Abstract
Background:
Prospective studies have suggested higher factor VIII (FVIII) levels is an independent risk factor for coronary heart disease (CHD) and stroke. However, limited information, including on genetic and epigenetic contributors to FVIII variation, is available specifically among African Americans (AAs), who have higher FVIII levels than Europeans.
Objectives:
We measured FVIII levels in ~3,400 AAs from the community-based Jackson Heart Study and assessed genetic, epigenetic, and epidemiological correlates of FVIII, as well as incident cardiovascular disease (CVD) associations.
Methods:
We assessed cross-sectional associations of FVIII with CVD risk factors as well as incident CHD, stroke, heart failure, and mortality associations. We additionally assessed associations with TOPMed whole genome sequencing data and an epigenome-wide methylation array.
Results:
Our results confirmed associations between FVIII and risk of incident CHD events and total mortality in AAs; mortality associations were largely independent of traditional risk factors. We also demonstrate an association of FVIII with incident heart failure, independent of B-type natriuretic peptide. Two genomic regions were strongly associated with FVIII (ABO and VWF). The index variant at VWF is specific to individuals of African descent and is distinct from the previously reported European VWF association signal. Epigenome-wide association analysis showed significant FVIII associations with several CpG sites in the ABO region. However, after adjusting for ABO genetic variants, ABO CpG sites were not significant.
Conclusions:
Larger sample sizes of AAs will be required to discover additional genetic and epigenetic contributors to FVIII phenotypic variation, which may have consequences for CVD health disparities.
Keywords: factor VIII, coagulation, thrombosis, GWAS, African American
INTRODUCTION
Coagulation factor VIII (FVIII) circulates bound to von Willebrand factor (VWF) and serves as a co-factor for factor IX-mediated activation of factor X, which ultimately generates a fibrin blood clot. Mutations of the factor VIII gene (F8) result in low levels of FVIII and the hereditary X-linked bleeding disorder hemophilia A [1] . Conversely, higher basal levels of FVIII are a risk factor for primary and recurrent venous thromboembolism (VTE) [2]. FVIII is an acute phase protein and levels tend to correlate with other inflammation biomarkers as well as traditional cardiovascular disease (CVD) risk factors such as age, body mass index (BMI), and diabetes [3]. Nonetheless, in some studies, higher FVIII levels were an independent risk factor for arterial thrombotic disease such as myocardial infarction (MI) or stroke [4-9], as well as overall mortality[10]. More recently, instrumental variable or Mendelian randomization analyses of FVIII have suggested FVIII levels may be causally related to both CHD and VTE risk [11].
Cardiovascular diseases, including MI, ischemic stroke, venous thromboembolic disease (VTE), and heart failure (HF) disproportionately affect African Americans (AAs) [12, 13]. FVIII levels are higher among AAs than individuals of European ancestry (EAs) [14, 15] and may be a stronger risk factor for VTE in AAs than EAs [16, 17], but the role of FVIII as a risk factor for CVD outcomes has been less well-studied among AAs [18].
Genetic factors contribute to inter-individual variation in FVIII levels, with heritability estimates in the range of 40-60% [19-21]. A major determinant is ABO blood group [19], but familial aggregation of high FVIII levels persists even after adjustment for ABO [22]. Through genome-wide association studies (GWAS) and exome studies, several additional FVIII associated loci have been discovered, though these studies were conducted primarily in individuals of European descent [11, 23-25].
Compared to traditional GWAS, whole genome sequencing (WGS) assesses genetic variants (both coding and non-coding) in the lower frequency range, as well as African population-specific variants poorly represented on genotyping arrays and current imputation reference panels [26]. Epigenetic factors can also influence complex traits such as FVIII level, but association of DNA methylation at a genome-wide scale with FVIII levels in population-based samples has not been previously examined. To further characterize the epidemiologic, genetic, and epigenetic correlates of FVIII, and the relationship of FVIII to CVD risk in AAs, we performed a series of analyses in ~3,400 AAs from the Jackson Heart Study (JHS).
METHODS
The Jackson Heart Study (JHS)
Between 2000 and 2004, JHS recruited 5,306 AA participants from the Jackson, Mississippi metropolitan area. A range of measures, including traditional and putative CVD risk factors, health behaviors, detailed demographic, socioeconomic and sociocultural factors, medication use, anthropometry, blood pressure, assessments of kidney function and diabetes, and biochemical analytes, were obtained at the baseline JHS examination and in subsequent clinic visits [23]. The current analysis is confined to 3,493 individuals who had FVIII measured as part of the JHS ancillary study “Thrombosis Genetics in African Americans” and gave consent that allows genetic research (Figure S1). Computed tomography (CT), ultrasound, and echocardiographic imaging data collection, reading, and quality control in JHS for assessment of carotid IMT, Left ventricular mass index (LVMI), LV hypertrophy (LVH), Ankle brachial index (ABI), coronary artery calcification (CAC) and abdominal aortic calcification (AAC), have been previously described [27-29]. All-cause mortality and incident coronary heart disease (CHD) and stroke events were adjudicated from the beginning of the study through 2014, while adjudication of incident HF events began in 2005 [30]. Overall CHD includes fatal CHD, myocardial infarction, coronary artery bypass surgery, or angioplasty. Hard CHD includes fatal CHD and myocardial infarction. Strokes were defined according to the World Health Organization definition and include both ischemic and hemorrhagic subtypes. Individuals with a prior history of stroke or CHD (prior to 2000) or HF (prior to 2005) or who did not consent to medical record abstraction were excluded from incident event analyses. Median follow-up for mortality is ~14 years, ~12 years for stroke and CHD, and ~10 years for HF.
Laboratory measurements
FVIII antigen level (as the percent of pooled normal plasma) was measured at the University of Vermont using EDTA plasma from the JHS baseline exam and a sandwich ELISA (Affinity Biologicals). Values above the upper limit of detection were set to 800%. High-sensitivity C-reactive protein (CRP), total cholesterol, high-density lipoprotein cholesterol (HDLc) and triglycerides (TG), serum creatinine and B-type natriuretic peptide (BNP) were measured as previously described.
Statistical Analysis of FVIII with CVD risk factors and outcomes
Cross-sectional associations of FVIII with baseline JHS participant characteristics and with measures of subclinical CVD were assessed using generalized estimating equations (GEE), to account for familial correlation, with FVIII as the independent variable and covariate adjustment for age and sex, and baseline characteristics and subclinical CVD measures treated as dependent variables. Effect estimates were reported per standard deviation (SD) change in FVIII. AAC, CAC, LVMI, cIMT, CRP, TG, and BMI (in JHS) were natural log(LN)-transformed prior to analysis. Cox proportional hazards models with sandwich variance estimator, to account for relatedness in the sample, were used to calculate hazard ratios and 95% confidence intervals (CI) for covariate-adjusted associations with all-cause mortality and incident CVD events. Associations were reported both using FVIII as a continuous trait (transformed as a z-score) and as a categorical variable divided into FVIII quartiles. All associations were assessed using SAS 9.3. Heritability of FVIII was estimated using a subset of 1,578 related JHS individuals from 433 families, adjusting for age and sex [30].
Whole genome sequencing (WGS) and FVIII association analysis
Eligible JHS participants underwent ~30X WGS at the Northwest Genomics Center at University of Washington through the NHLBI Trans-Omics for Precision Medicine (TOPMed) project. Details of the sequencing, variant calling, and quality control (QC) protocols used in TOPMed are described at https://www.nhlbiwgs.org/data-sets. Regression of inverse normalized FVIII on genotype was adjusted for age, sex, and the first ten principal components (PCs) for global ancestry. We used a linear mixed model approach to account for familial relationships, as implemented in SAIGE on the University of Michigan ENCORE server (https://encore.sph.umich.edu) [35]. We included 31,176,270 single nucleotide variants and small indels with sequence depth >10 and minor allele count >20. A significance threshold of 1 × 10−9 was used for single variant analyses [31]. To assess the number of distinct signals at a given locus, we performed step-wise conditional regression analysis.
We also performed genome-wide gene-based testing for rare variants with inverse-normalized FVIII using the mixed model SMMAT aggregate association testing method (adjusting for potential relatedness using an estimated kinship matrix and covariates) using Wu weights for each variant [32]. We aggregated variants for association tests grouping variants by gene and restricting to variants of MAF<1% that are either loss of function or missense and predicted to be pathogenic on the basis of a FATHMM-XF coding score>0.5 [33]. We use a Bonferroni correction for the number of tested genes containing more than one polymorphic variant (n= 18,750, p< 2.67 × 10−6).
Global and local ancestry estimation and admixture mapping
We utilized an earlier version of TOPMed WGS (freeze 5b, September 2017) to estimate global and local ancestry among JHS participants (n=2,958). Using as a reference panel 37 African, 35 European, and 20 Native American individuals with phased sequence data (for chromosomes 1-22) [34], we used RFMix version 1.5.4 [35] to infer the number of alleles inherited from each ancestral population (African, European, Native American). To estimate the overall admixture proportions for each JHS participant, we calculated the genome-wide average local ancestry. We used GENESIS [36] to perform admixture mapping using a linear mixed model, investigating each ancestral group (African, European, and Native American) separately, adjusting for age, sex and overall admixture proportions as fixed effects. To account for relatedness, we included ancestry-adjusted kinship estimates as a random effect. We used the genome-wide p-value significance threshold of 5.5×10−6, as estimated using the test statistic simulation approach described in [37].
Epigenome-wide analysis
Illumina Methylation EPIC array data (containing over 850,000 CpG methylation sites) was generated using blood samples collected at the JHS baseline exam. Methylation β values (the ratio of intensities between methylated and un-methylated alleles) were normalized with respect to background color intensity using the normal-exponential out-of-band (NOOB) preprocessing method in the R package minfi[38]. Cell counts (granulocytes, monocytes, natural killer, CD4+ T lymphocytes, naïve CD8+ T lymphocytes, exhausted cytotoxic CD8+ T cells (defined as CD8 positive CD28 negative CD45R negative), and plasma blasts) were estimated according to the method of Houseman et al and Horvath et al [39, 40]. Methylation β values were adjusted for important batch covariates (sample batch, plate, and plate position) using Combat as implemented in the sva and ChAMP R packages. Epigenome-wide association analysis (EWAS) was performed using ln transformed FVIII as the dependent variable, adjusted for age, sex, the first ten ancestry PCs from Affymetrix 6.0 GWAS array data, and estimated cell counts (n=1670). EWAS was performed using linear mixed models in R. For top CpGs, we adjusted for family structure as a random effect in the R package lmer; this was done for presented results. Effect sizes were based on Pearson correlation coefficients. We performed sensitivity analyses adjusting top CpGs for potential confounders of methylation levels (BMI, smoking, and socioeconomic status (SES) (as represented by income)), in a reduced sample size of n=1430. To assess statistical significance of findings, we used a genome-wide significance threshold of P = 3.6 × 10−8 [41].We removed lead CpGs which overlap common SNPs in African populations from 1000 Genomes, based on suggested masking from http://zwdzwd.github.io/InfiniumAnnotation [42].
RESULTS
Association of FVIII with cardiovascular risk factors and subclinical CVD
Of the 3,493 JHS participants included in the current analysis, the mean age was 55.6 years (range 21- 93), 38% were male, 13% were current smokers, 54% were obese, 57% had hypertension, 23% had diabetes, 11% had a prior history of CVD, and 2.9% were taking anticoagulant medication. FVIII levels ranged between 16% and 800% (median 135%, mean 145%, SD 59%). FVIII levels were strongly correlated with age and were higher in women (mean 149%, SD 60%) than men (mean 139%, SD 57%) (Table 1). One male participant had circulating levels (FVIII=16%) compatible with a diagnosis of mild hemophilia A, but bleeding history in this individual is unknown.
Table 1.
Associations between factor VIII and cardiovascular disease risk factors in the Jackson Heart Study (JHS), reported as the difference in the listed variable per standard deviation higher factor VIII. The mean (SD) or, for dichotomous variables, %, for each variable (untransformed) is also listed.
| Trait | Mean (SD) or % | N | β | Standard Error |
p-value |
|---|---|---|---|---|---|
| Age (years) | 55.59 (12.80) | 3493 | 2.91 | 0.25 | <0.0001 |
| Male sex | 37.79% | 3493 | −0.17 | 0.04 | <0.0001 |
| Current smoker | 13.31% | 3463 | −0.07 | 0.06 | 0.25 |
| Ln BMI (kg/m2) | 31.89 (7.31) | 3486 | 0.02 | 0.004 | <0.0001 |
| Waist (cm) | 101.2 (16.27) | 3486 | 2.04 | 0.29 | <0.0001 |
| Systolic BP (mmHg) | 127.37 (16.61) | 3487 | 0.32 | 0.29 | 0.26 |
| Diastolic BP (mmHg) | 75.77 (8.75) | 3487 | −0.18 | 0.14 | 0.20 |
| Fasting glucose (mg/dL) | 90.45 (8.86) | 2591 | 0.47 | 0.17 | 0.01 |
| Total Cholesterol (mg/dL) | 199.21 (40.62) | 3238 | 1.07 | 0.79 | 0.18 |
| LDLc (mg/dL) | 126.49 (36.94) | 3205 | 0.35 | 0.68 | 0.61 |
| HDLc (mg/dL) | 51.64 (14.78) | 3237 | −1.03 | 0.29 | 0.0003 |
| Triglycerides (mg/dL) | 107.57 (82.18) | 3238 | 0.07 | 0.01 | <0.0001 |
| C-reactive protein (mg/dL) | 0.53 (0.98) | 3487 | 0.20 | 0.02 | <0.0001 |
| Hypertension | 57.34% | 3493 | 0.10 | 0.05 | 0.01 |
| Diabetes | 23.15% | 3491 | 0.28 | 0.05 | <0.0001 |
Models (other than those for age and sex) are adjusted for age and sex.
Abbreviations: SD=standard deviation; HDLc=high density lipoprotein cholesterol; LDLc=low density lipoprotein cholesterol; BP=blood pressure.
BMI, C-reactive protein, and triglycerides were natural log transformed.
Fasting glucose was only tested in those without diabetes at visit 1.
In analyses adjusted for age and sex), higher BMI, larger waist circumference, higher TG, higher CRP, lower HDLc, higher fasting glucose, diabetes, and hypertension were each significantly associated with higher FVIII (all P<0.01) (Table 1). In a multivariate regression model containing terms for all CVD risk factors significantly associated with FVIII (P<0.01) (see Table 1), age, diabetes, triglycerides, and CRP remained strongly associated with higher FVIII levels (all P<0.001). As shown in Table 2, among subclinical disease outcomes available in JHS, higher FVIII was nominally associated with LVH (P=0.01). There was no evidence of association of FVIII with ABI, cIMT, LVMI, continuous or dichotomous AAC or CAC (all P>0.05).
Table 2.
Associations between factor VIII and subclinical disease measures in the Jackson Heart Study (JHS), reported as the difference in the listed variable per standard deviation higher FVIII. The mean (SD) or, for dichotomous variables, %, for each variable is also listed.
| Trait | Mean (SD) or % | N | β | Standard Error | p-value |
|---|---|---|---|---|---|
| Carotid IMT (mm) | 0.73 (0.19) | 3318 | 0.0001 | 0.003 | 0.97 |
| CAC (Agatston score) | 167.8 (506.93) | 1938 | 0.08 | 0.06 | 0.17 |
| AAC (Agatston score) | 895.77 (1629.07) | 1937 | 0.02 | 0.07 | 0.75 |
| Any CAC | 48.86% | 1938 | 0.003 | 0.06 | 0.96 |
| Any AAC | 66.39% | 1937 | −0.004 | 0.07 | 0.95 |
| Ankle-brachial index | 1.21 (0.17) | 3100 | −0.003 | 0.003 | 0.32 |
| LVMI (g/m2) | 36.31 (9.79) | 2233 | 0.003 | 0.006 | 0.58 |
| Left ventricular hypertrophy | 7.84% | 2233 | 0.17 | 0.07 | 0.01 |
Carotid IMT, CAC, AAC, and LVMI were natural log transformed prior to analysis. Any CAC and any AAC are reported as dichotomous variables (AAC>0 vs. AAC=0). AAC and CAC measures are from visit 2, not visit 1 when FVIII was measured. Models are adjusted for age and sex.
Abbreviations: IMT=intima media thickness; CAC=Coronary artery calcium; AAC=Abdominal aortic calcium; ABI=Ankle-brachial index; LVMI=Left ventricular mass index
Association of FVIII with incident clinical outcomes
In Cox models minimally adjusted for age and sex, continuous FVIII level was significantly associated with overall CHD, hard CHD, HF, and death (all P ≤ 0.05), but not with stroke (Table 3). These associations were somewhat attenuated upon adjustment for traditional CVD risk factors, including CRP (Table 3). In the model adjusted for CVD risk factors including CRP, the p-values for association with HF and mortality remained significant (both P < 0.05), and the association with hard CHD was marginally significant (P = 0.05). The hazard ratios (HR) per standard deviation increase in FVIII were 1.15 (95% confidence interval (CI) 1.03 – 1.28), 1.16 (1.08 – 1.25), and 1.15 (1.00 – 1.33) for HF, mortality, and hard CHD, respectively. When the FVIII association with HF was additionally adjusted for BNP, the HR remained significant (1.14; 95% CI 1.02 – 1.28; P=0.02). When the FVIII association with mortality was additionally adjusted for BNP, white blood count (WBC), and estimated glomerular filtration rate (eGFR), this association also remained significant (HR 1.15; 95% CI 1.06 – 1.24 per SD unit; P = 0.001).
Table 3:
Association between factor VIII and risk of clinical outcomes in the Jackson Heart Study
| Model 1 | Model 2 | Model 3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outcome | N | Events | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |||
| Stroke | 2990 | 110 | 1.09 | 0.92 | 1.30 | 0.31 | 1.06 | 0.87 | 1.28 | 0.57 | 1.02 | 0.84 | 1.24 | 0.87 |
| Overall CHD | 2899 | 147 | 1.13 | 1.00 | 1.28 | 0.05 | 1.06 | 0.93 | 1.22 | 0.37 | 1.05 | 0.92 | 1.21 | 0.46 |
| Hard CHD | 2905 | 101 | 1.22 | 1.08 | 1.39 | 0.001 | 1.16 | 1.01 | 1.35 | 0.04 | 1.15 | 1.00 | 1.33 | 0.05 |
| Heart Failure | 2755 | 190 | 1.21 | 1.10 | 1.32 | <0.0001 | 1.15 | 1.03 | 1.28 | 0.01 | 1.15 | 1.04 | 1.28 | 0.01 |
| Mortality | 3172 | 559 | 1.22 | 1.14 | 1.30 | <0.0001 | 1.18 | 1.10 | 1.27 | <0.0001 | 1.16 | 1.08 | 1.25 | <0.0001 |
Abbreviations: CHD=coronary heart disease; HR=hazard ratio; CI=confidence interval; CRP=C-reactive protein.
Hazard ratios are reported per standard deviation of factor VIII. Only individuals with complete covariates for all models are included. Model 1: Adjusted for age, sex; Model 2: Model 1 + BMI, blood pressure medications, type 2 diabetes, systolic blood pressure, total cholesterol, high-density lipoprotein cholesterol, current smoking; Model 3: Model 2 + CRP
When analyzed according to FVIII quartiles (Table S1), there again was a significant linear increase in risk of hard CHD, HF, and death in the minimally-adjusted model (P≤0.05), but only HF and mortality remained significant in the fully adjusted model. Individuals in the upper quartile of FVIII among JHS participants had an estimated 2.35-fold increased risk of HF (1.44 – 3.84) and 1.97-fold increased risk of death (1.50 – 2.59), compared to those in the bottom quartile.
Genetic association analysis of FVIII
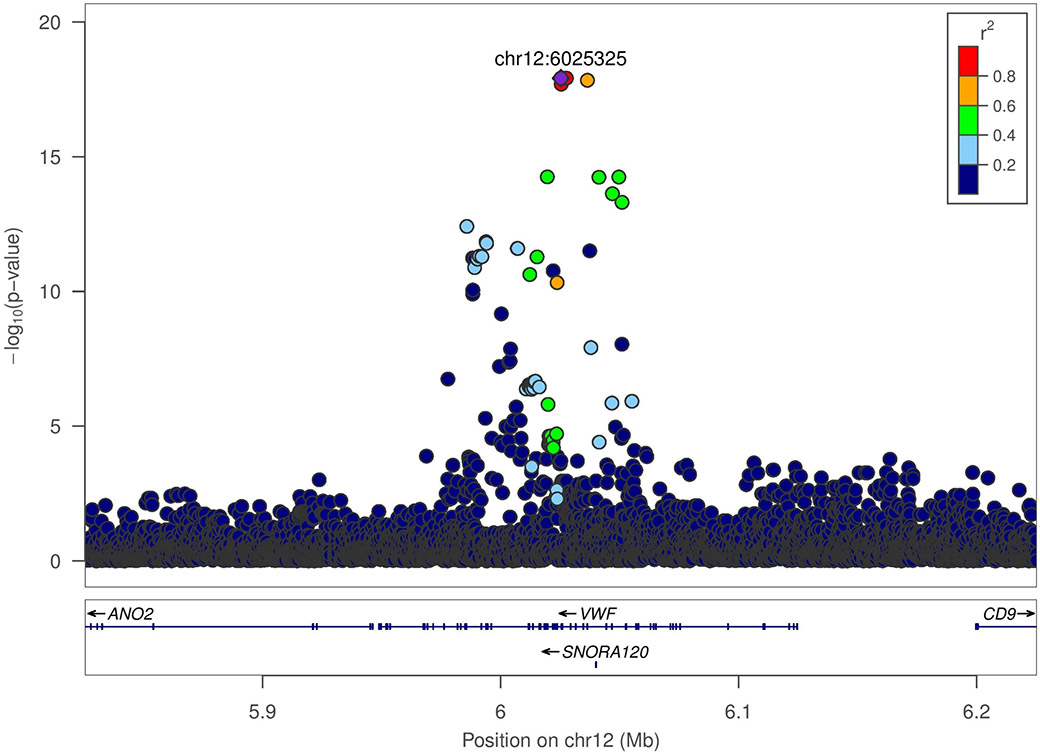
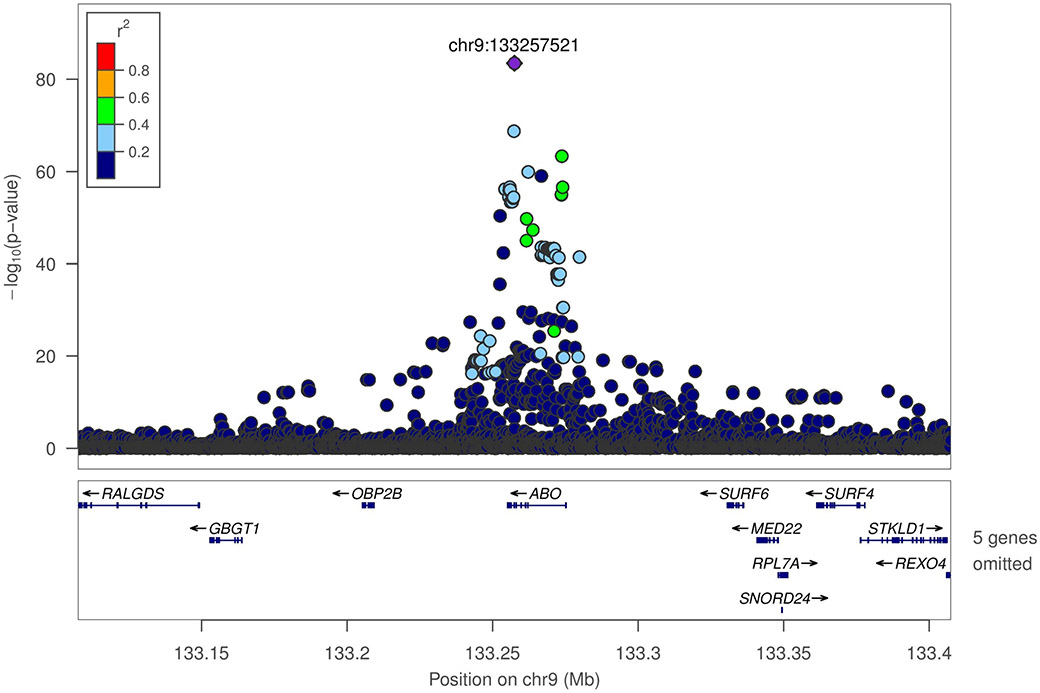
Adjusting for age and sex, heritability (h2) of inverse normalized natural log transformed FVIII was estimated as 0.47 (standard error (SE) = 0.06, p= 2.69 × 10−18) in JHS. In WGS-based association analysis of 3,349 JHS TOPMed participants, two genomic regions were strongly associated with FVIII (Figure S2). On chromosome 9q34, there was a broad association peak containing 312 genome-wide significant variants (P<1 × 10−9) centered on the ABO gene (index variant chr9:133257521_T/TC or rs8176719, MAF of 0.29, associated with 0.52 ± 0.03 SD unit higher FVIII, P=5 × 10−84) (Figure 1A). The effect allele of the 1 base pair (bp) indel index variant corresponds to the non-O allele of the ABO blood group. Upon conditional analysis adjusting for the O/non-O rs8176719 variant, the next strongest independent signal was chr9:133255669_CG/C_rs56392308 (MAF=0.064, P= 4.5 × 10−15) associated with 0.46 ± 0.05 lower FVIII, another 1 bp indel which encodes the A2 allele (ABO*A2.06). After a second round of conditional analysis adjusting for both O/non-O and A2 indels, there was a residual marginally significant signal at chr9:133264269_C/T (rs41302905 or rs141515001) associated with lower FVIII (MAF=0.003; P=5×10−5). This variant is part of an extended haplotype which includes two missense variants, rs41302905 and rs55876802, which together comprise part of several O2 alleles (ABO*O.02).
Figure 1.
LocusZoom plots for ABO and VWF loci, with linkage disequilibrium calculated in Jackson Heart Study participants with TOPMed freeze 6 data.
A. VWF locus (rs115708869)
b. ABO locus (rs8176719)
The second FVIII-associated region located on chromosome 12p13.31 contained 33 genome-wide significant variants (P<1 × 10−9) centered around the VWF gene (Figure 1B). The peak association signal at the VWF locus included nine single nucleotide variants in near complete LD with one another (rs115708869, rs142033986, rs115364369, rs184911391, rs74731445, rs76100694, rs114537734, rs114018824, rs57950734; MAF of ~0.11 for each) that were associated with 0.34 ± 0.05 SD unit lower FVIII (P=1-2 × 10−18). Based on 1000 Genomes allele frequencies, each of these nine SNV are considerably more prevalent among African (MAF ~0.15) compared to non-African populations (MAF<0.001). Eight of the nine SNVs are intronic, whereas rs57950734 encodes the missense variant p.His817Gln, which was previously associated with lower FVIII in a VWF gene-based association analysis conducted in AAs [43]. Upon conditional regression analysis of FVIII in JHS adjusting for the index VWF variant rs115708869, the strongest remaining association was chr12:6044584_A/G or rs216294 (MAF=0.05, P=1 × 10−5) with the minor allele associated with higher FVIII.
In gene-based rare variant association analysis of FVIII, there were no associations that reached the genome-wide significance threshold of < 2.7 × 10−6 (Figure S3, Table S2).
We further compared our single variant WGS-based FVIII association results in JHS AAs to previously published FVIII GWAS or exome-based association analyses that have been performed predominantly in European ancestry individuals (Table 4). At the ABO locus, the O/non-O variant rs8176719 variant most strongly associated with FVIII in our WGS-based analysis in AAs is in moderate LD (r2=0.82 in EUR, r2=0.51 in AFR in 1000 Genomes phase 1 data) with the sentinel variant rs687289 reported in the largest existing FVIII GWAS (n~36,000 multi-ethnic individuals, of whom ~70% were European ancestry) [11]. As expected, rs687289 is also strongly associated with FVIII in JHS AAs (P=1.8 × 10−50). By contrast, the sentinel variant at the VWF locus reported in [11] (rs2238109; MAF=0.39; β=0.026; P=3.5E−24) was only nominally associated with FVIII in JHS AAs (MAF=0.42; β=0.06; P=0.019). Of other VWF coding variants previously reported to be associated with FVIII (or VWF) levels [48], we observed nominally significant and directionally consistent associations with FVIII for rs1063856 (Thr789Ala), rs216321 (Ala852Gln), and rs2229446 (Arg2185Gln) in our JHS AA samples. Two other variants (at STXBP5 and ST3GAL4) reported in [11] had p-values ~0.06 in JHS and the estimated beta coefficients were directionally consistent (Table 4). We were unable to replicate a previously reported MAT1A rs2236568 variant suggestively associated with FVIII in AAs [24].
Table 4.
Significant loci from previous genome-wide association studies or exome-wide association studies, with results from Jackson Heart Study (JHS) (N=3,348). Effect sizes from previous studies are reported in study-specific units (often but not always for inverse normalized Factor VIII) and should only be compared for direction of effect.
| rsID | Closest gene(s) |
Chr | Position (Build 38) |
Effect allele |
Other allele |
Effect allele frequency |
β | P | Effect allele frequency |
β | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sabater-Lleal M et al., Circulation, 2019 (N=up to 46,354; multi-ethnic; genome-wide association study, includes n= 4,500 African Americans) |
JHS (N=3,348) | ||||||||||
| rs548630 | FCHO2, TMEM171, TNPO1 | 5 | 73110832 | A | C | 0.49 | −0.016 | 2.1 x 10−10 | 0.75 | −0.016 | 0.56 |
| rs9390460 | STXBP5 | 6 | 147373198 | T | C | 0.47 | −0.019 | 2.2 x 10−15 | 0.51 | −0.045 | 0.065 |
| rs9271597 | HLA region | 6 | 32623514 | A | T | 0.41 | −0.015 | 1.4 x 10−08 | 0.33 | −0.041 | 0.11 |
| rs4276643 | SCARA5 | 8 | 27946082 | T | C | 0.66 | −0.023 | 1.3 x 10−19 | 0.54 | −0.036 | 0.13 |
| rs10102164 | SOX17, RP1 | 8 | 54509054 | A | G | 0.19 | 0.019 | 2.4 x 10−09 | 0.18 | 0.033 | 0.30 |
| rs687289 | ABO | 9 | 133261703 | A | G | 0.36 | 0.15 | 1.9 x 10−770 | 0.41 | 0.37 | 1.8 x 10−50 |
| 9:13930481 | LINC00583, NFIB | 9 | 13930481 | Del | Ins | 0.85 | −0.032 | 2.7 x 10−10 | 0.97 | −0.009 | 0.90 |
| rs35458154 | ST3GAL4 | 11 | 126426930 | A | G | 0.03 | 0.048 | 3.1 x 10−08 | 0.005 | 0.33 | 0.065 |
| rs4981022 | STX2 | 12 | 103756096 | A | G | 0.69 | 0.025 | 3.0 x 10−20 | 0.69 | 0.023 | 0.39 |
| rs2238109 | VWF | 12 | 6044801 | A | T | 0.39 | 0.026 | 3.5 x 10−24 | 0.58 | 0.059 | 0.019 |
| rs4904820 | TCN2 | 14 | 91852591 | A | G | 0.49 | 0.014 | 1.8 x 10−08 | 0.69 | 0.007 | 0.80 |
| rs150926226 | TMLHE, F8 | X | 155491696 | C | G | 0.62 | 0.017 | 3.3 x 10−09 | 0.43 | −0.001 | 0.98 |
| Huffman JE et al., Blood, 2015 (N=28,291; multi-ethnic; Exome-chip), results displayed are from multi- ethnic analysis, including n=6 079 African Americans, KATNB1 significant in African Americans only |
JHS (N=3,348) | ||||||||||
| rs7962217 | VWF | 12 | 5952393 | T | C | 0.046 | 5.16 | 2.5 x 10−13 | 0.02 | 0.15 | 0.13 |
| rs41276738 | VWF | 12 | 6034812 | T | C | 0.0040 | −16.89 | 2.2 x 10−13 | 0.001 | −1.02 | 0.001 |
| rs141041254 | STAB2 | 12 | 103759154 | A | G | 0.00087 | 26.81 | 2.1 x 10−8 | Not in JHS results (minor allele count <20) | ||
| rs1800291 | F8 | X | 154930010 | C | G | 0.27 | −1.73 | 8.20 x 10−8 | 0.33 | 0.008 | 0.71 |
| rs142508811 | KATNB1 | 16 | 57754931 | T | C | 0.00027 | 39.36 | 4.80 x 10−4 | 0.001 | 0.696 | 0.06 |
| Tang W et al., Am J Hematol, 2015 (N=20,941; multi-ethnic; IBC Illumina iSELECT array, African American results from n= 5,020 displayed here) |
JHS (N=3,348) | ||||||||||
| rs8176693 | ABO | 9 | 133262254 | T | C | 0.1 | 37.24 | 2.51 × 10−114 | 0.11 | 0.65 | 1.17× 10−60 |
| rs2236568 | MAT1A | 10 | 80276167 | C | A | 0.24 | 5.28 | 1.69 × 10−6 | 0.23 | 0.003 | 0.93 |
| rs2229446 | VWF | 12 | 5993906 | T | C | 0.19 | −9.47 | 1.95 × 10−20 | 0.21 | −0.21 | 1.44× 10−12 |
| rs1800380 | VWF | 12 | 6029429 | T | C | 0.3 | 5.72 | 5.62 × 10−11 | 0.32 | 0.06 | 0.02 |
| rs4764482 | VWF | 12 | 6060567 | C | T | 0.2 | −5.74 | 8.12 × 10−8 | 0.19 | −0.04 | 0.19 |
Admixture analysis of FVIII
In a subset of n=2,958 JHS participants with genome-wide ancestry estimates and FVIII measurements, each percentage point higher of age- and sex-adjusted African ancestry was associated with 28%±10% higher mean FVIII (P=0.006). After additional adjustment for other FVIII correlates (BMI, triglycerides, CRP, diabetes, hypertension) and SES, the association of African ancestry proportion with FVIII was no longer significant (β = 13% ± 11%; P=0.21). We performed an admixture mapping scan in JHS using genome-wide local-ancestry estimates for African and European ancestry. None of the regions tested reached the empirically-derived significance threshold of P<5.5 × 10−6 (Figure S4).
Epigenome-wide association analysis of FVIII
EWAS identified 30 genome-wide significant CpGs (P <3.6 × 10−8) associated with FVIII. Nine of these CpG sites are located in or near ABO (Table 5). The most significant CpG was cg21160290 in ABO (Pearson’s correlation coefficient = 0.19, P = 5.21 × 10−22) located in the ABO 3’ UTR. Following additional adjustment for potential environmental or lifestyle confounders of methylation levels (BMI, smoking, and SES), the strong signal near ABO remained significant, with the same lead CpG (cg21160290), whereas most of the other CpG associations became non-significant after these adjustments. (Table S3). Some of the CpG associations were likely attenuated due to reduced sample size (n= 1430) due to missing covariate data but others may have been confounded by BMI, smoking, or SES. Finally, when we conditioned all CpGs within 1 Mb on each side of cg21160290 on our lead ABO genetic association signal rs8176719 in n= 1,657 individuals overlapping the WGS and EWAS datasets; the EWAS signal in the ABO region was markedly attenuated (cg21160290 p=0.06); this result is unsurprising given the high correlation between ABO SNP genotypes and CpG methylation beta values (r=0.58 for cg21160290 and rs8176719). Non-ABO CpG associations were not further examined, as they were not coincident with a genetic signal, and require further replication in additional African American cohorts.
Table 5.
Top CpG sites associated with factor VIII levels in Jackson Heart Study (N=1,670), with a p-value < 3.6 × 10−8. Models are adjusted for age, sex, cell counts, and 10 ancestry principal components, as well as for family as a random effect.
| CpG | Chr | Position (Build 37) |
Position (Build 38) |
Gene | Pearson Correlation |
p-value |
|---|---|---|---|---|---|---|
| cg21160290 | 9 | 136149941 | 133274525 | ABO | 0.19 | 5.21 x 10−22 |
| cg12020464 | 9 | 136131183 | 133255796 | ABO | −0.20 | 1.94 x 10−21 |
| cg22535403 | 9 | 136150032 | 133274616 | ABO | 0.19 | 2.35 x 10−21 |
| cg24267699 | 9 | 136151359 | 133275943 | ABO | 0.16 | 1.44 x 10−15 |
| cg14440550 | 9 | 136131118 | 133255731 | ABO | −0.17 | 7.54 x 10−15 |
| cg11879188 | 9 | 136149908 | 133274492 | ABO | 0.13 | 3.78 x 10−13 |
| cg13660174 | 9 | 136238392 | 133371516 | SURF4 | 0.14 | 1.36 x 10−11 |
| cg09376613 | 10 | 102087334 | 100327577 | PKD2L1 | 0.14 | 8.57 x 10−11 |
| cg06015525 | 12 | 57872123 | 57478340 | ARHGAP9 | 0.13 | 9.29 x 10−10 |
| cg14209264 | 11 | 64382444 | 64614972 | NRXN2 | 0.13 | 1.48 x 10−09 |
| cg26657675 | 16 | 85575407 | 85541801 | 0.12 | 1.65 x 10−09 | |
| cg17980786 | 3 | 32933637 | 32892145 | TRIM71 | 0.13 | 1.73 x 10−09 |
| cg02650017 | 17 | 47301614 | 49224252 | PHOSPHO1 | −0.13 | 2.91 x 10−09 |
| cg07793033 | 16 | 85256423 | 85222817 | 0.12 | 3.19 x 10−09 | |
| cg24044988 | 16 | 30197947 | 30186626 | CORO1A | 0.13 | 4.14 x 10−09 |
| cg06495135 | 22 | 40336939 | 39940935 | GRAP2 | 0.13 | 4.68 x 10−09 |
| cg06388937 | 10 | 104406651 | 102646894 | TRIM8 | 0.13 | 6.63 x 10−09 |
| cg13506600 | 9 | 136150361 | 133274945 | ABO | 0.12 | 7.81 x 10−09 |
| cg03315921 | 22 | 18243630 | 17760864 | BID | 0.12 | 8.27 x 10−09 |
| cg03044066 | 8 | 130047260 | 129035014 | 0.12 | 9.01 x 10−09 | |
| cg04883291 | 9 | 136126473 | 133251086 | 0.12 | 9.35 x 10−09 | |
| cg06192883 | 15 | 52554171 | 52261974 | MYO5C | 0.13 | 9.90 x 10−09 |
| cg18645241 | 21 | 43022102 | 41601942 | −0.12 | 1.08 x 10−08 | |
| cg07023538 | 10 | 71719637 | 69959881 | 0.14 | 1.12 x 10−08 | |
| cg26098679 | 1 | 23681008 | 23354515 | 0.13 | 1.47 x 10−08 | |
| cg06646796 | 14 | 24104741 | 23635532 | DHRS2 | 0.13 | 1.99 x 10−08 |
| cg16248756 | 7 | 127795594 | 128155542 | 0.13 | 9.60 x 10−09 | |
| cg07817261 | 20 | 47566479 | 48949942 | ARFGEF2 | 0.13 | 3.50 x 10−08 |
| cg00964361 | 20 | 30459158 | 31871355 | DUSP15 | 0.12 | 3.53 x 10−08 |
| cg19748455 | 17 | 76274856 | 78278775 | LOC100996291 | −0.13 | 3.53 x 10−08 |
Relationship of ABO and VWF FVIII-associated variants to clinical CVD outcomes
There was no evidence of association for either ABO rs8176719 or VWF rs115708869 with incident CHD, stroke, HF, or mortality in JHS (Table S4), though these analyses have limited statistical power due to the relatively small number of incident events. We used the Cerebrovascular Disease and Cardiovascular Disease Knowledge Portals to access summary statistics from larger GWAS analyses for stroke and CVD. African specific VWF lead variant rs115708869 was not present in CAD, stroke, or heart failure GWAS summary statistics. rs8176719 was also not present in summary statistics, but moderately correlated variant rs687289 (the lead from the largest FVIII GWAS [11]; r2=0.51 in 1000G AFR with rs8176719) was associated with coronary artery disease (p=4.76 × 10−6, OR=1.04, n=184,305) in the CARDIoGRAMplusC4D analysis [44], with ischemic stroke in the MEGASTROKE analysis (p=2.67 × 10−4, OR=1.03, n= 521,612) [45], and with heart failure in European UK Biobank participants (p=2.38 × 10−5, OR=1.08, n=394,156) [46]. Further analysis is needed to clarify the association of the multiple signals at the ABO locus with cardiovascular events, and to disentangle FVIII mediated effects from pleiotropic effects of ABO genetic variation on lipid and glycemic measures.
DISCUSSION
In a large prospective community-based study of AAs, we confirmed the association of FVIII with clinical events including hard CHD and total mortality. These FVIII associations were largely independent of traditional risk factors, including other inflammation biomarkers. We also demonstrate an association of FVIII with incident HF, independent of BNP. In WGS-based association analysis, we observed two genomic regions strongly associated with FVIII in AAs, the ABO region on chromosome 9q34 and the VWF gene on chromosome 12p13.31. The association signal at the VWF locus includes the African ancestral coding variant p.His817Gln and is distinct from the previously reported European VWF association signal for FVIII. At the ABO locus, there were at least two conditionally-independent association signals (O/non-O allele the A2 allele) in our AA sample.
FVIII and CVD risk in AAs
The role of FVIII as a risk factor for incident CHD [4-7] and stroke [8, 9] has been suggested in several prospective studies of healthy middle-aged and older adults, though it remains less clear whether the FVIII association is independent of other CVD risk factors [47]. FVIII is strongly correlated with atherosclerosis-related risk factors such as age, BMI, diabetes and other coagulation and inflammatory biomarkers [3]. Results from our cross-sectional analyses in JHS confirm age, diabetes, triglycerides, and CRP as the major correlates of higher FVIII in AAs. In a previous race-stratified analysis from REGARDS, FVIII was found to be associated with risk of overall and hard CHD, and to a lesser extent stroke, independent of traditional CVD risk factors and CRP in both EAs and AAs [18]. Among REGARDS AAs, the HR per SD unit increase in FVIII were 1.65 (1.28 – 2.13), 1.64 (1.28 – 2.11), and 1.38 (1.15 – 1.66) for hard CHD, overall CHD, and stroke, respectively. The weaker associations with incident CVD events observed in JHS may reflect the smaller numbers of cases compared to REGARDS, FVIII assay heterogeneity, or FVIII measurement error.
The stronger associations of FVIII with mortality compared to associations with incident CVD in JHS are consistent with the findings in several other multi-ethnic studies [52,53], with an approximately two-fold increased risk of mortality comparing the bottom quartile to the upper quartile. Some of these prior studies included individuals of both European and African ancestry, but our analysis is the first to demonstrate the association of FVIII with mortality in AAs. In MESA, FVIII was also associated with cancer-specific mortality [47]. The reason for the stronger relationship of FVIII to morality compared to incident CVD is unclear. FVIII is an acute phase reactant, and thus, similar to CRP and fibrinogen, may reflect a low-grade chronic inflammatory state characteristic of various chronic diseases including not only atherosclerosis but cancer and other age-related diseases.
The associations of FVIII with LVH and HF have not been reported. A study of British men found no association of FVIII with HF [48]. Nonetheless, hypercoagulability is a general feature of HF [49, 50], and arterial and venous thrombotic events are a common complication of patients with HF [51]. FVIII has also been associated with increased risk of incident atrial fibrillation [52], an important risk factor for HF. Several coagulation markers have been correlated with N-terminal-pro hormone BNP (NT-proBNP), suggesting increased coagulation activity may be related to neurohormonal activation and cardiac stress [53]. Given the importance of HF in CVD health disparities, further study of FVIII as a predictor of HF is warranted, particularly as the association observed in JHS was independent of adjustment for other risk factors and biomarkers such as BNP and renal function.
Genetics of FVIII in AAs
Our results show that the overall contribution of genetic factors to phenotypic variation in FVIII in AAs is similar to that previously reported in EAs (h2~50%) [19-21]. We were also able to confirm and extend the association of ABO and VWF variants to FVIII in AAs. In particular, the 1 bp deletion variants that define O/non-O and A2 alleles were directly genotyped in our analysis through WGS, thereby allowing us to “fine-map” these two ABO groups as the strongest determinants of FVIII (as opposed to correlated non-coding proxy variants). Our results are consistent with previous studies based on targeted genotyping or haplotype imputation showing O and A2 blood group alleles associated with lower FVIII [54] and that ABO accounts for ~10% of the variability of FVIII phenotypic variance in otherwise unselected individuals [55]. The effects of ABO blood groups on FVIII appear to be mainly mediated by VWF levels; lower VWF levels in O-group subjects are due to shorter VWF survival, mainly attributable to faster clearance [56-58]. A smaller, direct or VWF-independent ABO influence on FVIII has also been reported [54].
Epigenetic association analyses may identify novel mechanisms that contribute to regulation of hemostasis and thrombosis. Changes in gene expression and methylation of F8 and other inflammation-related genes were associated with SES [59] and neighborhood characteristics [60], suggesting that such epigenetic changes may mediate the effects of environmental or psycho-social stressors on CVD. By performing EWAS for FVIII in a subset of AAs from JHS, we identified several candidate loci, including differentially methylated CpG sites in the 3’UTR of ABO, which were also correlated with O vs non-O variant rs8176719. ABO gene transcription is dependent on differential DNA methylation of promoter and 3’ flanking regions [61, 62], and the non-O/O variant rs8176719 has been strongly associated both with ABO gene expression across a variety of tissues and with methylation at the same CpG sites [63]. Finally, it is important to note that our EWAS was performed using peripheral whole blood DNA. As methylation is a tissue-specific process, whole blood DNA methylation marks might not be a good proxy for methylation status at more biologically relevant cells, such as endothelial cells where FVIII is mainly expressed.
In contrast to the ABO locus, where the FVIII-associated variants appear to be largely consistent between AAs and EAs, we observed very little association with the previously reported European VWF rs2238109 variant associated with FVIII. Instead, the main association signal at the VWF locus in our AA sample is highly specific to AAs. Of the nine variants that comprise this AA-specific association signal, the most likely causal variant is rs57950734 which encodes p.His817Gln. This variant has been previously associated with lower FVIII in a targeted coding sequence analysis of VWF among ~4500 AAs [43]. FVIII circulates bound to VWF, which protects FVIII from early degradation. The VWF p.His817Gln variant, located within the VWF D′ domain within the FVIII binding region, is preferentially associated with lower FVIII (and has little effect on plasma VWF levels) [43]. This variant has also been reported in several patients with type 2N von Willebrand disease in which patients’ experience bleeding due to the inability of FVIII to appropriately bind to VWF [66]. Moreover, in vitro, the p.His817Gln amino acid substitution results in significantly lower FVIII binding capacity [64], suggesting it is the causal variant for lower FVIII observed in the JHS sample.
Paradoxically, the VWF p.His817Gln variant is common in AAs, but associated with lower FVIII. On average, FVIII levels are higher in AAs compared to EAs, yet we were unable to identify any African ancestral genetic factors that account for these differences through our WGS-based association analyses or through a complementary genome-wide local ancestry-based admixture scan in JHS. Genome-wide African admixture proportion was not associated with FVIII in JHS or in other studies[65], upon adjustment for other FVIII correlates including SES. We also did not observe any genome-wide significant association signal at the F8 structural gene locus. Of 37 missense variants within F8 identified by WGS in JHS with an allele frequency of 0.1% or greater, only rs1800297 showed suggestive evidence of association with higher/lower FVIII (P=0.06). The rs1800297 variant, which encodes a B-domain substitution D1241E, has been previously associated with FVIII levels in Europeans [68]. There was no association of FVIII with the common African rs1800291 missense variant (p.M2257V) in JHS, (P=0.71), consistent with the hypothesis that this is likely a benign African-derived variant [68,69]. Together, these observations suggest that non-genetic, epigenomic, or environmental factors may have a major contribution to the inter-ethnic FVIII phenotypic differences.
Mechanistic and causal relationships between FVIII, ABO, and CVD risk
While very large studies have established that ABO blood group, particularly O vs. non-O is associated with risk of VTE, CHD, and stroke [66-69], the mechanism is most clearly established for VTE, where both FVIII and VWF are important mediators. For arterial thrombotic diseases, the mechanistic relationships are less established, as ABO alleles are pleiotropically associated with many other potential CVD risk factors and mediators, particularly LDL cholesterol [70]. In JHS, we did not observe any association between O vs. non-O and risk of CHD, stroke, HF, or mortality, but our sample size is orders of magnitude smaller than those assessed in European studies. In a multi-ethnic analysis from REGARDS, ABO blood type did not account for the higher stroke risk among AAs [71].
The association of FVIII with VTE and the protection from CHD in individuals with genetically low FVIII (hemophilia A) [72] and hemophilia carriers [73] support a direct causal role of FVIII in arterial thrombosis. Other observational data have suggested that the association of FVIII with CVD risk appears to be independent of ABO blood group [4] but not necessarily independent of VWF [6]. A limitation of our study is the lack of availability of measured VWF (which is highly correlated with FVIII) in JHS; therefore a direct comparison of risk assessment was not possible. Recently, instrumental variable or Mendelian randomization analyses of FVIII using results of recent FVIII GWAS in Europeans have suggested FVIII levels may be causally related to both CHD and VTE risk, as the risk estimates were only modestly attenuated upon adjustment for VWF levels, whereas VWF (but not FVIII) was causally related to ischemic stroke [11]. The disparate association of the African VWF p.His817Gln variant with lower FVIII (but not VWF) levels [43, 64, 74] makes it a potentially attractive genetic instrument to further assess the causal role of FVIII in AAs. We were unable to observe any significant association with clinical events in JHS, but our power to detect such an association was likely limited by the small number of CVD cases and length of follow-up. Another limitation of our study is that VTE events have not been adjudicated in JHS; therefore, we were unable to assess the risk of our main FVIII-associated genetic variants with VTE risk. It will be important to perform genetic discovery in even larger samples of AAs to more comprehensively characterize the distinct and shared genetic determinants of FVIII and to assess whether the potential causal relationship of FVIII with CVD, and also the novel association of FVIII with heart failure/LVH, can be extended to broader AA populations.
Supplementary Material
ESSENTIALS.
Higher factor VIII (FVIII) levels are a known cardiovascular disease risk factor.
We here examine the epidemiological and genetic correlates of FVIII in African Americans.
FVIII was associated with incident heart failure, mortality, and coronary heart disease.
Genetic variants at ABO and VWF, as well as methylation at ABO, associated with FVIII.
Acknowledgements
This work was supported by a grant from the National Heart, Lung, and Blood Institute (NHLBI) (R01HL132947) to APR and LAL. LMR is supported by T32 HL129982. JGW is supported by U54GM115428 from the National Institute of General Medical Sciences.
The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201300049C and HHSN268201300050C), Tougaloo College (HHSN268201300048C), and the University of Mississippi Medical Center (HHSN268201300046C and HHSN268201300047C) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS.
This research project is supported by cooperative agreement U01 NS041588 co-funded by NINDS and the National Institute on Aging (NIA), NIH, Department of Health and Human Service. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the NIA. Representatives of the NINDS were involved in the review of the manuscript but were not directly involved in the collection, management, analysis or interpretation of the data. The authors thank the other investigators, the staff, and the participants of the REGARDS study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at http://www.regardsstudy.org Additional funding was provided by an investigator-initiated grant-in-aid K08-HL096841 from the National Heart, Lung, and Blood Institute National Institutes of Health, Department of Health and Human Service, Bethesda, MD, USA. Representatives from the National Heart, Lung, and Blood Institute did not have any role in the design and conduct of the study, the collection, management, analysis, and interpretation of the data, or the preparation or approval of the manuscript.
Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for “NHLBI TOPMed: The Jackson Heart Study” (phs000964) was performed at the University of Washington Northwest Genomics Center (HHSN268201100037C). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
Appendix
Membership in the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium and the TOPMed Hematology & Hemostasis Working Group is listed in the supplemental material.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Samuelson Bannow B, Recht M, Negrier C, Hermans C, Berntorp E, Eichler H, Mancuso ME, Klamroth R, O'Hara J, Santagostino E, Matsushita T, Kessler C. Factor VIII: Long-established role in haemophilia A and emerging evidence beyond haemostasis. Blood reviews. 2019; 35: 43–50. 10.1016/j.blre.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins PV, Rawley O, Smith OP, O'Donnell JS. Elevated factor VIII levels and risk of venous thrombosis. British journal of haematology. 2012; 157: 653–63. 10.1111/j.1365-2141.2012.09134.x. [DOI] [PubMed] [Google Scholar]
- 3.Kamphuisen PW, Eikenboom JC, Bertina RM. Elevated factor VIII levels and the risk of thrombosis. Arterioscler Thromb Vasc Biol. 2001; 21: 731–8. [DOI] [PubMed] [Google Scholar]
- 4.Meade TW, Cooper JA, Stirling Y, Howarth DJ, Ruddock V, Miller GJ. Factor VIII, ABO blood group and the incidence of ischaemic heart disease. Br J Haematol. 1994; 88: 601–7. [DOI] [PubMed] [Google Scholar]
- 5.Zakai NA, Katz R, Jenny NS, Psaty BM, Reiner AP, Schwartz SM, Cushman M. Inflammation and hemostasis biomarkers and cardiovascular risk in the elderly: the Cardiovascular Health Study. J Thromb Haemost. 2007; 5: 1128–35. 10.1111/j.1538-7836.2007.02528.x. [DOI] [PubMed] [Google Scholar]
- 6.Rumley A, Lowe GD, Sweetnam PM, Yarnell JW, Ford RP. Factor VIII, von Willebrand factor and the risk of major ischaemic heart disease in the Caerphilly Heart Study. Br J Haematol. 1999; 105: 110–6. [PubMed] [Google Scholar]
- 7.Folsom AR, Wu KK, Rosamond WD, Sharrett AR, Chambless LE. Prospective study of hemostatic factors and incidence of coronary heart disease: the Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 1997; 96: 1102–8. [DOI] [PubMed] [Google Scholar]
- 8.Folsom AR, Rosamond WD, Shahar E, Cooper LS, Aleksic N, Nieto FJ, Rasmussen ML, Wu KK. Prospective study of markers of hemostatic function with risk of ischemic stroke. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Circulation. 1999; 100: 736–42. [DOI] [PubMed] [Google Scholar]
- 9.Smith FB, Lee AJ, Fowkes FG, Price JF, Rumley A, Lowe GD. Hemostatic factors as predictors of ischemic heart disease and stroke in the Edinburgh Artery Study. Arterioscler Thromb Vasc Biol. 1997; 17: 3321–5. [DOI] [PubMed] [Google Scholar]
- 10.Yap ES, Timp JF, Flinterman LE, van Hylckama Vlieg A, Rosendaal FR, Cannegieter SC, Lijfering WM. Elevated levels of factor VIII and subsequent risk of all-cause mortality: results from the MEGA follow-up study. J Thromb Haemost. 2015; 13: 1833–42. 10.1111/jth.13071. [DOI] [PubMed] [Google Scholar]
- 11.Sabater-Lleal M, Huffman JE, de Vries PS, Marten J, Mastrangelo MA, Song C, Pankratz N, Ward-Caviness CK, Yanek LR, Trompet S, Delgado GE, Guo X, Bartz TM, Martinez-Perez A, Germain M, de Haan HG, Ozel AB, Polasek O, Smith AV, Eicher JD, Reiner AP, Tang W, Davies NM, Stott DJ, Rotter JI, Tofler GH, Boerwinkle E, de Maat MPM, Kleber ME, Welsh P, Brody JA, Chen MH, Vaidya D, Soria JM, Suchon P, van Hylckama Vlieg A, Desch KC, Kolcic I, Joshi PK, Launer LJ, Harris TB, Campbell H, Rudan I, Becker DM, Li JZ, Rivadeneira F, Uitterlinden AG, Hofman A, Franco OH, Cushman M, Psaty BM, Morange PE, McKnight B, Chong MR, Fernandez-Cadenas I, Rosand J, Lindgren A, Gudnason V, Wilson JF, Hayward C, Ginsburg D, Fornage M, Rosendaal FR, Souto JC, Becker LC, Jenny NS, Marz W, Jukema JW, Dehghan A, Tregouet DA, Morrison AC, Johnson AD, O'Donnell CJ, Strachan DP, Lowenstein CJ, Smith NL. Genome-Wide Association Transethnic Meta-Analyses Identifies Novel Associations Regulating Coagulation Factor VIII and von Willebrand Factor Plasma Levels. Circulation. 2019; 139: 620–35. 10.1161/circulationaha.118.034532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zakai NA, McClure LA, Judd SE, Safford MM, Folsom AR, Lutsey PL, Cushman M. Racial and regional differences in venous thromboembolism in the United States in 3 cohorts. Circulation. 2014; 129: 1502–9. 10.1161/circulationaha.113.006472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carnethon MR, Pu J, Howard G, Albert MA, Anderson CAM, Bertoni AG, Mujahid MS, Palaniappan L, Taylor HA Jr., Willis M, Yancy CW. Cardiovascular Health in African Americans: A Scientific Statement From the American Heart Association. Circulation. 2017; 136: e393–e423. 10.1161/cir.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 14.Conlan MG, Folsom AR, Finch A, Davis CE, Sorlie P, Marcucci G, Wu KK. Associations of factor VIII and von Willebrand factor with age, race, sex, and risk factors for atherosclerosis. The Atherosclerosis Risk in Communities (ARIC) Study. Thromb Haemost. 1993; 70: 380–5. [PubMed] [Google Scholar]
- 15.Lutsey PL, Cushman M, Steffen LM, Green D, Barr RG, Herrington D, Ouyang P, Folsom AR. Plasma hemostatic factors and endothelial markers in four racial/ethnic groups: the MESA study. J Thromb Haemost. 2006; 4: 2629–35. 10.1111/j.1538-7836.2006.02237.x. [DOI] [PubMed] [Google Scholar]
- 16.Roberts LN, Patel RK, Chitongo P, Bonner L, Arya R. African-Caribbean ethnicity is associated with a hypercoagulable state as measured by thrombin generation. Blood Coagul Fibrinolysis. 2013; 24: 40–9. 10.1097/MBC.0b013e32835a07fa. [DOI] [PubMed] [Google Scholar]
- 17.Patel RK, Ford E, Thumpston J, Arya R. Risk factors for venous thrombosis in the black population. Thromb Haemost. 2003; 90: 835–8. 10.1160/th03-05-0311. [DOI] [PubMed] [Google Scholar]
- 18.Zakai NA, Judd SE, Kissela B, Howard G, Safford MM, Cushman M. Factor VIII, Protein C and Cardiovascular Disease Risk: The REasons for Geographic and Racial Differences in Stroke Study (REGARDS). Thromb Haemost. 2018; 118: 1305–15. 10.1055/s-0038-1655766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orstavik KH, Magnus P, Reisner H, Berg K, Graham JB, Nance W. Factor VIII and factor IX in a twin population. Evidence for a major effect of ABO locus on factor VIII level. Am J Hum Genet. 1985; 37: 89–101. [PMC free article] [PubMed] [Google Scholar]
- 20.de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001; 357: 101–5. 10.1016/s0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 21.Souto JC, Almasy L, Borrell M, Gari M, Martinez E, Mateo J, Stone WH, Blangero J, Fontcuberta J. Genetic determinants of hemostasis phenotypes in Spanish families. Circulation. 2000; 101: 1546–51. [DOI] [PubMed] [Google Scholar]
- 22.Kamphuisen PW, Lensen R, Houwing-Duistermaat JJ, Eikenboom JC, Harvey M, Bertina RM, Rosendaal FR. Heritability of elevated factor VIII antigen levels in factor V Leiden families with thrombophilia. Br J Haematol. 2000; 109: 519–22. [DOI] [PubMed] [Google Scholar]
- 23.Huffman JE, de Vries PS, Morrison AC, Sabater-Lleal M, Kacprowski T, Auer PL, Brody JA, Chasman DI, Chen MH, Guo X, Lin LA, Marioni RE, Muller-Nurasyid M, Yanek LR, Pankratz N, Grove ML, de Maat MP, Cushman M, Wiggins KL, Qi L, Sennblad B, Harris SE, Polasek O, Riess H, Rivadeneira F, Rose LM, Goel A, Taylor KD, Teumer A, Uitterlinden AG, Vaidya D, Yao J, Tang W, Levy D, Waldenberger M, Becker DM, Folsom AR, Giulianini F, Greinacher A, Hofman A, Huang CC, Kooperberg C, Silveira A, Starr JM, Strauch K, Strawbridge RJ, Wright AF, McKnight B, Franco OH, Zakai N, Mathias RA, Psaty BM, Ridker PM, Tofler GH, Volker U, Watkins H, Fornage M, Hamsten A, Deary IJ, Boerwinkle E, Koenig W, Rotter JI, Hayward C, Dehghan A, Reiner AP, O'Donnell CJ, Smith NL. Rare and low-frequency variants and their association with plasma levels of fibrinogen, FVII, FVIII, and vWF. Blood. 2015; 126: e19–29. 10.1182/blood-2015-02-624551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang W, Cushman M, Green D, Rich SS, Lange LA, Yang Q, Tracy RP, Tofler GH, Basu S, Wilson JG, Keating BJ, Weng LC, Taylor HA, Jacobs DR Jr., Delaney JA, Palmer CD, Young T, Pankow JS, O'Donnell CJ, Smith NL, Reiner AP, Folsom AR. Gene-centric approach identifies new and known loci for FVIII activity and VWF antigen levels in European Americans and African Americans. American journal of hematology. 2015; 90: 534–40. 10.1002/ajh.24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, Hayward C, Rudan I, Sabater-Lleal M, Bis JC, de Maat MP, Rumley A, Kong X, Yang Q, Williams FM, Vitart V, Campbell H, Malarstig A, Wiggins KL, Van Duijn CM, McArdle WL, Pankow JS, Johnson AD, Silveira A, McKnight B, Uitterlinden AG, Aleksic N, Meigs JB, Peters A, Koenig W, Cushman M, Kathiresan S, Rotter JI, Bovill EG, Hofman A, Boerwinkle E, Tofler GH, Peden JF, Psaty BM, Leebeek F, Folsom AR, Larson MG, Spector TD, Wright AF, Wilson JF, Hamsten A, Lumley T, Witteman JC, Tang W, O'Donnell CJ. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010; 121: 1382–92. 10.1161/circulationaha.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Taliun SAG, Corvelo A, Gogarten SM, Kang HM, Pitsillides AN, LeFaive J, Lee S-b, Tian X, Browning BL, Das S, Emde A-K, Clarke WE, Loesch DP, Shetty AC, Blackwell TW, Wong Q, Aguet F, Albert C, Alonso A, Ardlie KG, Aslibekyan S, Auer PL, Barnard J, Barr RG, Becker LC, Beer RL, Benjamin EJ, Bielak LF, Blangero J, Boehnke M, Bowden DW, Brody JA, Burchard EG, Cade BE, Casella JF, Chalazan B, Chen Y-DI, Cho MH, Choi SH, Chung MK, Clish CB, Correa A, Curran JE, Custer B, Darbar D, Daya M, Andrade Md, DeMeo DL, Dutcher SK, Ellinor PT, Emery LS, Fatkin D, Forer L, Fornage M, Franceschini N, Fuchsberger C, Fullerton SM, Germer S, Gladwin MT, Gottlieb DJ, Guo X, Hall ME, He J, Heard-Costa NL, Heckbert SR, Irvin MR, Johnsen JM, Johnson AD, Kardia SLR, Kelly T, Kelly S, Kenny EE, Kiel DP, Klemmer R, Konkle BA, Kooperberg C, Köttgen A, Lange LA, Lasky-Su J, Levy D, Lin X, Lin K-H, Liu C, Loos RJF, Garman L, Gerszten R, Lubitz SA, Lunetta KL, Mak ACY, Manichaikul A, Manning AK, Mathias RA, McManus DD, McGarvey ST, Meigs JB, Meyers DA, Mikulla JL, Minear MA, Mitchell B, Mohanty S, Montasser ME, Montgomery C, Morrison AC, Murabito JM, Natale A, Natarajan P, Nelson SC, North KE, O’Connell JR, Palmer ND, Pankratz N, Peloso GM, Peyser PA, Post WS, Psaty BM, Rao DC, Redline S, Reiner AP, Roden D, Rotter JI, Ruczinski I, Sarnowski C, Schoenherr S, Seo J-S, Seshadri S, Sheehan VA, Shoemaker MB, Smith AV, Smith NL, Smith JA, Sotoodehnia N, Stilp AM, Tang W, Taylor KD, Telen M, Thornton TA, Tracy RP, Berg DJVD, Vasan RS, Viaud-Martinez KA, Vrieze S, Weeks DE, Weir BS, Weiss ST, Weng L-C, Willer CJ, Zhang Y, Zhao X, Arnett DK, Ashley-Koch AE, Barnes KC, Boerwinkle E, Gabriel S, Gibbs R, Rice KM, Rich SS, Silverman E, Qasba P, Gan W, Papanicolaou GJ, Nickerson DA, Browning SR, Zody MC, Zöllner S, Wilson JG, Cupples LA, Laurie CC, Jaquish CE, Hernandez RD, O’Connor TD, Abecasis GR. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019: 563866 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter MA, Crow R, Steffes M, Rock W, Heilbraun J, Evans G, Skelton T, Jensen R, Sarpong D. Laboratory, reading center, and coordinating center data management methods in the Jackson Heart Study. Am J Med Sci. 2004; 328: 131–44. [DOI] [PubMed] [Google Scholar]
- 28.Taylor HA Jr., Wilson JG, Jones DW, Sarpong DF, Srinivasan A, Garrison RJ, Nelson C, Wyatt SB. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethnicity & disease. 2005; 15: S6-4–17. [PubMed] [Google Scholar]
- 29.Liu J, Fox CS, Hickson D, Sarpong D, Ekunwe L, May WD, Hundley GW, Carr JJ, Taylor HA. Pericardial adipose tissue, atherosclerosis, and cardiovascular disease risk factors: the Jackson heart study. Diabetes care. 2010; 33: 1635–9. 10.2337/dc10-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998; 62: 1198–211. 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulit SL, de With SA, de Bakker PI. Resetting the bar: Statistical significance in whole-genome sequencing-based association studies of global populations. Genetic epidemiology. 2017; 41: 145–51. 10.1002/gepi.22032. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Huffman JE, Brody JA, Wang C, Lee S, Li Z, Gogarten SM, Sofer T, Bielak LF, Bis JC, Blangero J, Bowler RP, Cade BE, Cho MH, Correa A, Curran JE, de Vries PS, Glahn DC, Guo X, Johnson AD, Kardia S, Kooperberg C, Lewis JP, Liu X, Mathias RA, Mitchell BD, O'Connell JR, Peyser PA, Post WS, Reiner AP, Rich SS, Rotter JI, Silverman EK, Smith JA, Vasan RS, Wilson JG, Yanek LR, Redline S, Smith NL, Boerwinkle E, Borecki IB, Cupples LA, Laurie CC, Morrison AC, Rice KM, Lin X. Efficient Variant Set Mixed Model Association Tests for Continuous and Binary Traits in Large-Scale Whole-Genome Sequencing Studies. Am J Hum Genet. 2019; 104: 260–74. 10.1016/j.ajhg.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018; 34: 511–3. 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, Skoglund P, Lazaridis I, Sankararaman S, Fu Q, Rohland N, Renaud G, Erlich Y, Willems T, Gallo C, Spence JP, Song YS, Poletti G, Balloux F, van Driem G, de Knijff P, Romero IG, Jha AR, Behar DM, Bravi CM, Capelli C, Hervig T, Moreno-Estrada A, Posukh OL, Balanovska E, Balanovsky O, Karachanak-Yankova S, Sahakyan H, Toncheva D, Yepiskoposyan L, Tyler-Smith C, Xue Y, Abdullah MS, Ruiz-Linares A, Beall CM, Di Rienzo A, Jeong C, Starikovskaya EB, Metspalu E, Parik J, Villems R, Henn BM, Hodoglugil U, Mahley R, Sajantila A, Stamatoyannopoulos G, Wee JT, Khusainova R, Khusnutdinova E, Litvinov S, Ayodo G, Comas D, Hammer MF, Kivisild T, Klitz W, Winkler CA, Labuda D, Bamshad M, Jorde LB, Tishkoff SA, Watkins WS, Metspalu M, Dryomov S, Sukernik R, Singh L, Thangaraj K, Paabo S, Kelso J, Patterson N, Reich D. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016; 538: 201–6. 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013; 93: 278–88. 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conomos MP GS, Brown L, Chen H, Rice K, Sofer T, Thornton T, Yu C. . GENESIS: GENetic EStimation and Inference in Structured samples (GENESIS): Statistical methods for analyzing genetic data from samples with population structure and/or relatedness., 2019.
- 37.Grinde KE, Brown LA, Reiner AP, Thornton TA, Browning SR. Genome-wide Significance Thresholds for Admixture Mapping Studies. Am J Hum Genet. 2019; 104: 454–65. 10.1016/j.ajhg.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortin JP, Triche TJ Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017; 33: 558–60. 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. J Infect Dis. 2015; 212: 1563–73. 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012; 13: 86 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saffari A, Silver MJ, Zavattari P, Moi L, Columbano A, Meaburn EL, Dudbridge F. Estimation of a significance threshold for epigenome-wide association studies. Genet Epidemiol. 2018; 42: 20–33. 10.1002/gepi.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic acids research. 2017; 45: e22 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnsen JM, Auer PL, Morrison AC, Jiao S, Wei P, Haessler J, Fox K, McGee SR, Smith JD, Carlson CS, Smith N, Boerwinkle E, Kooperberg C, Nickerson DA, Rich SS, Green D, Peters U, Cushman M, Reiner AP. Common and rare von Willebrand factor (VWF) coding variants, VWF levels, and factor VIII levels in African Americans: the NHLBI Exome Sequencing Project. Blood. 2013; 122: 590–7. 10.1182/blood-2013-02-485094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJF, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015; 47: 1121–30. 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen W-M, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu F-C, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee J-M, Lemmens R, Leys D, Lewis CM, Lin W-Y, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT, Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M, Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen W-M, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, Hoed Md, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu F-C, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee J-M, Lemmens R, Leys D, Lewis CM, Lin W-Y, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O’Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Amin N, Aparicio HS, Arnett DK, Attia J. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics. 2018; 50: 524–37. 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aragam Krishna G, Chaffin M, Levinson Rebecca T, McDermott G, Choi Seung H, Shoemaker MB, Haas Mary E, Weng L-C, Lindsay Mark E, Smith JG, Newton-Cheh C, Roden Dan M, London B, null n, Wells Quinn S, Ellinor Patrick T, Kathiresan S, Lubitz Steven A, Bloom Heather L, Dudley Samuel C, Shalaby Alaa A, Weiss R, Gutmann R, Saba S. Phenotypic Refinement of Heart Failure in a National Biobank Facilitates Genetic Discovery. Circulation. 2019; 139: 489–501. 10.1161/CIRCULATIONAHA.118.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folsom AR, Delaney JA, Lutsey PL, Zakai NA, Jenny NS, Polak JF, Cushman M. Associations of factor VIIIc, D-dimer, and plasmin-antiplasmin with incident cardiovascular disease and all-cause mortality. American journal of hematology. 2009; 84: 349–53. 10.1002/ajh.21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wannamethee SG, Whincup PH, Papacosta O, Lennon L, Lowe GD. Associations between blood coagulation markers, NT-proBNP and risk of incident heart failure in older men: The British Regional Heart Study. International journal of cardiology. 2017; 230: 567–71. 10.1016/j.ijcard.2016.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JH, Shah P, Tantry US, Gurbel PA. Coagulation Abnormalities in Heart Failure: Pathophysiology and Therapeutic Implications. Curr Heart Fail Rep. 2016; 13: 319–28. 10.1007/s11897-016-0308-6. [DOI] [PubMed] [Google Scholar]
- 50.Jug B, Vene N, Salobir BG, Sebestjen M, Sabovic M, Keber I. Procoagulant state in heart failure with preserved left ventricular ejection fraction. Int Heart J. 2009; 50: 591–600. [DOI] [PubMed] [Google Scholar]
- 51.Zannad F, Stough WG, Regnault V, Gheorghiade M, Deliargyris E, Gibson CM, Agewall S, Berkowitz SD, Burton P, Calvo G, Goldstein S, Verheugt FW, Koglin J, O'Connor CM. Is thrombosis a contributor to heart failure pathophysiology? Possible mechanisms, therapeutic opportunities, and clinical investigation challenges. Int J Cardiol. 2013; 167: 1772–82. 10.1016/j.ijcard.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Alonso A, Tang W, Agarwal SK, Soliman EZ, Chamberlain AM, Folsom AR. Hemostatic markers are associated with the risk and prognosis of atrial fibrillation: the ARIC study. Int J Cardiol. 2012; 155: 217–22. 10.1016/j.ijcard.2010.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mongirdiene A, Kursvietiene L, Kasauskas A. The coagulation system changes in patients with chronic heart failure. Medicina (Kaunas). 2010; 46: 642–7. [PubMed] [Google Scholar]
- 54.Song J, Chen F, Campos M, Bolgiano D, Houck K, Chambless LE, Wu KK, Folsom AR, Couper D, Boerwinkle E, Dong JF. Quantitative Influence of ABO Blood Groups on Factor VIII and Its Ratio to von Willebrand Factor, Novel Observations from an ARIC Study of 11,673 Subjects. PLoS ONE. 2015; 10: e0132626 10.1371/journal.pone.0132626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campos M, Buchanan A, Yu F, Barbalic M, Xiao Y, Chambless LE, Wu KK, Folsom AR, Boerwinkle E, Dong JF. Influence of single nucleotide polymorphisms in factor VIII and von Willebrand factor genes on plasma factor VIII activity: the ARIC Study. Blood. 2012; 119: 1929–34. 10.1182/blood-2011-10-383661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallinaro L, Cattini MG, Sztukowska M, Padrini R, Sartorello F, Pontara E, Bertomoro A, Daidone V, Pagnan A, Casonato A. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood. 2008; 111: 3540–5. 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- 57.Albanez S, Ogiwara K, Michels A, Hopman W, Grabell J, James P, Lillicrap D. Aging and ABO blood type influence von Willebrand factor and factor VIII levels through interrelated mechanisms. J Thromb Haemost. 2016; 14: 953–63. 10.1111/jth.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casari C, Lenting PJ, Wohner N, Christophe OD, Denis CV. Clearance of von Willebrand factor. J Thromb Haemost. 2013; 11 Suppl 1: 202–11. 10.1111/jth.12226. [DOI] [PubMed] [Google Scholar]
- 59.Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SL, Shively CA, Seeman TE, Liu Y, Diez Roux AV. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics. 2015; 10: 958–69. 10.1080/15592294.2015.1085139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith JA, Zhao W, Wang X, Ratliff SM, Mukherjee B, Kardia SLR, Liu Y, Roux AVD, Needham BL. Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis. Epigenetics. 2017; 12: 662–73. 10.1080/15592294.2017.1341026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kominato Y, Hata Y, Takizawa H, Tsuchiya T, Tsukada J, Yamamoto F. Expression of human histo-blood group ABO genes is dependent upon DNA methylation of the promoter region. The Journal of biological chemistry. 1999; 274: 37240–50. 10.1074/jbc.274.52.37240. [DOI] [PubMed] [Google Scholar]
- 62.Sano R, Nakajima T, Takahashi K, Kubo R, Yazawa S, Kominato Y. The 3' flanking region of the human ABO histo-blood group gene is involved in negative regulation of gene expression. Legal medicine (Tokyo, Japan). 2011; 13: 22–9. 10.1016/j.legalmed.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, Zheng J, Duggirala A, McArdle WL, Ho K, Ring SM, Evans DM, Davey Smith G, Relton CL. Systematic identification of genetic influences on methylation across the human life course. Genome biology. 2016; 17: 61 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kroner PA, Foster PA, Fahs SA, Montgomery RR. The defective interaction between von Willebrand factor and factor VIII in a patient with type 1 von Willebrand disease is caused by substitution of Arg19 and His54 in mature von Willebrand factor. Blood. 1996; 87: 1013–21. [PubMed] [Google Scholar]
- 65.Lutsey PL, Wassel CL, Cushman M, Sale MM, Divers J, Folsom AR. Genetic admixture is associated with plasma hemostatic factor levels in self-identified African Americans and Hispanics: the Multi-Ethnic Study of Atherosclerosis. J Thromb Haemost. 2012; 10: 543–9. 10.1111/j.1538-7836.2012.04663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, Yang SH, Xu H, Li JJ. ABO blood group system and the coronary artery disease: an updated systematic review and meta-analysis. Scientific reports. 2016; 6: 23250 10.1038/srep23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He M, Wolpin B, Rexrode K, Manson JE, Rimm E, Hu FB, Qi L. ABO blood group and risk of coronary heart disease in two prospective cohort studies. Arteriosclerosis, thrombosis, and vascular biology. 2012; 32: 2314–20. 10.1161/atvbaha.112.248757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasan SK, Rostgaard K, Majeed A, Ullum H, Titlestad KE, Pedersen OB, Erikstrup C, Nielsen KR, Melbye M, Nyren O, Hjalgrim H, Edgren G. ABO Blood Group and Risk of Thromboembolic and Arterial Disease: A Study of 1.5 Million Blood Donors. Circulation. 2016; 133: 1449–57; discussion 57. 10.1161/circulationaha.115.017563. [DOI] [PubMed] [Google Scholar]
- 69.Williams FM, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, Traylor M, Bevan S, Dichgans M, Rothwell PM, Sudlow C, Farrall M, Silander K, Kaunisto M, Wagner P, Saarela O, Kuulasmaa K, Virtamo J, Salomaa V, Amouyel P, Arveiler D, Ferrieres J, Wiklund PG, Ikram MA, Hofman A, Boncoraglio GB, Parati EA, Helgadottir A, Gretarsdottir S, Thorsteinsdottir U, Thorleifsson G, Stefansson K, Seshadri S, DeStefano A, Gschwendtner A, Psaty B, Longstreth W, Mitchell BD, Cheng YC, Clarke R, Ferrario M, Bis JC, Levi C, Attia J, Holliday EG, Scott RJ, Fornage M, Sharma P, Furie KL, Rosand J, Nalls M, Meschia J, Mosely TH, Evans A, Palotie A, Markus HS, Grant PJ, Spector TD. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Annals of neurology. 2013; 73: 16–31. 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y, Chen C, Ke X, Xiong L, Shi Y, Li J, Tan X, Ye S. Analysis of circulating cholesterol levels as a mediator of an association between ABO blood group and coronary heart disease. Circulation Cardiovascular genetics. 2014; 7: 43–8. 10.1161/circgenetics.113.000299. [DOI] [PubMed] [Google Scholar]
- 71.Zakai NA, Judd SE, Alexander K, McClure LA, Kissela BM, Howard G, Cushman M. ABO blood type and stroke risk: the REasons for Geographic And Racial Differences in Stroke Study. J Thromb Haemost. 2014; 12: 564–70. 10.1111/jth.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosendaal FR, Briet E, Stibbe J, van Herpen G, Leuven JA, Hofman A, Vandenbroucke JP. Haemophilia protects against ischaemic heart disease: a study of risk factors. British journal of haematology. 1990; 75: 525–30. [DOI] [PubMed] [Google Scholar]
- 73.Sramek A, Kriek M, Rosendaal FR. Decreased mortality of ischaemic heart disease among carriers of haemophilia. Lancet (London, England). 2003; 362: 351–4. [DOI] [PubMed] [Google Scholar]
- 74.ISTH-SSC VWF Online Database (VWFdb)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.