Abstract
There is broad interest in engineering phenylalanine ammonia-lyase (PAL) for its biocatalytic applications in industry and medicine. While site-specific mutagenesis has been employed to improve PAL stability or substrate specificity, combinatorial techniques are poorly explored. Here, we report development of a directed evolution technique to engineer PAL enzymes. Central to this approach is a high-throughput enrichment that couples E. coli growth to PAL activity. Starting with the PAL used in the formulation of pegvaliase for PKU therapy, we report previously unidentified mutations that increase turnover frequency almost twofold after only a single round of engineering.
The ammonia lyase (AL) class and aminomutase (AM) class of enzymes have been the focus of decades of research and development for industrial and biomedical applications. Their prosthetic group, 4-methylideneimidazole-5-one (MIO), either catalyzes the transformation of an l-α-amino acid into the α,β-unsaturated carboxylic acid counterpart via the non-oxidative elimination of ammonia or into the spatially isometric β-amino acid, respectively1. Hence, application of MIO-enzymes in both directions has yielded intermediates for pharmaceuticals2,3, agrochemicals4-6, polymers7-9, and flavonoids2,10-12.
Phenylalanine ammonia lyase (PAL) has been of great interest as a treatment for the genetic disease phenylketonuria (PKU). Daily subcutaneous injection of a purified and PEGylated recombinant PAL from Anabaena variabilis (PEG-rAvPAL; Palynziq®, BioMarin Pharmaceutical Inc.) was approved by the US FDA in 2018 as an enzyme substitution therapy for PKU13. Concurrently, an orally administered engineered PAL-expressing probiotic Escherichia coli Nissle 1917 is currently under investigation by Synlogic Inc14. Other formulations of this enzyme are also being explored as therapeutics15,16 as well as for the production of low phenylalanine (phe) protein dietary supplementation for PKU17-20 and cancer 21-24 patients.
This significant interest has resulted in various efforts to improve enzyme properties. Structural and sequence homology between aromatic ALs and AMs has fuelled rational engineering efforts to alter or improve stability25-27, substrate specificity28, and enantioselectivity 29. However, application of combinatorial approaches that leverage evolutionary selection to search large sequence spaces for improved properties30 has not been well-explored for this class of enzymes31,32. Here, we developed a growth-based high-throughput enrichment scheme and screened a mutagenized PAL library to identify variants with improved kinetic properties. Core to this enrichment is the growth rescue of E. coli by PAL in minimal medium with phe, which cannot be used as the sole nitrogen source by K-12 strains33. Consequently, E. coli can only grow if PAL actively deaminates phe to release ammonium, a highly preferred nitrogen-source (Figure 1a). We executed our directed evolution technique using the A. variabilis PAL (DM-rAvPAL/AvPAL*)20, because of its clinical significance, and identified mutants with improved catalytic properties. The mutations identified here have not been previously reported as important for PAL catalytic activity, demonstrating the advantage of our approach for scanning unexplored sequence space over previous efforts.
Figure 1.
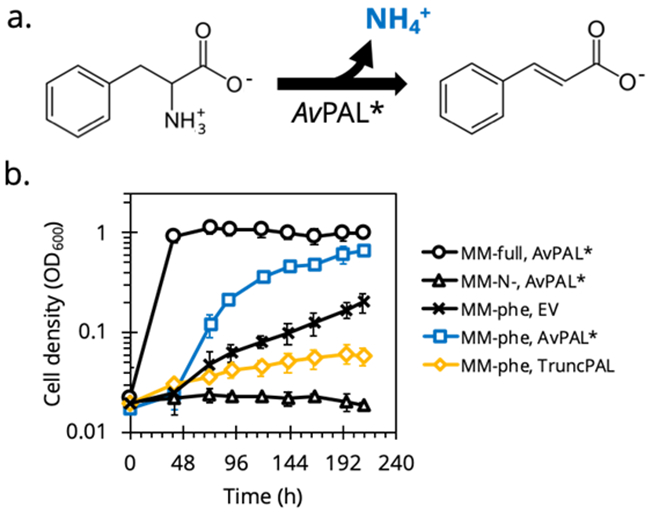
Initial study demonstrating growth-rescue of E. coli by PAL activity. (a.) Growth rescue relies on deamination of phenylalanine by PAL to form ammonium (NH4+), a preferred nitrogen source for E. coli. (b.) E. coli cells expressing active AvPAL* (□) in MMphe,init grow faster than wild-type cells (✕) or those expressing truncated inactive AvPAL* (◇). Cells grown in MMfull,init (○) and MMN-,init (△) as controls.
Initial growth studies demonstrated that AvPAL* could rescue growth of E. coli in phe selective minimal media (MMphe,init) with a ~70% biomass yield relative to complete minimal media (MMfull,init), demonstrating coupling between growth and enzyme activity (Figure 1b). However, controls strains had unexpected, albeit slow, growth. Overexpression of noncatalytic proteins such as green fluorescent protein (sfGFP) or a truncated AvPAL* (TruncPAL) decreased background growth (Figure 1b) but still adversely affected the dynamic range to reliably select for highly active PAL over inactive mutants or other suppressors, if left unoptimized. Phenylalanine metabolism under austere conditions, viz nitrogen starvation, has not been well studied, and transaminases (AspC, IlvE, TyrB, HisC) may have unreported promiscuous activity on phe20. Unfortunately, we observed no difference in the basal growth of E. coli in MMphe,init after deleting each transaminase (Figure S1) suggesting no single gene was responsible for basal growth.
Subsequently, we optimized conditions to increase growth rate and shorten the lag by testing different media formulations (carbon source, pH, strain background, culture volume, phe concentration, and the presence of an additional nitrogen source) (Figure 2a). We initially performed this optimization using MG1655, which grows poorly in minimal media because of inefficient pyrimidine utilization from a mutation in rph33. Switching to a strain with a corrected allele (MG1655rph+) shortened the lag phase by 18 h and culturing in glucose reduced the lag phase by another 24 h (Figure 2b).
Figure 2.
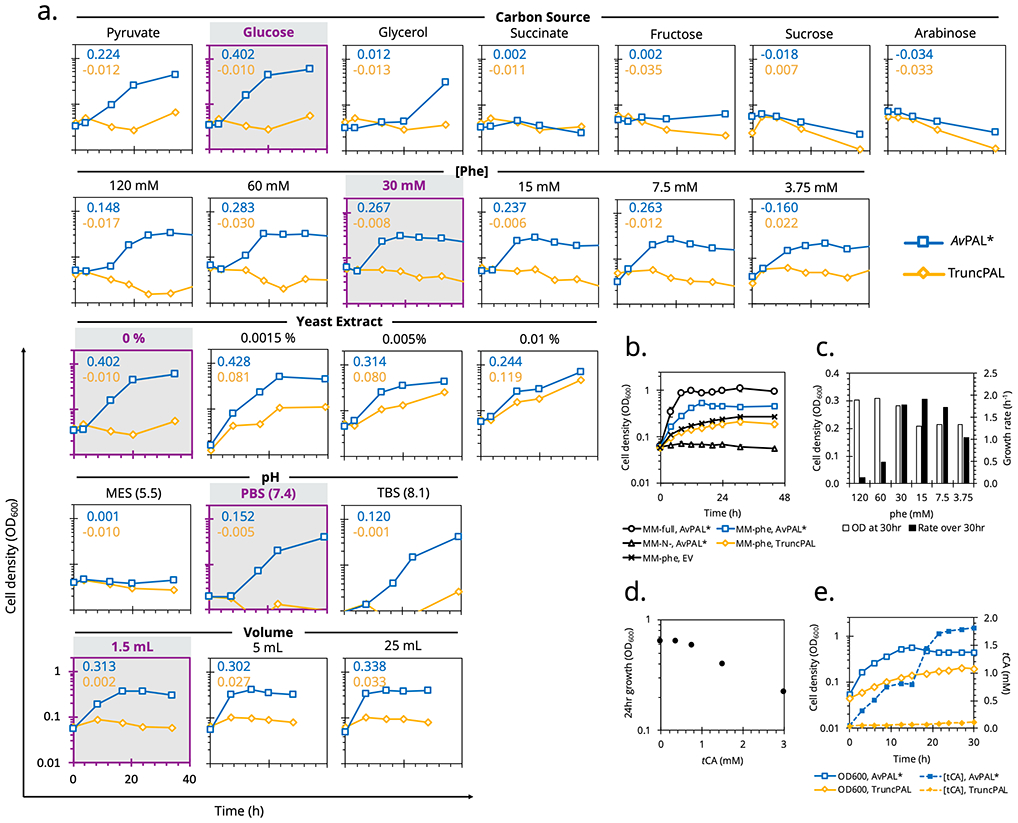
Optimizing conditions of growth-based PAL selection. (a.) E. coli MG1655(DE3)ΔendA,ΔrecA cells expressing AvPAL* (blue) or truncated AvPAL* (orange) were grown in MMphe under different conditions. Growth rates (OD600/day) are in the top right corner with optimum conditions shaded. (b.) The final optimized conditions allowed growth recovery in 12 h compared to 3 d previously. (c.) While optimizing the growth media, we observed that at phe > 30 mM, final biomass density decreased and lag time increased, suggesting toxicity due to rapid accumulation of tCA. At phe < 30 mM, final biomass densities dropped, and when ≤ 7.5 mM, growth rate also slowed, suggesting insufficient nitrogen to sustain growth. (d.) Media supplemented with tCA inhibited the growth of E. coli at concentrations ≥ 1 mM. (e.) AvPAL* expressing cells produce and secrete ~ 1 mM tCA before arresting growth. The subsequent bolus increase in tCA during death phase is likely due to cell lysis. All curves representative of duplicates with less than 10% error.
We found that not only was phe concentration important for optimal growth (Figure 2a,c), but that trans-cinnamic acid (tCA) was toxic to cells. Cells grown in MMfull,opt showed impaired growth when supplemented with 1.5 mM tCA (Figure 2c). When grown in MMphe,opt containing <30 mM phe, growth rate and biomass yield were reduced by low nitrogen availability. However, at phe >30 mM, tCA accumulated too quickly causing toxicity, and the cells not only experience a long lag but also quickly arrest growth (Figure 2c-e). The final optimized conditions in Figure 2e show that despite growth levelling out at OD600 0.3, the lag was virtually eliminated. Thus, we determined that subculturing the cells into fresh medium at OD600 0.2 would minimize tCA toxicity and basal growth — maximizing the difference between inactive and active PAL expressing cells. To validate enrichment occurs under these conditions, we created a mock library by transforming a 1:10 or 1:1000 mixture of AvPAL*-to-sfGFP-expressing. We measured cell fluorescence by flow-cytometry and observed decreasing fluorescence and increasing PAL activity at successive rounds of subculture in MMphe,opt (Figure S3).
After finalizing the conditions to enrich active PAL, we created a mutant library of 105 variants with an average error rate of 2.8 aa/protein. The entire library was grown in MMphe,opt over three rounds, subculturing each time at OD600 of 0.2 (Figure S2). We subsequently plated the cells on non-selective LB medium, picked fifteen random colonies and screened their lysates (Figure S3c) and purified protein (Figure 3a) for PAL activity. Eight of the fifteen, including M222L and L4P/G218S, showed 1.5- to 2-fold higher activity with the other three showing similar activity to parental AvPAL* (Figure 3a). Colonies with same sense or no mutation showed PAL activity similar to that of AvPAL*. This result suggests successful enrichment of higher activity mutants over lower/inactive mutants. E. coli expressing M222L and L4P/G218S mutants showed faster growth compared to AvPAL* in MMphe,opt, with all attaining the same OD600 at stationary phase (Figure 3b). The greater differences in growth profiles at early growth stages between mutants (M222L and L4P/G218S) and parental AvPAL* is consistent with the enrichment strategy of subculturing at low OD600. Furthermore, residues 218 and 222 are directly adjacent to the active site of AvPAL* and in close vicinity of the MIO-adduct20. Comparing the crystal structure of AvPAL* to these mutants shows potential changes in hydrogen bonding within the active site (Figure S4).
Figure 3.
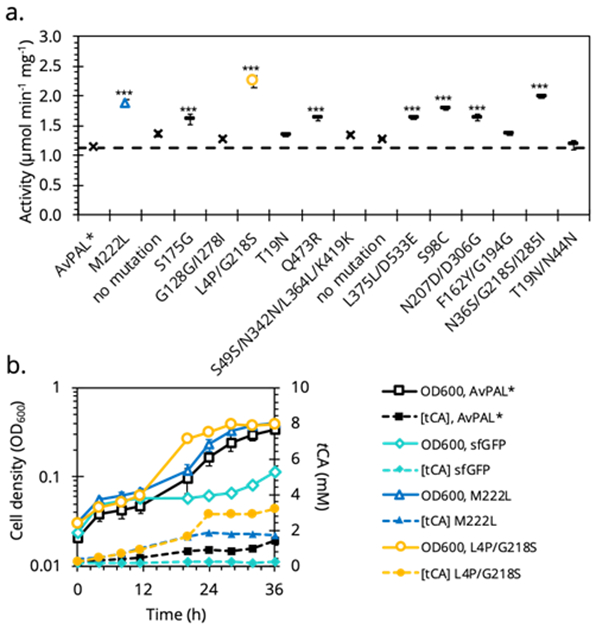
Identification of AvPAL* mutants (–) by growth-coupled enrichment. (a.) Purified PAL activity of 15 randomly picked colonies compared to wildtype. (b.) The growth profiles (solid) and tCA production (dotted) in MMphe,opt of select mutants (○ and △). (*** significant compared to wildtype or silent mutants (✕), p < 0.001)
Previous studies with AvPAL* have demonstrated that kinetic parameters, pH optimum, thermal and proteolytic stabilities are relevant to therapeutic efficacy for PKU enzyme-replacement therapy. The kcat of both the mutants was 70 – 80% higher than parental AvPAL* (Figure 4a) whereas the KM of M222L was similar to that of the parent and that of L4P/G218S was ~2.5× higher. Overall, the M222L mutant showed improved catalytic efficiency compared to AvPAL*, while L4P/G218S mutant showed a trade-off between turnover frequency and substrate “affinity”. AvPAL* is reported to have a pH optimum in the range of 7.5 – 8.520 and we observed similar results for both the mutants (Figure 4b), albeit with a slightly narrower optimal range. Temperature stability was assessed by subjecting the mutants to different temperatures for 1 h before measuring enzyme activity at optimal conditions (37 °C, pH 7.4). The enzymes remained stable from 37 °C to 55 °C and began a modest decrease in relative activity at 65 °C before denaturing at 80 °C (Figure 4c). The proteolytic stability was evaluated by incubating purified enzymes to trypsin. M222L was as trypsin-resistant as AvPAL* but L4P/G218S showed rapid loss of activity within five minutes (Figure 4d).
Figure 4.
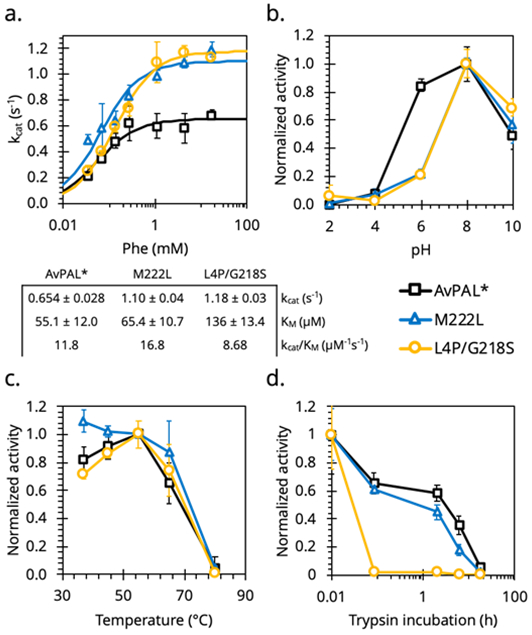
Biochemical characterization of PAL mutants. Two mutants showing higher than wildtype activity were characterized to establish (a.) kinetic parameters, (b.) pH optimum, (c.) temperature optimum, (d.) and resistance to protease degradation.
Our results show that the catalytic properties of this class of enzymes, which are important for both industrial and biomedical applications, can be engineered using directed evolution. Further, the large sequence space we rapidly searched to identify mutations at residues previously unrecognized as functionally important, serves as evidence of this technique’s strength. Since deamination activity serves as the foundation of technique, we offer this method as demonstration that may be applicable to other ALs as well. While the enzyme AvPAL* has specific implications as a therapy for PKU, several other ALs are under clinical investigation to treat disorders and biosynthesize industry chemicals. Our method could further direct AL temperature and pH stability or proteolytic resistance. The improvements in turnover rates observed here are unprecedented in the literature, either through rational or combinatorial methods, and has tremendous translation potential, especially for PKU.
Supplementary Material
Footnotes
Conflicts of interest
Tufts University and all authors have applied for a patent.
References
- 1.Cooke HA, Christianson C. v. and Bruner SD, Current Opinion in Chemical Biology, 009, 13, 453–461. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Kim BG, Kim M and Ahn JH, Journal of Microbiology and Biotechnology, 2015, 25, 1442–1448. [DOI] [PubMed] [Google Scholar]
- 3.Walker KD, Klettke K, Akiyama T and Croteau R, Journal of Biological Chemistry, 2004, 279, 53947–53954. [DOI] [PubMed] [Google Scholar]
- 4.An J, Zhang M and Lu Q, Journal of Food Engineering, 2007, 78, 340–344. [Google Scholar]
- 5.Shin SY, Jung SM, Kim MD, Han NS and Seo JH, Enzyme and Microbial Technology, 2012, 51, 211–216. [DOI] [PubMed] [Google Scholar]
- 6.Hoagland RE, Weed Technology, 1996, 10, 651–674. [Google Scholar]
- 7.McKenna R and Nielsen DR, Food, Pharmaceutical and Bioengineering Division - Core Programming Topic at the 2011 AIChE Annual Meeting, 2011, 1, 340–350. [Google Scholar]
- 8.Verhoef S, Ruijssenaars HJ, de Bont JAM and Wery J, Journal of Biotechnology, 2007, 132, 49–56. [DOI] [PubMed] [Google Scholar]
- 9.Verhoef S, Wierckx N, Westerhof RGM, de Winde JH and Ruijssenaars HJ, Applied and Environmental Microbiology, 2009, 75, 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Wood K. v. and Morgan JA, Applied and Environmental Microbiology, 2005, 71, 2962–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Du G, Zhou J and Chen J, Metabolic Engineering, 2013, 16, 48–55. [DOI] [PubMed] [Google Scholar]
- 12.il Park N, Xu H, Li X, Kim YS, Lee MY and Park SU, Process Biochemistry, 2012, 47, 2575–2580. [Google Scholar]
- 13.Hydery T and Coppenrath VA, Drug Target Insights, , DOI: 10.1177/1177392819857089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castañeda MT, Adachi O and Hours RA, Journal of Industrial Microbiology and Biotechnology, 2015, 42, 1299–1307. [DOI] [PubMed] [Google Scholar]
- 15.US007560263B2, 2009, 2. [Google Scholar]
- 16.Shen R, Fritz RR and Abell CW, Cancer Research, 1977, 37, 1051–1056. [PubMed] [Google Scholar]
- 17.Chesters C, Wilding M, Goodall M and Micklefield J, Angewandte Chemie - International Edition, 2012, 51, 4344–4348. [DOI] [PubMed] [Google Scholar]
- 18.Bell SM, Wendt DJ, Zhang Y, Taylor TW, Long S, Tsuruda L, Zhao B, Laipis P and Fitzpatrick PA, PLOS ONE, 2017, 12, e0173269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F, Huang N, Zhou L, Cui W, Liu Z, Zhu L, Liu Y and Zhou Z, Applied Biochemistry and Biotechnology, 2017, 183, 699–711. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Gamez A, Archer H, Abola EE, Sarkissian CN, Fitzpatrick P, Wendt D, Zhang Y, Vellard M, Bliesath J, Bell SM, Lemontt JF, Scriver CR and Stevens RC, Journal of Molecular Biology, 2008, 380, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovelock SL and Turner NJ, Bioorganic and Medicinal Chemistry, 2014, 22, 5555–5557. [DOI] [PubMed] [Google Scholar]
- 22.Louie G. v., Bowman ME, Moffitt MCC, Baiga TJ, Moore BSS and Noel JP, Chemistry and Biology, 2006, 13, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang L and Moore BS, Journal of Bacteriology, 2005, 187, 4286–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartsch S, Wybenga GG, Jansen M, Heberling MM, Wu B, Dijkstra BW and Janssen DB, ChemCatChem, 2013, 5, 1797–1802. [Google Scholar]
- 25.Wohlgemuth R, Current Opinion in Microbiology, 2010, 13, 283–292. [DOI] [PubMed] [Google Scholar]
- 26.Wu B, Szymanski W, Wietzes P, de Wildeman S, Poelarends GJ, Feringa BL and Janssen DB, ChemBioChem, 2009, 10, 338–344. [DOI] [PubMed] [Google Scholar]
- 27.Turner NJ, Current Opinion in Chemical Biology, 2011, 15, 234–240. [DOI] [PubMed] [Google Scholar]
- 28.Wrenbeck EE, Azouz LR and Whitehead TA, Nature Communications, 2017, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flachbart LK, Sokolowsky S and Marienhagen J, ACS Synthetic Biology, 2019, 8, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reitzer LJ, in Escherichia coli and Salmonella: cellular and molecular biology, ed. Neidhardt FC, American Society for Microbiology, Washington, D.C., 1996, pp. 380–390. [Google Scholar]
- 31.Gelfand DH and Steinberg RA, Journal of Bacteriology, 1977, 130, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzmán GI, Utrilla J, Nurk S, Brunk E, Monk JM, Ebrahim A, Palsson BO and Feist AM, Proceedings of the National Academy of Sciences, 2015, 112, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen K, Journal of Bacteriology, 1993, 175, 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


