Abstract
Fibrous scaffolds fabricated via electrospinning are being explored to repair injuries within dense connective tissues. However, there is still much to be understood regarding the appropriate scaffold properties that best support tissue repair. In this study, the influence of the stiffness of electrospun fibers on cell invasion into fibrous scaffolds is investigated. Specifically, soft and stiff electrospun fibrous networks are fabricated from crosslinked methacrylated hyaluronic acid (MeHA), where the stiffness is altered via the extent of MeHA crosslinking. Meniscal fibrochondrocyte (MFC) adhesion and migration into fibrous networks is investigated, where the softer MeHA fibrous networks are easily deformed and densified through cellular tractions and the stiffer MeHA fibrous networks support ~50% greater MFC invasion over weeks when placed adjacent to meniscal tissue. When the scaffolds are sandwiched between meniscal tissues and implanted subcutaneously, the stiffer MeHA fibrous networks again support enhanced cellular invasion and greater collagen deposition after 4 weeks when compared to the softer MeHA fibrous networks. These results indicate that the mechanics and deformability of fibrous networks likely alter cellular interactions and invasion, providing an important design parameter towards the engineering of scaffolds for tissue repair.
Keywords: hydrogels, electrospinning, fibrous networks, cell migration, meniscus
Graphical Abstract
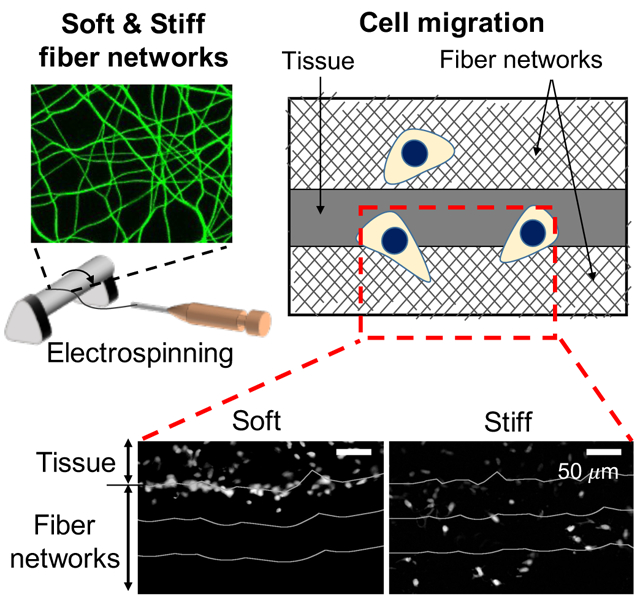
Electrospun fibrous hydrogels are being developed for meniscal tissue repair. To better understand the influence of scaffold biophysical properties on cellular invasion, fiber stiffness is varied. Softer networks are easily deformed by cells to densify fibers, whereas stiffer networks support increased cell invasion both by seeded cells and when placed adjacent to meniscus tissue.
1. Introduction
Biomaterial scaffolds are being designed to act as structural templates to support cellular colonization and new tissue formation in tissue repair strategies.[1–3] For example, fibrous scaffolds are often employed as implantable acellular materials to restore the function and organization of dense connective tissues (e.g., knee meniscus, tendon, cartilage), exploiting their porosity and similarities in structural features to adjacent tissues.[1,2,4] Specifically, nano- to micro-scale polymer fibers can be generated by electrospinning[5] and collected as either random or aligned fibrous networks to mimic the highly organized collagen fibers of these dense connective tissues. Recent advances in biomaterials design and electrospinning techniques have expanded the range of biophysical (e.g., alignment,[6,7] diameter[8,9] and stiffness[10,11]) and biochemical (e.g., adhesion ligand[10–12] and growth factor[13–15]) properties of scaffolds to better explore features that enable expedited tissue repair.
In this approach, the rapid cellularization of scaffolds is desired, so that invading cells are able to initiate matrix deposition and tissue repair as quickly as possible. One challenge to the use of electrospun fibers as a scaffold are the small pores that are formed when fibers are collected, particularly in aligned scaffolds, that can act as a barrier to cell invasion. For instance, the nuclei of adjacent host cells are often too large and too stiff to migrate through the small pores.[1,2] Attenuated cell migration might also favor new tissue formation at the scaffold interface, which could further act as a barrier to hinder cell migration into scaffolds.[16,17]
To address this issue, numerous strategies have been employed in scaffold engineering to increase scaffold porosity and pore size. For instance, porosity has been increased in scaffolds through the spinning of populations of sacrificial polyethylene oxide (PEO) fibers that can be readily dissolved away to retain only lower densities of stable fibers[15,18–20]. Additionally, approaches such as increased fiber diameters (even up to ~6 μm),[21,22] more directed fiber deposition through direct-write electrospinning,[23] and laser-assisted ablation techniques[24] have been utilized to increase scaffold porosity and cell infiltration. Further, either haptotactic[25] or chemotactic[15] gradients have been created through the depths of fibrous scaffolds to direct cellular migration.
Beyond these strategies to create additional porosity and paths for migration into scaffolds, the biophysical properties (e.g., stiffness) of fibers may also be important to increase cell invasion. Indeed, it has been reported that the stiffness of extracellular matrix (ECM) (e.g., collagen gel, Matrigel), adjusted by variation of gelation temperature and ECM concentration, alters the speed of 3D cell migration through the ECM.[26–28] Additionally, the increase of ECM stiffness reduced the number of nascent and retracting adhesions, and promoted the formation of stable adhesions, which promoted 3D cell migration.[26]
In addition to cell responses within ECM environments, synthetic hydrogel fibers with altered fiber mechanics have also been used to study the effects of fiber stiffness on cell behaviors. For instance, soft hydrogel fibers conjugated with RGD peptide (adhesion motif) led to increased focal adhesion formation,[11] cell spreading,[11,29] migration,[30] and proliferation[11] over stiff hydrogel fibers. The ability of cells to deform the softer fibers and increase local fiber density was implicated as a reason for these outcomes. Although these results imply that the stiffness of electrospun hydrogel fibers influence cell behaviors, little work has been completed to investigate how fiber mechanics influence cell invasion, particularly in the repair of dense connective tissues.
In this study, we developed a system to investigate cell invasion through fibrous scaffolds, fabricated through the electrospinning of hyaluronic acid (HA)-based hydrogels where fiber stiffness was varied, while other features such as fiber diameter, adhesion ligand (RGD peptide) density, pore size between fibers, and thickness of fibrous scaffolds were maintained as a constant. Specifically, we investigated how fiber stiffness influenced meniscal fibrochondrocyte (MFC) adhesion and invasion through these fibrous networks either as seeded cells or when migrating from adjacent meniscal tissue.
2. Results and discussion
2.1. Fabrication of soft and stiff hydrogel fibers
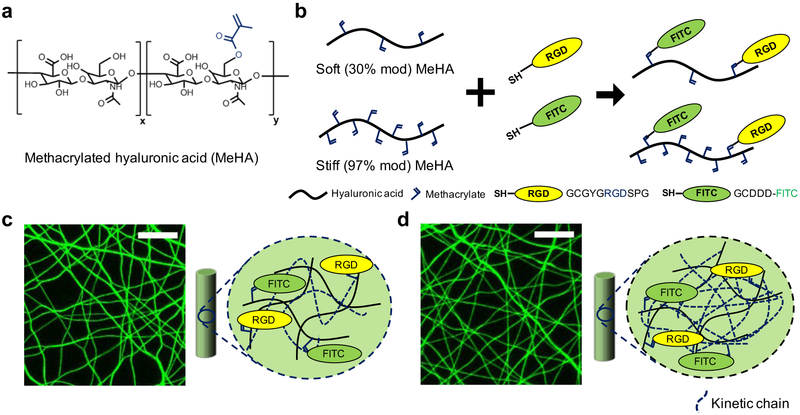
To study the effect of fiber stiffness on meniscal fibrochondrocyte (MFC) migration into dense fibrous networks, methacrylated hyaluronic acid (MeHA) was obtained by modifying the primary hydroxyl group of sodium hyaluronic acid (HA) with methacrylates (Figure 1a). For soft and stiff hydrogel fibers, MeHA macromers either with ~30% (soft) or ~97% (stiff) of methacrylate modification were prepared and conjugated with thiolated fluorescein (for fiber visualization) and RGD peptides (for cell attachment) via a Michael addition reaction between thiol and methacrylate groups (Figure 1b).[29] The peptide-conjugated MeHA macromers were electrospun, stabilized with ultraviolet (UV) light, and hydrated in phosphate buffered saline (PBS).
Figure 1.
Overview of material design for the fabrication of soft and stiff fibrous networks. (a) Hydroxyl groups on hyaluronic acid (HA) were modified with methacrylates to form MeHA at various modification levels (soft: ~30%, stiff: ~97%). (b) Michael type addition reactions were used to conjugate RGD (yellow) and FITC (green) peptides on MeHA. Representative maximum projection images of suspended and hydrated electrospun fibers and schematic showing fiber composition for (c) soft and (d) stiff fibrous networks. Scale bar is 20 μm. Dashed lines indicate kinetic chains generated by methacrylate radical polymerization.
When both soft and stiff fibers were collected under identical electrospinning conditions, the diameters of hydrated soft fibers were ~50% greater than that of stiff fibers due to enhanced swelling given the differences in crosslink density.[29] As differences in fiber diameter could influence cell behaviors, as well as the density of RGD in the fibers and the pore sizes between fibers, this was not acceptable to probe our question related to the influence of fiber mechanics on cellular invasion. To address this, the electrospinning parameters (distance between a needle and the collector, ejection rate of macromer solution) were altered to achieve the same diameter, RGD density, and pore size of hydrated fibrous networks fabricated from the low and high modified macromers.[29] As a result, soft and stiff MeHA single fibers with Young’s modulus (measured from hydrated single fibers via atomic force microscope using three-point bending analysis) of ~0.12 and ~2 GPa, respectively, were obtained, while maintaining the diameters of the two groups at ~830 nm, similar RGD density (indicated by fluorescence intensity of fibers), and pore areas (measured from fluorescent images of fibrous networks) of ~20 μm2 for both hydrated fibers (Figure 1c,d). Notably, the Young’s moduli of these MeHA single fibers are similar to that reported for various single fibrous proteins such as collagens (0.16 ~ 14.7 GPa).[31,32] Although we do not believe it is a concern due to the highly hydrated nature of the fibers, no studies were conducted to examine any differences in fiber hydrophobicity based on different levels of MeHA modification.
2.2. MFC behavior on fibrous networks
Before studying the influence of fiber stiffness on MFC migration into the fibrous hydrogel networks, we first investigated whether the differences in stiffness of soft and stiff fibers influenced MFC adhesion and spreading. MFCs were seeded on thin layers of soft and stiff fibrous networks collected over polydimethylsiloxane (PDMS) wells (Figure 2a and S1a), and the local fiber densities under cells and the cell spreading area were assessed after 1 and 24 hours of culture (Figure 2b, Figure S2). It was shown previously that cells on soft fibrous hydrogel networks increased the local fiber density by deforming adjacent fibers, which in turn promoted cell spreading and focal adhesion formation.[11,29] In contrast, cell spreading on stiff fibrous networks was significantly attenuated as the stiffer fibers reduced the ability of cells to deform their local fibers.
Figure 2.
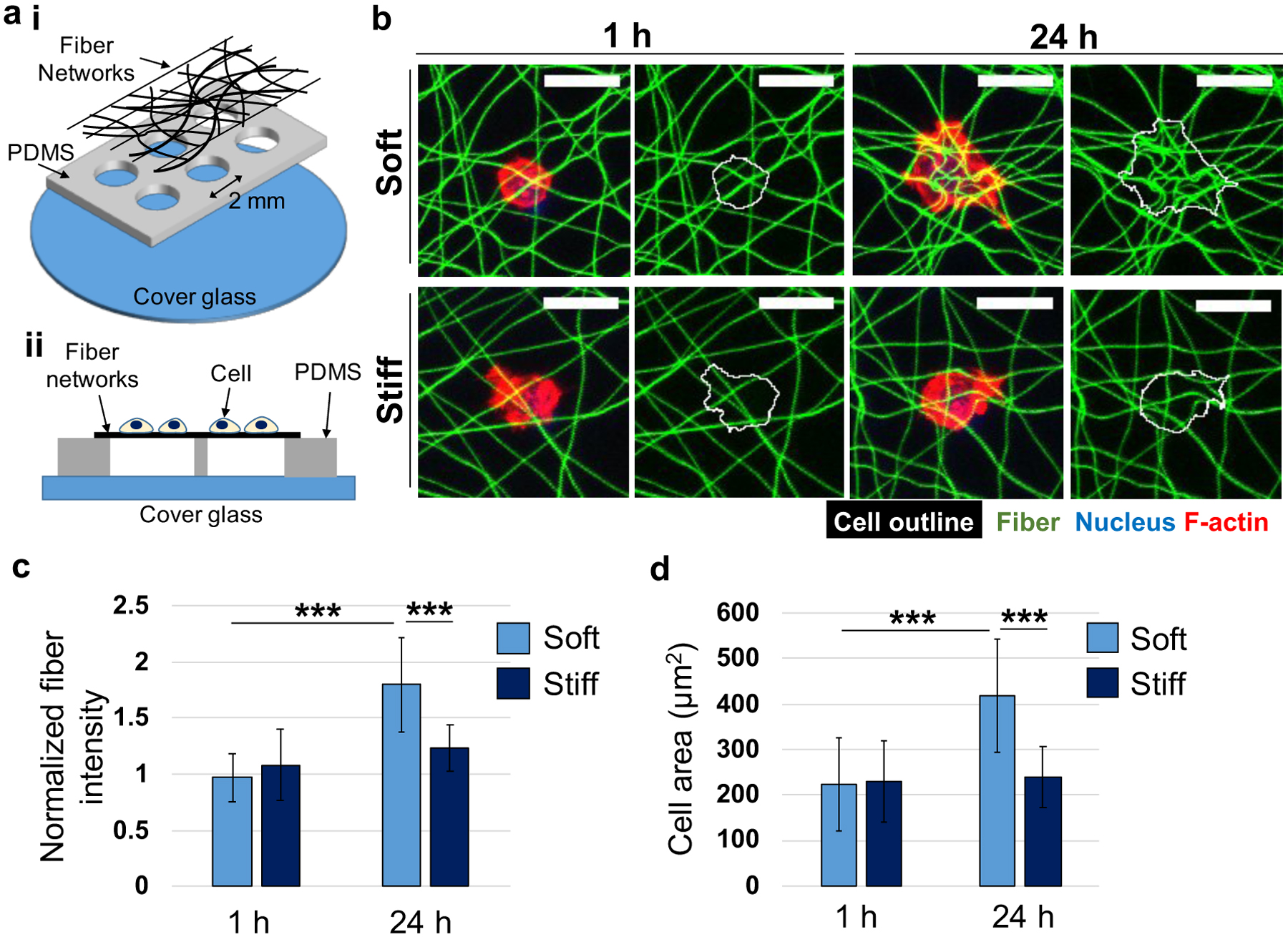
Meniscal fibrochondrocyte (MFC) behavior on thin layers of soft and stiff fibrous networks. (a) Schematic of PDMS wells containing suspended thin fibrous networks, viewed from the (i) top or (ii) side. MFCs were seeded on top of the PDMS wells. (b) Representative maximum projection images of MFCs cultured on soft and stiff fibrous networks (green) for 1 or 24 h. Cells were labeled for nucleus (blue) and F-actin (red) and an outline around each cell was drawn (white). Scale bar is 20 μm. Quantification of (c) normalized fiber intensity (fiber density) at cellular regions and (d) MFC spreading area. Soft, 1 h: n = 24; Stiff, 1 h: n = 19; Soft, 24 h: n = 23; Stiff, 24 h: n = 13 cells. Two-way ANOVA with Tukey’s post hoc testing. ***p < 0.001.
After 1 hour of culture on soft and stiff fibrous networks, the fiber density was not significantly different within the cellular region (~1.0 and ~1.1 for soft and stiff fibrous networks, respectively) (Figure 2c), which is likely too early to see significant deformation of the fibrous networks. However, the normalized intensity of soft fibrous networks after 24 hours was ~1.9 and ~1.4 times higher than that of soft fibrous networks after 1 hour and stiff fibrous networks after 24 hours, respectively, indicating that the density of soft fibrous networks was increased by MFC-mediated fiber deformation, while stiff fibrous networks were less deformed. Similarly, MFC spreading (as measured through cell spread area) on soft and stiff fibrous networks were not significantly different after 1 hour of culture. However, cell spreading on soft fibrous networks increased ~1.9 times between 1 hour and 24 hours of culture, whereas the spreading did not significantly change on stiff fibrous networks over time (Figure 2d).
In addition to cell behaviors on thin layers of fibrous networks, cells on thick layers of soft and stiff MeHA fibrous networks were investigated, moving towards systems with the potential for cell invasion. For this experiment, MFCs were seeded on thick layers of hydrated MeHA fibrous networks attached to PDMS wells (Figure 3a) and observed after 1 and 24 hours of culture. The thickness of hydrated fibrous networks was the same for both fibrous network mechanics (~134 μm) when measured 24 hours after hydration to reach equilibrium swelling; this was achieved by adjusting the time of electrospinning (Figure 3b).
Figure 3.
MFC behavior on thick layers of soft and stiff fibrous networks. (a) Schematic of PDMS wells containing suspended thick fibrous networks, viewed from the (i) top or (ii) side. MFCs were seeded on top of the PDMS wells. (b) Quantification of thickness of hydrated fibrous networks after 24 hours of swelling. NS: not significant. Representative (c) 3D reconstructed and (d) cross-section images of MFCs cultured on soft and stiff fibrous networks (green) for 1 or 24 h. Cells were stained for nuclei (blue) and F-actin (red). Scale bars are (c) 10 μm and (d) 20 μm. White arrows and dotted line in d indicate approximate location of MFCs and top surface of fibrous networks, respectively. (e) Quantification of percentage of MFCs within fibrous networks. n = 4 wells. Two-way ANOVA with Tukey’s post hoc testing. ***p < 0.001.
The 3D reconstructed and cross-sectional images showed that MFCs placed on top of soft fibrous networks after 1 hour were later found within fibrous networks after 24 hours, while MFCs were maintained primarily on the surface of stiff fibrous networks during the 24 hour culture period (Figure 3c,d). Statistical analysis confirmed that the concentration of cells within soft fibrous networks after 24 hours of culture was ~5.0 and ~9.5 times greater than that within soft and stiff fibrous networks after 1 hour, respectively, and was ~3.86 greater than that within stiff fibrous networks after 24 hours (Figure 3e). It is likely that the MFC-mediated deformation of local soft fibers resulted in the pulling of the soft fibers around cells during 24 hours of culture, such that cells appeared to reside within the network at the final time point. Taken together, MFCs on fibrous networks with altered stiffness showed distinct responses, allowing us to investigate the influence of fiber stiffness on MFC migration into fibrous networks.
2.3. MFC migration into fibrous networks
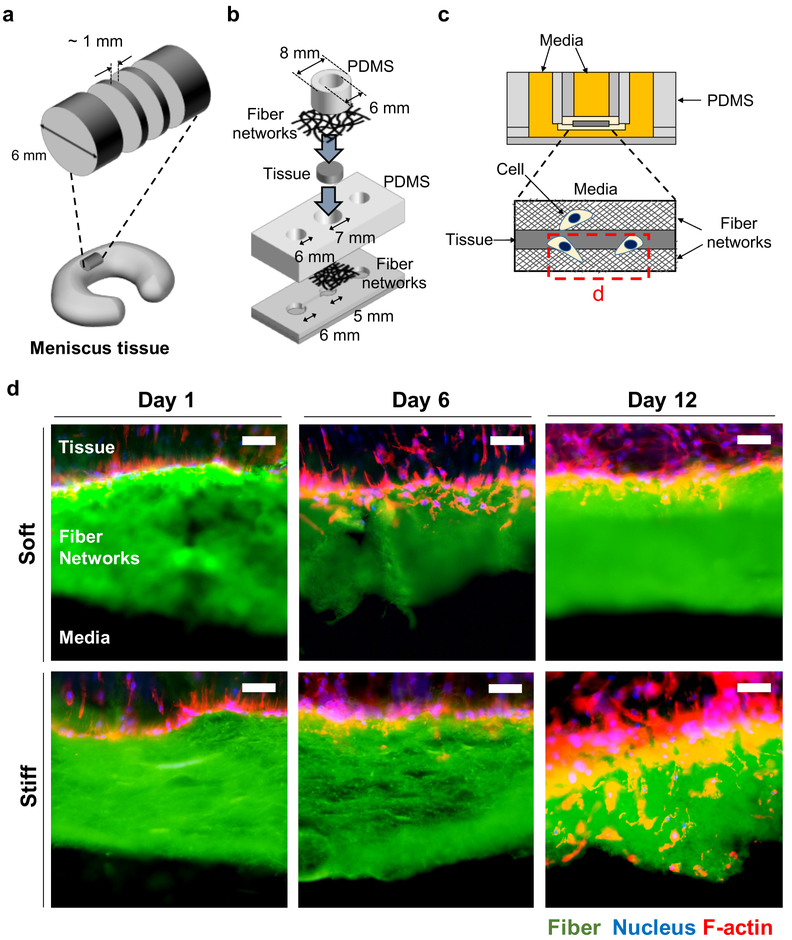
To study the influence of fiber stiffness on cell migration through these fibrous networks, MFC migration from a native meniscal tissue into soft and stiff MeHA fibrous networks was imaged and analyzed. Meniscal tissue explants were isolated with ~1 mm thickness (Figure 4a) and sandwiched between soft and stiff MeHA fibrous networks within a custom-designed cell migration chamber (Figure 4b) and cultured for 1, 6 and 12 days before analysis of MFC invasion (Figure 4c). Representative images of fibrous networks and MFCs (labeled by nucleus and F-actin) showed that cells that were located at the interface between the tissue and soft fibrous networks on day 1 had migrated into the fibrous networks before day 6, while cells adjacent to and encountering stiff fibrous networks migrated through the networks after day 6 (Figure 4d).
Figure 4.
Cell migration chamber to study MFC migration into thick fibrous networks. (a-c) Schematic of (a) slicing meniscal tissue, (b) cell migration chamber sandwiching a tissue slice, and (c) cross-section view of assembled migration chamber. Red dotted box in c indicates acquisition position of images in d. (d) Representative cryo-sectioned images of tissues sandwiched between soft and stiff fibrous networks (green). Cells were fixed on days 1, 6, and 12, and stained for nuclei (blue) and F-actin (red). Scale bar is 50 μm.
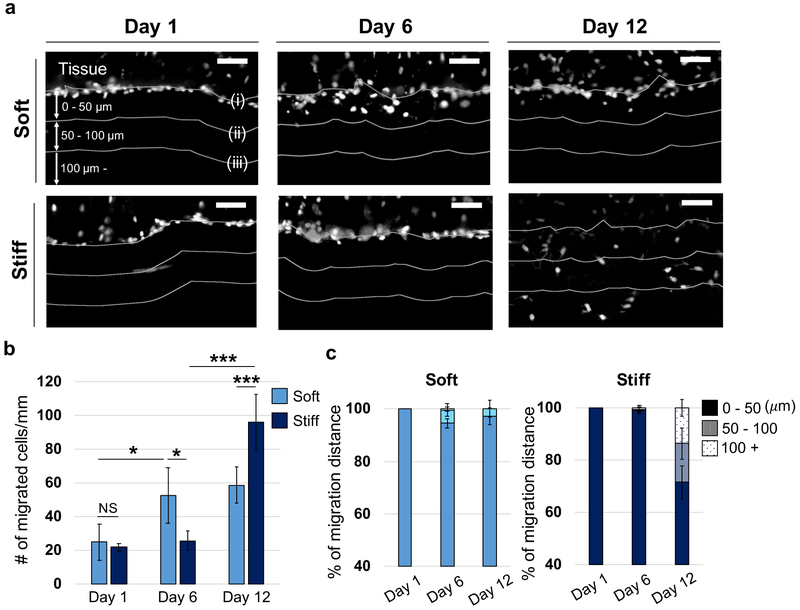
For the quantitative analysis of the influence of fiber stiffness on cell migration, cryo-sections and nuclear images were utilized to analyze MFC density and migration distance within soft and stiff MeHA fibrous networks (Figure 5a). There was no statistical difference between thicknesses of soft and stiff fibrous networks at each time point investigated (Figure S3). In soft fibrous networks, MFC density (number of cells migrated through fibrous networks, normalized by the length of tissue-fiber interface) on day 6 was ~2.1 times greater than the density on day 1, and there were no statistical differences in the density on day 12 (Figure 5b). In contrast, in stiff fibrous networks, MFC density on day 6 was similar to the density on day 1, but increased ~3.7 times on day 12. Additionally, the density within stiff fibrous networks was ~1.6 times greater than that within soft fibrous networks on day 12. Similar to the cell density, the fraction of cells that migrated further than 50 μm from the tissue-fiber interface was significantly increased within soft and stiff fibrous networks on days 6 and 12, respectively (Figure 5c). The fraction of cells that migrated further than 50 μm within stiff fibrous networks was also ~10.0 times greater than that within soft fibrous networks on day 12.
Figure 5.
Quantification of MFC migration into thick fibrous networks using cell migration chambers. (a) Representative images of nuclei (white) in cryo-sectioned tissues sandwiched between soft and stiff fibrous networks. MFCs were fixed on days 1, 6 and 12. White lines indicate (i) interface between tissue and fibrous network, and distances of (ii) 50 or (iii) 100 μm away from the interface. Scale bar is 50 μm. (b) Quantification of number of cells that migrated into the fibrous networks per length of the tissue-fiber interface. n = 5 tissue constructs. Two-way ANOVA with Tukey’s post hoc testing. NS: not significant, *p < 0.05, ***p < 0.001. (c) Quantification of cell migration distance into fibrous networks (reported as percentage of 3 groups (0–50 μm, 50–100 μm, 100 μm-)) across the different days. n = 5 tissue constructs.
Taken together, more MFCs eventually migrated further within stiff fibrous networks than soft networks, although migration from the fibrous interface was slower. It is likely that the softer fibrous networks allowed rapid MFC invasion into the fibrous networks at the interface via MFC-mediated deformation of local fibers, as seen in Figures 2 and 3, but then these increased fiber densities became a hindrance for further migration through the fibrous environments. Indeed, while MFCs were deforming and recruiting soft fibrous networks, local fibers could be clustered or coiled,[11,29] to decrease porosity. Additionally, increased adhesive ligand (RGD peptide) density with fiber recruitment, could also slow down the mobility of MFCs. Although efficient cell migration requires intermediate adhesive ligand density for the balance of cell adhesion and detachment on surrounding fibers,[33,34] the detachment process could be delayed with increased ligand density. In contrast, the stiffer fibrous networks were not significantly deformed by the invading MFCs, allowing cell migration with maintained porosity and ligand density.
2.4. Fibrous scaffolds for a meniscal tissue repair model
The preceding in vitro MFC migration assay indicates that the stiff MeHA fibrous networks allowed more cell invasion than soft MeHA fibrous networks on day 12 (Figure 5b). To verify if this concept is observed in vivo and whether it also leads to differences in the deposition of matrix within scaffolds, soft and stiff MeHA fibrous networks used throughout the MFC migration study were employed as fibrous scaffolds for a meniscal tissue repair study.[15,35] Specifically, soft and stiff MeHA fibrous scaffolds were inserted into partially transected cylindrical meniscus tissues and these repair constructs were implanted subcutaneously in athymic rats and assessed after 4 weeks for location of cell nuclei and deposition of collagen types I and II (Figure 6a,b). Cell density and collagen intensity within fibrous scaffolds were normalized to the cell density and collagen intensity at control regions of the tissue (100 μm width and 200 μm away from scaffolds)[15] to minimize the influence of heterogeneity in cellularity across tissue samples (Figure S4). There was no statistical difference between the thicknesses of implanted soft and stiff fibrous scaffolds (Figure S5).
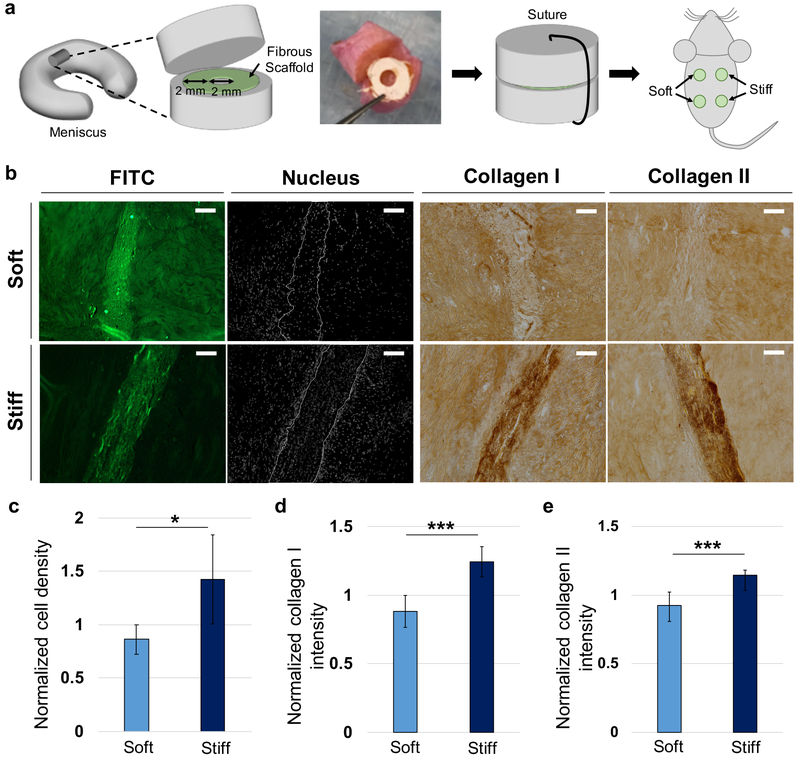
Figure 6.
Application of fibrous networks as scaffolds for meniscal repair. (a) Schematic for implantation of meniscal tissue construct containing soft and stiff fibrous scaffolds. Scaffolds concentrically cut with biopsy punches were inserted into the transected area of meniscal tissue. After suturing, tissue constructs were subcutaneously implanted into athymic rats. (b) Representative images of fibrous scaffold (FITC, green), nuclei (white) and collagen types I and II (dark brown) after 4 weeks of implantation. Scale bar is 100 μm. Quantification of normalized (c) cell density, (d) collagen type I staining intensity, and (e) collagen type II staining intensity within fibrous scaffolds at 4 weeks. n = 6 tissue constructs. Unpaired t-tests. *p < 0.05, ***p < 0.001.
After 4 weeks, the cell density within stiff fibrous scaffolds was ~1.66 times greater than that within soft fibrous scaffolds, implying that stiff scaffolds permitted more cell invasion than soft scaffolds (Figure 6c). Similarly, the normalized intensity of collagen type I and II within stiff fibrous scaffolds was ~1.41 and ~1.23 times higher than that within soft scaffolds, respectively (Figure 6d,e). These trends were similar to those observed in MFC migration within the cell migration chambers, although there are likely differences in migration signals between the assays. MFCs in the cell migration chamber likely entered the fibrous networks to follow gradients of nutrients and oxygen from cell culture media, while MFCs in the meniscus tissue repair study actively migrate into fibrous scaffolds in the presence of diverse chemical signals or complex interactions with adjacent cells.
It has been reported previously with ECM fibers that 3D migration requires a balance of contractility with ECM stiffness to stabilize adhesions.[26] Further, cell migration along fiber-like micropatterns requires stable adhesions.[36] In our synthetic system, adhesion and migration is likely a balance between the ligand density and fiber stiffness, which may change over time with cellular contraction, particularly in soft deformable fibers. On the other hand, the scaffold porosity must also support the passage of cells and particularly their nuclei. Thus, in this study, the stiffer fibrous networks likely met this threshold or exhibited enough deformation for MFC migration, while also minimizing fiber deformation and densification that could impede migration. Overall, the optimal biophysical parameters to support cell invasion and dense connective tissue repair are likely a delicate balance of these factors. In our example, the stiffer fibrous scaffolds would be more useful in meniscal tissue repair strategies than soft fibrous scaffolds due to their higher cellularity and matrix deposition within these scaffolds.
It is important to note that the appropriate biophysical parameters may be directly related to the cells investigated and the material system used. For example, the Young’s moduli of the soft and stiff MeHA fibrous scaffolds (population of MeHA fibers, thickness of ~134 μm) used in this study were previously determined through indentation testing to be ~0.3 and ~2.1 MPa, respectively.[29] This is higher than measured values of juvenile bovine meniscus tissues (~50 kPa),[16] although it is difficult to compare these measurements and to understand the important mechanical properties in the cell microenvironment, particularly when they change over time with densification by cells. It should also be noted that mechanical loading of the scaffolds, which may occur upon implantation of a mechanically active tissues such as the meniscus, may also alter outcomes as this could influence parameters such as scaffold porosity. This study also used HA in fiber fabrication, as HA is involved in diverse biological processes (e.g., proliferation, wound healing),[37,38] is important in cartilage development[39] and meniscal injury,[40,41] and can be chemically modified to tailor biochemical and biophysical properties. For example, HA fibers can be conjugated with adhesive ligands,[10,25,29,42] processed with varied fiber mechanics,[10,29] and incorporate enzymatic[43] and hydrolytic[15] degradation mechanisms. Thus, there is much diversity that can be engineered into scaffolds for tissue repair, and our findings implicate fiber stiffness as a parameter that should be considered.
3. Conclusion
Electrospun fibrous scaffolds have been employed to repair dense connective tissues, exploiting their structural similarities to tissues to allow cellularization. To better understand how scaffold biophysical properties influence the invasion of MFCs into scaffolds, we fabricated soft and stiff fibrous networks from hyaluronic acid with the same fiber diameter, RGD peptide density, pore size, and scaffold thickness. The stiffness of the fibers influenced cell behavior, with increased cell spreading and cell-mediated densification of fibers observed when MFCs were seeded on top of soft fibrous scaffolds, which then reduced MFC invasion into scaffolds from adjacent meniscal tissue. When implanted subcutaneously between pieces of meniscal tissue, cells invaded into both soft and stiff scaffolds after 4 weeks; however, cellularity and collagen deposition were greater in the stiffer scaffold. This suggests that fiber mechanics contributes to the behavior of cells towards the colonization of scaffolds for tissue repair. Somewhat counterintuitively, softer fibers allow for greater fiber deformability, but this can actually reduce cellular invasion if they increase fiber recruitment to reduce local porosity. Ultimately, this work provides additional information towards our understanding of the design of scaffolds for connective tissue repair.
4. Experimental Section
Materials:
Hydrogel macromers and thiolated peptides were prepared as described previously.[29] Briefly, methacrylated hyaluronic acid (MeHA, Figure 1a) was obtained by methacrylate esterification with the primary hydroxyl group of sodium hyaluronic acid (HA, 75 kDa, Lifecore Biomedical). The degree of methacrylate modification (Soft: ~30%, stiff: ~97%) was altered through the amount of methacrylic anhydride introduced during synthesis and was determined via 1H NMR (Bruker, 360 MHZ). Thiolated fluorescein peptides (GCDDD-carboxyfluorescein) were synthesized using standard FMOC chemistry and thiolated RGD peptides (GCGYGRGDSPG) were purchased from Genscript. Michael addition reactions between thiol groups from peptides and methacrylate groups from MeHA were employed to conjugate both fluorescent and RGD peptides to the MeHA macromers. After 4 days of dialysis at 4 °C, peptide-conjugated MeHA macromers were isolated by freezing and lyophilizing.
Fabrication of fibrous networks:
MeHA fibrous networks were obtained by electrospinning solutions of 4 wt% MeHA, 3.5 wt% polyethylene oxide (PEO, 900 kDa, Sigma), and 0.05 wt% Irgacure 2959 (Ciba Specialty Chemicals) in deionized water.[29] The distance between the needle and collector and the ejection rate of the polymer solution were varied to achieve a diameter of ~830 nm for soft and stiff MeHA fibers, while voltages (needle: +26–27 kV, collector: −3–4 kV, deflector: +6–8 kV) were maintained constant. To stabilize the fibers, the networks were purged with nitrogen and the top and bottom of the scaffolds were exposed to ultraviolet light (15 mW cm−2, 320–390 nm, Omnicure S1500, Excelitas Technologies) for 20 min each.
Meniscal fibrochondrocyte (MFC) isolation and culture:
To isolate MFCs, juvenile bovine menisci (2~3 months, Animal Technologies Inc.) were dissected, minced into ~1 mm3 cubes, and placed in tissue culture plates containing Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) with 10% fetal bovine serum and 1% penicillin, streptomycin, fungizone (PSF).[44] Minced tissues were incubated at 37 °C and MFCs gradually emerged from the tissue segments over 2 weeks. MFCs at passage 3 were used throughout the experiments.
Cell behaviors on thin and thick fibrous networks:
To assess MFC behaviors on thin scaffolds, soft and stiff MeHA fibers were directly collected on methacrylated polydimethylsiloxane (PDMS) (DOW Corning) wells (2 mm in diameter) during electrospinning and exposed to light as above for 20 minutes (Figure 2a). The networks were hydrated in PBS for 2 days, seeded with 5 × 104 MFCs, and analyzed. To assess MFC behaviors on thick scaffolds, ~0.9 ml and ~1.75 ml polymer solution for soft and stiff fibers, respectively, were electrospun on foil to obtain thicknesses of ~134 μm. After light exposure to the top and bottom of the fibrous networks for 20 minutes each, scaffolds were cut with a 10 mm biopsy punch and attached to PDMS wells (5 mm in diameter) via a thin layer of PDMS cured at interfaces (overnight at 37 °C) (Figure 3a). The networks were hydrated on the PDMS wells in PBS for 2 days, seeded with 2.5 × 104 MFCs, and analyzed. Cells on thin and thick fibrous networks were fixed after 1 or 24 hours of culture at 37 °C and stained with Hoechst 33342 (5 μg ml−1, Invitrogen) and rhodamine phalloidin (1:200, Invitrogen) to visualize nuclei and F-actin, respectively.
Fluorescence images of nuclei, F-actin and fibers were acquired using a confocal microscope (Leica TCS SP5). To analyze fiber deformation, Image J was used to determine the fluorescence signal of cellular and control (acellular) regions on maximum projection images of fibers (Figure S2). The signal of the cellular regions was normalized to the signal of the control regions to account for the heterogeneity in distributions of fibers. Additionally, maximum projection images of F-actin were analyzed to obtain cell spreading areas via Image J. To quantify cells within fiber networks, confocal images of nuclei, F-actin and fibers were 3D reconstructed and used to determine whether cells were located within or above the fiber networks. Specifically, cells with fibers over their nuclei were regarded as cells located within fiber networks.
MFC migration assay using cell migration chambers:
Cell migration chambers consisting of top and bottom parts were fabricated to observe cell migration into fibrous networks from meniscal tissue (Figure 4a). For the bottom parts, two layers of PDMS blocks containing holes and a channel were cast using 3D printed molds purchased from Proto Labs (Accura SL 5530 with post-thermal curing process). To assemble the PDMS blocks and fibrous networks, interfaces were coated with a thin layer of a mixture of PDMS base and curing agent and cured overnight at 37 °C. For the top parts, 6 mm diameter PDMS wells were punched with biopsy punches (Integra Miltex, Kai Medical) and attached to fibrous networks via a thin layer of PDMS cured at the interfaces. Juvenile bovine menisci were vertically cut with ~4 mm thickness and horizontally punched with 6 mm biopsy punches. The tissue constructs in cylindrical shape were sliced to ~1mm thickness, sandwiched between the top and bottom parts of the cell migration chamber, and the reservoirs were filled with cell culture media. Tissue constructs were fixed on days 1, 6 and 12, sequentially immersed in 30% sucrose, a mixture of 30% sucrose and optimum cutting temperature (OCT, Sakura Finetek) compound (1:1 ratio), and OCT compound for one day at 4 °C, and frozen with 2-methylbutane (Sigma) and liquid nitrogen. The frozen tissues were sectioned to 30 μm thickness and stained with Hoechst 33342 (5 μg ml−1, Invitrogen) and rhodamine phalloidin (1:200, Invitrogen) for nucleus and F-actin imaging, respectively. For consistency, cells migrating into fibrous networks at the bottom were imaged and analyzed in the study. For analysis, the interfaces between meniscus tissues and fibrous scaffolds were determined by fluorescence images of the scaffolds, and used to draw additional lines (50 or 100 μm away from the interfaces) on fluorescent images of nuclei via Image J. Nuclei on the fluorescent images were analyzed to quantify cell location and migration distance.
Meniscal tissue repair model study:
To investigation migration into fibrous scaffolds in vivo, a meniscal tissue repair model previously developed was utilized.[15,35] All of the animal studies were approved by the Animal Care and Use Committee of the Corporal Michael Crescenz VA Medical Center. Meniscal tissue constructs were cut into cylindrical shapes (diameter: ~8 mm, height: ~4 mm), partially transected at ~2 mm, and fibrous scaffolds concentrically cut using 2 and 6 mm biopsy punches were inserted into the transected meniscus tissue and secured in place using absorbable sutures (Y497G, Ethicon) (Figure 6a). Four tissue constructs containing soft or stiff fibrous scaffolds were subcutaneously implanted into athymic rats (RH-Foxn1rnu, 8–10 week-old, ~300 g, Envigo). Rats were euthanized via CO2 asphyxiation at week 4 and tissue constructs containing scaffolds were retrieved. Tissue constructs were paraffin embedded, sectioned to 10 μm thickness and kept at 4 °C. To image nuclei, sectioned tissues were deparaffinized and stained with Hoechst 33342 (5 μg ml−1, Invitrogen). To visualize collagen type I and II deposition, sectioned tissues were deparaffinized, treated with hyaluronidase (300 μg ml−1) and proteinase K for 60 min and 4 min, respectively, and antibodies applied for collagen type I (10 μg ml−1, Millipore) or II (10 μg ml−1, Developmental Studies Hybridoma Bank, University of Iowa) overnight at 4 °C. The next day, tissues were sequentially incubated with a secondary antibody, streptavidin HRP, and chromogen reagent for 10 min. Image J was used to determine the scaffolds and control (100 μm width and 200 μm away from the scaffold) regions, and analyze the number of nuclei and intensity of collagen types I and II (Figure S4). For cell density and collagen intensity, the number of nuclei and intensity of collagen within the scaffold regions were normalized to the number of nuclei and intensity of collagen within control tissue regions, respectively, to minimize the heterogeneity in cellularity across the samples.
Fluorescence imaging:
To analyze MFC behaviors on thin and thick fibrous networks, fibrous networks and MFCs were imaged at 25x on a Leica TCS SP5 confocal microscope with 1 μm step size of z-stacks. Imaging of MFCs, fiber networks (or fibrous scaffolds) and collagen type I and II from the cell migration assay was performed at 20x on an Olympus BX51 epifluorescence microscope.
Analysis of thickness of fibrous networks/scaffolds:
Fluorescent images of cryo-sectioned fibrous networks after 1, 6, and 12 days of the MFC migration assay within cell migration chambers or sectioned fibrous scaffolds after 4 weeks of implantation were analyzed to obtain their thicknesses. Thicknesses of 5 regions of each fibrous networks/scaffolds were measured via Image J and averaged as the representative thickness values for the sample. 5 or 6 replicates individual samples were analyzed for each condition.
Statistical analysis:
Graphpad Prism 8 software was utilized for statistical analysis throughout the experiments. Data are presented as mean ± standard deviation. Significance in data comparison was determined by two-way ANOVA with Tukey’s post hoc testing or unpaired t-tests, where a P value < 0.05 was considered statistically significant.
Supplementary Material
5. Acknowledgements
The authors thank Dr. Michelle Kwon for assistance in immunohistochemistry. This work was supported by the National Science Foundation through the Center for Engineering MechanoBiology STC (CMMI: 15-48571) and the UPenn MRSEC program (DMR-1120901), as well as through the National Institutes of Health (R01 AR056624, F32 DK117568).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Kwang Hoon Song, Department of Bioengineering, University of Pennsylvania, 210 South 33rd Street, Philadelphia, PA 19104, USA; Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA.
Su-Jin Heo, Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA; McKay Orthopaedic Research Laboratory, Department of Orthopaedic Surgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Translational Musculoskeletal Research Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA.
Ana P. Peredo, Department of Bioengineering, University of Pennsylvania, 210 South 33rd Street, Philadelphia, PA 19104, USA Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA; McKay Orthopaedic Research Laboratory, Department of Orthopaedic Surgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Translational Musculoskeletal Research Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA.
Matthew D. Davidson, Department of Bioengineering, University of Pennsylvania, 210 South 33rd Street, Philadelphia, PA 19104, USA Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA.
Robert L. Mauck, Department of Bioengineering, University of Pennsylvania, 210 South 33rd Street, Philadelphia, PA 19104, USA Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA; McKay Orthopaedic Research Laboratory, Department of Orthopaedic Surgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Translational Musculoskeletal Research Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA.
Jason A. Burdick, Department of Bioengineering, University of Pennsylvania, 210 South 33rd Street, Philadelphia, PA 19104, USA Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA USA; Translational Musculoskeletal Research Center, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA.
References
- [1].Mauck RL, Burdick JA, Ann. Biomed. Eng 2015, 43, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qu F, Guilak F, Mauck RL, Nat. Rev. Rheumatol 2019, 15, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Place ES, Evans ND, Stevens MM, Nat. Mater 2009, 8, 457. [DOI] [PubMed] [Google Scholar]
- [4].Liu W, Thomopoulos S, Xia Y, Adv. Healthc. Mater 2012, 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sill TJ, von Recum HA, Biomaterials 2008, 29, 1989. [DOI] [PubMed] [Google Scholar]
- [6].Park SH, Kim MS, Lee B, Park JH, Lee HJ, Lee NK, Jeon NL, Suh KY, ACS Appl. Mater. Interfaces 2016, 8, 2826. [DOI] [PubMed] [Google Scholar]
- [7].Ray WZ, Siewe DY, Liu W, MacEwan MR, Xie J, Xia Y, ACS Nano 2010, 4, 5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Levorson EJ, Raman Sreerekha P, Chennazhi KP, Kasper FK, Nair SV, Mikos AG, Biomed. Mater 2013, 8, 014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim BS, Park KE, Kim MH, You HK, Lee J, Park WH, Int. J. Nanomedicine 2015, 10, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim IL, Khetan S, Baker BM, Chen CS, Burdick JA, Biomaterials 2013, 34, 5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA, Chen CS, Nat. Mater 2015, 14, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim TG, Park TG, Tissue Eng 2006, 12, 221. [DOI] [PubMed] [Google Scholar]
- [13].Kim IL, Pfeifer CG, Fisher MB, Saxena V, Meloni GR, Kwon MY, Kim M, Steinberg DR, Mauck RL, Burdick JA, Tissue Eng. Part A 2015, 21, 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sahoo S, Ang LT, Goh JCH, Toh SL, J. Biomed. Mater. Res. - Part A 2010, 93, 1539. [DOI] [PubMed] [Google Scholar]
- [15].Qu F, Holloway JL, Esterhai JL, Burdick JA, Mauck RL, Nat. Commun 2017, 8, 1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Q, Wang C, Han B, Qu F, Qi H, Li CY, Mauck RL, Han L, J. Biomech 2018, 72, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Qu F, Li Q, Wang X, Cao X, Zgonis MH, Esterhai JL, Shenoy VB, Han L, Mauck RL, Sci. Rep 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Baker BM, Gee AO, Metter RB, Nathan AS, Marklein RA, Burdick JA, Mauck RL, Biomaterials 2008, 29, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Baker BM, Shah RP, Silverstein AM, Esterhai JL, Burdick JA, Mauck RL, Proc. Natl. Acad. Sci 2012, 109, 14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Phipps MC, Clem WC, Grunda JM, Clines GA, Bellis SL, Biomaterials 2012, 33, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Z, Cui Y, Wang J, Yang X, Wu Y, Wang K, Gao X, Li D, Li Y, Zheng XL, Zhu Y, Kong D, Zhao Q, Biomaterials 2014, 35, 5700. [DOI] [PubMed] [Google Scholar]
- [22].Soliman S, Sant S, Nichol JW, Khabiry M, Traversa E, Khademhosseini A, J. Biomed. Mater. Res. - Part A 2011, 96 A, 566. [DOI] [PubMed] [Google Scholar]
- [23].Lee J, Jang J, Oh H, Jeong YH, Cho DW, Mater. Lett 2013, 93, 397. [Google Scholar]
- [24].Lee BLP, Jeon H, Wang A, Yan Z, Yu J, Grigoropoulos C, Li S, Acta Biomater 2012, 8, 2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sundararaghavan HG, Burdick JA, Biomacromolecules 2011, 12, 2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM, Nat. Commun 2015, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wolf K, te Lindert M, Krause M, Alexander S, te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P, J. Cell Biol 2013, 201, 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P, Proc. Natl. Acad. Sci 2006, 103, 10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davidson MD, Song KH, Lee M-H, Llewellyn J, Du Y, Baker BM, Wells RG, Burdick JA, ACS Biomater. Sci. Eng 2019, 5, 3899. [DOI] [PubMed] [Google Scholar]
- [30].Wang WY, Davidson CD, Lin D, Baker BM, Nat. Commun 2019, 10, 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guthold M, Liu W, Sparks EA, Jawerth LM, Peng L, Falvo M, Superfine R, Hantgan RR, Lord ST, Cell Biochem. Biophys 2007, 49, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang L, Van Der Werf KO, Koopman BFJM, Subramaniam V, Bennink ML, Dijkstra PJ, Feijen J, J. Biomed. Mater. Res. - Part A 2007, 82, 160. [DOI] [PubMed] [Google Scholar]
- [33].Lautscham LA, Kämmerer C, Lange JR, Kolb T, Mark C, Schilling A, Strissel PL, Strick R, Gluth C, Rowat AC, Biophys. J 2015, 109, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Singh SP, Schwartz MP, Lee JY, Fairbanks BD, Anseth KS, Biomater. Sci 2014, 2, 1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Qu F, Lin J-MG, Esterhai JL, Fisher MB, Mauck RL, Acta Biomater 2013, 9, 6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM, J Cell Sci 2012, 125, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Burdick JA, Prestwich GD, Adv. Mater 2011, 23, H41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim IL, Mauck RL, Burdick JA, Biomaterials 2011, 32, 8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].DeLise AM, Fischer L, Tuan RS, Osteoarthr. Cartil 2000, 8, 309. [DOI] [PubMed] [Google Scholar]
- [40].Suzuki Y, Takeuchi N, Sagehashi Y, Yamaguchi T, Itoh H, Iwata H, Arch. Orthop. Trauma Surg 1998, 117, 303. [DOI] [PubMed] [Google Scholar]
- [41].Dernek B, Kesiktas FN, Duymus TM, Diracoglu D, Aksoy C, J. Phys. Ther. Sci 2017, 29, 1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wade RJ, Bassin EJ, Gramlich WM, Burdick JA, Adv. Mater 2015, 27, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wade RJ, Bassin EJ, Rodell CB, Burdick JA, Nat. Commun 2015, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mauck RL, Martinez-Diaz GJ, Yuan X, Tuan RS, Anat. Rec 2007, 290, 48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.