Abstract
The ability for biological cells to produce mechanical forces is important for the development, function, and homeostasis of tissue. The measurement of cellular forces is not a straightforward task because individual cells are microscopic in size and the forces they produce are at the nanonewton scale. Consequently, studies in cell mechanics rely on advanced biomaterials or flexible structures that permit one to infer these forces by the deformation they impart on the material or structure. Here, we review the scientific progression on the use of deformable materials and deformable structures to measure cellular forces. We summarize the findings and insights made possible with these approaches in the field of cell mechanics.
1. Introduction
Biological cells are dynamic entities as they are able to crawl, contract, and probe their physical surroundings through the generation of cytoskeletal forces.[1] Their ability to generate these forces is essential to the development, function, and maintenance of tissue for it provides a mechanism for cells to migrate to areas of need or pull together to provide shape and integrity to a tissue.[2,3] Failure to produce an appropriate degree of force can lead to abnormalities such as developmental defects, poor tissue function, or diseased states.[4,5] The wide interest in cellular forces and their importance in cell biology has fueled the development of tools that can measure these forces in individual cells and in multicellular structures.
Cellular forces arise from regions of tension within the cytoskeleton that are transmitted to points of adhesion with the extracellular matrix (ECM) or between neighboring cells. Cytoskeletal tension is due to the motor activity of bipolar myosin filaments as it binds to two or more actin filaments, causing them to move past each other in an antiparallel fashion.[6] If a cell is in free suspension or unconnected to another material, the movement of cytoskeletal filaments due to actin-myosin interactions causes the shape of a cell to contract. However, when a cell is attached to structures within the ECM or to adjacent cells, the movement of cytoskeletal filaments is restrained, but a contractile force is still present. By definition, the tension in the cytoskeleton produces cellular forces that act on the surroundings of the cell (cell-matrix forces) or at the junctions between cells (cell-cell forces). These forces can range from the scale of piconewtons to several hundreds of nanonewtons depending on the size and maturation of the adhesion site as well as the degree of actin-myosin activity within a cell.
The tools used to measure cellular forces, often termed traction force microscopy (TFM), rely on a fundamental principle of physics attributed to Robert Hooke, who was also the first to record the discovery of cells in Micrographia in 1665.[7] In accordance with Hooke’s law for elasticity and springs, one can quantify the amount of force applied to an elastic material or structure by observing the deformation that is causes. However, the measurement of cellular forces is a difficult task because of their microscopic size and transmission of force to multiple points of adhesion that are nanoscale. Moreover, cells are living entities that respond to physical stimuli through a process known as mechanotransduction, which can be defined as the generation of biological responses by the effect of applied forces or the mechanical properties of the microenvironment.[8] Perhaps one of the most fascinating and simultaneously challenging aspects of studying cellular forces is that they can change in response to the mechanical properties of the tool itself. The adaptability of cells to the tools used to measure them makes it difficult to characterize the natural state of cells. Thus, one of the major goals of cell mechanics is not only to measure cellular forces, but also to identify the mechanism by which cells sense, interpret, and respond to physical stimuli.
In this review, we provide an overview of the deformable materials and structures used to measure cellular forces during migration and contraction. In both sections, we highlight how these studies have helped to reveal how cellular forces are regulated and what roles they play in the biological function of cells. We close with a future outlook on the emerging directions in cell mechanics.
2. Deformable materials
The first approaches to measure cellular forces were performed on deformable materials. It is common with these approaches to seed cells onto the surface of a substrate that has adhesive ligands for cellular attachment. As cell generate cell-matrix forces at their locations of adhesion, they cause the substrate to deform. Since a cell’s adhesions are often distributed within the cell-substrate interface, they can be described as tractions, i.e. force per unit area. The magnitude and direction of cellular forces are inferred by the amount of deformation at its surface (Figure 1A).
Figure 1:
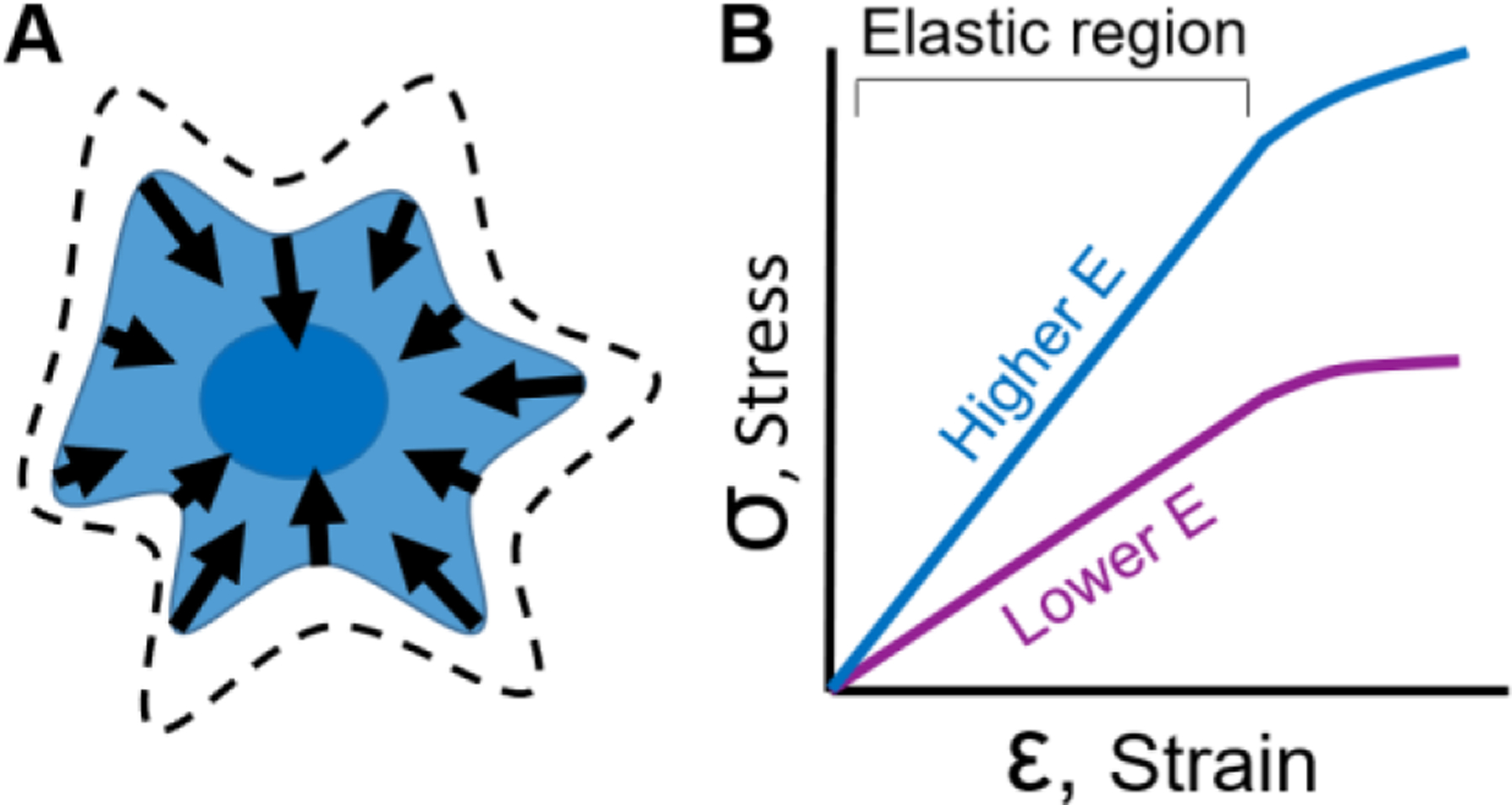
Cellular forces can be measured using deformable materials. (A) A cell grown on the surface of a deformable material produces contractile forces that causes displacements on the surface of the material. Dashed lines represent the projected cell area before contraction. Arrows indicate the location, direction, and magnitude of the forces produced by a cell. (B) To measure cellular forces, elastic materials are often used which have a linear relationship between applied stress and observed strain. The slope of this linear relationship is referred to as the modulus of elasticity or Young’s Modulus (E). To achieve the same amount of strain in a soft (lower E, purple line) or stiff material (higher E, blue line), more stress needs to be applied to the stiff material. Once the Young’s Modulus of a material is known, one can measure deformation caused by cellular forces and infer the magnitude and direction of those forces.
To infer cellular forces from material deformation, a key material relationship must be characterized: the proportion in which applied forces cause deformations to a material. This relationship is usually described as stress versus strain (Figure 1B) where stress (σ) is the applied force divided by the area over which it is applied and strain (ε) is the ratio change in distance between points on a surface. Knowing the stress versus strain behavior of a material enables one to calculate the modulus of elasticity (E) or Young’s Modulus, which is the slope of the stress versus strain data in its linear elastic region (Figure 1B). It is ideal to use materials whose chemistry and manufacturing process lead to stable and consistent material properties. Materials historically and commonly used include polysiloxane (silicone), polyacrylamide, native ECM, and engineered gels, which will be described in the sections below.
2.1. Polysiloxane deformation
The first work to measure cellular forces involved seeding cells on a thin membrane of polysiloxane, more commonly known as silicone rubber. As a cell contracted or migrated, the membrane deformed to the point that it buckled or wrinkled (Figure 2A).[9] This technique was used to compare forces of different cell types, noting that fibroblasts produced the most wrinkles (Figure 2B), while macrophages were less contractile and cause no wrinkles on the surface.[10] The authors of this seminal work made their deformable substrates by exposing a film of polysiloxane fluid to a flame in order to crosslink a thin membrane at the surface of the polymer. Composed of a silicon-oxygen backbone, polysiloxane is considered to be biologically inert and non-toxic. Additionally, the material is linearly elastic within the range of forces produced by cells. Polysiloxanes are also relatively unsusceptible to problems due to biochemical exchange, such as shrinkage. Perhaps their greatest advantage is their transparency to optical and fluorescence microscopy, which make it possible to observe the cells as well as the wrinkles they cause.[9]
Figure 2:
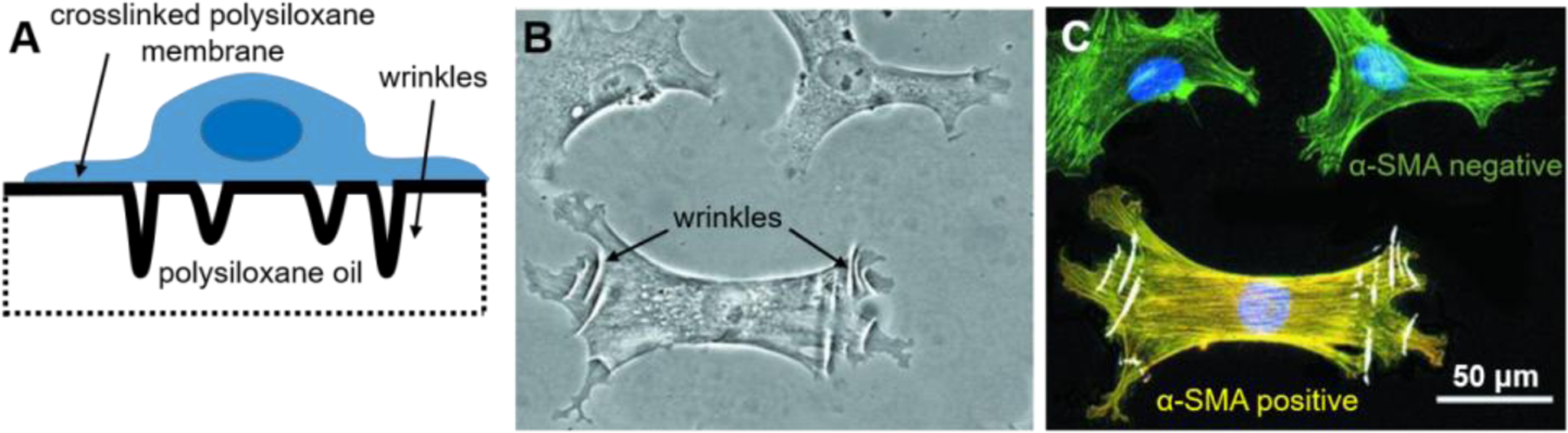
The first seminal method to study cellular forces involved silicone wrinkling. (A) Cells were seeded on a polysiloxane fluid that was briefly exposed to a flame to crosslink a thin surface. Cellular forces deformed the membrane and caused to buckle or wrinkle. (B) Wrinkles in the silicone membrane can be visualized under light microscopy. (C) When used in conjunction with fluorescent microscopy, the magnitude of cellular forces can be correlated with the expression of biomarkers. For example, a fibroblast that is positive for α-smooth muscle actin (SMA) (yellow) create cellular forces that produce visible wrinkles, unlike those that are negative for α-SMA (green). B and C are adapted under the terms of the Creative Commons Attribution 3.0 Unported.[14] Copyright 2001, The American Society for Cell Biology.
Others expand on the original wrinkling method by using UV irradiation to reduce the stiffness of the polysiloxane membranes, making it more permissible to wrinkling at a lower range of cellular forces.[11,12] This method was used to measure cellular forces during cytokinesis[11] and to examine the dynamics of cellular forces during cell locomotion[12]. Silicone wrinkling has also been used to assess the regulation of myosin activity by caldesmon[13] and to identify α-smooth muscle actin as a biomarker of contractile myofibroblasts[14] (Figure 2B–C). This wrinkling method was seminal to the field and was a simple method to adopt by others, requiring only silicone oil and a flame, but also necessitated a fair degree of skill to produce membranes that were consistent in thickness and stiffness between substrates.
In order to gain more quantitative data on cellular forces, a method was developed in which wrinkling is avoided and latex beads are embedded into the top surface of a polysiloxane membrane to visually track the deformations in the substrate (Figure 3A).[15] Specifically, latex beads were brushed into the liquid surface of polysiloxane and then a glow discharge device was used to form a crosslinked membrane that encapsulated the beads. Beads in an undeformed, zero-displacement state (before cellular deformation) were imaged as a reference image (Figure 3B). Keratocytes then migrated into an area and displaced the beads. Bead displacement was calculated by comparing an image of displaced beads to the reference image of the zero-displacement state (Figure 3C–D). Using this technique, it was observed that the largest forces occurred at the marginal edges of the cell, not at the extended front edge.[15–17] In addition, this technique facilitated the first quantitative data of the magnitude (length of arrow) and direction (orientation of arrow) of cellular forces.[15–18]
Figure 3:
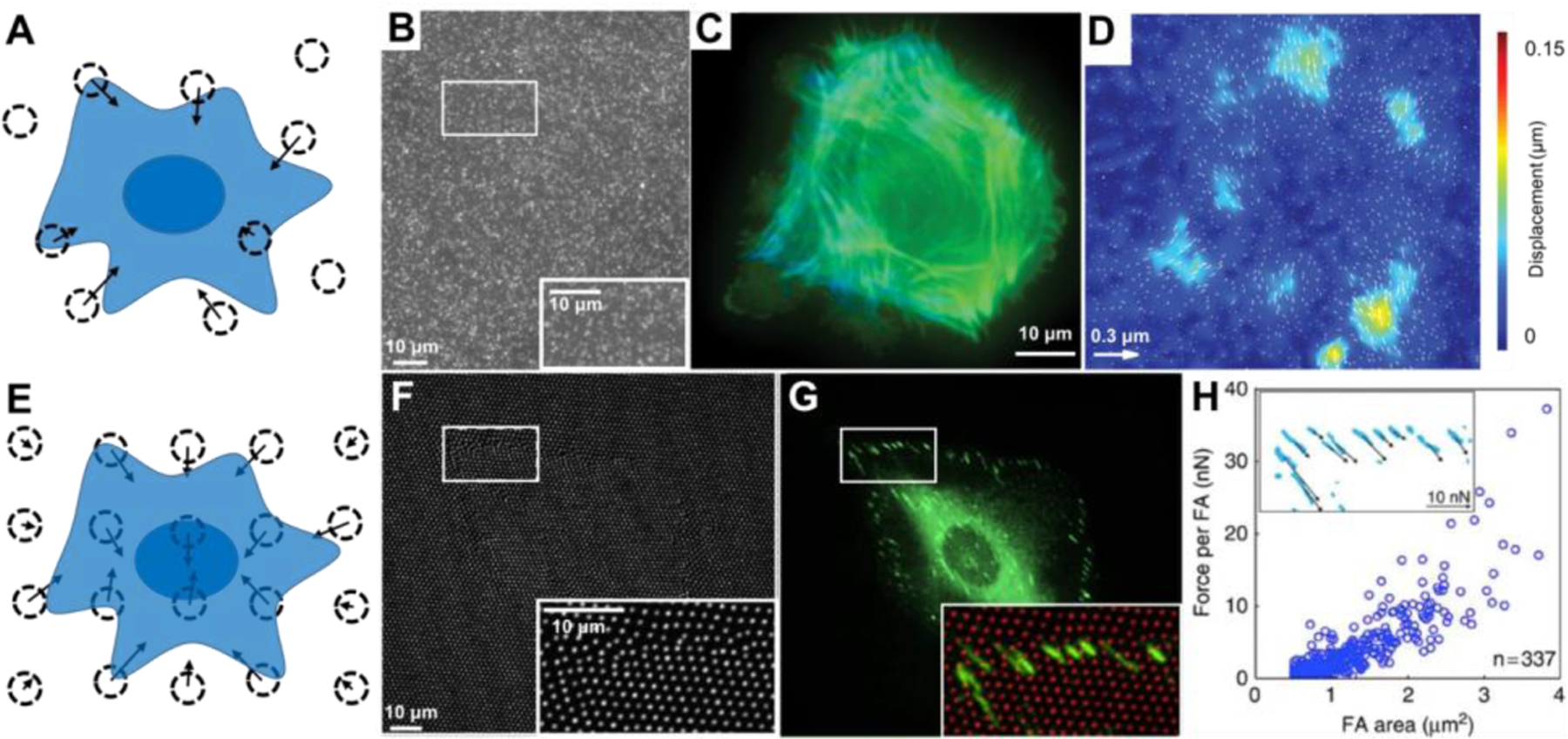
Cellular forces have been measured by the displacement of beads embedded in polysiloxane. (A) Surface displacements can be calculated from the movement of beads randomly embedded in the material. Dotted circles indicate the bead location after cellular displacement and the arrows indicate the magnitude and direction of their displacement. (B-D) Fluorescent microscopy allows for the analysis of bead displacements (white dots in B) and spatially arrangement of actin filaments (green and blue in C, where blue is the actin closer to the surface). Bead displacement vectors can be calculated (arrows in D) and overlaid on a colormap to visualize areas of large displacements (colors in D, units of colormap are μm.). (E) Beads can also be placed in a non-random orthogonal or hexagonal pattern. (F-H) Fluorescent microscopy allows for imaging the displaced position of the bead (white dots in F and red dots in the inset in G) along with fluorescent microscopy of their focal adhesions (FA) by paxillin (green in G) to examine the relationship between FA area and cellular forces (graph in H, inset in H shows zoom region of FAs with force vectors indicated location, direction, and magnitude). B-D are adapted under the terms of Creative Commons Attribution 4.0 International License.[18] Copyright 2011, Public Library of Sciences. F-H are adapted under the terms of Creative Commons Attribution 4.0 International License.[19] Copyright 2016, Springer Nature.
To avoid the challenge of taking an image of the substrate in its undeformed, zero-displacement state as a reference image, a pattern of beads or markers have been used for reference-free traction force microscopy[20] (Figure 3E). Because this method involves the elimination of the reference image, it is termed “reference-free.” Rather than generating randomly spaced markers, reference-free traction force microscopy involves creating an orthogonal or hexagonal pattern of beads or visual markers and measuring their displacement from the grid. The pattern reduces the technical challenge of documenting the zero-displacement location of the markers by either 1) imaging the substrate before adding cells or 2) removing cells and re-imaging. This method was used to understand the relationship of focal adhesion size and force generation (Figure 3F–H).[19,20]
2.2. Polyacrylamide deformation
After over a decade and a half since the first seminal work using polysiloxane, a new material was advanced to measure cellular forces: polyacrylamide gels.[21,22] Polyacrylamide is a hydrogel polymer composed of acrylamide (-CH2CHCONH2-) units. The authors of this work chose it because it is elastic over a wide range of cellular forces and its stiffness can be tailored by changing the amount of cross-linker (bis-acrylamide). Similar to polysiloxane, polyacrylamide has optical qualities that permit optical and immunofluorescent microscopy and its surface can be coated with one or more types of ECM proteins to permit cellular adhesion.[22]
Early studies with polyacrylamide found that the stiffness of a substrate can cause a mechanotransduction response in cells that affects the extent to which they are able to spread, how fast they can migrate, and their ability to form focal adhesions that are strong and stable.[21] Initially, non-fluorescent latex beads were embedded into the polyacrylamide gels and used to measure cellular forces[22], and later fluorescent beads were used.[23] Reference-free traction force microscopy, which overcomes the computational challenge of imaging twice and back calculating deformation, was first conducted on polysiloxane[20] and can also be employed on polyacrylamide.[24] In addition to patterning latex markers, adhesion molecules on the surface can be controlled by patterning extracellular matrix proteins to simultaneously measure cellular forces while also characterizing the effect of adhesion molecules on cellular forces.[25,26] To increase throughput, markers can be spaced such that the cell binds to a location over two markers, pulling them together, and simplifying the analysis to increase throughput.[27] Ultimately, polyacrylamide deformation for traction force microscopy has been used for a variety of applications and for a variety of cell types[28–38] and many would agree that it is currently the most widely used traction force microscopy method.
2.3. Native ECM and engineered gel deformation
Since cells live in a 3D environment in vivo, there is a strong interest in the use of deformable materials that can be used for measuring cellular forces and within which cells can be cultured. Materials derived from native ECM such as collagen can be used to make a hydrogel. Unlike polysiloxane or polyacrylamide, these native ECM gels permit the diffusion of gas and nutrients for cell survival. For example, epithelial cells and tissues were embedded in type I collagen with fluorescent markers.[39] Cellular forces were inferred from bead displacement to find that single cells generate tension (pulling) in 3D and that multicellular tissues can cause compression (pushing).[39] Additionally, biopolymer gels composed of collagen, fibrin, or matrigel were used to measure cellular forces in 3D and to show that breast carcinoma cells generate constant force during exposure to varying collagen concentrations and matrix stiffnesses.[40] This somewhat contradicts findings on 2D platforms in which stiffer substrates induced more cell spreading and more mature (spread and organized) focal adhesions.[41,42]
While ECM proteins provide a physiologically relevant environment to cells, they often behave in a non-elastic manner, making them mechanically complex. Additionally, seeded cells may degrade, synthesize, and/or remodel native proteins, leading to unclear interpretations about how material deformation relates to cellular force. To avoid these complications, engineered synthetic gels were created that are mechanically stable (mechanical properties are minimally by cells) but that also promote cell adhesion. As an example, a polyethylene glycol (PEG)-based hydrogel with proteolytically degradable domains and adhesive domains to facilitate cell invasion and spreading was used to measure cellular forces in 3D.[43] This showed that cells in 3D probe the extracellular environment with long, slender cellular extensions that produce large tractions.[43] Using a similar PEG hydrogel functionalized with arginine-glycine-aspartic acid-serine (RGDS)[44], cellular force measurements were conducted where cells are seeded on top of a substrate (such that the cell is in a 2D, not fully encapsulated environment) and displacement of markers is measured in 3D (to reveal 3D cellular forces); this is referred to as 2.5D.[45] This revealed out-of-plane forces at the focal adhesions, which have not been observed in 2D measurements.[45]
As will be discussed in Section 3.4, it is also possible to conduct measurements of multiple cells that act as a tissue while embedded in native ECM or engineered gel. This is distinct from the methods discussed in this section in that rather than measuring cell-induced deformation of a material (by visualizing wrinkles or marker displacement), this involves measuring the deformation of a structure.
3. Deformable Structures
A deformable material can be used to craft a deformable structure, commonly shaped as a beam or cantilever. When a cell applies a force on the structure, it causes the structure to bend or deflect, which results in a visible displacement at the point where the force is applied. According to Hooke’s law, the displacement (δ) can be used to measure the amount of force (F) by
| (1) |
where the spring constant (k) of the structure is empirically tested or mathematically determined by the geometry of the structure and the modulus of elasticity of its material. Deformable structures have been constructed with a range of different shapes and size in order to suit a particular application or type of cell; these devices include microposts, silicon cantilevers, nanoposts, tissue posts, and fibrous matrices.[46]
3.1. Microposts
Microposts are an array of cantilevers made from polydimethylsiloxane (PDMS) that can be used to measure cellular forces by quantifying their deflections (Figure 4A).[47] Based on the close spacing between the posts, a cell can spread out and attach to the tips of multiple posts, allowing one to measure its forces at several locations simultaneously since microposts are able to deflect independently from their neighbors.[48] Based upon the amount of deflection at the tip of a micropost and its direction, one can calculate the force generated by a cell using Hooke’s law (Equation 1), where the spring constant is k = 3πEd⁴/64L3, E is the modulus of elasticity of PDMS, d is the diameter, and L is the length of the post (Figure 4B). Notably, this method is a direct calculation of force and overcomes some of the mathematical challenges associated with measuring forces using traction force microscopy.[49,50]
Figure 4.
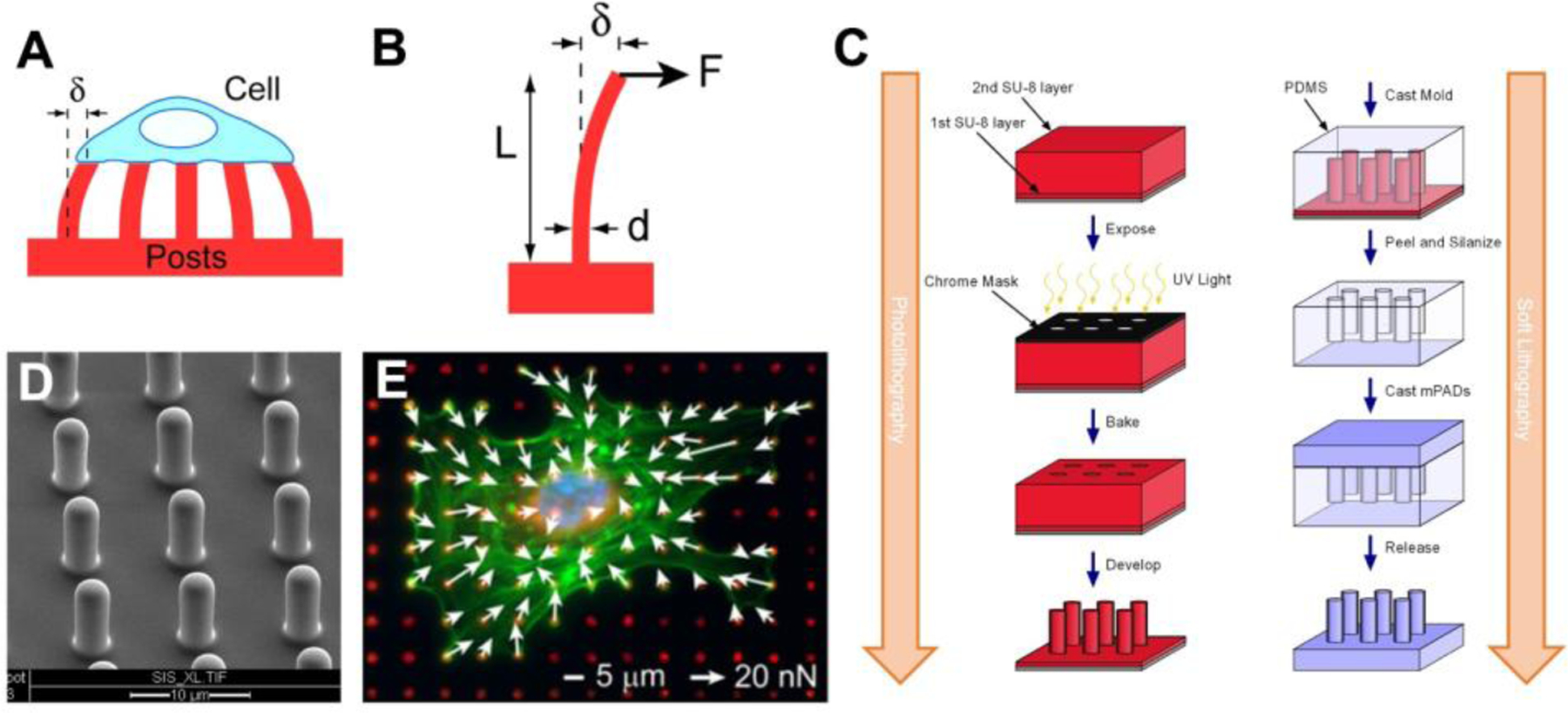
Cellular forces can be measured using microposts. (A) Cells deflect the microposts as they contract and the amount of force can be calculated from the deflection of the posts (δ). (B) Each post is a cantilever beam of length (L) and diameter (d) that deflects in proportion to the force applied at its tip (F). (C) Using photolithography techniques, a master structure of the microposts is made using SU-8 photoresist on a silicon wafer by exposing it to UV light through a chrome mask, baking it to crosslink the SU-8, and then using a solvent to develop the final master structure by removing the unexposed SU-8. Next, soft lithography techniques are used create a negative mold of the microposts by casting them in PDMS, then fluorosilanizing the mold and using it to cast the final PDMS structure. (D) An array of PDMS microposts imaged by scanning electron microscopy. (E) Cellular forces are measured by quantifying the defection of the posts which have been labeled with a fluorescent dye (red). White arrows indicate direction and magnitude of the forces, green is F-actin staining, and blue marks the nucleus. C is adapted with permission.[51] Copyright 2007, Elsevier. E is adapted with permission.[52] Copyright 2012, Elsevier.
Microposts are manufactured using photolithography to build a master structure and soft lithography to produce the final structures in PDMS (Figure 4C). Multiple PDMS substrates can be generated from the same master with sequential casting of negative PDMS molds, which ensures that the dimensions of the microposts are similar between experiments (Figure 4D). Alternatively, PDMS microposts can be created by directly casting from a silicon master that has holes instead of posts.[53] After casting, the spring constant of the microposts can be estimated by the length and diameter of the final structures and the modulus of elasticity of PDMS.[54,55] Calibrating the spring constant can also be determined empirically, e.g. using atomic force microscopy (AFM), but often these direct measurements are difficult to perform due to the size and scale of the microposts.[55]
To quantify cell forces, one typically confines the adhesion of the cells to the tips of the microposts. Arrays of microposts are prepared for cell seeding by stamping fibronectin or another ECM protein on the tips of the posts to support the formation of focal adhesions. The sides of each post are typically treated with Pluronic F-127 to prevent protein adsorption and thereby limit adhesion. Depending on the application, alternative techniques have been used to encourage cell adhesion, such as gold-tipped microposts[56] or sacrificial layers.[57] Using optical or fluorescence microscopy approaches, the deflection of the micropost can be recorded for fixed and stained samples (Figure 4E) or with live-cell imaging (Figure 5A). Image analysis routines are used to track the distance between the centroid of each micropost and its undeflected position (Figure 5B) in order to determine its deflection (Figure 5C).[53] In this manner, it is possible to measure the magnitude and direction of the force at each post by equation 1 for a single image (Figure 4E) or for video data (Figure 5D).[58]
Figure 5.
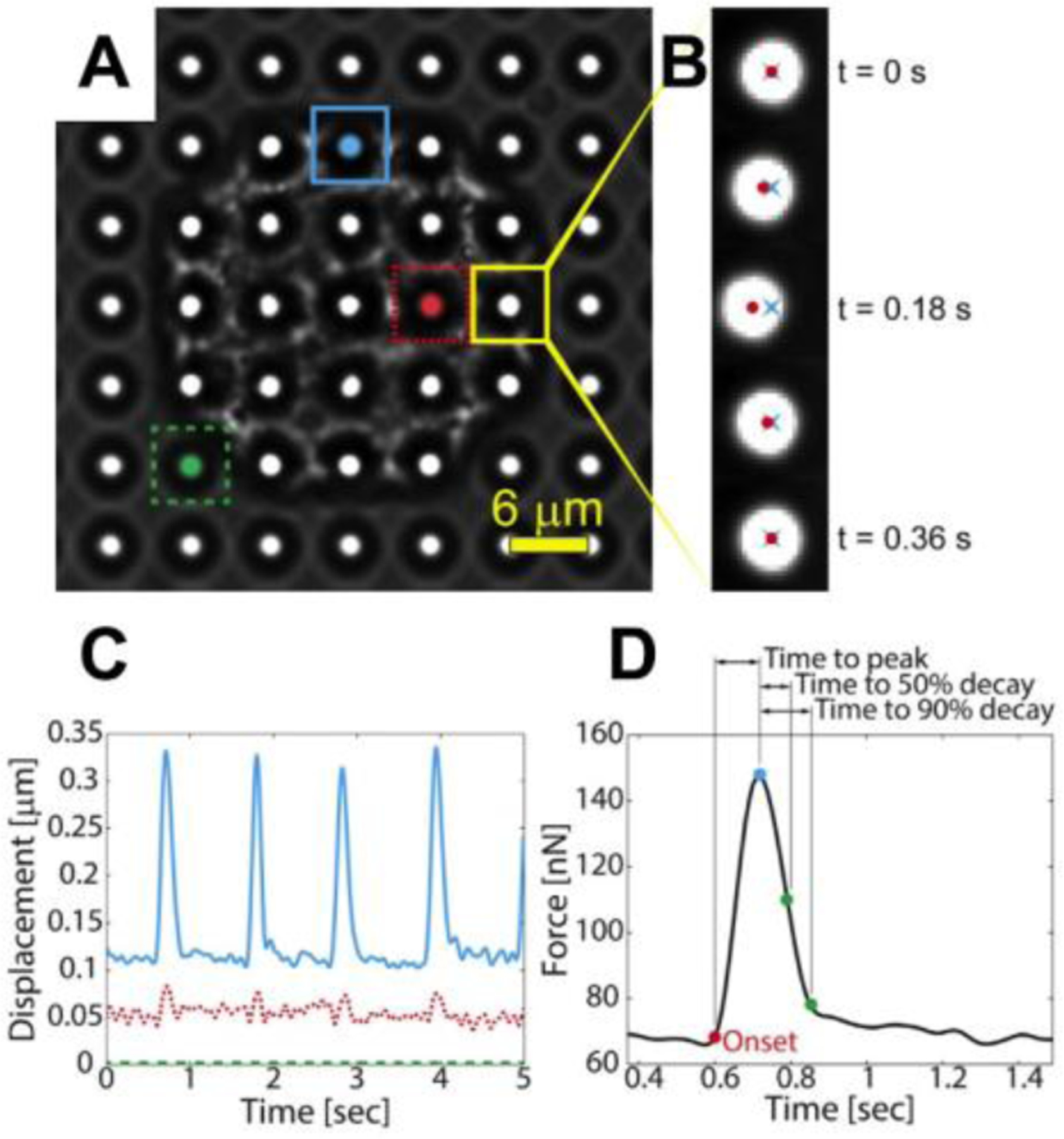
Quantification of cellular forces by tracking the displacement of the microposts. (A) A single cardiomyocyte is shown on microposts. (B) The displacement of a post is tracked over time by determining the distance from the centroid of the tip of the post (red dot) to its undeflected position (blue “x”). (C) The dynamics if a cell’s contractile forces can be monitored as shown by the displacement vs. time and (D) force vs. time graphs. Posts near the boundary of the cell (blue) tend to deflect more than posts near the center of the cell (red), while posts where the cell is not attached do not deflect (green). Reproduced with permission.[59] Copyright 2016, Elsevier.
Microposts can be a versatile tool for measuring cellular forces. With immunofluorescent techniques, they have been used to examine signaling pathways that regulate cell-matrix forces and focal adhesion size[47,60,61] Using optical microscopy for live cell imaging, they can be used to quantify the dynamics of twitch contractions in neonatal rat cardiomyocytes[57,59,62–64] and pluripotent stem cell-derived cardiomyocytes.[57,65–68] Microposts can also be used to measure the forces of platelet aggregates that form on the tips of microposts in an effort to understand the mechanisms that regulate thrombus formation.[69–71] The versatility of microposts is further detailed in the following sections that highlight a number of studies on mechanotransduction cues by altering the geometry of the posts, changing the stiffness or topography of the extracellular environment, or by applying an external force.
3.1.1. Controlling extracellular stiffness of microposts
Cells form linkages known as focal adhesions with their extracellular environment, creating tension within the cytoskeleton and exerting a force on the surrounding environment. During cell migration, these forces permit a cell to probe its environment and acquire feedback via its integrin receptors and mechanotransduction proteins, allowing the cell to adjust its behavior in response to mechanical cues in its extracellular environment.[72] In fact, cell-matrix forces are largely dependent on substrate stiffness; typically, larger forces are observed on stiffer posts.[73] The correlation between substrate stiffness and force has been observed not only at the cellular level, but also at the molecular level using microposts and a vinculin-based molecular tension sensor that uses Förster resonance energy transfer (FRET).[74] By changing the mechanical properties of the microenvironment – height, diameter, shape, and spacing of microposts – one can examine how the extracellular environment impacts differentiation, growth, and migration.
To mimic changes in the extracellular environment, the stiffness of the posts can be altered by changing their height and diameter. Varying the stiffness of posts is used to study cellular response to mechanical stimuli, such as stem cell differentiation and cell growth. For example, mesenchymal stem cells, which can become cartilage, bone, muscle, or fat cells, have been observed to “feel” stiffness and differentiate accordingly.[75] Longer posts are softer, mimicking fat, while short pillars are more rigid, like bone. Substrate stiffness in conjunction with other factors cues stem cell differentiation. Additionally, by utilizing posts of ellipsoidal cross-sectional shape to create higher stiffness in the direction of its major axis, cells have been observed to grow in the direction in which their microenvironment is more rigid.[76] Likewise, cells favor migration and growth towards stiffer posts in studies using adjacent posts with differentiable height, spacing and diameter.[77–79]
3.1.2. Regulating cell shape, configuration, and migration on microposts
As demonstrated by micropost studies, biochemical cues also drive cell behavior. By strategically stamping fibronectin in a pattern on the posts, one can control the direction of cell migration, or the ability of cells to spread and form cell-cell contact. For example, by confining cells to spread within a confined area (Figure 6 A–D), cell-matrix forces were found to be dependent on spread area as well as substrate stiffness (Figure 6 E).[52] The mechanotransduction effect of spread area and stiffness also influenced the size of focal adhesions within a cells, confirming that force and adhesion size are closely related.
Figure 6.
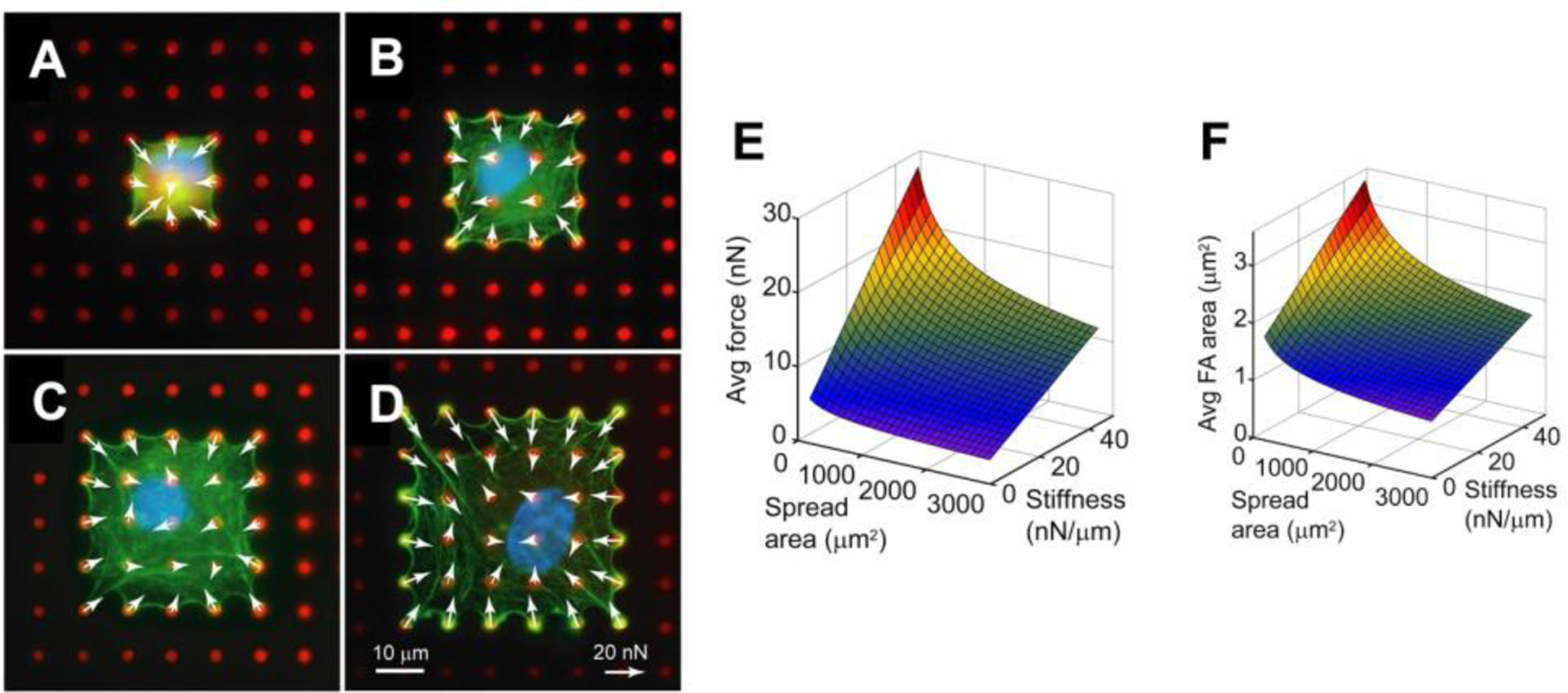
Human pulmonary artery endothelial cells confined to areas of (A) 441 μm2, (B) 900 μm2, (C) 1521 μm2, and (D) 2304 μm2. The red dots are microposts and white arrow represent force vectors. Surface plots indicate that the (E) average cell-matrix force and (F) average focal adhesion area has an inverse relationship with spread area and stiffness. Adapted with permission.[52] Copyright 2012, Elsevier.
The ability for cells to generate cell-cell forces may regulate tissue organization and structure.[80] These forces act perpendicular to the surface of cell-cell contact, forming an adherens junction which relies on myosin-dependent tension. Stamping fibronectin in a bowtie pattern on the tips of microposts can constrain cell-cell contact to one interface (Figure 7 A–D). The size of the adherens junctions between a pair of endothelial cells was observed to be in proportion to the amount of cell-cell force acting at the junction, i.e. tugging force (Figure 7E).[80] Modulation of cytoskeletal tension through actin, myosin, or microtubules had a direct effect on junction size (Figure 7 E).[80] However, when cell-cell forces in epithelial cells were measured using TFM, a correlation with the size of adherens junctions was not observed.[81] Instead, there was a correlation between cell-matrix and cell-cell forces. Microposts were later used to measure cell-cell forces at desmosomes in cancer cells and a correlation between cell-cell force and junction size was observed.[82] These observations indicate that cell-cell junctions are sensitive to cellular forces, but it may not be common to all cell types or experimental conditions.
Figure 7.
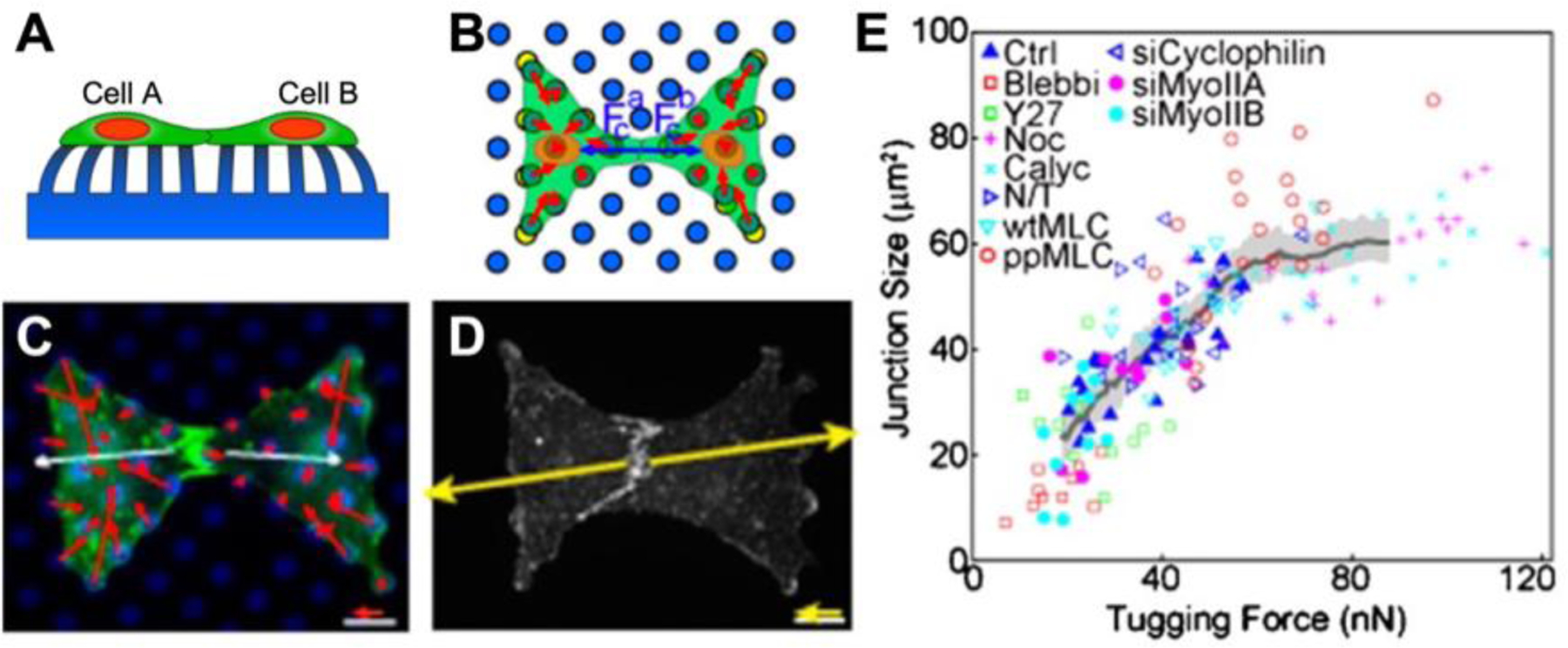
Measuring cellular forces with microposts and bowties. (A) Cells produce both forces with their extracellular environment (cell-matrix force) and forces with other cells (cell-cell force). (B) As indicated by the red arrows, the vector sum of cell-matrix forces exerted by a cell is zero. Fc (blue arrows) is the cell-cell force exerted by each cell. The cell-cell force is equal in magnitude but opposite in direction to the vector sum of a cell-matrix force. Cell A pulls on cell B with a cell-cell force equal and opposite to the force cell B exerts on cell A. Cells were constrained to a bowtie pattern by stamping fibronectin on the tips of select microposts. (C) Red arrows indicate cell-matrix forces. (D) Yellow arrows indicate cell-cell forces, i.e. tugging force. (E) A scatter plot showing adherens junction size vs. cell-cell (tugging) force under conditions that affect cytoskeletal tension. Adapted under the terms of PNAS License to Publish.[80] Copyright 2010, National Academy of Sciences.
A monolayer, as seen in the epithelium and endothelium, is a single layer of cells growing side by side, encompassing multiple cell-cell force interactions. Monolayers are difficult to study because they involve complex cell patterns, have many cell-cell interactions, and migrate collectively.[83] In an epithelial monolayer, the largest cellular forces were observed in the border cells.[84] At a distance close to one cell length from the edge, internal cells had forces half as strong as the border cells.[84] Furthermore, the endothelium is responsible for regulating the transmigration of molecules and leukocytes between the blood and tissue.[85] The transmigration process involves rolling adhesion, firm adhesion, and penetration. Using microposts, the cellular forces were observed to increase during firm adhesion, with the cells directly in contact with the leukocytes increasing the most.[85] Monolayers exposed to shear flow are also of interest to study changes in cellular forces and cell-cell forces in response to mechanical stimuli.[86] Microposts can be built into a flow chamber designed to deliver laminar and disturbed flow. Compared to static conditions, endothelial cellular forces increased under laminar flow, but decreased under disturbed flow.[86]
Cellular migration aids in embryotic development, tissue repair, and immune response, rendering it a necessary process to maintain homeostasis.[87] Studying cellular forces during distinctive stages of migration or in different environments may clarify the mechanical mechanisms regulating pathological functions. During one-dimensional cell migration, a cell protrudes forward, adheres to a substrate, contracts, and releases its rear. Fibroblasts migrating along a line of microposts revealed cellular contraction does not cause rear release. Rather, focal adhesion disassembly at the rear is independent of contraction.[87] In another study focused on unidirectional cell migration, computational models validated experimental micropost results.[88] As shown by micropost deflection, cells exhibit higher forces at their boundary and lower forces at their center with the highest force at the trailing edge. Studying cells on microposts arrays with variable spacing between the posts revealed that cells exhibited smaller forces on high density areas (more posts underneath) and lower forces on low density areas (less posts underneath). Additionally, cells tend to migrate towards increasing micropost densities.[88]
Similarly, cells encounter complex microenvironments during 3D cell migration. Cells were studied on microposts with different surface coatings and post spacings to mimic physical confinement.[89] Cells traveled faster on the tops of microposts, while cells squeezing between microposts were slower. To further investigate how cells respond to a confined space, they were covered with a second set of microposts. When the spacing between the top and bottom microposts was reduced such that cells could adhere to both sets of microposts, it was found that they migrated at a higher speed.[89] In another study, cell migration towards a chemotactic stimulus was observed inside a microfluidic device consisting of microposts embedded in channels of varying cross-sectional area.[90] Average cellular force decreased as the cross-sectional area decreased (more confined). Inhibiting or activating myosin- did not affect cell forces in confined environments, but did have an impact on cells in channels with a larger cross-sectional area.[90]
3.1.3. Force applied to microposts
The microposts previously discussed are unable to deflect without the influence of cellular forces, mimicking a static extracellular environment. However, forces applied by the extracellular environment act as mechanical signals that influence cellular function. For example, cells respond to topographical cues in their microenvironment by rearranging their cytoskeleton to migrate along these cues, while mechanical cues, such as uniaxial strain, have a competing effect on cell alignment as cells orient themselves perpendicular to the direction of strain.[91] Implications of rival cues on engineered tissues can be better understood by observing a cell’s behavior due to influences from the topographical and mechanical environment.[91] Manipulating traditional micropost arrays can recreate the mechanical stimuli found in vivo. These mechanical cues include actuation (magnetic microposts), stretch (vacuum chamber), and flow (microfluidics).
With magnetic microposts, a magnetic field acting on cobalt nanowires or iron particles dispersed throughout microposts causes the posts to bend and exert an external force on the cells (Figure 8A–F).[92–94] Magnetic microposts are fabricated in a manner similar to Figure 4C with an additional step to load the negative mold with magnetic material. Either cobalt nanowires (Figure 8C and 8D) or iron micro-/nanoparticles (Figure 8E and 8F) are integrated into the PDMS.[93,94] This technique has also been used with dry micro-/nanoparticles, meaning the particles are poured into the negative mold without the aid of a solution instead of pre-mixing magnetic particles into PDMS.[95] A magnetic field is applied to the microposts by sliding magnets positioned on opposite sides of the micropost array. The magnetic field is larger when the magnets are closer together and smaller when the magnets are farther apart, thus controlling the induced torque. Cellular forces are calculated using Equation 1 where the spring constant is k = 3πEd⁴/64(L³-Lw3) and Lw is the length of the nanowire or region of embedded magnetic particles.[92]
Figure 8.
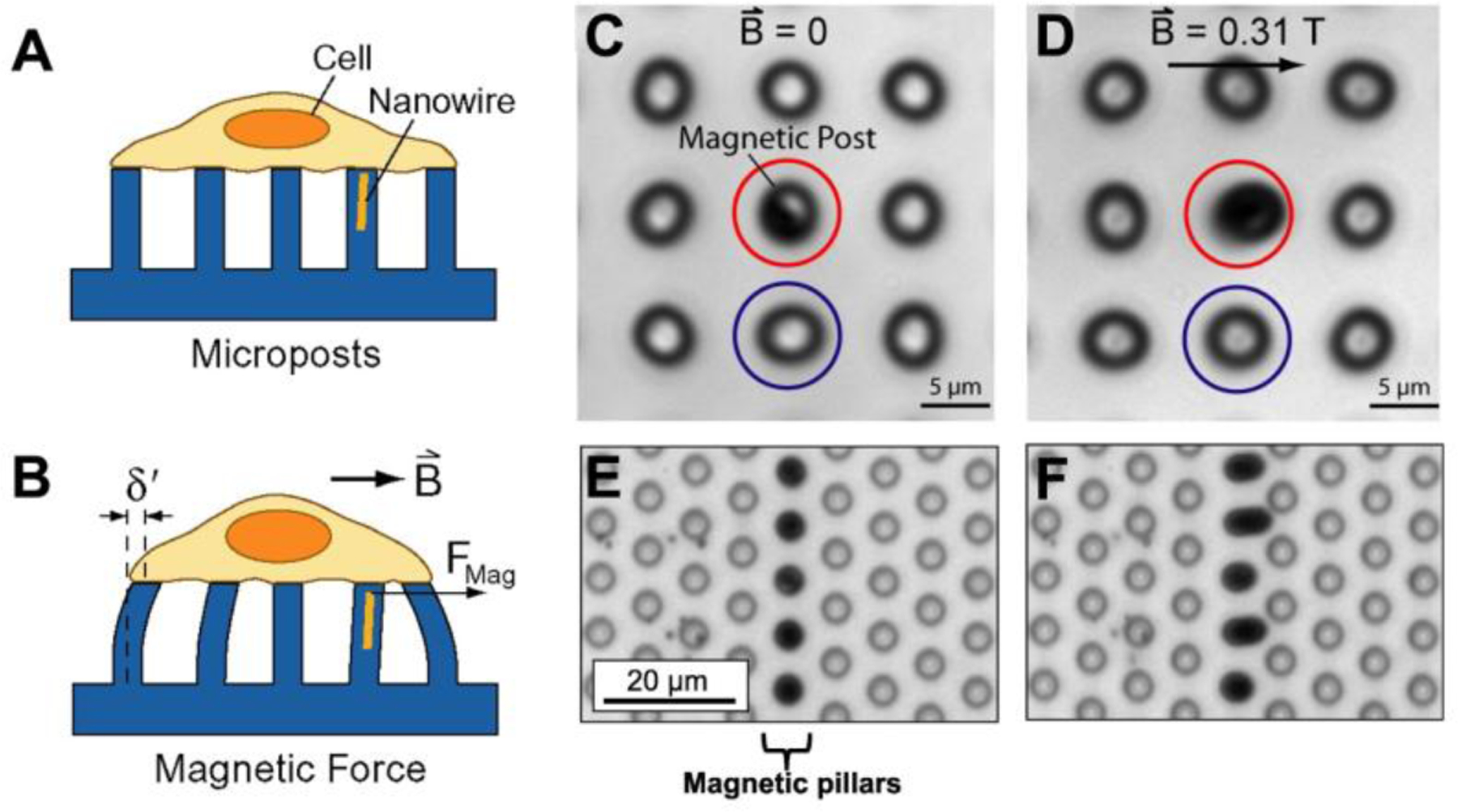
Magnetic microposts can be used to apply force to cells and quantify their response. (A) Cells adhere to the tops of magnetic microposts and contract, deflecting the posts. (B) Applying a magnetic field B causes an external force Fmag on the cell. The cell adjusts its environment by altering its force, which can be determined by measuring δ’. There are two types of magnetic microposts: those with nanowires and those with embedded particles. (C) Magnetic post with nanowire (red) surrounded by non-magnetic posts (blue). (E) Posts with magnetic particles (dark, center column) surrounded by non-magnetic posts (transparent). (D and F) Applying a magnetic field causes the magnetic post to deflect and the non-magnetic posts remain stationary. A and B are adapted with permission.[92] Copyright 2008, AIP Publishing LLC. C and D are Adapted under the terms of PNAS License to Publish.[93] Copyright 2007, National Academy of Sciences. E and F are adapted with permission.[94] Copyright 2017, Elsevier.
Various studies with magnetic microposts have revealed changes in cellular contractility, focal adhesions, and spread area in response to the magnitude and frequency of applied forces. Interestingly, cellular force response has been observed to vary based on location within a cell. Cells exhibited increased focal adhesion size at the site of actuation and decreased cellular forces at the peripheral adhesions of cells.[93] With smooth muscle cells, contractility increased quickly then leveled off for posts externally adherent to the cells, while internal posts experienced a steady increase in contractility.[96] Additionally, cells exhibited differing responses depending on initial spread area and pretension. Cell with low pretension showed decreased cellular forces and increased area after stimulation, while the cells with elongated bodies and high pretension exhibited increased force, but decreased spread area.[97] Moreover, actuation frequency impacted the contractile responsiveness of cells, with cells favoring actuation frequencies closest to heartrate frequencies.[96] Taken together, these studies exemplify the usefulness of magnetic microposts in understanding mechanotransduction.
In addition to magnetics, external forces can be applied to microposts integrated within a stretchable membrane via a vacuum chamber.[98] This technique allows single focal adhesions to be monitored. In an effort to understand pressure-induced vascular disease progression during sustained stretching, vascular smooth muscle cells have been observed to increase their contractility first, then relinquish their contractility gradually, largely due to periphery focal adhesions.[98]
Cells in vivo experience forces due to shear flow. Tools combining microfluidics and microposts help to quantify cell forces in response to shear stress.[86] After platelets are activated, they contract to form strong, stable blood clots (Figure 9D). A microfluidic device with embedded blocks and posts was created to measure platelet forces under flow (Figure 9A and 9B).[99] During flow, the blocks created a shear gradient which induced platelet aggregation. The deflection of the accompanying post (Figure 9C) indicated platelet forces which can be used to identify healthy, functioning platelets, or platelets of trauma patient requiring a blood transfusion (Figure 9E and 9F).[99] Another device combined microfluidics and magnetically actuated microposts to apply shear stress to blood clots as they form.[100] The posts were actuated to maintain constant deflection during platelet aggregation, documenting elasticity of the blood clot over time.[100]
Figure 9.
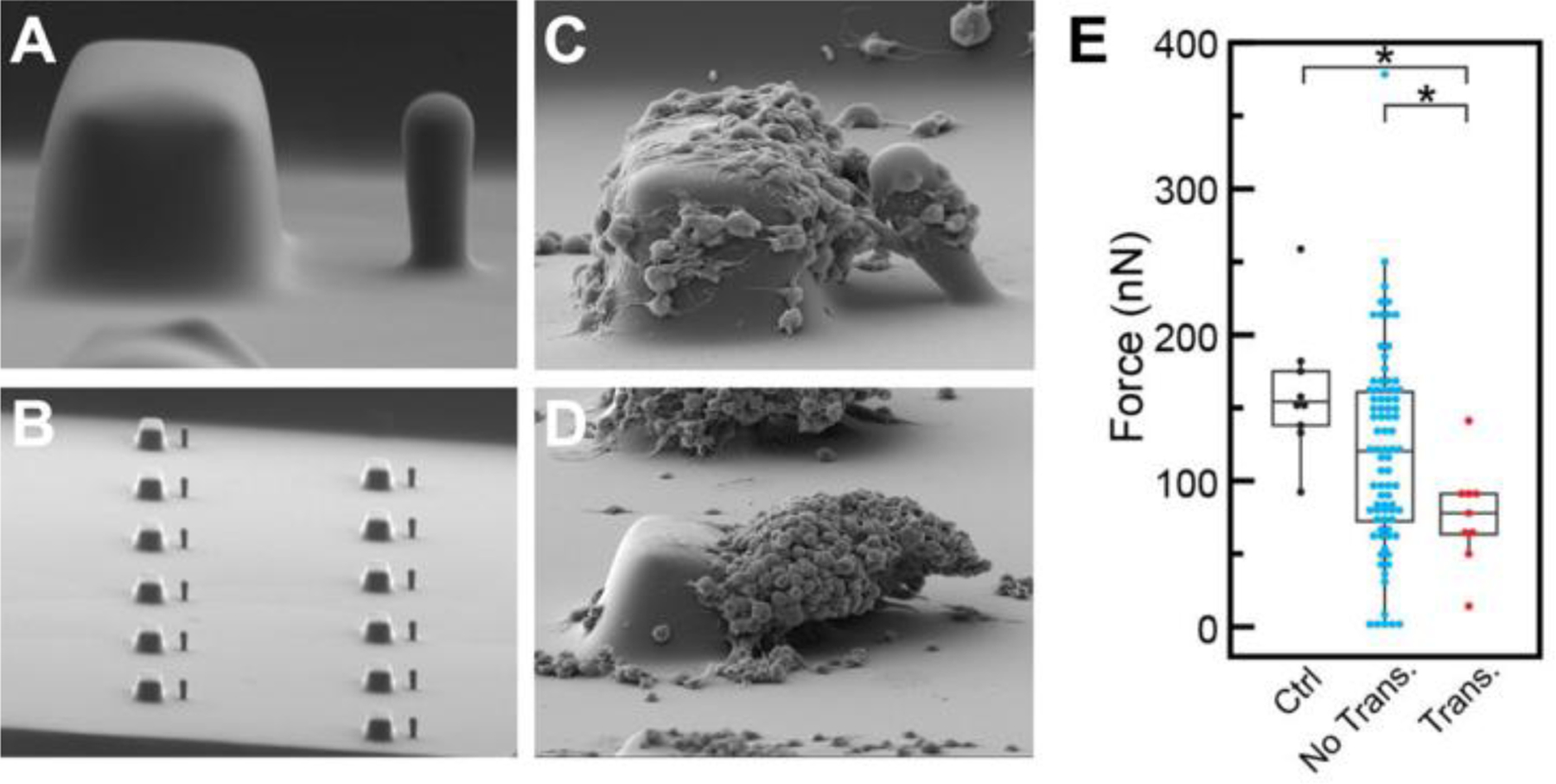
Blocks and posts embedded into a microfluidic device can determine platelet forces and bleeding risk in trauma patients. (A) A single block and post. (B) Many blocks and posts within a channel. (C) Platelet forces are determined by deflection of the post. (D) Block and post after platelet aggregate has formed. (E) In comparison to trauma patients not requiring a blood transfusion (blue), mean platelet force was significantly decreased in trauma patients requiring a transfusion (red). Healthy platelets reported the strongest forces (black). Adapted under the terms of Creative Commons Attribution 4.0 International License.[99] Copyright 2019, Springer Nature.
3.2. Silicon cantilevers
Microelectromechanical systems (MEMS) are microscale devices composed of electrical and mechanical elements that include sensors and/or actuators.[101] They consist typically of structures made from crystalline silicon material due to its advantageous mechanical properties[102,103] and its compatibility with micromachining techniques that can deposit, pattern, or etch this material with nanoscale precision.[104] MEMS was used as one of the first approaches to measure cellular forces with a deformable structure.[105] Specifically, a device was fabricated with a multitude of cantilevers on a silicon wafer. Each cantilever was horizontal in orientation and was contained within its own cavity underneath the surface of the wafer, but with the tip of the cantilever extending out to the surface through an opening. As a cell crawled across the surface of the device, it made contact with the tip of the cantilever and displaced it, which was measured optically.[105] In this manner, it was found that migrating cells not only generate forces at their leading edge, but have substantially larger, retrograde forces at their trailing edge. MEMS devices have also measured cell forces in embryonic Drosophia axons by measuring deflection of cantilevers optically relative to a fixed reference beam.[106] This MEMS device is a series of flexible silicon beams attached to a rigid probe which can be inserted into a biological environment to simultaneously exert forces and measure biological response. Using this system, it was observed that after release of externally applied tension to the Drosophia neurons, the cells contracted and generated forces to restore tension, demonstrating an influence of mechanical tension to neuronal behavior.[106]
Atomic force microscopy (AFM) is a versatile experimental technique used to push and pull on molecules, cells, tissues, or other materials on the nanoscale. Cantilever deflection is measured from laser deflection onto a photosensitive detector. Similar to aforementioned deformable structures, force is calculated from deflection, the cantilever dimensions, and the known stiffness of the cantilever. On the cellular scale, AFM can be used to measure mechanics such as cell stiffness or cellular response to force by pushing on the cell with an AFM cantilever.[107–109] More similar to aforementioned measurements examining forces exerted by cells (as opposed to externally exerting forces on cells), it is also possible to adhere cells between a surface and an AFM cantilever and measure cell contraction (the cell pulling the cantilever toward the surface).[110] In a system with an AFM combined with a “side-view” microscope, contraction of human bone osteosarcoma epithelial single cells was examined. It was observed that cells contract laterally at the midpoint before contracting vertically, suggesting directional cytoskeletal remodeling or contraction.[110] AFM cantilevers are often composed of silicon, usually silicon or silicon nitride. Similar to MEMS, these materials are used because they are mechanically well-characterized and structures with micron dimensional tolerances can be created using micromachining.[111] After being machined from silicon, cantilevers are often coated with thin metal layers such as gold for the purposes of functionalization and/or increased reflectance for deflection measurements.[112,113] A similar AFM method was used to measure mechanics and contraction dynamics of individual platelets. This work found that platelets contract immediately after binding to fibrinogen and complete contraction within 15 minutes, generating higher forces in stiffer microenvironments.[114] While this technique is challenging in throughput (one cell measured at a time) and cost (both the high cost of the AFM and the disposable cantilevers), it is capable of providing cell contraction forces temporarily and cell adhesion forces with simultaneous cell visualization in a variety of microenvironments.
3.3. Nanoposts
Nanoposts are smaller versions of microposts; they allow for higher spatial resolution of cell forces because there are more posts under each cell. However, fabrication of posts at a nanoscale level has its challenges. The casting process for nanopost arrays causes surface cracks to accumulate on negative mold which become more severe with each replication.[115] These cracks are not due to peeling the negative mold away from the nanoposts (mechanical stress), but rather thermal stresses that fracture the PDMS. To combat thermally induced stresses, it is recommended that lower temperatures are used during fabrication and negative molds are only reused a few times.[115] Furthermore, master molds are often treated with silane vapor to prevent PDMS from permanently bonding to its surface.[116] This silane coating increases the geometry of the posts on the master, consequently reducing the dimensions of the nanoposts due to the double-casting process. As a result, silane deposition time should be monitored or the coating can adversely affect nanoscale dimensions of PDMS structures.[116]
A single cell covers multiple nanoposts, permitting cellular forces to be measured at multiple points on a cell (Figure 10A, 10B, 10C). For this reason, platelets have been studied on nanoposts to determine the role of the surface receptor GPIb-IX-V complex, which interacts with a protein called von Willebrand factor (VWF), in forming a hemostatic plug.[71] In blocking the receptors GPIbα and αIIbβ3, the GPIb-IX-V complex was determined to transmit a significant portion of cytoskeletal forces to VWF and the extracellular environment (Figure 10D).[71] Nanoposts have also been used to understand the mechanism cardiomyocytes use to probe matrix stiffness.[117] It was determined protein kinase C signaling activates non-muscle myosin and muscle myosin contractions, which work together to sense extracellular matrix rigidity.[117]
Figure 10.
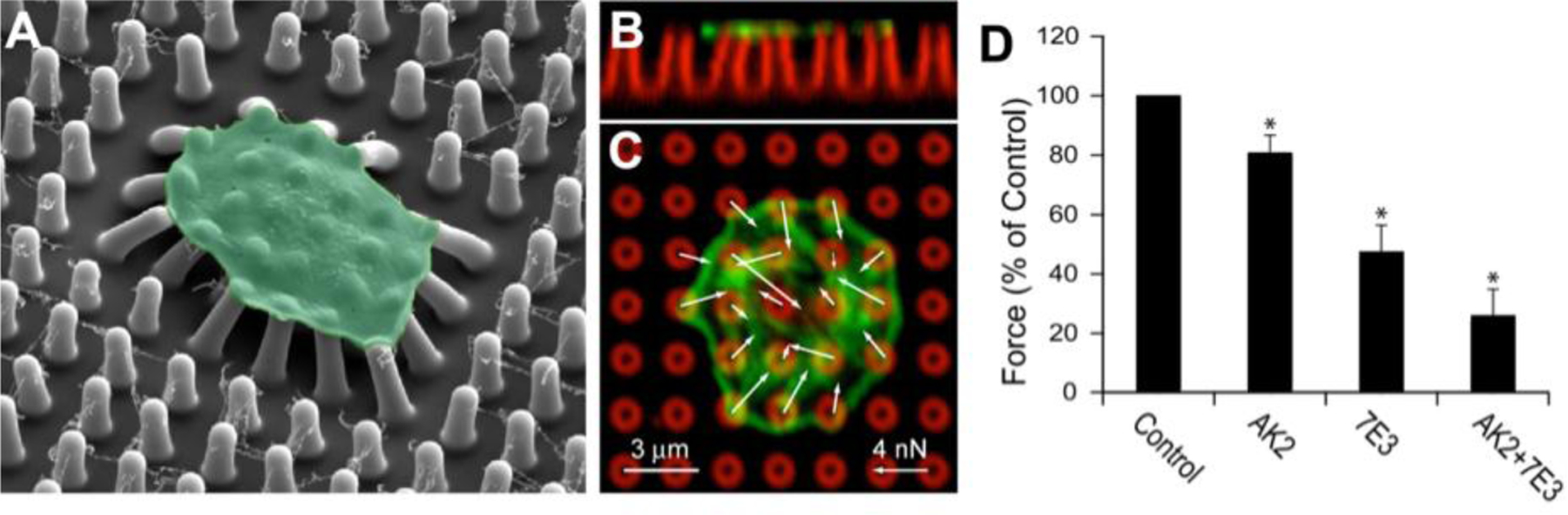
In addition to microposts and the block and post microfluidic device, platelets have been studied on nanoposts. (A) Nanoposts allow forces from a single platelet to be measured at many different locations. The image from scanning electron microscopy depicts several nanoposts under a single platelet (green). Side view (B) and top view (C) of a platelet on nanoposts. The white arrows represent force vectors, the green is an F-actin label, the red is the posts. (D) Platelet force was inhibited when platelets were treated with antibodies AK2, 7E3 and AK2 and 7E3 to block GPIbα, αIIbβ3, and both receptors respectively to examine how they contribute to the transmission of platelet forces. Adapted with permission.[71] Copyright 2016, Elsevier.
Cell-matrix forces have been studied on microposts and nanoposts to compare the cellular response on posts of differing sizes.[118] Nanoposts illuminate more details regarding cellular forces that may be overlooked on microposts and more closely resemble continuous deformable materials.[118] Ideally, cellular forces measured using nanoposts should yield similar values to those measured using TFM. Although platelets have been found to spread in response to topography[119], nanoposts are small enough to induce a cell spread area comparable to protein-coated flat surfaces.[71]
3.4. Tissue posts
Tissues within hydrogels that are at the millimeter to hundreds of micrometers in length can be formed between the tips of two or more PDMS cantilevers, typically larger than microposts, to mimic a 3D environment while also measuring their contractile forces.[120] The first incarnation of this approach was a device engineered to study the effects of drugs on electrically-stimulated muscle tissue over long periods of time.[121] Around the same time, microfabricated tissue gauges (or μTUGs) that were used to study the maturation of engineered tissues and monitor their response to biochemical, electrical, and mechanical stimuli.[122] Shortly thereafter, a fibrin-based method with cardiomyocytes was developed to measure their dynamic twitch contractions as an engineered heart tissue.[123] In both instances, the PDMS cantilevers have a T-shaped cap at their tips that prevent the tissues from pulling off the ends when the cantilevers are highly bent. Forces produced by the tissues are calculated using Equation 1 where the spring constant is k = Ewt³/6a2(3L-a) , and w is the width of the cantilever beam, t is the thickness, and a is the length from the bottom of the cantilever to the centroid of the top portion of the “T”. These tissue posts can be modified by changing the stiffness, altering the geometry, adding electrodes, or employing magnetic actuation to more closely model a cell’s natural environment.[124]
Various experiments with tissue gauges have revealed their capabilities. One application of tissue gauges is to study asthma by exposing airway smooth muscle tissue to contractile and relaxant agents.[125] Moreover, by integrating carbon electrodes into traditional μTUGs, the effects of electrical stimulation on developing cardiac microtissues can be explored. Cardiac tissues matured on stiffer posts and exposed to electrical stimulation established superiority in both structure (cell alignment) and function (force generation).[126,127] Furthermore, to study the effect of extracellular matrix remodeling on tissue structure and forces, μTUGs can be used. Contractility was found to be dependent on post stiffness, while protein expression and extracellular matrix stiffening was correlated to mechanical stress.[122] Additionally, μTUGs are an appropriate tool for high-throughput monitoring of drug-induced effects on various types of cells.[122,126] However, PDMS is prone to absorbing small hydrophobic molecules, which may upset the outcome of experiments, especially in drug screening studies.[128]
A similar device used to study engineered heart tissues consists of one rigid post and one flexible post (Figure 11A).[129] In a study with pluripotent stem cell derived cardiomyocytes, the flexible post contained an embedded magnet in its tip (Figure 11B). A giant magnetoresistive (GMR) sensor below the device recorded voltage changes as the force of the beating tissue deflected the post and magnet. The deflection recorded by the sensor was consistent with the optical deflection measurements (Figure 11C). The voltage was used to calculate the tissue beating frequency and twitch force in real-time.[129] Another study aimed to understand the role of afterload, or systolic load, in maturing engineered heart tissues.[130] This device still had one flexible post and one rigid post; however, the flexible post stiffness was adjustable with braces that varied the effective length of the posts (Figure 11D). The longer the brace, the shorter the post, and the higher the stiffness and afterload. Overall, increased afterload was determined to aid in the maturation of hiPSC-CMs (Figure 11E).[130] A similar system utilizing a piezo-bending actuator in one post, produces automated force measurements that could aid in cardiac repair and drug screening.[131]
Figure 11.
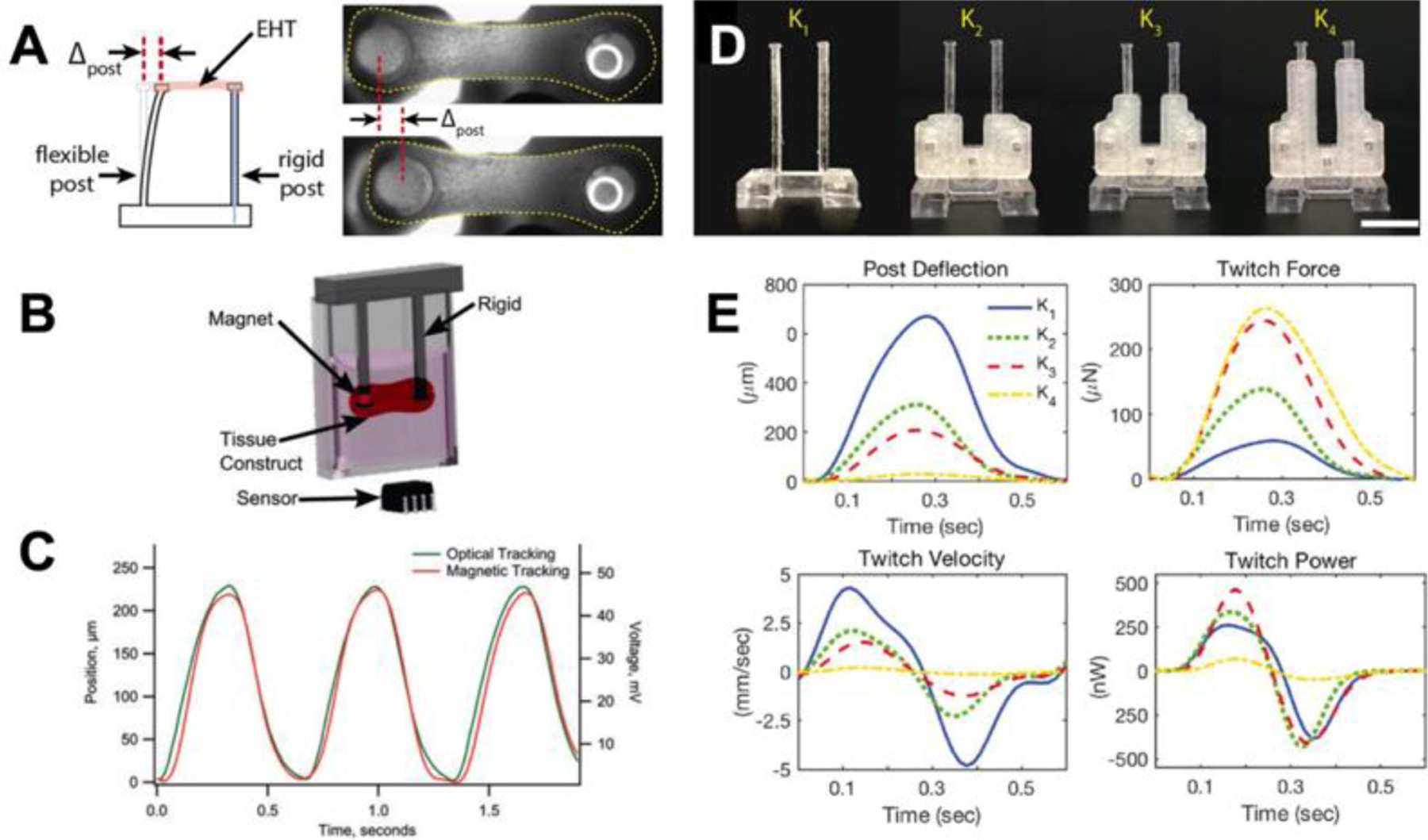
Human cardiomyocytes can be studied as a single cell on microposts or as a tissue on tissue posts. (A) To study engineered heart tissues, one post remains rigid, while the other is flexible to measure deflection. (B) In some cases, post deflection is measured by a GMR sensor which senses the movement of an embedded magnet inside the flexible post. (C) The graph shows correlation between the deflection measured optically and the deflection reported by the GMR sensor. (D) Tissue posts with braces to vary effective length and modify stiffness. (E) Post deflection, twitch force, twitch velocity, and twitch power for various post lengths. A, D, and E are adapted with permission.[130] Copyright 2018, Elsevier. B and C are adapted under the terms of Creative Commons Attribution 4.0 International Public License.[129] Copyright 2016, Mary Ann Liebert, Inc. publishers.
Tissue posts can be used in conjunction with other techniques and tools, including FRET and microfluidics. One experiment combined μTUGs with FRET to study the fibronectin matrix within collagen-based microtissues during wound healing.[132] These tools allowed the changes in spatial distributions and shape of fibronectin within the tissue to be measured.[132] Tissue posts can also be integrated into microfluidics, creating a device capable of monitoring clot mechanics to illustrate how shear flow and biochemical stimuli impact clot stiffening.[133]
3.5. Wires and fibers
A device called Biowire provides the structural and electrical stimulation needed to mature cardiomyocytes into cardiac tissue. The first-generation Biowire is fabricated using soft lithography techniques in a manner similar to microposts with a piece of sterile surgical silk suture placed in the center channel of the PDMS.[134] The second-generation Biowire II platform consists of two elastomer-based wires laid over a shallow chamber made of polystyrene (Figure 12A and 12B).[135,136] Both Biowire and Biowire II are placed inside an electrical stimulation chamber to mature the heart cells.[134]
Figure 12.
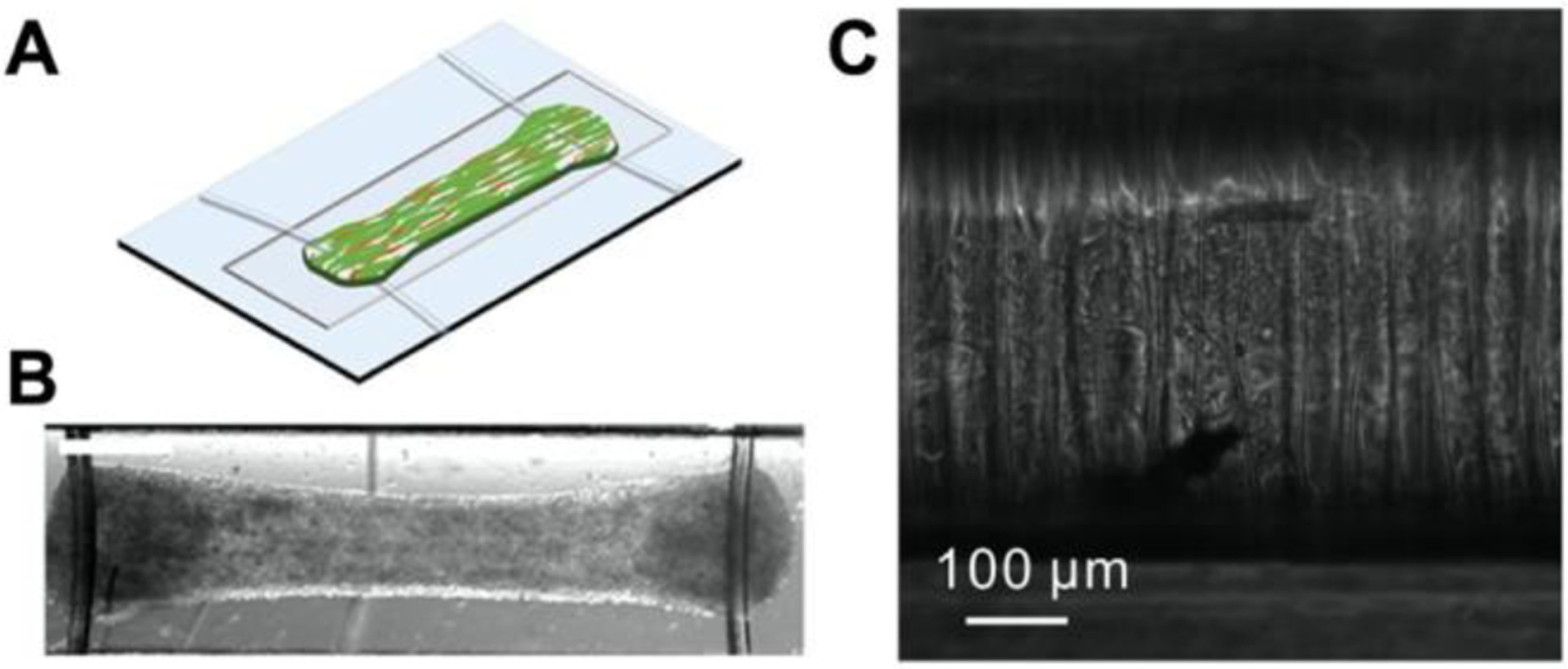
Fibrous platforms are used to mature stem cells into cardiac tissue. (A-B) Biowire II consists of a tissue suspended between two wires. (C) Cardiac tissue on a filamentous matrix. A and B are adapted under the terms of ASC AuthorChoice License.[135] Copyright 2019, American Chemical Society. C is adapted with permission.[137] Copyright 2014, Elsevier.
Differentiated cardiomyocytes from human pluripotent stem cells (hPSC) can be used to engineer heart tissues. However, differentiated cells must first be matured into adult cardiomyocytes. Biowire presents the architectural structure and electrical stimulation needed to develop mature heart cells. On the original Biowire platform, cells aligned themselves along the suture of the 3D environment and, after several weeks of exposure to electrical frequency, the cells displayed traits more consistent with adult cardiomyocytes than the control group.[138] Furthermore, Biowire offers a high-throughput, high-content screening platform and has been used to investigate the effects of kinase inhibitors on cardiac tissue in addition to the side effects are cardiovascular risk of new drugs.[139,140] The Biowire II platform uses the deflection of the wires to measure systolic and diastolic forces of cardiac tissue in real time.[135] Biowire II has been used to monitor the progression of cardiac fibrosis at the interface of fibrotic and healthy tissue, while simultaneously functioning as a drug screening platform.[135]
Filamentous matrices, or a matrix of synthetic parallel fibers, are fabricated using two-photon polymerization (TPP) and can be employed to examine cardiomyocytes (Figure 12C).[137] The spacing of the fibers regulates the structural alignment of cardiomyocytes, while the fiber diameter is altered to mimic cardiac tissue stiffness.[141] This platform has been used to monitor cardiac tissue from patients with long QT syndrome type 3 (LQT3) and tissues with a sacromeric mutation. A filamentous matrix allows the contractile defects of tissues and their vulnerability to topographical cues (stiffness) or environmental stressors (pharmaceuticals) to be quantified.[137,141]
Aside from cardiomyocytes, filamentous matrices are suitable for other applications. Fibers can mimic the extracellular matrix to study cell emergence from monolayers, wound gap closure, and cell migration.[142] On matrices with large interfiber spacing, cells were observed to break away on their own or in small groups, while multiple groups of cells emerged collectively on matrices with small interfiber spacing.[142] Furthermore, fibrous scaffolds constructed using spinneret-based tunable engineered parameters (STEP) technique were used to study how various mechanical and chemical stimuli impact cellular behavior, including cell migration.[143]
To better understand tissue remodeling, recent work has utilized a fibrillar collagen matrix to study macrophage migration towards an attractive force source and in response to deformation fields produced by fibroblasts.[144] This approach reveals that long-distance mechanical signals may drive cell migration over structural cues.
4. Conclusion and Future Outlook
In this review, we discussed the measurement of cellular forces using deformable materials including polysiloxane, polyacrylamide, native ECM, and engineered gels as well as deformable structures composed of polydimethylsiloxane, silicon, or printable polymers. Through all these techniques, deformation (material or structural) is measured and used to calculate force from the known properties such as material stiffness and/or structural dimensions. Over the last three decades, approaches to measure cellular forces have been invented and improved, facilitating characterization of the strength of cellular forces directionally and temporarily during cell processes such as migration or maturation, in healthy or diseased states, and in the presence or absence of key cellular markers or external stimuli. Using different approaches to measure force and a wide variety of cell types can help to elucidate the common features in cell mechanics and increase our confidence in establishing robust biomechanical relationships for cells. For example, the effect of stiffness on cellular force generation has been examined by measuring bead displacement on multiple materials[18,21] as well as on structures.[47] Additionally, the observation that cellular forces increase as function of focal adhesion size has been corroborated with multiple methods.[19,20,47] While each method has its strengths and drawbacks, collectively these methods work symbiotically to elucidate biophysical relationships and increase confidence that these relationships are not an artifact of the observation method.
Despite the improvement in these techniques and the vast knowledge gained from them, some challenges prevent accessibility of these techniques to all fields and research groups. Material wrinkling, the earliest method to characterize cell forces, facilitated the first method to comparison of cell forces but are ultimately non-quantitative.[9,10] Later methods facilitated quantification and the first force maps indicating cell force magnitude and direction[15,16] but are challenging and time-consuming because they require cell removal to obtain a reference image. Others do not require a reference image, but the manufacturing of each substrate requires clean room preparation[20] or individual printing of each reference point.[19] Ultimately this means that each technique has one or more challenges including high cost (particularly start-up cost), manufacturing complexity or difficulty, imaging temporal limitations, quantification or computational analysis challenges, and/or throughput. Traction force microscopy methods can also be confounded by other variables; for example, substrate topography and microenvironmental stiffness that affect cell function.
Looking to the future, an ideal system may increase throughput and decrease technical challenges involved in system manufacturing, data collection, and/or data analysis. This system may involve techniques in which system manufacturing is less expensive, increased throughput (i.e. more cells measured per unit or reduced manufacturing time per unit), and/or requires only widely available, inexpensive equipment. It would also be desirable to use simple image acquisition approaches to capture material or structural deformation and fast computational tools to measure cellular forces with accuracy. Additionally, a system should be versatile to a variety of cell types (varied sizes, strengths, functions) and compatible with other cellular assays, i.e. immunofluorescence microscopy, RNA-seq, etc., to understand how cell mechanics relates to cell function. Finally, an ideal system could both measure cellular forces and not inadvertently affect their behavior, unless by intention, in order to facilitate our understanding of mechanobiology.
Acknowledgements
This work was supported by the National Science Foundation (CMMI-1661730, CMMI-1824792), Wings for Life Foundation (WFL-US-28-17), and National Institutes of Health (EB028094, HL147462).
AUTHOR BIOGRAPHIES

Ava M. Obenaus earned her B.S. in mechanical engineering and her M.E. in aerospace and mechanical engineering from Illinois Institute of Technology. She is currently a Ph.D. student in the Department of Mechanical Engineering at the University of Washington. Her research interests include thrombus formation under blood flow and microfluidics.

Molly Y. Mollica earned her B.S. in biomedical engineering and her M.S. in mechanical engineering from Ohio State University. She is currently a Ph.D. student in the Department of Bioengineering at the University of Washington. Her research interests include thrombosis mechanics, mechanobiology, and DNA origami nanotechnology.

Nathan J. Sniadecki earned his B.S. in mechanical engineering from the University of Notre Dame and M.S. and Ph.D. in mechanical engineering from the University of Maryland at College Park. He was postdoctoral fellow at Johns Hopkins University and the University of Pennsylvania. He is currently a Professor and Associate Chair in the Department of Mechanical Engineering and an Adjunct Professor in the Department of Bioengineering at the University of Washington. His research interests include cell mechanics, mechanobiology, and bio-MEMs.
References
- [1].Fletcher DA, Mullins RD, Nature 2010, 463, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Janmey PA, McCulloch CA, Annu. Rev. Biomed. Eng 2007, 9, 1. [DOI] [PubMed] [Google Scholar]
- [3].Guck J, Lautenschläger F, Paschke S, Beil M, Integr. Biol 2010, 2, 575. [DOI] [PubMed] [Google Scholar]
- [4].Miller CJ, Davidson LA, Nat. Rev. Genet 2013, 14, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kai FB, Laklai H, Weaver VM, Trends Cell Biol. 2016, 26, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Murrell M, Oakes PW, Lenz M, Gardel ML, Nat. Rev. Mol. Cell Biol 2015, 16, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hooke R, Micrographia, 1665. [Google Scholar]
- [8].Katta S, Krieg M, Goodman MB, Annu. Rev. Cell Dev. Biol 2015, 31, 347. [DOI] [PubMed] [Google Scholar]
- [9].Harris AK, Wild P, Stopak D, Science (80-. ). 1980, 208, 177. [DOI] [PubMed] [Google Scholar]
- [10].Harris AK, Stopak D, Wild P, Nature 1981, 290, 249. [DOI] [PubMed] [Google Scholar]
- [11].Burton K, Taylor DL, Lett. to Nat 1997, 385, 450. [DOI] [PubMed] [Google Scholar]
- [12].Burton K, Park JH, Taylor DL, Mol. Biol. Cell 1999, 10, 3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Helfman DM, Levy ET, Berthier C, Shtutman M, Riveline D, Grosheva I, Lachish-Zalait A, Elbaum M, Bershadsky AD, Mol. Biol. Cell 1999, 10, 3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C, Mol. Biol. Cell 2001, 12, 2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K, J. Cell Biol 1994, 127, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oliver T, Dembo M, Jacobson K, Cell Motil. Cytoskeleton 1995, 31, 225. [DOI] [PubMed] [Google Scholar]
- [17].Dembo M, Oliver T, Ishihara A, Jacobson K, Biophys. J 1996, 70, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gutierrez E, Tkachenko E, Besser A, Sundd P, Ley K, Danuser G, Ginsberg MH, Groisman A, PLoS One 2011, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bergert M, Lendenmann T, Zündel M, Ehret AE, Panozzo D, Richner P, Kim DK, Kress SJP, Norris DJ, Sorkine-Hornung O, et al. , Nat. Commun 2016, 7, DOI 10.1038/ncomms12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al. , Nat. Cell Biol 2001, 3, 466. [DOI] [PubMed] [Google Scholar]
- [21].Pelham R, Wang L-L, Proc. Natl. Acad. Sci. U. S. A 1997, 94, 13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y-L, Pelham RJ, in Methods, 1998, pp. 489–496. [DOI] [PubMed] [Google Scholar]
- [23].Munevar S, Wang YL, Dembo M, Biophys. J 2001, 80, 1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Polio SR, Rothenberg KE, Stamenović D, Smith ML, Acta Biomater. 2012, 8, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Polio SR, Parameswaran H, Canović EP, Gaut CM, Aksyonova D, Stamenović D, Smith ML, Integr. Biol. (United Kingdom) 2014, 6, 357. [DOI] [PubMed] [Google Scholar]
- [26].Tseng P, Di Carlo D, Adv. Mater 2014, 26, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Myers DR, Qiu Y, Fay ME, Tennenbaum M, Chester D, Cuadrado J, Sakurai Y, Baek J, Tran R, Ciciliano JC, et al. , Nat. Mater 2017, 16, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Califano JP, Reinhart-King CA, Cell. Mol. Bioeng 2010, 3, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kandow CE, Georges PC, Janmey PA, Beningo KA, Methods Cell Biol. 2007, 83, 29. [DOI] [PubMed] [Google Scholar]
- [30].Wang HB, Dembo M, Hanks SK, Wang YL, Proc. Natl. Acad. Sci. U. S. A 2001, 98, 11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Maskarinec SA, Franck C, Tirrell DA, Ravichandran G, Proc. Natl. Acad. Sci. U. S. A 2009, 106, 22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Oakes PW, Patel DC, Morin NA, Zitterbart DP, Fabry B, Reichner JS, Tang JX, Blood 2009, 114, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rape AD, Guo W, Wang Y, Biomaterials 2011, 32, 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Reinhart-King CA, Dembo M, Hammer DA, Biophys. J 2005, 89, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ribeiro AJS, Denisin AK, Wilson RE, Pruitt BL, Methods 2016, 94, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shiu YT, Li S, Marganski WA, Usami S, Schwartz MA, Wang YL, Dembo M, Chien S, Biophys. J 2004, 86, 2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stroka KM, Aranda-Espinoza H, Cell Motil. Cytoskeleton 2009, 66, 328. [DOI] [PubMed] [Google Scholar]
- [38].Tay CY, Wu YL, Cai P, Tan NS, Venkatraman SS, Chen X, Tan LP, NPG Asia Mater. 2015, 7, 1. [Google Scholar]
- [39].Gjorevski N, Nelson CM, Biophys. J 2012, 103, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Steinwachs J, Metzner C, Skodzek K, Lang N, Thievessen I, Mark C, Münster S, Aifantis KE, Fabry B, Nat. Methods 2016, 13, 171. [DOI] [PubMed] [Google Scholar]
- [41].Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D, Biophys. J 2004, 86, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pelham RJ, Wang YL, Proc. Natl. Acad. Sci. U. S. A 1997, 94, 13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS, Nat. Methods 2010, 7, 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Miller JS, Shen CJ, Legant WR, Baranski JD, Blakely BL, Chen CS, Biomaterials 2010, 31, 3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS, Proc. Natl. Acad. Sci. U. S. A 2013, 110, 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yang MT, Fu J, Wang YK, Desai RA, Chen CS, Nat. Protoc 2011, 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS, Proc. Natl. Acad. Sci 2002, 100, 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim D, Wong PK, Park J, Levchenko A, Y. Sun, 2009, 203. [DOI] [PubMed] [Google Scholar]
- [49].Butler JP, Toli-Nørrelykke IM, Fabry B, Fredberg JJ, Am. J. Physiol. - Cell Physiol 2002, 282, 595. [DOI] [PubMed] [Google Scholar]
- [50].Sabass B, Gardel ML, Waterman CM, Schwarz US, Biophys. J 2008, 94, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sniadecki NJ, Chen CS, Methods Cell Biol. 2007, 83, 313. [DOI] [PubMed] [Google Scholar]
- [52].Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ, Biophys. J 2012, 103, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li B, Xie L, Starr ZC, Yang Z, Lin JS, Wang JHC, Cell Motil. Cytoskeleton 2007, 64, 509. [DOI] [PubMed] [Google Scholar]
- [54].Fuard D, Tzvetkova-Chevolleau T, Decossas S, Tracqui P, Schiavone P, Microelectron. Eng 2008, 85, 1289. [Google Scholar]
- [55].Cheng Q, Sun Z, Meininger GA, Almasri M, Rev. Sci. Instrum 2010, 81, 2008. [DOI] [PubMed] [Google Scholar]
- [56].Ghassemi S, Rossier O, Sheetz MP, Wind SJ, Hone J, J. Vac. Sci. Technol. B Microelectron. Nanom. Struct 2009, 27, 3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Taylor RE, Kim K, Sun N, Park SJ, Sim JY, Fajardo G, Bernstein D, Wu JC, Pruitt BL, Biomed. Microdevices 2013, 15, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Desai RA, Yang MT, Sniadecki NJ, Legant WR, Chen CS, J. Vis. Exp 2007, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Beussman KM, Rodriguez ML, Leonard A, Taparia N, Thompson CR, Sniadecki NJ, Methods 2016, 94, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Al-Rekabi Z, Wheeler MM, Leonard A, Fura AM, Juhlin I, Frazar C, Smith JD, Park SS, Gustafson JA, Clarke CM, et al. , J. Cell Sci 2016, 129, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Al-Rekabi Z, Fura AM, Juhlin I, Yassin A, Popowics TE, Sniadecki NJ, Cell Adhes. Migr 2019, 13, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ, J. Biomech. Eng 2014, 136, 051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao Y, Zhang X, Sensors Actuators, A Phys. 2006, 125, 398. [Google Scholar]
- [64].Broughton K, Russell B, Biomech. Model Mechanobiol 2015, 14, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yang X, Rodriguez M, Leonard A, Sun L, Fischer K, Wang Y, Ritterhoff J, Zhao L, Kolwicz S, Pabon L, et al. , Stem Cell Reports 2019, 13, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zaunbrecher RJ, Abel AN, Beussman K, Leonard A, von Frieling-Salewsky M, Fields PA, Pabon L, Reinecke H, Yang X, Macadangdang J, et al. , Circulation 2019, CIRCULATIONAHA.119.039521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Miklas H, Clark JW, Levy EC, Detraux S, Leonard D, Beussman A, Showalter K, Hofsteen M, Yang P, Macadangdang X, Raftery J, Madan D, Kim A, Murry D-H, Fiehn C, Sniadecki O, Wang N, Ruohola-Baker Y, Prep. 2019, DOI 10.1038/s41467-019-12482-1. [DOI] [Google Scholar]
- [68].Yang X, Rodriguez M, Pabon L, Fischer K, Reinecke H, Regnier M, Sniadecki N, Ruohola-Baker H, Murry C, J. Mol. Cell. Cardiol 2014, 72, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Feghhi S, Tooley WW, Sniadecki NJ, J. Biomech. Eng 2016, 138, 104506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liang XM, Han SJ, Reems JA, Gao D, Sniadecki NJ, Lab Chip 2010, 10, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Feghhi S, Munday AD, Tooley WW, Rajsekar S, Fura AM, Kulman JD, López JA, Sniadecki NJ, Biophys. J 2016, 111, 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR, Science (80-. ). 2003, 302, 1704. [DOI] [PubMed] [Google Scholar]
- [73].Cheng Q, Sun Z, Meininger G, Almasri M, Sensors Actuators, B Chem 2013, 188, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sarangi BR, Gupta M, Doss BL, Tissot N, Lam F, Mège RM, Borghi N, Ladoux B, Nano Lett. 2017, 17, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Buxboim A, Discher DE, Nat. Methods 2010, 7, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Saez A, Anon E, Ghibaudo M, Du Roure O, Di Meglio JM, Hersen P, Silberzan P, Buguin A, Ladoux B, J. Phys. Condens. Matter 2010, 22, DOI 10.1088/0953-8984/22/19/194119. [DOI] [PubMed] [Google Scholar]
- [77].Ghassemi S, Biais N, Maniura K, Wind SJ, Sheetz MP, Hone J, J. Vac. Sci. Technol. B Microelectron. Nanom. Struct 2008, 26, 2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wei J, Shi J, Wang B, Tang Y, Tu X, Roy E, Ladoux B, Chen Y, Microelectron. Eng 2016, 158, 22. [Google Scholar]
- [79].Kim EJ, Fleischman AJ, Muschler GF, Roy S, Biomed. Microdevices 2013, 15, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS, Proc. Natl. Acad. Sci 2010, 107, 9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Maruthamuthu V, Sabass B, Schwarz US, Gardel ML, Proc. Natl. Acad. Sci 2011, 108, 4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Broussard JA, Yang R, Huang C, Nathamgari SSP, Beese AM, Godsel L, Hegazy M, Lee S, Zhou F, Sniadecki N, et al. , Mol. Biol. Cell 2017, 28, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chen T, Saw TB, Mège R-M, Ladoux B, J. Cell Sci. 2018, 131, jcs218156. [DOI] [PubMed] [Google Scholar]
- [84].du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B, Proc. Natl. Acad. Sci 2005, 102, 2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liu Z, Sniadecki NJ, Chen CS, Cell. Mol. Bioeng 2010, 3, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ting LH, Jahn JR, Jung JI, Shuman BR, Feghhi S, Han SJ, Rodriguez ML, Sniadecki NJ, Am. J. Physiol. Circ. Physiol 2012, 302, H2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Han SJ, Rodriguez ML, Al-Rekabi Z, Sniadecki NJ, Cell Adhes. Migr 2016, 10, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wang Z, Geng Y, J. Biomech. Eng 2015, 137, 031006. [DOI] [PubMed] [Google Scholar]
- [89].Hui J, Pang SW, Biosci. Rep 2019, 39, BSR20181596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Raman PS, Paul CD, Stroka KM, Konstantopoulos K, Lab Chip 2013, 13, 4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tamiello C, Bouten CVC, Baaijens FPT, Sci. Rep 2015, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Sniadecki NJ, Lamb CM, Liu Y, Chen CS, Reich DH, Rev. Sci. Instrum 2008, 79, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, Kirschner SB, Liu Y, Reich DH, Chen CS, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Nagayama K, Inoue T, Hamada Y, Matsumoto T, J. Biomech 2017, 65, 194. [DOI] [PubMed] [Google Scholar]
- [95].Khademolhosseini F, Chiao M, J. Microelectromechanical Syst 2013, 22, 131. [Google Scholar]
- [96].Lin YC, Kramer CM, Chen CS, Reich DH, Nanotechnology 2012, 23, DOI 10.1088/0957-4484/23/7/075101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Nagayama K, Inoue T, Hamada Y, Sugita S, Matsumoto T, Biomed. Microdevices 2018, 20, DOI 10.1007/s10544-018-0328-y. [DOI] [PubMed] [Google Scholar]
- [98].Mann JM, Lam RHW, Weng S, Sun Y, Fu J, Lab Chip 2012, 12, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ting LH, Feghhi S, Taparia N, Smith AO, Karchin A, Lim E, John AS, Wang X, Rue T, White NJ, et al. , Nat. Commun 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Rutering J, Ilmer M, Recio A, Coleman M, Vykoukal J, Alt E, Orleans N, Nat. Rev Drug Discov 2016, 5, 1. [Google Scholar]
- [101].Grayson ACR, Shawgo RS, Johnson AM, Flynn NT, Li Y, Cima MJ, Langer R, Proc. IEEE 2004, 92, 6. [Google Scholar]
- [102].Petersen KE, Micromechanics MEMS Class. Semin. Pap. to 1990 1997, 70, 58. [Google Scholar]
- [103].Hopcroft MA, Nix WD, Kenny TW, J. Microelectromechanical Syst 2010, 19, 229. [Google Scholar]
- [104].Madou MJ, Fundamentals of Microfabrication, 2002. [Google Scholar]
- [105].Galbraith CG, Sheetz MP, Proc. Natl. Acad. Sci. U. S. A 1997, 94, 9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jagannathan R, Alireza T, Saif MTA, Biophys. J 2010, 99, 3208.21081068 [Google Scholar]
- [107].Haase K, Pelling AE, Soc JR. Interface 2015, 12, DOI 10.1098/rsif.2014.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Polacheck WJ, Chen CS, Nat. Methods 2016, 13, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Petersen NO, McConnaughey WB, Elson EL, Proc. Natl. Acad. Sci 1982, 79, 5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chaudhuri O, Parekh SH, Lam WA, Fletcher DA, Nat. Methods 2009, 6, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tortonese M, Eng. Med. Biol 1997. [DOI] [PubMed] [Google Scholar]
- [112].Kirmizis D, Logothetidis S, Int. J. Nanomedicine 2010, 5, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Alonso JL, Goldmann WH, Life Sci. 2003, 72, 2553. [DOI] [PubMed] [Google Scholar]
- [114].Lam WA, Chaudhuri O, Crow A, Webster KD, De Li T, Kita A, Huang J, Fletcher DA, Nat. Mater 2011, 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Tooley WW, Feghhi S, Han SJ, Wang J, Sniadecki NJ, J. Micromechanics Microengineering 2011, 21, DOI 10.1088/0960-1317/21/5/054013. [DOI] [Google Scholar]
- [116].Ting LH, Feghhi S, Han SJ, Rodriguez ML, Sniadecki NJ, J. Nanotechnol. Eng. Med 2012, 2, 041006. [Google Scholar]
- [117].Pandey P, Hawkes W, Hu J, Megone WV, Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, et al. , Dev. Cell 2018, 44, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, Sheetz MP, Hone J, Proc. Natl. Acad. Sci 2012, 109, 5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sandmann R, Henriques SSG, Rehfeldt F, Köster S, Soft Matter 2014, 10, 2365. [DOI] [PubMed] [Google Scholar]
- [120].Elson EL, Genin GM, Interface Focus 2016, 6, DOI 10.1098/rsfs.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vandenburgh H, Shansky J, Benesch-Lee F, Barbata V, Reid J, Thorrez L, Valentini R, Crawford G, Muscle and Nerve 2008, 37, 438. [DOI] [PubMed] [Google Scholar]
- [122].Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS, Proc. Natl. Acad. Sci 2009, 106, 10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Hansen A, Eder A, Bönstrup M, Flato M, Mewe M, Schaaf S, Aksehirlioglu B, Schwörer A, Uebeler J, Eschenhagen T, Circ. Res 2010, 107, 35. [DOI] [PubMed] [Google Scholar]
- [124].Ramade A, Legant WR, Picart C, Chen CS, Boudou T, Microfabrication of a Platform to Measure and Manipulate the Mechanics of Engineered Microtissues, Elsevier Inc, 2014. [DOI] [PubMed] [Google Scholar]
- [125].West AR, Zaman N, Cole DJ, Walker MJ, Legant WR, Boudou T, Chen CS, Favreau JT, Gaudette GR, Cowley EA, et al. , Am. J. Physiol. Cell. Mol. Physiol 2013, 304, L4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS, Tissue Eng. Part A 2012, 18, 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Truitt R, Mu A, Corbin EA, Vite A, Brandimarto J, Ky B, Margulies KB, JACC Basic to Transl. Sci 2018, 3, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Toepke MW, Beebe DJ, Lab Chip 2006, 6, 1484. [DOI] [PubMed] [Google Scholar]
- [129].Bielawski KS, Leonard A, Bhandari S, Murry CE, Sniadecki NJ, Tissue Eng. - Part C Methods 2016, 22, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Leonard A, Bertero A, Powers JD, Beussman KM, Bhandari S, Regnier M, Murry CE, Sniadecki NJ, J. Mol. Cell. Cardiol 2018, 118, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Mannhardt I, Warncke C, Trieu HK, Müller J, Eschenhagen T, J. Tissue Eng. Regen. Med 2019, 13, 3. [DOI] [PubMed] [Google Scholar]
- [132].Legant WR, Chen CS, Vogel V, Integr. Biol. (United Kingdom) 2012, 4, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Chen Z, Lu J, Zhang C, Hsia I, Yu X, Marecki L, Marecki E, Asmani M, Jain S, Neelamegham S, et al. , Nat. Commun 2019, 10, 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sun X, Nunes SS, J. Vis. Exp 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wang EY, Rafatian N, Zhao Y, Lee A, Lai BFL, Lu RX, Jekic D, Davenport Huyer L, Knee-Walden EJ, Bhattacharya S, et al. , ACS Cent. Sci 2019, 5, 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Zhao Y, Wang EY, Davenport LH, Liao Y, Yeager K, Vunjak-Novakovic G, Radisic M, Zhang B, Adv. Healthc. Mater 2019, 8, 1. [DOI] [PubMed] [Google Scholar]
- [137].Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, Conklin BR, Grigoropoulos CP, Healy KE, Biomaterials 2014, 35, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sun X, Nunes SS, Methods 2016, 101, 21. [DOI] [PubMed] [Google Scholar]
- [139].Conant G, Ahadian S, Zhao Y, Radisic M, Sci. Rep 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Xiao Y, Zhang B, Liu H, Miklas JW, Gagliardi M, Pahnke A, Thavandiran N, Sun Y, Simmons C, Keller G, et al. , Lab Chip 2014, 14, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Ma Z, Huebsch N, Koo S, Mandegar MA, Siemons B, Boggess S, Conklin BR, Grigoropoulos CP, Healy KE, Nat. Biomed. Eng 2018, 2, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sharma P, Ng C, Jana A, Padhi A, Szymanski P, Lee JSH, Behkam B, Nain AS, Mol. Biol. Cell 2017, 28, 2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Sheets K, Wunsch S, Ng C, Nain AS, Acta Biomater. 2013, 9, 7169. [DOI] [PubMed] [Google Scholar]
- [144].Pakshir P, Alizadehgiashi M, Wong B, Coelho NM, Chen X, Gong Z, Shenoy VB, McCulloch C, Hinz B, Nat. Commun 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]


