Abstract
Objective
We previously reported that children exposed to secondhand smoke (SHS) that carried variants in the NAT1 gene had over two-fold higher hair cotinine levels. Our objective was to determine if NAT1 polymorphisms confer increased risk for developing asthma in children exposed to SHS.
Methods
White participants in the Cincinnati Childhood Allergy and Air Pollution Study (n= 359) were genotyped for 10 NAT1 variants. Smoke exposure was defined by hair cotinine and parental report. Asthma was objectively assessed by spirometry and methacholine challenge. Findings were replicated in the Genomic Control Cohort (n=638).
Results
Significant associations between 5 NAT1 variants and asthma were observed in the CCAAPS exposed group compared to none in the unexposed group. There was a significant interaction between NAT1 rs13253389 and rs4921581 with smoke exposure (p=0.02, p=0.01) and hair cotinine level (p=0.048, p=0.042). Children wildtype for rs4921581 had increasing asthma risk with increasing hair cotinine level, whereas those carrying the NAT1 minor allele had an increased risk of asthma regardless of cotinine level. In the GCC, 13 NAT1 variants were associated with asthma in the smoke-exposed group, compared to 0 in the unexposed group, demonstrating gene-level replication.
Conclusions
Variation in the NAT1 gene modifies asthma risk in children exposed to secondhand-smoke. To our knowledge, this is the first report of a gene-environment interaction between NAT1 variants, smoke exposure, cotinine levels, and pediatric asthma. NAT1 genotype may have clinical utility as a biomarker of increased asthma risk in children exposed to smoke.
Keywords: NAT1, secondhand smoke exposure, children, asthma
INTRODUCTION
Cigarette smoke is a major contributor to illness in children(1). Commonly referred to as secondhand smoke (SHS), inhaled byproducts of cigarette smoking exacerbate 400,000 to 1,000,000 cases of childhood asthma yearly(2–4). Moreover, SHS exposure induce new cases of asthma in children and adolescents. Still, 35% of children in the United States live in homes where residents or visitors smoke regularly(5). Thus, despite well-known risks associated with SHS exposure, a substantial population of children are chronically exposed to SHS, resulting in an accumulation of toxic chemicals and metabolites that have the potential to cause disease. Nicotine and cotinine levels are used as a proxy for systemic exposure to over 4700 chemicals present in SHS(6). Despite the regular use of biomarkers to determine systemic SHS exposures, a large amount of inter-individual variability exists(7–10). Much of this variation has been ascribed to factors such as race, age, and gender, but genetic variation also offers a sizable contribution(6, 11). Many genes are involved in nicotine metabolism, some of which are highly polymorphic and alter the conversion of nicotine to cotinine. Our previously published findings demonstrate that polymorphisms in N-acetyltransferase 1 (NAT1), a xenobiotic metabolism gene, result in higher levels of cotinine in hair samples from SHS-exposed children participating in the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS)(12). Specifically, children exposed to SHS that carried the minor allele A of rs13253389 in NAT1 had over two-fold higher hair cotinine levels than those that were homozygous for the major allele. This finding suggests that NAT1 regulates metabolism of nicotine to cotinine.
The NAT1 gene is part of a family of ayrlamine N-acetyltransferases that are xenobiotic conjugating enzymes. These enzymes are responsible for acetylation of arylamines and arylhydrazines, O-acetylation of N-arylhydroxylamines, and N,O acetyl transfer of N-hydroxamic acid(13). NAT1 also functions in the acetylation of heterocyclic and aromatic amines, which are found in many substances including cigarette smoke(14),(15). The NAT genes present a crucial role in the detoxification and activation reactions of numerous xenobiotics originating not only from tobacco-derived aromatic and heterocyclic amine carcinogens but also from drug metabolism(16). Genetic polymorphisms in the NAT1 genes have been associated with “slow acetylator” and “fast acetylator” phenotypes, which exhibit reduced or enhanced metabolism of xenobiotic compounds, respectively(13). NAT1 polymorphism has been shown to increase the risk of multiple sclerosis(17), colorectal cancer(18) and lung and upper aerodigestive cancer(19) in smokers.
Given our observation that NAT1 polymorphisms result in significant differences in hair cotinine levels among SHS-exposed children and the association between SHS and respiratory disease, we hypothesized that having NAT1 polymorphisms would confer increased risk for developing asthma. We genotyped white children participating in CCAAPS as well as the Genomic Control Cohort (GCC) for single nucleotide polymorphisms (SNPs) in NAT1. We then evaluated associations between NAT1 SNPs, SHS exposure and asthma.
METHODS
Discovery Cohort
Population:
Subjects for the discovery population were obtained from participants in CCAAPS, a birth cohort of 762 infants born to atopic parent(s) between 2001 and 2003 in Cincinnati, Ohio and northern Kentucky(20). Infants were identified by birth records and eligible parents: 1) had at least one allergy symptom and 2) were skin prick test (SPT) positive to at least one aeroallergen(20). Children were examined annually at ages 1, 2, 3, 4 and 7 years of age for development of asthma and allergy symptoms, asthma, allergic rhinitis, eczema and food allergy. Medial history and environmental exposures are collected, and the children also underwent skin prick testing to 15 aeroallergens and milk and egg at each visit. Analyses were limited to whites because of differences in nicotine metabolism in whites compared to African-Americans(10), as well as differences in genetic allele frequencies between the races. White CCAAPS children with available genotyping data from our custom Illumina Array(12) and hair cotinine data available from the age 2 or 4 exam were included (n=359). Informed consent and parental permission was obtained from all parents. This study was approved by the Institutional Review Board at the University of Cincinnati.
SHS Exposure Determination:
SHS exposure histories were obtained using our validated panel of four questions(21). Parents were asked about the mothers smoking habits, the number of smokers living in the child’s home, the number of hours per day the child is around SHS at any location and SHS exposures in the car. Children whose parents responded positively to any of these four questions during any clinical exam were defined as SHS exposed; those answering no to all questions were defined as unexposed. Hair samples were obtained by cutting approximately 40 strands of hair in the occipital region of the scalp. Samples were adjusted for weight and analyzed for cotinine level via radioimmunoassay with a limit of detection of 0.02ng/mg. Cotinine values from hair samples taken at ages 2 and 4 were averaged; if only one sample was available then its value was utilized.
Asthma Determination:
In CCAAPS, asthma was defined at age 7 by reported symptoms and objective measures of lung function(22). Pulmonary function was determined at age 7 via spirometry testing according to ATS-ERS guidelines(23). Each child participant completed at least 4 acceptable maneuvers after the spirometers were verified for volume accuracy. Children were defined as having asthma if the parent reported asthma symptoms (tight or clogged chest or throat in the past 12 months, difficulty breathing or wheezy after exercise, wheezing or whistling in the chest in the previous 12 months, or a previous doctor’s diagnosis of asthma) and the child demonstrated either significant airway reversibility (>12% increase in FEV1) or a positive methacholine challenge test result(22). Cases were defined as children that were determined to have asthma at the age 7 visit; controls did not have a determination of asthma. Allergic asthma was defined as those that had a positive skin prick test to an aeroallergen at any age. Asthma severity was assessed by forced expiratory volume (FEV1).
DNA Isolation and Genotyping:
The CCAAPS cohort had banked buccal and/or saliva samples available; if both samples were available, saliva samples were used. DNA was isolated from buccal cells collected via cytology brush or saliva and using the Zymo Research Genomic DNA II Kit™ (Orange, CA) or Oragene DNA kit (DNA Genotek Inc.). Genotypes were assigned using GenomeStudio’s genotyping module. Genotyping was performed using a custom Illumina GoldenGate assay. The custom assay was populated with genes related to nicotine and xenobiotic metabolism and oxidative stress. SNPs with reported minor allele frequencies (MAF) ≥10% in the CEU population that captured the common genetic variation were selected using a coefficient of determination (r2) of 0.8 in HapMap (http://hapmap.ncbi.nlm.nih.gov/). In the CCAAPS population, one of the NAT1 SNPs had a MAF of 8.6% but was retained in the analyses. For this analysis, NAT1 SNPs that failed Hardy Weinberg Equilibrium (p < 0.0001) or had missing call rates greater than 10% were excluded.
Replication Cohort
Population:
White subjects for the replication population were obtained from participants in the GCC, a CCHMC-funded cohort of 1020 children representative of the Greater Cincinnati population with available SNP array data. Participants were invited to participate in the GCC without respect to any disease status. Participants in the GCC filled out questionnaires regarding medical history, including asthma and allergy symptoms and asthma, allergic rhinitis and eczema diagnosis by a physician. Blood samples were collected and DNA was extracted. In these analyses, participants were restricted to whites.
SHS Exposure and Asthma Determination:
SHS exposure was determined by parent report using the same four question screener as the discovery CCAAPS population. GCC asthma cases were defined by parental report of doctor-diagnosed asthma. Controls were participants without parental report of asthma. Allergic asthma was defined as those with parental report of eczema, hay fever or environmental allergies. Since asthma is often not diagnosed until later in childhood, we restricted the controls to GCC participants ≥7 years of age to minimize misclassification.
DNA Isolation and Genotyping:
Genomic DNA was extracted from blood samples using Manual PerfectPure DNA Blood Kit. Genotyping was performed on the Illumina OMNI5 platform. SNPs that failed Hardy Weinberg Equilibrium (p < 0.0001) or had missing call rates greater than 10% were excluded. All NAT1 SNPs had a MAF ≥10% (n=60).
Statistical Analyses
Principal component analyses was performed in EIGENSTRAT(24) using 100 ancestry-informative markers (AIMs) included on the CCAAPS chip, to account for potential population stratification. To address multiple testing, we determined the average pairwise linkage disequilibrium (LD, as measured by r2) for all SNP combinations and calculated the Bonferroni correction using Simple Interactive Statistical Analyses Software. The LD-adjusted Bonferroni corrected p-value for CCAAPS was 0.01. Associations between asthma and NAT1 SNPs were tested using logistic regression in all subjects as well as in subjects exposed and non-exposed to SHS. Additionally, interaction analyses between NAT1 polymorphism and hair cotinine level was conducted in the CCAAPS population. Interaction analyses were not held to our multiple testing correction criteria since the analyses were stratified, therefore the interaction was considered significant at p<0.05. All models were adjusted for sex and were carried out in SAS 9.4 (SAS, Cary, NC). In the GCC, models were also adjusted for age.
RESULTS
Demographics of Subjects
Of the 359 white children included from the CCAAPS cohort, 13.4% were asthmatic, 79.1% had allergic asthma and the mean FEV1 was 93 ± 13%. The asthma rate in the GCC cohort (n=638) was slightly higher at 15.5% (66.7% had allergic asthma), but this is due to the restriction of the controls to those ≥7 years of age (Table 1). We previously reported that hair cotinine was higher in children with parental reported smoke exposure than those without (0.139 vs. 0.058, p<0.0001)(12). Parental reported SHS exposure and hair cotinine levels were significantly higher in CCAAPS children with asthma compared to non-asthmatics (p=0.02 and p=0.0006, respectively, Table 1). SHS exposure was not associated with asthma in the GCC (p=1.00). The proportion of males did not differ significantly between groups in either population. White CCAAPS children included in this analysis did not differ from the 237 white CCAAPS children not included with respect to sex, asthma diagnosis or SHS exposure (data not shown).
Table 1:
Population Description and Sample Size in White Discovery and Replication Populations.
| CCAAPS | GCC | |||||
|---|---|---|---|---|---|---|
| Asthma (N=48) |
No Asthma (N=311) |
p-value | Asthma (N=99) |
No Asthma (N=539) |
p-value | |
| Males, N (%) | 30 (62.5) | 168 (54.0) | 0.27 | 48 (48.5) | 267 (49.5) | 0.93 |
| Parental report SHS, N (%) | 24 (50.0) | 102 (32.8) | 0.02 | 20 (20.2) | 108 (20.0) | 1.00 |
| % aeroallergen SPT+, N (%) | 30 (79.2) | 239 (77.9) | 0.72 | n/a† | ||
| Hair cotinine ng/mg (mean(SD)) | 0.15 (0.21) | 0.08 (0.13) | 0.0006 | n/a † | ||
| Age (mean) | 7 | 7 | n/a* | 11.9 | 12.6 ‡ | 0.13 |
All children in the CCAAPS birth cohort are evaluated for asthma at the age 7 clinical exam.
These variables are not available for the GCC.
GCC controls were restricted to those that were ≥7 years of age.
Genetic Associations with Asthma and SHS Exposure in CCAAPS
In our previous report, children carrying 1 or 2 copies of the minor allele of NAT1 rs13253389 had ~2-fold higher cotinine levels than children without the minor allele(12). We first tested whether this SNP was associated with asthma in the context of SHS exposure. Indeed, children with parental reported SHS exposure that carried one or two copies of the minor rs13253389 allele had a higher prevalence of asthma (18.9% and 44.4%, respectively) than children with only the major allele (10.1%, p=0.007, Supplemental Table 1). In contrast, asthma prevalence was not associated with rs13253389 in children not SHS exposed (Supplemental Table 1).
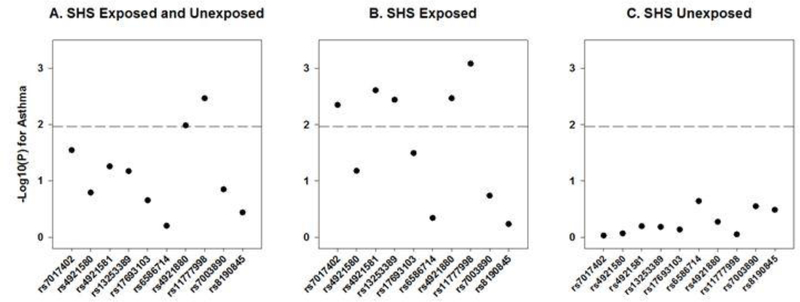
We then evaluated the 10 NAT1 SNPs and examined associations with asthma (Fig 1, Supplemental Table 2). In the whole population, only one of the NAT1 SNPs reached statistical significance at our LD-adjusted Bonferroni p-value threshold of 0.01 (Fig 1a, Supplemental Table 2). After stratifying by parental-reported SHS exposure, five of the 10 SNPs significantly associated with asthma in the exposed group (Fig 1b, Supplemental Table 2). Importantly, CCAAPS children without parental-reported SHS exposure did not demonstrate any significant genetic associations between NAT1 and asthma (Fig 1c, Supplemental Table 2).
Figure 1:
Association of NAT1 SNPs with asthma stratified by secondhand smoke exposure in white CCAAPS children. Gray line indicates LD-adjusted Bonferroni corrected p=0.01.
We next conducted interaction analyses of the SNPs with SHS exposure. To mitigate the potential for recall bias associated with the parental-reported SHS exposure, we examined both SNP*parental-reported SHS exposure as well as NAT1*hair cotinine interaction in subjects with parental-reported SHS exposure. Cotinine is an internal dose marker for SHS exposure, and levels of this biomarker in patient hair represent a quantitative measure of chemicals that may be present in SHS. Two SNPs had a significant SNP*parental-reported SHS interaction (Table 2). Restricting the analysis to children with parental-reported SHS exposure, 4 SNPs had significant SNP*hair cotinine level interaction. Two SNPs, rs4921581 and rs13253389, showed significant interaction with both parental-reported SHS and hair cotinine level (Table 2). We previously reported that these SNPs are in strong LD(12). While both SNPs were significantly associated with asthma, rs4921581 had a larger effect size (OR 2.80 vs OR 2.67, Supplemental Table 2).
Table 2.
Interaction analyses of NAT1 SNPs, Parental-Reported SHS Exposure and Hair Cotinine Level with Asthma in White CCAAPS Children.
| SNP | Major/Minor allele | MAF | SNP*SHS Interaction p-value | SNP*Hair Cotinine Interaction* |
|---|---|---|---|---|
| rs7017402 | G/A | 0.123 | 0.079 | 0.959 |
| rs4921580 | C/G | 0.126 | 0.135 | 0.007 |
| rs4921581 | G/A | 0.333 | 0.014 | 0.042 |
| rs13253389 | G/A | 0.344 | 0.020 | 0.048 |
| rs17693103 | G/T | 0.179 | 0.112 | 0.568 |
| rs6586714 | G/A | 0.086 | 0.188 | 0.115 |
| rs4921880 | A/T | 0.227 | 0.101 | 0.989 |
| rs11777998 | G/C | 0.123 | 0.051 | 0.060 |
| rs7003890 | -G/A | 0.442 | 0.885 | 0.030 |
| rs8190845 | G/A | 0.126 | 0.861 | 0.157 |
Only children with parental-reported SHS exposure.
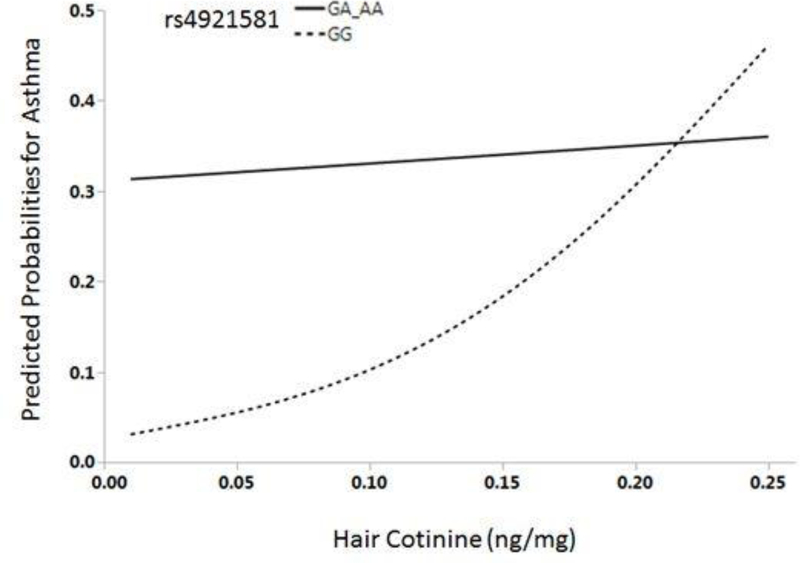
We then looked at the interactive effect of NAT1 rs4921581 genotype and hair cotinine level on asthma risk graphically. At the highest levels of hair cotinine (>0.25ng/mg), we did not observe an effect of the rs4921581 SNP on asthma risk. Only 12 of the 126 children had hair cotinine >0.25ng/mg; they were subsequently removed from this analysis. In the remaining 114 children, we observed a differential risk by genotype for asthma (Fig 2). In children with the GG genotype, asthma risk increases with increasing levels of hair cotinine (Fig 2). In contrast, the risk of asthma in children with the AA or GA genotype is high even at the lowest levels of hair cotinine, suggesting that even low levels of SHS exposure in children with at least one A allele are detrimental.
Figure 2:
Predicted probabilities of asthma by hair cotinine levels stratified by NAT1 rs4921581 genotype in white CCAAPS children.
Replication of the association of NAT1, SHS exposure and asthma in the GCC
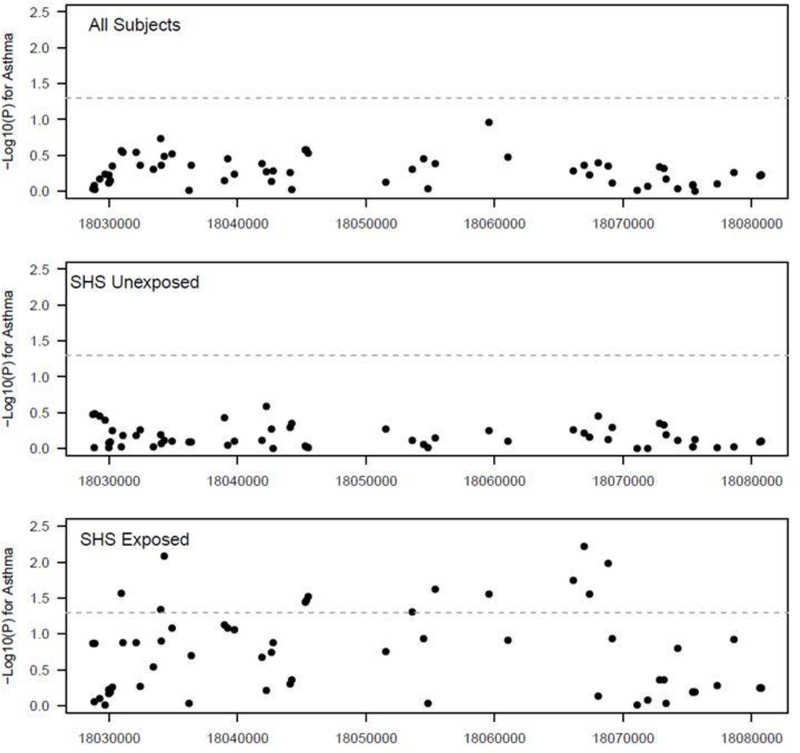
In order to replicate of our findings, we evaluated parental report of SHS exposure and asthma as well as NAT1 genotype in the GCC population. We had genotyping information on 60 NAT1 SNPs. Five of our 10 NAT1 SNPs genotyped in the CCAAPS population were included in the 60 available for the GCC. When all GCC subjects were evaluated, none of the NAT1 SNPs reached nominal significance (Fig 3). After stratification for parental-reported SHS exposure, we observed a significant asthma association for 13 NAT1 SNPs in the exposed group (p<0.05), compared to none in the unexposed group (Fig 3). Although the 13 significant SNPs in the GCC did not overlap with the 5 significant SNPs in the CCAAPS population, this demonstrates gene-level replication.
Figure 3:
Association of NAT1 SNPs with asthma stratified by SHS exposure in white GCC children. Gray line indicates p=0.05.
DISCUSSION
Our data demonstrate that NAT1 genetic variation interacts with SHS exposure to increase the risk of asthma in children. Children exposed to SHS who carried the risk allele had a high risk of asthma regardless of exposure level, while asthma risk in children without the variant was positively correlated with hair cotinine level. At higher levels of hair cotinine, the asthma risk reached the same level for children with and without the variant, suggesting that the genetic effect may be masked at higher levels of exposure. Taken together, these findings suggest that NAT1 variation plays a role in modifying asthma likelihood in children exposed to SHS. To our knowledge, this is the first report of a gene-environment interaction between NAT1 genetic polymorphisms, SHS exposure, and pediatric asthma.
Our group previously demonstrated that the NAT1 rs13253389 SNP was associated with increased hair cotinine levels, which may be attributed to altered nicotine metabolism. In our current analysis, this SNP exhibited a statistically significant association with a diagnosis of asthma, but only in the context of SHS exposure. These results suggest that NAT1 genetic variation alters the metabolism of multiple xenobiotic compounds present in SHS, not just nicotine, which promote airway hyper responsiveness in exposed children.
Genetic polymorphism in NAT1 confers measurable differences in its detoxifying function. Slow and fast acetylator NAT1 phenotypes types have previously been identified. In adults, slow acetylators have a 2.5-fold increased risk of diisocyanate-induced asthma, suggesting that slow acetylators cannot effectively neutralize toxic diisocyanates and their metabolic byproducts(25). Variation in a NAT1 tagging SNP has been associated with a 70% decreased risk of diisocyanate-associated asthma compared to asymptomatic controls in diisocyanate-exposed workers(26). Similarly, in SHS-exposed children, NAT1 variation may subject them to accumulation of numerous toxic compounds, including diisocyanates(27). Inadequate inactivation of harmful compounds may stimulate increased mucous production, constrict the smooth muscle of the airway, or promote accumulation of inflammatory cells, all physiological hallmarks in asthma development.
In addition to its role in detoxifying harmful substances via acetylation, NAT1 inactivates cysteinyl leukotrienes (CysLTs)(28). CysLTs are inflammatory lipid mediators that play a crucial role in airway narrowing as well as mediation of hyper-responsiveness to histamine that are both associated with asthma, recurrent episodes of coughing, wheezing, and breathlessness(29),(30),(31). Consequentially, by inactivating CysLTs and modulating histamine activity, NAT1 may inhibit their pro-inflammatory mechanism and alter the occurrence or severity of asthma(32).
NAT enzymes have been demonstrated in bronchial epithelial cells, including Clara cells and type II pneumocytes, suggesting that inhaled aromatic amine pollutants, such as those present in SHS, may undergo NAT-dependent biotransformation in lung epithelium(33).
Furthermore, exposure of lung cells to high levels of oxidants, such as H2O2 or peroxynitrite, impairs NAT1-dependent cellular biotransformation of aromatic amines, possibly compromising detoxification pathways. These findings suggest a contribution of NAT enzyme function on respiratory disease outcomes and how NAT alteration may result in deleterious pollutant effects on health.
The most significant NAT1 tagging SNP in this study, rs4921581, is located within an intron. While it is possible that rs4921581 is directly involved in the observed phenotype, this is unlikely since it is a tagging SNP and did not replicate in the GCC, even though other variants in NAT1 did exhibit association. Further, we investigated whether any of our significant SNPs were cis-expression quantitative trait loci (cis-eQTLs) using the Genotype-Tissue Expression (GTEx) project(34) but there was no evidence that any of our NAT1 SNPs affect expression levels. Using HaploReg v4.1(35), we identified that rs4921581 is predicted to alter a regulatory motif as well as histone sites in blood and immune cells, therefore it may have a regulatory function. Further, some of our NAT1 SNPs were either associated with an eQTL or in LD with a SNP associated with an eQTL in whole blood, but none of these SNPs were top-associated cis-eQTLs (false discovery rate <0.5)(36). Therefore, our SNP is most likely in linkage disequilibrium with an undetermined causal variant.
NAT1 genetic variants with known effects on enzyme activity levels have been previously described. An alteration in the consensus polyadenylation site within the NAT1*10 allele(37–39) and a G445A transition (Val149Ile, rs4987076) in the NAT1*11 allele are both associated with increased enzymatic activity(40), while a G560A substitution (Arg187Gln, rs4986782) and C559T base substitution (Arg187Stop) are associated with significantly decreased NAT1 activity(14, 40). Additionally, NAT1*17, *1, and *22 have been associated with reduced NAT1 enzymatic activity, but their frequency in whites is so low that these are probably of little consequence at the population level(41, 42). Further sequencing and biologic studies are needed to determine causal NAT1 variants in our population.
The strengths of this study include the comprehensive phenotyping of the CCAAPS subjects, specifically the objective asthma diagnosis and available hair cotinine levels, as well as the availability of the GCC as a replication population from the same geographical region. Study participants were all white children in the Greater Cincinnati area; consequently, future studies will be needed in order to verify the generalizability of our results. Although our discovery cohort was modestly sized, our similar findings in the GCC replication cohort provide replication and support our findings. Due to power concerns given our modest sample size, we did not adjust for other potential confounders such as air pollution and allergen exposure or evaluate asthma severity and sub-phenotypes. Larger, future studies are necessary to disentangle these effects. Because we used a tagging SNP selection approach, there may be other NAT1 SNPs with associations with hair cotinine and asthma diagnosis which were not directly tested. Given our gene-level rather than SNP-level replication, to establish which variants are contributing in a causal manner it will be necessary to first evaluate all variants in the region, and then perform mechanistic studies to demonstrate metabolic or pathophysiologic relevance.
Conclusion
Our data supports that NAT1 SNP rs4921581 is associated with increased risk of asthma, specifically in SHS-exposed children. These findings may have direct clinical utility, as knowledge of NAT1 genotype can potentially inform caregivers about asthma risk when SHS-exposure status is known. Future studies examining the mechanisms underlying the associations between NAT1 polymorphism, SHS exposure, and asthma may lead to the development of novel and efficacious pharmacologic therapies.
Supplementary Material
Acknowledgments
The authors thank the clinic staff for their efforts in study coordination, recruitment, data management, and data collection. We also thank all of the CCAAPS and GCC participants and families for their time and commitment. We also thank Angela Sadler for editorial assistance.
Abbreviations
- AIMs
ancestry-informative markers
- CCAAPS
Cincinnati Childhood Allergy and Air Pollution Study
- GCC
Genomic Control Cohort
- Cis-eQTLs
cis-expression quantitative trait loci
- GTEx
Genotype-Tissue Expression
- LD
linkage disequilibrium
- MAF
minor allele frequencies
- NAT1
N-acetyltransferase
- SHS
secondhand smoke
- SNPs
single nucleotide polymorphisms
- SPT
skin prick test
Footnotes
Declaration of Interests
The authors report no conflicts of interest.
References
- 1.Department of Health and Human Services, Centers for Disease Control and Prevention,, Coordinating Center for Health Promotion,, National Center for Chronic Disease Prevention and Health Promotion,, Office on Smoking and Health,. The Health Consequences of Involuntary Exposure to Tobacco Smoke: A Report of the Surgeon General. Atlanta, GA. : 2006. [PubMed] [Google Scholar]
- 2.Moritsugu KP. The 2006 Report of the Surgeon General: the health consequences of involuntary exposure to tobacco smoke. Am J Prev Med. 2007;32(6):542–3. Epub 2007/05/30. doi: 10.1016/j.amepre.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 3.NTP (National Toxicology Program). Report on Carcinogens, Thirteenth Edition. Research Triangle Park, NC. : 2014.
- 4.ACS. Secondhand Smoke and Children Fact Sheet. Atlanta, GA: American Cancer Society, 2006 Contract No.: January 11, 2007. [Google Scholar]
- 5.Schuster MA, Franke T, Pham CB. Smoking patterns of household members and visitors in homes with children in the United States. Arch Pediatr Adolesc Med. 2002;156(11):1094–100. [DOI] [PubMed] [Google Scholar]
- 6.Benowitz NL, Hukkanen J, Jacob P 3rd, Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009(192):29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benowitz NL, Jacob P 3rd, Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56(5):483–93. [DOI] [PubMed] [Google Scholar]
- 8.Benowitz NL, Jacob P 3rd, Individual differences in nicotine kinetics and metabolism in humans. NIDA Res Monogr. 1997;173:48–64. [PubMed] [Google Scholar]
- 9.Benowitz NL, Perez-Stable EJ, Herrera B, Jacob P 3rd, Slower metabolism and reduced intake of nicotine from cigarette smoking in Chinese-Americans. Journal of the National Cancer Institute. 2002;94(2):108–15. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Stable EJ, Herrera B, Jacob P 3rd, Benowitz NL Nicotine metabolism and intake in black and white smokers. Jama. 1998;280(2):152–6. [DOI] [PubMed] [Google Scholar]
- 11.Mwenifumbo JC, Tyndale RF. Molecular genetics of nicotine metabolism. Handb Exp Pharmacol. 2009(192):235–59. [DOI] [PubMed] [Google Scholar]
- 12.LeMasters GK, Khurana Hershey GK, Sivaprasad U, Martin LJ, Pilipenko V, Ericksen MB, et al. N-acetyltransferase 1 polymorphism increases cotinine levels in Caucasian children exposed to secondhand smoke: the CCAAPS birth cohort. The pharmacogenomics journal. 2015;15(2):189–95. doi: 10.1038/tpj.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sim E, Walters K, Boukouvala S. Arylamine N-acetyltransferases: from structure to function. Drug Metab Rev. 2008;40(3):479–510. Epub 2008/07/22. doi: 10.1080/03602530802186603. [DOI] [PubMed] [Google Scholar]
- 14.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9(1):29–42. Epub 2000/02/10. [PubMed] [Google Scholar]
- 15.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. Epub 2002/09/28. [DOI] [PubMed] [Google Scholar]
- 16.Khlifi R, Messaoud O, Rebai A, Hamza-Chaffai A. Polymorphisms in the human cytochrome P450 and arylamine N-acetyltransferase: susceptibility to head and neck cancers. Biomed Res Int. 2013;2013:582768. Epub 2013/10/24. doi: 10.1155/2013/582768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briggs FB, Acuna B, Shen L, Ramsay P, Quach H, Bernstein A, et al. Smoking and risk of multiple sclerosis: evidence of modification by NAT1 variants. Epidemiology. 2014;25(4):605–14. Epub 2014/03/15. doi: 10.1097/EDE.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen M, Autrup H, Olsen A, Tjonneland A, Overvad K, Raaschou-Nielsen O. Prospective study of NAT1 and NAT2 polymorphisms, tobacco smoking and meat consumption and risk of colorectal cancer. Cancer Lett. 2008;266(2):186–93. Epub 2008/03/29. doi: 10.1016/j.canlet.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 19.McKay JD, Hashibe M, Hung RJ, Wakefield J, Gaborieau V, Szeszenia-Dabrowska N, et al. Sequence variants of NAT1 and NAT2 and other xenometabolic genes and risk of lung and aerodigestive tract cancers in Central Europe. Cancer Epidemiol Biomarkers Prev. 2008;17(1):141–7. Epub 2008/01/18. doi: 10.1158/1055-9965.EPI-07-0553. [DOI] [PubMed] [Google Scholar]
- 20.LeMasters GK, Wilson K, Levin L, Biagini J, Ryan P, Lockey JE, et al. High prevalence of aeroallergen sensitization among infants of atopic parents. J Pediatr. 2006;149(4):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biagini Myers JM, Khurana Hershey GK, Deka R, Burkle JW, Levin LS, Bernstein DI, et al. Asking the right questions to ascertain early childhood secondhand smoke exposures. J Pediatr. 2012;160(6):1050–1. Epub 2012/04/13. doi: S0022–3476(12)00243–0 [pii] 10.1016/j.jpeds.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reponen T, Vesper S, Levin L, Johansson E, Ryan P, Burkle J, et al. High environmental relative moldiness index during infancy as a predictor of asthma at 7 years of age. Ann Allergy Asthma Immunol. 2011;107(2):120–6. doi: 10.1016/j.anai.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Asthma Education and Prevention Program (NAEPP) NHLBI. Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. NIH Publication No. 07–4051. National Institutes of Health, National Heart, Lung and Blood Institute, 2007. [Google Scholar]
- 24.Narayanaswamy CRaDR. J Royal Statistical Society Series C. 1991;40(2):309–16. [Google Scholar]
- 25.Wikman H, Piirila P, Rosenberg C, Luukkonen R, Kaaria K, Nordman H, et al. N-Acetyltransferase genotypes as modifiers of diisocyanate exposure-associated asthma risk. Pharmacogenetics. 2002;12(3):227–33. Epub 2002/04/03. [DOI] [PubMed] [Google Scholar]
- 26.Yucesoy B, Kissling GE, Johnson VJ, Lummus ZL, Gautrin D, Cartier A, et al. N-Acetyltransferase 2 Genotypes Are Associated With Diisocyanate-Induced Asthma. J Occup Environ Med. 2015;57(12):1331–6. Epub 2015/12/08. doi: 10.1097/JOM.0000000000000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agency USEP. Health and Environmental Effects Profile for Methyl Isocyanate. Cincinnati, OH: 1986.
- 28.Kim JM, Park BL, Park SM, Lee SH, Kim MO, Jung S, et al. Association analysis of N-acetyl transferase-2 polymorphisms with aspirin intolerance among asthmatics. Pharmacogenomics. 2010;11(7):951–8. Epub 2010/07/07. doi: 10.2217/pgs.10.65. [DOI] [PubMed] [Google Scholar]
- 29.Frieri M. Asthma concepts in the new millennium: update in asthma pathophysiology. Allergy Asthma Proc. 2005;26(2):83–8. Epub 2005/06/24. [PubMed] [Google Scholar]
- 30.Bai TR, Knight DA. Structural changes in the airways in asthma: observations and consequences. Clinical science (London, England : 1979). 2005;108(6):463–77. Epub 2005/05/18. doi: 10.1042/cs20040342. [DOI] [PubMed] [Google Scholar]
- 31.Elias JA, Zhu Z, Chupp G, Homer RJ. Airway remodeling in asthma. J Clin Invest. 1999;104(8):1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M, Muller J, Leier I, Jedlitschky G, Ball HA, Moore KP, et al. Metabolism of cysteinyl leukotrienes in monkey and man. Eur J Biochem. 1990;194(1):309–15. Epub 1990/11/26. [DOI] [PubMed] [Google Scholar]
- 33.Dairou J, Petit E, Ragunathan N, Baeza-Squiban A, Marano F, Dupret JM, et al. Arylamine N-acetyltransferase activity in bronchial epithelial cells and its inhibition by cellular oxidants. Toxicology and applied pharmacology. 2009;236(3):366–71. Epub 2009/03/03. doi: 10.1016/j.taap.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Gibson G. Human genetics. GTEx detects genetic effects. Science. 2015;348(6235):640–1. doi: 10.1126/science.aab3002. [DOI] [PubMed] [Google Scholar]
- 35.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badawi AF, Hirvonen A, Bell DA, Lang NP, Kadlubar FF. Role of aromatic amine acetyltransferases, NAT1 and NAT2, in carcinogen-DNA adduct formation in the human urinary bladder. Cancer Res. 1995;55(22):5230–7. Epub 1995/11/15. [PubMed] [Google Scholar]
- 38.Bell DA, Badawi AF, Lang NP, Ilett KF, Kadlubar FF, Hirvonen A. Polymorphism in the N-acetyltransferase 1 (NAT1) polyadenylation signal: association of NAT1*10 allele with higher N-acetylation activity in bladder and colon tissue. Cancer Res. 1995;55(22):5226–9. Epub 1995/11/15. [PubMed] [Google Scholar]
- 39.Zenser TV, Lakshmi VM, Rustan TD, Doll MA, Deitz AC, Davis BB, et al. Human N-acetylation of benzidine: role of NAT1 and NAT2. Cancer Res. 1996;56(17):3941–7. Epub 1996/09/01. [PubMed] [Google Scholar]
- 40.Deitz AC, Doll MA, Hein DW. A restriction fragment length polymorphism assay that differentiates human N-acetyltransferase-1 (NAT1) alleles. Analytical biochemistry. 1997;253(2):219–24. Epub 1997/12/31. doi: 10.1006/abio.1997.2379. [DOI] [PubMed] [Google Scholar]
- 41.Cascorbi I, Roots I, Brockmoller J. Association of NAT1 and NAT2 polymorphisms to urinary bladder cancer: significantly reduced risk in subjects with NAT1*10. Cancer Res. 2001;61(13):5051–6. Epub 2001/06/30. [PubMed] [Google Scholar]
- 42.Wikman H, Thiel S, Jager B, Schmezer P, Spiegelhalder B, Edler L, et al. Relevance of N-acetyltransferase 1 and 2 (NAT1, NAT2) genetic polymorphisms in non-small cell lung cancer susceptibility. Pharmacogenetics. 2001;11(2):157–68. Epub 2001/03/27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.