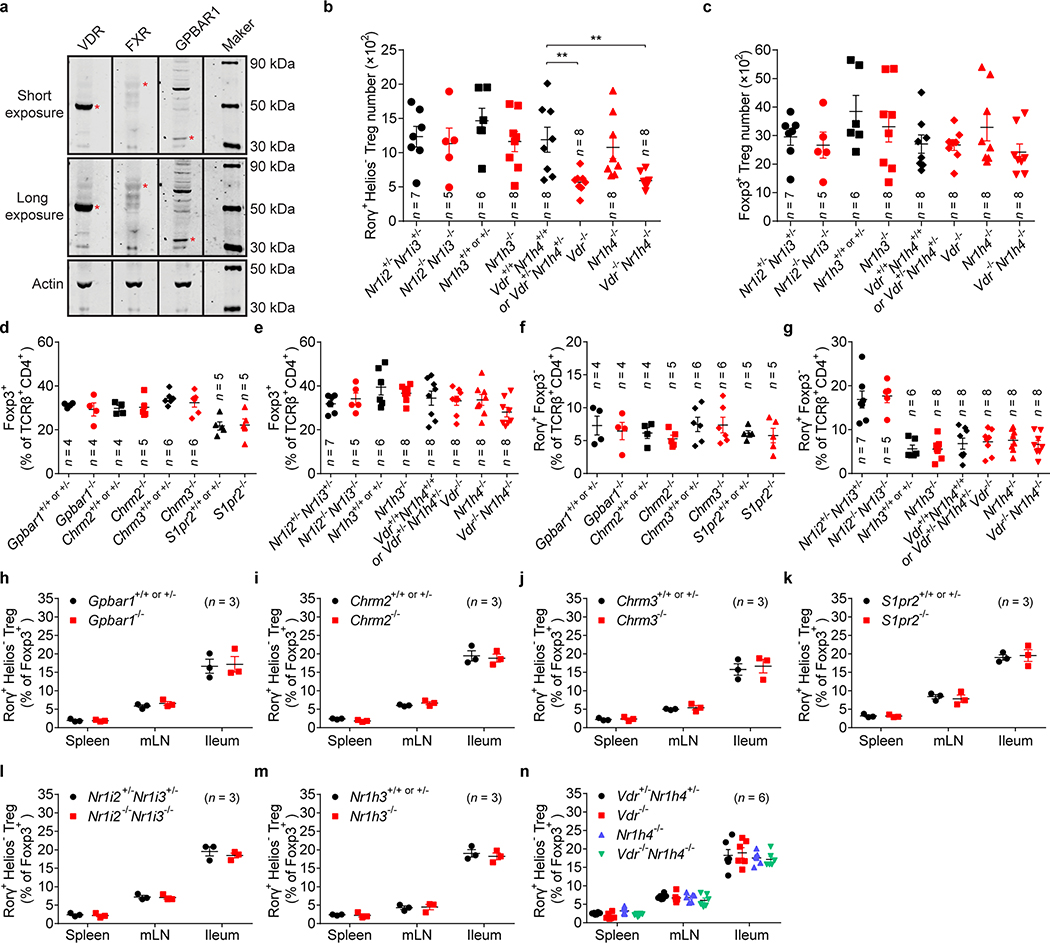
Extended Data Fig. 6. The impact of bile acid receptor deficiency on Tregs or Th17 cells in gut and peripheral lymphoid organs.
(a) Protein expression of VDR, FXR (NR1H4), and GPBAR1 in the colonic tissue of SPF C57BL/6J mice was analyzed by western blot. The red asterisks indicate the corresponding molecular weight of VDR (53 kDa), FXR (69 kDa), and GPBAR1 (33 kDa). For gel source data, see Supplementary Figure 1. (b, c) Absolute numbers of RORγ+Helios– in the colonic Foxp3+CD4+TCRβ+ Treg population (b) and of Foxp3+ Tregs in the CD4+TCRβ+ population (c) from mice deficient in nuclear receptors (Nr1i2–/–Nr1i3–/–, Nr1h3–/–, Vdr–/–, Nr1h4–/–, and Vdr–/–Nr1h4–/–) and their littermate controls. (d, e) Frequencies of Foxp3+ in the CD4+TCRβ+ cell population from mice deficient in G protein-coupled receptors (Gpbar1–/–, Chrm2–/–, Chrm3–/–, and S1pr2–/–) and their littermate controls (d) and from mice deficient in nuclear receptors (Nr1i2–/–Nr1i3–/–, Nr1h3–/–, Vdr–/–, Nr1h4–/–, and Vdr–/–Nr1h4–/–) and their littermate controls (e). (f, g) Frequencies of RORγ+Foxp3– in the CD4+TCRβ+ cell population from mice described in d, e. (h-n) Tregs in the spleen, mesenteric lymph node (mLN), and ileum from the indicated mice were analyzed. Frequencies of RORγ+Helios– in the Foxp3+CD4+TCRβ+ Treg population from Gpbar1–/– (h), Chrm2–/– (i), Chrm3–/– (j), S1pr2–/– (k), Nr1i2–/–Nr1i3–/– (l), Nr1h3–/– (m), and Vdr–/–, Nr1h4–/–, and Vdr–/–Nr1h4–/– (n) mice and their littermate controls are shown. Data are representative of two or three independent experiments in a, d-n, or are pooled from two or three independent experiments in b, c. n represents biologically independent animals. Bars indicate mean ± SEM values. ∗∗p < 0.01 in one-way analysis of variance followed by the Bonferroni post hoc test in b.