Abstract
The subcellular localization of proteins is very important for characterizing its function in a cell. Accurate prediction of the subcellular locations in computational paradigm has been an active area of interest. Most of the work has been focused on single localization prediction. Only few studies have discussed the multi-target localization, but have not achieved good accuracy so far; in plant sciences, very limited work has been done. Here we report the development of a novel tool Plant-mSubP, which is based on integrated machine learning approaches to efficiently predict the subcellular localizations in plant proteomes. The proposed approach predicts with high accuracy 11 single localizations and three dual locations of plant cell. Several hybrid features based on composition and physicochemical properties of a protein such as amino acid composition, pseudo amino acid composition, auto-correlation descriptors, quasi-sequence-order descriptors and hybrid features are used to represent the protein. The performance of the proposed method has been assessed through a training set as well as an independent test set. Using the hybrid feature of the pseudo amino acid composition, N-Center-C terminal amino acid composition and the dipeptide composition (PseAAC-NCC-DIPEP), an overall accuracy of 81.97 %, 84.75 % and 87.88 % is achieved on the training data set of proteins containing the single-label, single- and dual-label combined, and dual-label proteins, respectively. When tested on the independent data, an accuracy of 64.36 %, 64.84 % and 81.08 % is achieved on the single-label, single- and dual-label, and dual-label proteins, respectively. The prediction models have been implemented on a web server available at http://bioinfo.usu.edu/Plant-mSubP/. The results indicate that the proposed approach is comparable to the existing methods in single localization prediction and outperforms all other existing tools when compared for dual-label proteins. The prediction tool will be a useful resource for better annotation of various plant proteomes.
Keywords: Artificial intelligence, machine learning, multi-location, prediction tool, protein science, subcellular localization, web server
In order for various cellular processes to be carried out in a cell, the correct identification of subcellular localization of a protein is crucial. Determining the localization of proteins through experiments can be laborious, expensive and time-consuming. Computational prediction is thus essential to decipher protein function and faster genome annotation. It also aids the identification of drug targets. We present here a software, Plant-mSubP, which is a publicly available web platform at http://bioinfo.usu.edu/Plant-mSubP/. Plant-mSubP uses artificial intelligence to effectively predict the protein localizations (11 single- and three dual-target) inside the plant cell. This resource can assess a high number of proteins in order to find their desired location in a cell, and thus will be useful in better annotation of various plant proteomes.
Background
The cell is a three-dimensional space composed of several compartments, having different physicochemical environment and function. For efficient functioning, the cell’s functional machinery - protein needs to be present at specific cellular compartments. Improper localization of proteins may result in disease and cell death (Park et al. 2011; Mer and Andrade-Navarro 2013). Therefore, subcellular location is an essential attribute in the functional characterization of proteins (Casadio et al. 2008). In recent years, knowledge about protein subcellular localization has earned enormous attention due to its important roles in elucidating protein functions, identifying drug targets and many more (Chou and Cai 2005). Thus, predicting the subcellular localization of protein is an important issue in proteomics. Since biochemical experiments are expensive and time-consuming, computational approaches gained an attention in prediction of subcellular localization. Several in silico approaches have been proposed to predict the subcellular localization. In one of the approach as reported in Lin et al. (2011) the motifs recognized by the sorting proteins and receptors of the protein transport machinery to move protein products from the cytosol to other subcellular localizations. This method is limited by the knowledge of sorting signals and absence of known motifs. In Adelfio et al. (2013), they used the sequence homology feature to proteins of experimentally verified localizations with the assumption that similar proteins target similar localizations. There are many deviations of this rule which may mislead the prediction (e.g. the proteins of the Lsg1 family of GTPases). Further, in some other methods, it uses protein sequence features such as amino acid composition, dipeptide composition, pseudo amino acid composition based on the assumption that the physicochemical properties of the protein residues may somehow be coupled to the physicochemical properties of the environment where the protein performs its function. Therefore, the differences in environments will be engraved in the protein amino acid compositions (Nakashima and Nishikawa 1994; Nielsen et al. 1997; Emanuelsson et al. 2000; Chou 2001; Höglund et al. 2006; Mak et al. 2008). The advantage of this approach is that it can be applied to any set of compartments and proteins, provided there is enough availability of data. Several approaches have been developed on annotation-based methods. Recently, Gene Ontology (GO)-based features have gained popularity for the prediction of protein subcellular locations (Chou and Cai 2004; Wan et al. 2011, 2012a, b, 2013; Mei 2012). Also, a combination of GO, composition and evolutionary features have been successfully used. To date, the GO-based features have shown better accuracy in predicting the subcellular localizations in both single- and multi-label localizations (Chou and Shen 2006). Although it shows superior results, it has several bottlenecks.
The set of distinct GO terms derived from a given data set may not be representative for other data sets; means the generalization capabilities of the predictors may be weakened when new GO terms outside the predefined GO term set are found in the test proteins. The GO term set also varies from species to species. Although the GO-based model looks promising, there are no specific classes defined for the multi-located proteins. Since overall actual accuracy is the most desired measure in multi-located classes, the existing GO-based models do not show up the actual accuracy of the multi-class proteins which is misleading the accuracy performance. In addition to this, in the existing multi-target approaches, there have been no report of comparing the performances of different data sets, e.g. how the models developed from single-label proteins differ from the models developed on a combined set of single- and multi-target proteins data set, or the models developed from multi-target protein data sets only.
Most of the existing methods are limited to the prediction of single-location proteins. These methods generally exclude the occurrences of multi-label proteins. But the fact is, multi-location proteins exist that can simultaneously reside at, or move between, two or more different subcellular localizations (Chou and Shen 2007; Chou et al. 2011; Lin et al. 2011; Xiao et al. 2011; Wu et al. 2012). Recently, several multi-label predictors such as Plant-mPLoc (Chou and Shen 2010a), Virus-mPLoc (Shen and Chou 2010), iLoc-Plant (Wu et al. 2011), iLoc-Virus (Xiao et al. 2011), HybridGO-Loc (Wan et al. 2014), Y-Loc (Briesemeister et al. 2010) and mGOASVM (Wan et al. 2012a) have been proposed. These predictors use the GO information and have demonstrated superiority over existing methods. Some other methods are based on predicting the transit peptides; Sperschneider et al. (2017) proposed a web tool, LOCALIZER for predicting plant and effector protein localization to three classes; chloroplast, mitochondria and nuclei. Chen et al. (2017) proposed a method to identify the peroxisome subcellular locations in plants. BUSCA (Savojardo et al. 2018) combines different computational tools to predict signal and transit peptides, GPI anchors and transmembrane domains. It has one module available for plant proteins but no option for predicting dual- or multi-label proteins.
Many subcellular predictors have been developed especially for specific species (Kaundal and Raghava 2009; Kaundal et al. 2013). A different promising approach has been proposed based on account amino acid composition at different levels of amino acid exposure (Emanuelsson et al. 2000). Efficient feature representation of a protein is a very important aspect of subcellular localization (Chou and Shen 2007). Hence there is a demand to accurately predict the subcellular localizations efficiently which further helps in the correct annotation of various proteomes.
In literature, dual targeting of a multitude of proteins has been described for native plant proteins (Peeters and Small 2001; Silva-Filho 2003; Mackenzie 2005; Mitschke et al. 2009). Also, protein folding, post-translational modification and protein–protein interactions can be involved in determining the targeting of proteins with multiple sites of action (Karniely and Pines 2005; Mitschke et al. 2009). It has been seen that various amino acid features significantly contribute to the dual targeting of localizations (Mitschke et al. 2009).
In this paper, we propose a simple and efficient predictor tool based on the sequence features. It can be used to classify single- and dual-label proteins subcellular localization. The system predicts the 11 single localizations (cell membrane, cell wall, plastid, cytoplasm, endoplasmic reticulum, extracellular, Golgi apparatus, mitochondrion, nucleus, peroxisome and vacuole) and three dual-localized protein classes (cytoplasm-nucleus, mitochondrion-plastid and cytoplasm-Golgi apparatus). Various sequence-based features of a protein sequence viz. amino acid composition (AAC), dipeptide composition (DIPEP), pseudo amino acid composition (PseAAC), N-terminal-Center-C-terminal (NCC) composition, physicochemical properties, Composition and Transition, and Quasi-sequence-order-based methods, and a range of hybrid features are explored in a machine learning framework to develop diverse prediction models for better confidence and reliability.
Implementation
For the development of any useful sequence-based statistical predictor for a biological system as reported in a series of recent publications (Chou et al. 2011; Wu et al. 2011; Lin et al. 2013; Chen et al. 2018; Feng et al. 2019), one should implement the 5-step rules (Chou et al. 2011) such as (i) construction of a valid benchmark data set to train and test the predictor; (ii) mathematical representation of biological sequence samples which will reflect their intrinsic correlation with the target to be predicted; (iii) an algorithm (or engine) for performing prediction operation; (iv) cross-validation tests to objectively evaluate the anticipated accuracy of the predictor; and (v) a user-friendly web server/tool for the predictor that is easily accessible to the public. We have implemented our best-performing prediction models on the publicly available tool, Plant-mSubP and is freely accessible on the web.
Data sets generation
To develop an efficient prediction system, it is important to first gather data sets of known subcellular localization and extract diverse relevant features out of it for use in the training and testing of machine learning classifiers. The protein sequences of all the plants were extracted from the UniProt database release 2018_02 (http://www.uniprot.org) using [keywords: SUBCELLULAR LOCATION AND reviewed: yes]. Sequence annotations marked as ‘PROBABLE’, ‘POSSIBLE’ and ‘BY SIMILARITY’ were discarded. This resulted in 16 494 unique sequences of proteins, annotated to 14 different single- and dual-label subcellular localizations as detailed in Table 1.
Table 1.
Distribution of subcellular localization classes (single- and dual-located) for all plant data from UniProt database release 2018_02 in the training data set and independent testing data set. *About 10 % of sequences from the original training data set were kept separate for independent testing. In total, 16 494 plant protein sequences were found after applying the filters [viridiplantae AND annotation:(type: location confidence: experimental)].
| Type | Subcellular location | # sequences retrieved | # sequences after redundancy check (30 % cut-off) | *Training data set | Training data set (sequences length > 50) | Independent data set (sequences length > 50) |
|---|---|---|---|---|---|---|
| Single label | Plastid | 11 302 | 2979 | 2678 | 2468 | 248 |
| Cytoplasm | 739 | 403 | 361 | 351 | 40 | |
| Extracellular | 237 | 186 | 166 | 140 | 14 | |
| Nucleus | 734 | 636 | 571 | 568 | 63 | |
| Mitochondrion | 759 | 537 | 481 | 447 | 52 | |
| Cell membrane | 1256 | 927 | 830 | 829 | 92 | |
| Golgi apparatus | 277 | 229 | 204 | 204 | 23 | |
| Endoplasmic reticulum | 393 | 320 | 285 | 280 | 29 | |
| Vacuole | 260 | 198 | 176 | 176 | 20 | |
| Peroxisome | 80 | 63 | 57 | 57 | 06 | |
| Cell wall | 52 | 47 | 42 | 37 | 05 | |
| Dual label | Mito-plastid | 141 | 133 | 118 | 118 | 13 |
| Cyto-nucleus | 210 | 196 | 175 | 170 | 20 | |
| Cyto-Golgi | 54 | 38 | 34 | 34 | 04 | |
| Total | 16 494 | 6892 | 6178 | 5879 | 629 |
After reducing the sequence identity with a cut-off of <30 % using BlastClust, a total of 6892 proteins were left for further use. This was done within the class as well as across the classes. About 10 % of these data, i.e. 714 sequences, were kept separate for independent testing; thus, a total of remaining 6178 proteins constituted our initial training data set (column 5, Table 1). Testing on independent data sets that are not used during the machine learning model development has been reported to be the best benchmark to test the performance of various prediction modules. Further, to remove any potential fragments, 5879 sequences were extracted out of the 6178 proteins by filtering those sequences whose length was greater than 50 (column 6, Table 1) and were used in the training/testing of various machine learning algorithms. Similarly, in the independent test data, out of 714 proteins, 629 sequences were extracted whose length was greater than 50 (column 7, Table 1).
Feature representation methods
With the explosive growth of biological sequences, one of the most important and difficult problems in computational biology is the expression of a biological sequence with a discrete model or a vector, yet retaining sequence-order information or key pattern characteristics. In this work, the following diverse features have been used:
Amino acid composition (AAC)
Dipeptide composition (DIPEP)
Pseudo amino acid composition (PseAAC)
Terminal-based N-Center-C (NCC) amino acid composition
The above four features have been explained in detail in our previous studies on the identification and characterization of various plastid types (Kaundal et al. 2013), and so not discussed here. In the current study, we wanted to explore these features to see if they could predict the multi-target protein localizations as well. In addition, we extracted and implemented the following diverse features from protein sequences to achieve high accuracy.
Physicochemical property-based composition
The physicochemical properties of amino acids are successfully used for prediction of protein function, structure and subcellular localizations with various alterations. In literature, it has been shown that the physicochemical properties such as acidic, basic, hydrophobicity, hydrophilicity, neutral and atomic composition play an important role in the residing the protein the cellular compartment. Compositions of amino acids of these classes are calculated as a feature to represent the protein. Thus, each protein is represented by a 26-dimensional feature vector.
Composition and Transition
A protein sequence can be represented and categorized into three classes according to its attributes (Dubchak et al. 1999), where each amino acid in the sequence is encoded as 1, 2 or 3 depending on the class that it belongs to. The attributes used here are hydrophobicity, normalized van der Waals volume, polarity and polarizability. The corresponding classification for each attribute is listed in Table 2.
Table 2.
Group attributes and classification of various amino acids in a protein, as defined in Dubchak et al. (1999).
| Group 1 | Group 2 | Group 3 | |
|---|---|---|---|
| Hydrophobicity | Polar | Neutral | Hydrophobicity |
| R, K, E, D, Q, N | G, A, S, T, P, H, Y | C, L, V, I, M, F, W | |
| Normalized van der Waals volume | 0–2.78 | 2.95–4.0 | 4.03–8.08 |
| G, A, S, T, P, D, C | N, V, E, Q, I, L | M, H, K, F, R, Y, W | |
| Polarity | 4.9–6.2 | 8.0–9.2 | 10.4–13.0 |
| L, I, F, W, C, M, V, Y | P, A, T, G, S | H, Q, R, K, N, E, D | |
| Polarizability | 0–1.08 | 0.128–0.186 | 0.219–0.409 |
| G, A, S, D, T | C, P, N, V, E, Q, I, L | K, M, H, F, R, Y, W | |
| Charge | Positive | Neutral | Negative |
| K, R | A, N, C, Q, G, H, I, L, M, F, P, S, T, W, Y, V | D, E | |
| Secondary structure | Helix | Strand | Coil |
| E, A, L, M, Q, K, R, H | V, I, Y, C, W, F, T | G, N, P, S, D | |
| Solvent accessibility | Buried | Exposed | Intermediate |
| A, L, F, C, G, I, V, W | R, K, Q, E, N, D | M, S, P, T, H, Y |
After this classification, three types of descriptors: composition (C), Transition (T) and Distribution (D) are calculated.
Composition (CTDC)
The composition is defined as the global percentage for each of the encoded classes in a protein sequence.
| (1) |
where nr is the number of amino acids of type r in the encoded sequence; N is the length of the sequence.
Transition (CTDT)
Transition is defined as each of the changes between classes for the encoded sequences, a transition from class 1 to 2 is the percent frequency with which 1 is followed by 2 or 2 is followed by 1 in the encoded sequences.
| (2) |
where nrs, nsr are the numbers of dipeptide encoded as rs and sr in the sequence; N is the length of the sequence.
Quasi-sequence-order descriptors (QSO)
The QSO descriptors are derived from the distance matrix between the 20 amino acids. Based on the definitions and figures used in protr package (Xiao et al. 2015) for the equations originally described in Chou (2000), a quasi-sequence-order descriptor can be defined for each of the amino acids as:
| (3) |
where fr is the normalized occurrence for amino acid type i and N is a weighting factor (w = 0.1). These are the first 20 quasi-sequence-order descriptors. The other 30 quasi-sequence-order are defined as:
Support Vector Machines
Support Vector Machine (SVM) is a machine learning technique first introduced by Cortes and Vapnik (1995). It is a statistical learning theory based on optimization principle. This technique has been used in the field of image processing, speech processing, protein subcellular localization prediction, protein secondary structure prediction and many other areas. The main aim of SVM is to separate the training data by maximization the margin with maximum computing efficiency. Multi-class classification is implemented by using a series of binary classification. Many methods have been used for multi-class classification like Directed Acyclic Graph Support Vector Machines (DAGSVM), One-vs.-Rest (OvR) and One-vs.-One (OvO). Radial basis function (RBF) is a popular kernel widely used for classification. In our study, we have used OvR strategy which involves training a single classifier per localization class, with the samples of that class as positive samples and all other localization classes as negatives. Making decisions means then applying all classifiers to an unseen sample and predicting the label for which the corresponding classifier reports the highest confidence score. The idea here is to reduce the problem of multi-class classification to multiple binary classification problems.
Training/testing schema.
We have used 5-fold cross-validation technique for training/testing procedure, using the OvR strategy for decision-making. Here, the training data are divided into five parts. For development of model, four parts are combined to form a training set and fifth part is used as testing data set. This process is repeated five times by changing the training and testing data set. Finally, the models are tested on an independent data set called as validation set.
Evaluation parameters.
The evaluation of models is done based on following parameters.
(i) Sensitivity: It is defined as a percent of truly predicted true proteins,
| (4) |
Specificity: It is the percent of non-true correctly predicted as non-true proteins,
| (5) |
Accuracy: It is the percentage of correctly predicted proteins (true and non-true proteins),
| (6) |
Precision: It is the percentage of positive predictions those are correct calculated as:
| (7) |
(v) Rate of False Predictions (RFP): It is defined as the probability of false predictions percentage from the predictions set,
| (8) |
Error Rate (ER): ER is defined as the percentage of misclassified samples,
| (9) |
Matthews Correlation Coefficient (MCC): MCC is defined as the parameter for prediction of class. For perfect prediction, it is equal to 1 and 0 for random prediction. It is given by
| (10) |
where TP = True Positives, TN = True Negatives, FP = False Positives, and FN = False Negatives.
Results and Discussion
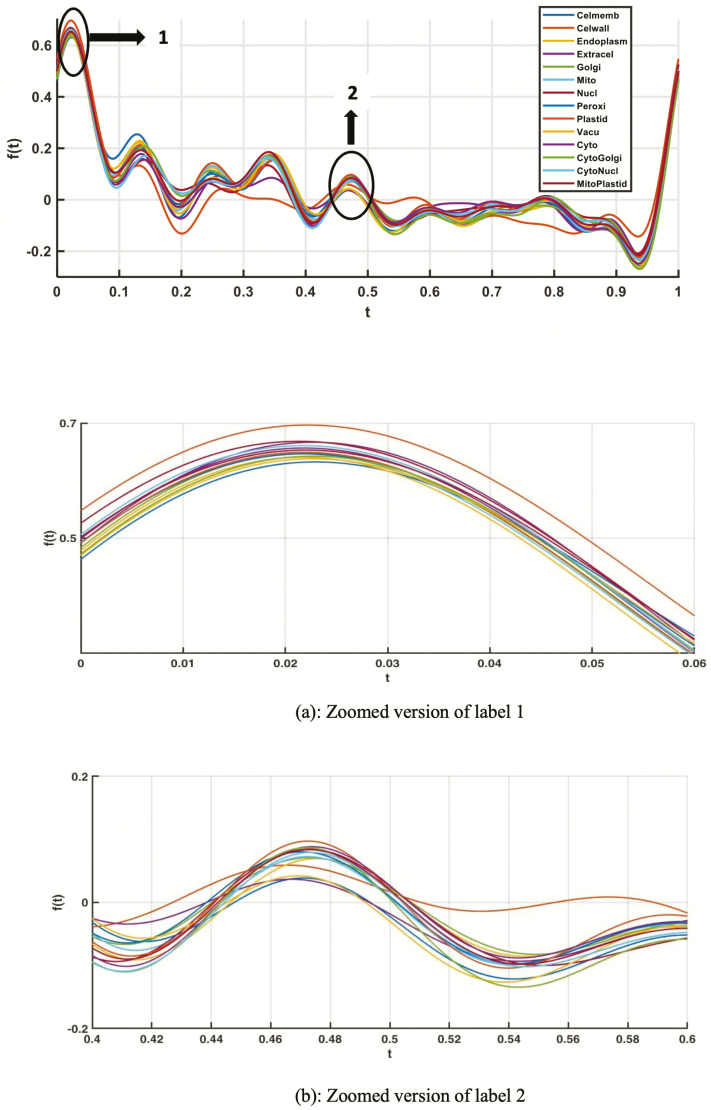
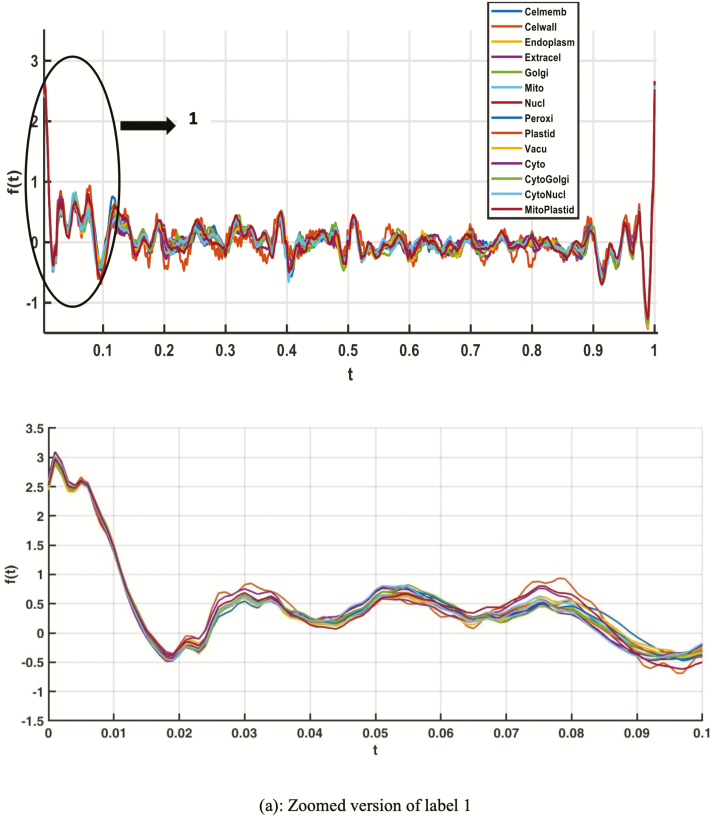
To assess the distinguishing capability of various protein features, we first studied the Andrews plot. Andrews plot is a method in high dimensional data to visualize the latent structure. It has been used to represent multivariate data. The Andrews plot of amino acid composition (AAC) and the PseAAC-NCC-DIPEP features is shown in Figs 1 and 2.
Figure 1.
Andrews plot of amino acid composition (AAC) feature for all the single- and dual-label localizations.
Figure 2.
Andrews plot of PseAAC-NCC-DIPEP feature for all the single- and dual-label localizations.
From the variations in the plots, it can be elucidated that the extracted features are capable to distinguish the different localization classes. This shows that composition-based models and other sequence features could be used in a machine learning framework to develop prediction models for classifying protein sequences of different subcellular localizations.
Five-fold cross-validation training/testing
In this study, the 5-fold cross-validation technique was used with SVM as the prediction model. The performance of various models was evaluated based on various statistical parameters as explained above. In a 5-fold cross-validation test in the training data set, the overall accuracy of the SVM model results is listed in Table 3. It shows that the PseACC-NCC-DIPEP model provides the superior result on all three types of data sets; single-label, single- and dual-label combined, and dual-label proteins. The PseAAC-NCC-DIPEP feature achieves an overall accuracy of 81.97 % on the single-label training set (Table 3a), 84.75 % on the single- and dual-label combined training data set (Table 3b) and 87.88 % on the dual-label only proteins data set (Table 3c). We did not see a significant difference in prediction performances across the data set types as depicted in Table 3a–c; for example, in the PseACC-NCC-DIPEP model, there is a marginal increase of 3.3 % accuracy of the dual-label model over the combined data set module. It is worth mentioning here that in our separate comparative analysis (results not shown), we also explored the use of Artificial Neural Networks (ANN) but achieved much lower overall accuracies in a 5-fold training/testing procedure as compared to the SVMs.
Table 3.
(a) Performance comparison by 5-fold cross-validation testing on the training data set of single-label proteins using SVMs; (b) Performance comparison of 5-fold cross-validation testing on the combined training data set (single- + dual-label) using SVMs; (c) Performance comparison of 5-fold cross-validation testing on the dual-localized training data set using SVMs. Bold values represents the best performance. RBF = radial basis function of SVM; C = regularization parameter.
| (a) | ||||
|---|---|---|---|---|
| Feature representation methods | Overall accuracy (%) (single-label data) | |||
| AAC (σ = 2, C = 10) | 73.65 | |||
| DIPEP (σ = 50, C = 500) | 77.56 | |||
| PseAAC (σ = 10, C = 500) | 75.49 | |||
| NCC (σ = 10, C = 50) | 74.36 | |||
| PseAAC-NCC-DIPEP (σ = 50, C = 500) | 81.97 | |||
| NCC-DIPEP (σ = 50, C = 500) | 81.18 | |||
| QSO (σ = 10, C = 500) | 73.25 | |||
| NCC-DIPEP-CTDC-CTDT- QSO (σ = 5, C = 30) | 80.42 | |||
| (b) | ||||
| Feature representation methods | Overall accuracy (%) (single- + dual-label data) | |||
| AAC (σ = 2, C = 10) | 68.48 | |||
| DIPEP (σ = 50, C = 500) | 74.59 | |||
| PseAAC (σ = 10, C = 500) | 71.87 | |||
| NCC (σ = 10, C = 50) | 70.74 | |||
| PseAAC-NCC-DIPEP (σ = 50, C = 500) | 84.75 | |||
| NCC-DIPEP (σ = 50, C = 500) | 83.96 | |||
| Physicochem [atomi + hydrophobicity, basic] | 73.21 | |||
| NCC-DIPEP-physicochem | 83.79 | |||
| Quasi-sequence-order descriptors | 54.38 | |||
| NCC-DIPEP-CTDC-CTDT- QSO | 60.02 | |||
| (c) | ||||
| Model | Kernel | C | Gamma | Overall accuracy (%) (dual- label data) |
| AAC | RBF | 10 | 0.001 | 76.64 |
| DIPEP | RBF | 10 | 0.001 | 82.29 |
| PseAAC | RBF | 10 | 0.001 | 77.63 |
| NCC | RBF | 10 | 0.001 | 86.02 |
| NCC-DIPEP | RBF | 10 | 0.001 | 87.57 |
| PseAAC-NCC-DIPEP | RBF | 10 | 0.001 | 87.88 |
Independent testing/benchmarking.
Next, we performed a test on the independent data sets, the 10 % data that were kept separate for testing (as in Table 1). The comparison results are reported in Table 4. As reported in previous studies (Chou and Elrod 1999; Kaundal and Raghava 2009; Kaundal et al. 2010, 2013; Tung et al. 2017), the best way to test the prediction performance of a particular tool is to test it on independent data sets, which have not been used in the process of training/testing of machine learning. From the results in Table 4a–c, it shows that the PseAAC-NCC-DIPEP feature is superior providing an overall accuracy of 64.36 % on the single-label data set, 64.84 % on the single- and dual-target combined data set and 81.08 % accuracy on the independent dual-target data set. This shows that the dual-target proteins might contain some specialized signals for dual targeting which are not well represented when we develop training classifiers on a combined data. The overall results show that PseAAC-NCC-DIPEP feature is superior in predicting the single- and dual-label subcellular localizations.
Table 4.
(a) Comparison of prediction results on an ‘independent data set’ based on models trained from single-label proteins using SVMs; (b) Comparison of prediction results on an ‘independent data set’ based on models trained from combined data set (single- + dual-label); (c) Comparison of prediction results on an ‘independent data set’ based on models trained from dual-label proteins data set. Bold values represents the best performance.
| (a) | ||||
|---|---|---|---|---|
| Feature representation methods | Accuracy (%) | |||
| AAC (σ = 2, C = 10) | 59.11 | |||
| DIPEP (σ = 50, C = 500) | 59.11 | |||
| PseAAC (σ = 10, C = 500) | 59.12 | |||
| NCC (σ = 10, C = 50) | 50.34 | |||
| PseAAC-NCC-DIPEP (σ = 50, C = 500) | 64.36 | |||
| NCC-DIPEP (σ = 50, C = 500) | 64.05 | |||
| QSO (σ = 10, C = 500) | 57.05 | |||
| NCC-DIPEP-CTDC-CTDT-QSO (σ = 5, C = 300) | 61.46 | |||
| (b) | ||||
| Feature representation methods | Accuracy (%) | |||
| AAC (σ = 2, C = 10) | 57.71 | |||
| DIPEP (σ = 50, C = 500) | 58.95 | |||
| PseAAC (σ = 10, C = 500) | 56.60 | |||
| NCC (σ = 10, C = 50) | 52.88 | |||
| PseAAC-NCC-DIPEP (σ = 50, C = 500) | 64.84 | |||
| NCC-DIPEP (σ = 50, C = 500) | 64.42 | |||
| Quasi-sequence-order descriptors | 58.94 | |||
| NCC-DIPEP-CTDC-CTDT-QSO | 38.49 | |||
| (c) | ||||
| Model | Kernel | C | Gamma | Accuracy (%) |
| AAC | RBF | 10 | 0.001 | 72.56 |
| DIPEP | RBF | 10 | 0.001 | 72.97 |
| PseAAC | RBF | 10 | 0.001 | 75.67 |
| NCC | RBF | 10 | 0.001 | 78.37 |
| NCC-DIPEP | RBF | 10 | 0.001 | 75.67 |
| PseAAC-NCC-DIPEP | RBF | 10 | 0.001 | 81.08 |
Comparison with other existing tools.
Further, we assessed the performance of our tool, Plant-mSubP with the existing tools for predicting both the single- and dual-label subcellular localizations. In literature, many methods have been reported to predict the subcellular localizations but most of them are for single-class proteins. In this paper, we have compared our method with the existing methods for plant subcellular localization such as YLoc (Briesemeister et al. 2010), Euk-mPloc (Chou and Shen 2010b) and iLoc-Plant (Wu et al. 2011) that were developed for multi-label proteins. The prediction results for the YLoc, Euk-mPloc and iLoc-Plant are assessed on the independent data set as created in Table 1. The comparison results are reported in Table 5. The results show that our proposed method is better than the three compared tools to predict the subcellular localizations, single- as well as dual-target proteins. We believe Plant-mSubP will be helpful in better annotation of the existing and novel plant proteomes.
Table 5.
Comparison of actual prediction accuracy of Plant-mSubP on an ‘independent data set’ with the existing web tools that support multi-label localizations. Actual accuracy is calculated (in percentage) as the ratio of number of localization samples correctly predicted divided by the total number of samples in the independent data set.
| Web tools | Prediction accuracy (%) (single- + dual-label data) | Prediction accuracy (%) (dual-label data) |
|---|---|---|
| YLoc | 34.35 | 35.89 |
| Euk-mPloc 2.0 | 53.5 | 44.86 |
| iLoc-Plant | 37.42 | 34.42 |
| Our method [Plant-mSubP] | 64.84 | 81.08 |
Tool development and availability
In various recent publications (Chou and Shen 2009; Kaundal and Raghava 2009; Kaundal et al. 2010, 2013; Chou 2011, 2013; Chou et al. 2011; Chen et al. 2018; Feng et al. 2019), it is demonstrated that user-friendly and openly accessible web tools represent the future direction for developing practically more useful computational tools.
From our analysis, the best-performing prediction algorithms were implemented on the web server called, Plant-mSubP (http://bioinfo.usu.edu/Plant-mSubP/). Its framework has been implemented using R, with the user interface and web server designed with the Shiny package. It has an intuitive interface in which the user can either upload a multi-FASTA format file or paste their sequences in a box. When the user submits a job, it will test the sanity of the sequences using protr R package; besides, it will check that the input sequences have a length more than 50 amino acids required to calculate N-Center-C terminal Composition features (Kaundal and Raghava 2009; Kaundal et al. 2010, 2013). The protein features extraction for Composition, Transition and Quasi-sequence-order descriptors are done using protr R package. Other features extraction was made with our in-house scripts in R. The web server currently supports a prediction workload up to a thousand sequences (1000).
Predictions methods implemented on the server were selected based on efficiency and fast-paced, including two options for a faster prediction (amino acid composition-based and dipeptide composition-based), two options for an accurate prediction using comprehensive hybrid features models (PseAAC-NCC-DIPEP and NCC-DIPEP-CTDC-CTDT-QSO) and a homology-search-based option (blastp). Support Vector Machines predictions were implemented using the e1071 R package. After the job submission, users can search throughout the results presented in an enriched table format or download a file with that information to be opened in a spreadsheet software (e.g. Excel); downloading the sequences alignments is also an option in case the user selects the homology-based BLAST approach for comparing the subcellular localization predictions with the machine-learned classifiers.
On the Plant-mSubP web server, we have also provided the links to download the sequences used to construct the predictions models (training sets) and the testing sequences used for independent test, separated by each subcellular localization class.
Conclusion
An accurate prediction of protein localization is a very critical step in any functional genome annotation process. Various experimental procedures such as large-scale phenotyping screens, microarray or RNA-Seq experiments, protein–protein interaction assays etc. all rely heavily on the subcellular localization information. It is, therefore, necessary to continuously expand our knowledge in this area and develop highly accurate prediction tools. Although some tools exist to predict single localization of the proteins, very few have been developed for dual-targeting proteins and have limited accuracy. Very limited work has been reported for plant proteins. In this paper, we have developed an integrated machine learning framework to accurately predict the subcellular localizations of protein targeting for both the single and dual locations in plants. Various features of proteins have been explored and found that the PseAAC-NCC-DIPEP feature is superior in predicting the subcellular localizations for both single- and dual-targeting proteins. Using an independent data set for each localization class, we have compared our method with the available sequence-based prediction tools that also support dual-location prediction and found that our method, Plant-mSubP outperforms the existing methods. We believe the web server should be helpful to the users in the correct annotation of various proteomes.
Availability and Requirements
Project name: Plant-mSubP
Project home page: http://bioinfo.usu.edu/Plant-mSubP/
Operating system(s): Linux
Programming language: R, Python, MATLAB
Other requirements: N/A
License: N/A
Any restrictions to use by non-academics: No restrictions to use this web tool
Acknowledgements
The authors acknowledge the support to this study from faculty start-up funds to R.K. from the Center for Integrated BioSystems/Department of Plants, Soils, and Climate, Utah State University. This research was also supported by the Utah Agricultural Experiment Station, Utah State University, and approved as journal paper number 9261. The authors also thank the anonymous referees for help in improving the research article.
Conflict of Interest
None declared.
Literature Cited
- Adelfio A, Volpato V, Pollastri G. 2013. SCLpredT: ab initio and homology-based prediction of subcellular localization by N-to-1 neural networks. SpringerPlus 2:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briesemeister S, Rahnenführer J, Kohlbacher O. 2010. YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Research 38:W497–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R, Martelli PL, Pierleoni A. 2008. The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Briefings in Functional Genomics & Proteomics 7:63–73. [DOI] [PubMed] [Google Scholar]
- Chen W, Feng P, Yang H, Ding H, Lin H, Chou KC. 2018. iRNA-3typeA: identifying three types of modification at RNA’s adenosine sites. Molecular Therapy - Nucleic Acids 11:468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Teng XL, Xiao XG. 2017. Subcellular localization of a plant catalase-phenol oxidase, AcCATPO, from amaranthus and identification of a non-canonical peroxisome targeting signal. Frontiers in Plant Science 8:1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC. 2000. Prediction of protein subcellular locations by incorporating quasi-sequence-order effect. Biochemical and Biophysical Research Communications 278:477–483. [DOI] [PubMed] [Google Scholar]
- Chou KC. 2001. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 43:246–255. [DOI] [PubMed] [Google Scholar]
- Chou KC. 2011. Some remarks on protein attribute prediction and pseudo amino acid composition. Journal of Theoretical Biology 273:236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC. 2013. Some remarks on predicting multi-label attributes in molecular biosystems. Molecular Biosystems 9:1092–1100. [DOI] [PubMed] [Google Scholar]
- Chou KC, Cai YD. 2004. Prediction of protein subcellular locations by GO-FunD-PseAA predictor. Biochemical and Biophysical Research Communications 320:1236–1239. [DOI] [PubMed] [Google Scholar]
- Chou KC, Cai YD. 2005. Predicting protein localization in budding yeast. Bioinformatics 21:944–950. [DOI] [PubMed] [Google Scholar]
- Chou KC, Elrod DW. 1999. Protein subcellular location prediction. Protein Engineering 12:107–118. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. 2006. Predicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic K-nearest neighbor classifiers. Journal of Proteome Research 5:1888–1897. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. 2007. Recent progress in protein subcellular location prediction. Analytical Biochemistry 370:1–16. [DOI] [PubMed] [Google Scholar]
- Chou K-C, Shen H-B. 2009. REVIEW : recent advances in developing web-servers for predicting protein attributes. Natural Science 01:63–92. [Google Scholar]
- Chou KC, Shen HB. 2010a. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, Shen HB. 2010b. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: euk-mPLoc 2.0. PLoS One 5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, Wu ZC, Xiao X. 2011. iLoc-Euk: a multi-label classifier for predicting the subcellular localization of singleplex and multiplex eukaryotic proteins. PLoS One 6:e18258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Machine Learning 297:273–297. [Google Scholar]
- Dubchak I, Muchnik I, Mayor C, Dralyuk I, Kim SH. 1999. Recognition of a protein fold in the context of the structural classification of proteins (SCOP) classification. Proteins 35:401–407. [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Feng P, Yang H, Ding H, Lin H, Chen W, Chou KC. 2019. iDNA6mA-PseKNC: identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 111:96–102. [DOI] [PubMed] [Google Scholar]
- Höglund A, Dönnes P, Blum T, Adolph HW, Kohlbacher O. 2006. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22:1158–1165. [DOI] [PubMed] [Google Scholar]
- Karniely S, Pines O. 2005. Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Reports 6:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal R, Raghava GP. 2009. RSLpred: an integrative system for predicting subcellular localization of rice proteins combining compositional and evolutionary information. Proteomics 9:2324–2342. [DOI] [PubMed] [Google Scholar]
- Kaundal R, Sahu SS, Verma R, Weirick T. 2013. Identification and characterization of plastid-type proteins from sequence-attributed features using machine learning. BMC Bioinformatics 14:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal R, Saini R, Zhao PX. 2010. Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiology 154:36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TH, Murphy RF, Bar-Joseph Z. 2011. Discriminative motif finding for predicting protein subcellular localization. IEEE/ACM Transactions on Computational Biology and Bioinformatics 8:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WZ, Fang JA, Xiao X, Chou KC. 2013. iLoc-Animal: a multi-label learning classifier for predicting subcellular localization of animal proteins. Molecular Biosystems 9:634–644. [DOI] [PubMed] [Google Scholar]
- Mackenzie SA. 2005. Plant organellar protein targeting: a traffic plan still under construction. Trends in Cell Biology 15:548–554. [DOI] [PubMed] [Google Scholar]
- Mak MW, Guo J, Kung SY. 2008. PairProSVM: protein subcellular localization based on local pairwise profile alignment and SVM. IEEE/ACM Transactions on Computational Biology and Bioinformatics 5:416–422. [DOI] [PubMed] [Google Scholar]
- Mei S. 2012. Multi-label multi-kernel transfer learning for human protein subcellular localization. PLoS One 7:e37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer AS, Andrade-Navarro MA. 2013. A novel approach for protein subcellular location prediction using amino acid exposure. BMC Bioinformatics 14:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Fuss J, Blum T, Höglund A, Reski R, Kohlbacher O, Rensing SA. 2009. Prediction of dual protein targeting to plant organelles. The New Phytologist 183:224–235. [DOI] [PubMed] [Google Scholar]
- Nakashima H, Nishikawa K. 1994. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. Journal of Molecular Biology 238:54–61. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. International Journal of Neural Systems 8:581–599. [DOI] [PubMed] [Google Scholar]
- Park S, Yang JS, Shin YE, Park J, Jang SK, Kim S. 2011. Protein localization as a principal feature of the etiology and comorbidity of genetic diseases. Molecular Systems Biology 7:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Small I. 2001. Dual targeting to mitochondria and chloroplasts. Biochimica et Biophysica Acta 1541:54–63. [DOI] [PubMed] [Google Scholar]
- Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R. 2018. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Research 46:W459–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. 2010. Virus-mPLoc: a fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. Journal of Biomolecular Structure & Dynamics 28:175–186. [DOI] [PubMed] [Google Scholar]
- Silva-Filho MC. 2003. One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Current Opinion in Plant Biology 6:589–595. [DOI] [PubMed] [Google Scholar]
- Sperschneider J, Catanzariti AM, DeBoer K, Petre B, Gardiner DM, Singh KB, Dodds PN, Taylor JM. 2017. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Scientific Reports 7:44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CH, Chen CW, Sun HH, Chu YW. 2017. Predicting human protein subcellular localization by heterogeneous and comprehensive approaches. PLoS One 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Mak MW, Kung SY. 2011. Protein subcellular localization prediction based on profile alignment and gene ontology. IEEE International Workshop on Machine Learning for Signal Processing: 1–6. [Google Scholar]
- Wan S, Mak MW, Kung SY. 2012a. mGOASVM: multi-label protein subcellular localization based on gene ontology and support vector machines. BMC Bioinformatics 13:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Mak MW, Kung SY. 2012b. GOASVM: protein subcellular localization prediction based on gene ontology annotation and SVM. ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings: 2229–2232. [Google Scholar]
- Wan S, Mak MW, Kung SY. 2013. GOASVM: a subcellular location predictor by incorporating term-frequency gene ontology into the general form of Chou’s pseudo-amino acid composition. Journal of Theoretical Biology 323:40–48. [DOI] [PubMed] [Google Scholar]
- Wan S, Mak MW, Kung SY. 2014. HybridGO-Loc: mining hybrid features on gene ontology for predicting subcellular localization of multi-location proteins. PLoS One 9:e89545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZC, Xiao X, Chou KC. 2011. iLoc-Plant: a multi-label classifier for predicting the subcellular localization of plant proteins with both single and multiple sites. Molecular Biosystems 7:3287–3297. [DOI] [PubMed] [Google Scholar]
- Wu ZC, Xiao X, Chou KC. 2012. iLoc-Gpos: a multi-layer classifier for predicting the subcellular localization of singleplex and multiplex Gram-positive bacterial proteins. Protein and Peptide Letters 19:4–14. [DOI] [PubMed] [Google Scholar]
- Xiao N, Cao DS, Zhu MF, Xu QS. 2015. protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 31:1857–1859. [DOI] [PubMed] [Google Scholar]
- Xiao X, Wu ZC, Chou KC. 2011. iLoc-Virus: a multi-label learning classifier for identifying the subcellular localization of virus proteins with both single and multiple sites. Journal of Theoretical Biology 284:42–51. [DOI] [PubMed] [Google Scholar]