Abstract
Objective
We evaluated the prevalence of pathogenic repeat expansions in replication factor C subunit 1 (RFC1) and disabled adaptor protein 1 (DAB1) in an undiagnosed ataxia cohort from North America.
Methods
A cohort of 596 predominantly adult-onset patients with undiagnosed familial or sporadic cerebellar ataxia was evaluated at a tertiary referral ataxia center and excluded for common genetic causes of cerebellar ataxia. Patients were then screened for the presence of pathogenic repeat expansions in RFC1 (AAGGG) and DAB1 (ATTTC) using fluorescent repeat-primed PCR (RP-PCR). Two additional undiagnosed ataxia cohorts from different centers, totaling 302 and 13 patients, respectively, were subsequently screened for RFC1, resulting in a combined 911 subjects tested.
Results
In the initial cohort, 41 samples were identified with 1 expanded allele in the RFC1 gene (6.9%), and 9 had 2 expanded alleles (1.5%). For the additional cohorts, we found 20 heterozygous samples (6.6%) and 17 biallelic samples (5.6%) in the larger cohort and 1 heterozygous sample (7.7%) and 3 biallelic samples (23%) in the second. In total, 29 patients were identified with biallelic repeat expansions in RFC1 (3.2%). Of these 29 patients, 8 (28%) had a clinical diagnosis of cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS), 14 had cerebellar ataxia with neuropathy (48%), 4 had pure cerebellar ataxia (14%), and 3 had spinocerebellar ataxia (10%). No patients were identified with expansions in the DAB1 gene (spinocerebellar ataxia type 37).
Conclusions
In a large undiagnosed ataxia cohort from North America, biallelic pathogenic repeat expansion in RFC1 was observed in 3.2%. Testing should be strongly considered in patients with ataxia, especially those with CANVAS or neuropathy.
Cerebellar ataxia is a heterogeneous genetic disorder characterized by inability to control balance and coordination. Roughly 50% of patients remain undiagnosed despite advanced genomic testing.1–4The most common genetic ataxias, as well as several rarer forms, are caused by nucleotide repeat expansions, which typically require targeted non–sequence-based testing to identify.5–8
Recent studies identified a recessive intronic (AAGGG) repeat expansion in replication factor C subunit 1 (RFC1) related to cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS) in Australia and the United Kingdom.9,10 In addition, this expansion may be responsible for up to 22% (33/150) of sporadic cerebellar ataxia and 63% (32/51) of ataxia associated with sensory neuropathy.9 Similarly, a dominant pathologic pentanucleotide (ATTTC) repeat insertion was identified within a normal (ATTTT) tandem repeat element in the intronic 5′ untranslated region of the disabled adaptor protein 1 (DAB1) gene causing spinocerebellar ataxia type 37 (SCA37) in patients from the southern Iberian Peninsula.11
To address the frequency of these repeat expansion disorders in North America, we assessed a large cohort of 596 patients from the United States with unidentified cerebellar ataxia. We identified biallelic RFC1 expansion in 1.5% (n = 9) and found no patients with a pathogenic DAB1 expansion. We further tested 2 additional cohorts from different centers (the larger of which consisted of approximately one-third samples from patients in Canada, with the remainder from the United States) and identified RFC1-mediated ataxia cases in 5.6% (17/302) and 23% (3/13), respectively, for a total prevalence of 3.2% (29/911).
Methods
Standard protocol approvals, registrations, and patient consents
Patients were enrolled at the University of California, Los Angeles (UCLA) Ataxia Center, clinically assessed for acquired causes of ataxia, and then considered for genetic causes.2 Only patients with negative testing for the common genetic ataxias (SCA1, SCA2, SCA3, SCA6, SCA7, and Friedreich ataxia) were included in this study. The majority (∼ two-thirds) were adult and sporadic onset. All patients consented for DNA collection for genetic analysis. Peripheral blood was collected from patients, and DNA was then isolated and purified using the Gentra Puregene Blood Kit (Qiagen) for genetic testing. The study methods used were approved by the UCLA Institutional Review Board. Additional DNA samples from 302 patients enrolled at the University of Chicago and 13 patients enrolled at the Brigham and Women's Hospital under institutional review board-approved procedures with identical inclusion criteria were subsequently tested as well. For the purpose of assessing disease prevalence, only probands from affected families were included in this study, and all pathogenic repeat expansions were confirmed in additional affected family members when available.
Repeat expansion testing
RFC1 gene repeat expansion analysis
Fluorescent repeat-primed PCR (RP-PCR) was performed to detect RFC1 pathogenic (AAGGG)n alleles using previously published primers9,10 with an optimized touchdown PCR protocol and Qiagen HotStarTaq. One primer set included the forward 5′FAM-ACTGACAGTGTTTTTGCCTGT-3′ primer, the anchor 5′-CAGGAAACAGCTATGACC-3′ primer, and the repeat 5′-CAGGAAACAGCTATGACCAAGGGAAGGGAAGGGAAGGGAAGGG-3′ primer that identifies the (AAGGG) repeats. A second primer set included the forward 5′FAM-TCAAGTGATACTCCAGCTACACCGT-3′ primer, the anchor 5′-CAGGAAACAGCTATGACC-3′ primer, and 3 repeat primers that identify the (AAGGG) repeats 5′-CAGGAAACAGCTATGACCAACAGAGCAAGACTCTGTTTCAAAAAAGGGAAGGGAAGGGAAGGGAA-3′, 5′-CAGGAAACAGCTATGACCAACAGAGCAAGACTCTGTTTCAAAAAGGGAAGGGAAGGGAAGGGAA-3′, and 5′-CAGGAAACAGCTATGACCAACAGAGCAAGACTCTGTTTCAAAAGGGAAGGGAAGGGAAGGGAA-3′. Fragment length analysis was performed using an Applied Biosystems 3730xl DNA Analyzer with Peak Scanner software (v. 2.0). To determine whether the genotype of samples with positive RP-PCR results was heterozygous or biallelic, standard PCR was performed using published primers,9 forward 5′-TCAAGTGATACTCCAGCTACACCGTTGC-3′ primer and the reverse 5′ GTGGGAGACAGGCCAATCACTTCAG-3′ primer. Observation of a band at or near 348 bp (wild-type size) corresponding to repeat sizes of less than approximately 60 repeats (approximately 650 bp) identified patients as heterozygotes. As an internal control to prevent false positives, standard PCR was also performed simultaneously on the same sample with primers designed to amplify a 282-bp band from the SPG11 gene (figure e-1, links.lww.com/NXG/A266).
For a proportion of samples at the University of Chicago, 3 separate reactions each using 100 ng of genomic DNA were performed to confirm the existence of a true biallelic (AAGGG) repeat expansion (figure e-2, links.lww.com/NXG/A267). First, a flanking PCR was performed using primers that surrounded the RFC1 region of interest. The flanking primers included a labeled forward primer9 (5′-/56-FAM/ACTGACAGTGTTTTTGCCTGT-3′) (10 μm) and a reverse primer9 (5′-GGCTGAGGCAGGAGATTCAC-3′) (10 μm). Second, RP‐PCR was performed to detect individual nonpathogenic (AAAAG) motifs. The primers for the (AAAAG) RP-PCR included the same forward primer as the flanking reaction, an M13 anchor primer (5′-CAGGAAACAGCTATGACC-3′) (10 μm) and an (AAAAG) specific primer (5′-CAGGAAACAGCTATGACC_AAAAGAAAAGAAAAGAAAAGAAAAG-3′) (1 μm). Both the flanking and (AAAAG) RP-PCR products were amplified using the Takara LA PCR kit in combination with a touchdown PCR. Third, to detect individual pathogenic (AAGGG) motifs, a PCR reaction was performed using the same forward primer as the flanking, the M13 anchor primer, and an (AAGGG) specific primer9 (5′-CAGGAAACAGCTATGACC_AAGGGAAGGGAAGGGAAGGGAAGGG-3′). To obtain full amplification of the expanded (AAGGG) motif, the Qiagen HotStarTaq chemistry was used with 400 μm of deoxyribonucleotide triphosphates using a standard PCR. All 3 products were loaded on an ABI3730xl DNA analyzer after denaturing with a 5% GS500 Rox/formamide mixture and subsequently analyzed using GeneMarker v2.6.0 (SoftGenetics Inc.).
To validate the assays performed at the different centers, a proportion of samples (>50%) determined to have heterozygous or biallelic AAGGG expansions were assessed at least 2 times by both expansion RP-PCR testing and standard PCR genotyping in 2 separate laboratories.
DAB1 gene repeat expansion analysis
RP-PCR was performed to detect expansion of the normal (ATTTT) DAB1 repeat region and for detection of the pathogenic (ATTTC)n insertion using published primers.11,12 Fragment length analysis was performed as described above.
Data availability
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Results
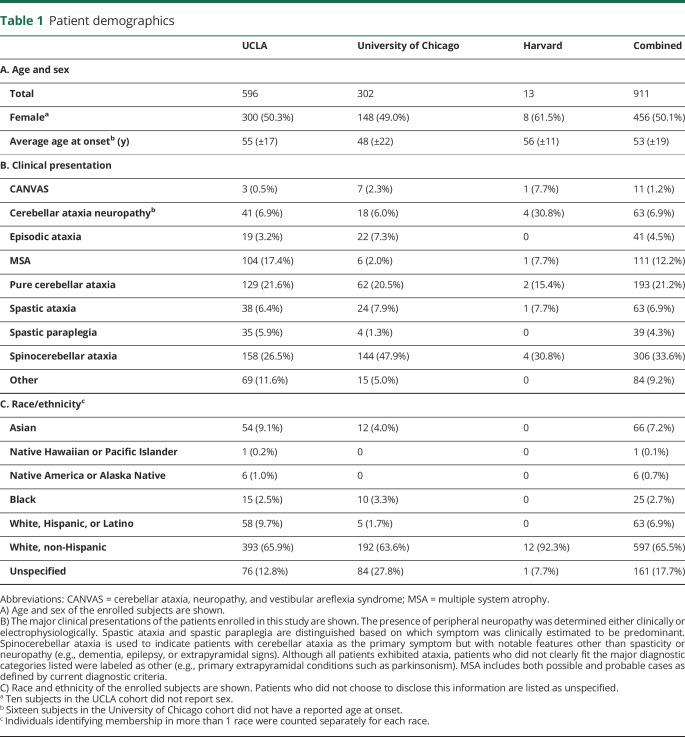
We examined the prevalence of repeat expansions in RFC1 and DAB1 in a large cohort from the tertiary referral Ataxia Center at UCLA. The demographics of this 596 subject cohort are described in table 1. Average age was 55 years, 50% of the patients were female, and 66% were white, non-Hispanic. The most common phenotypes were spinocerebellar ataxia (26.5%), pure cerebellar ataxia (21.6%), and multiple system atrophy (17.4%). To assess for the presence of pathogenic (AAGGG) repeat expansions in RFC1, fluorescent repeat-primed fragment analysis was performed and identified at least 1 expansion in 50 of 596 patients (8.4%, figure 1 and figure e-1, links.lww.com/NXG/A266). Standard PCR was used to genotype subjects for the presence of a heterozygous or a pathogenic biallelic expansion. Nine subjects (1.5%) were found with biallelic expansions. Of these patients, 3 subjects presented clinically with CANVAS (33%, 100% of phenotype), 5 had cerebellar ataxia with neuropathy (56%, 12% of phenotype), and 1 had spinocerebellar ataxia (11%, 0.6% of phenotype).
Table 1.
Patient demographics
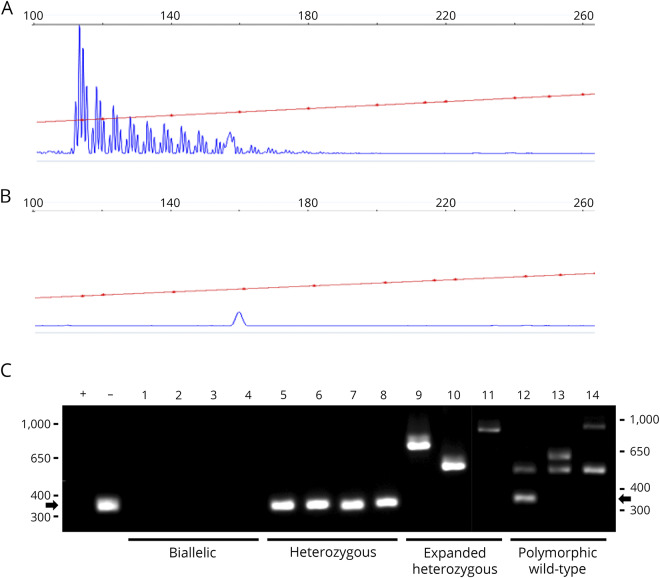
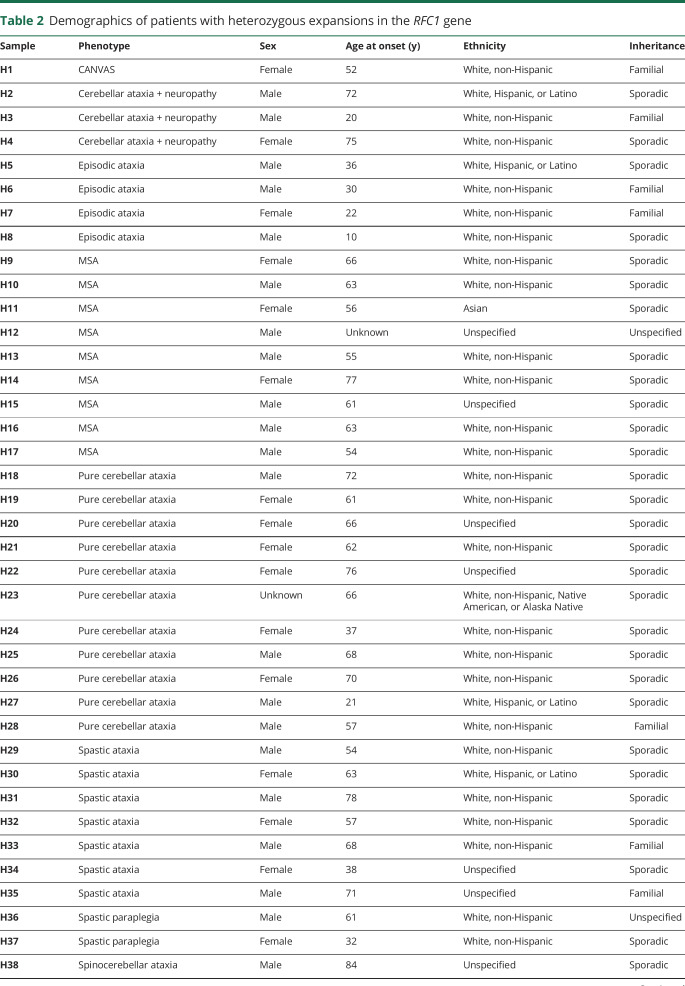
Figure 1. RFC1 expansion analysis.
Representative RP-PCR results from a patient with disease due to (A) biallelic expanded (AAGGG) pathogenic alleles or a control individual (B) with wild-type alleles. Samples with RP-PCR evidence of an expanded RFC1 allele were genotyped by standard PCR for biallelic expansion (C). Standard PCR allows categorization of individuals as biallelic with pathogenic expansions (no band, lanes 1-4), heterozygous wild-type (348 bp band, arrow, lanes 5-8), heterozygous with a non-pathogenic polymorphic expansion (variable sized bands, lanes 9-11), or wild-type with one or more non-pathogenic polymorphic expansion(s) (variable sized band(s), lanes 12-14). + = biallelic control; − = wild-type control; M = marker; RFC1 = replication factor C subunit 1.
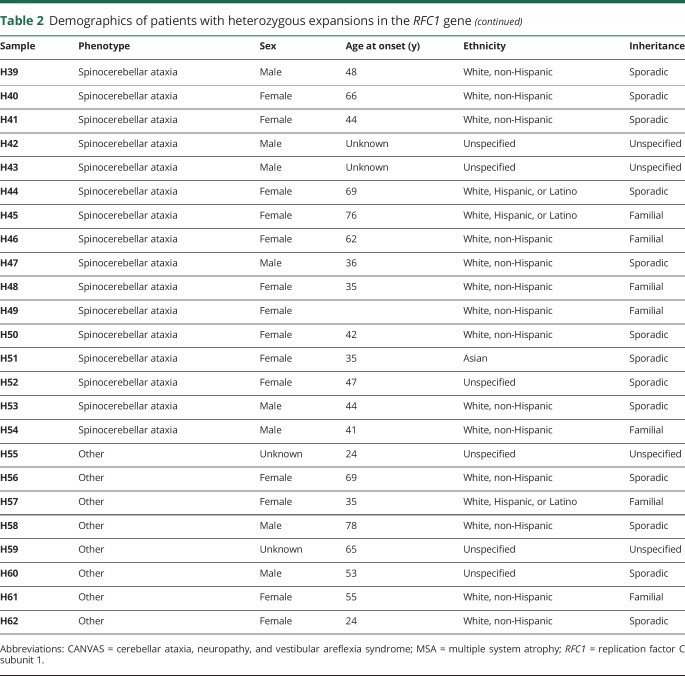
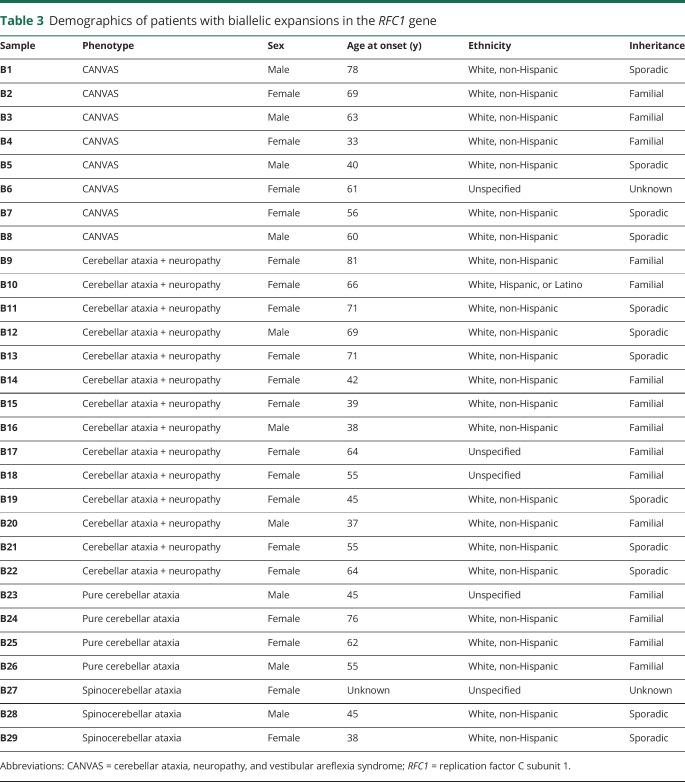
To validate these findings, we tested 2 additional cohorts from centers in different regions of the United States, the University of Chicago (UC) and Brigham and Women's Hospital affiliated with Harvard Medical School. Demographics were similar to the UCLA cohort (table 1). The larger UC cohort, consisting of both sporadic and familial cases with roughly one-third of subjects originating from Canada, showed heterozygous expansions in 20 individuals (6.6%) and pathogenic biallelic expansion in 17 patients (5.6%) (figure e-2, links.lww.com/NXG/A267). Four subjects had CANVAS (24%, 57% of phenotype), 7 had cerebellar ataxia with neuropathy (41%, 39% of phenotype), 4 had pure cerebellar ataxia (23.5%, 6.5% of phenotype), and 2 had spinocerebellar ataxia (12%, 1.4% of phenotype). In the smaller Harvard cohort, 1 individual showed heterozygous expansion (7.7%), and 3 of 13 patients (23%) had pathogenic biallelic expansions. Two of these had cerebellar ataxia with neuropathy (67%, 50% of phenotype), while 1 had CANVAS (33%, 100% of phenotype). Collectively, 62 individuals were found with heterozygous expansion (6.8%, 62/911, table 2), and 29 of 911 subjects showed pathogenic biallelic expansions across all cohorts for a total prevalence of 3.2% (29/911, table 3). Of the biallelic cases, the majority of the patients were white (24/29, 83%), 1 patient was Hispanic (1/29, 3.4%), and the race/ethnicity of 5 patients was not reported (17%). Twelve of the cases showed sporadic onset (41%). In total, 8 subjects had CANVAS (28%, 73% of phenotype), 14 had cerebellar ataxia with neuropathy (48%, 22% of phenotype), 4 had pure cerebellar ataxia (14%, 2.1% of phenotype), and 3 had spinocerebellar ataxia (10%, 1.0% of phenotype).
Table 2.
Demographics of patients with heterozygous expansions in the RFC1 gene
Table 3.
Demographics of patients with biallelic expansions in the RFC1 gene
For DAB1 repeat expansion analysis, 83/596 (13.9%) subjects showed an expanded ATTTT allele by RP-PCR analysis; however, none of these patients possessed the pathogenic ATTTC insertion, indicating that no patients within the cohort had SCA37.
Discussion
Overall, in a large undiagnosed ataxia cohort of 911 patients from 3 tertiary referral centers in the United States, biallelic pathogenic (AAGGG) repeat expansions in RFC1 were observed in 3.2% (n = 29, 95% CI 2.0%–4.3%) of patients from the United States (25/29, 86%) and Canada (4/29, 14%). The observation that the majority of cases (24/24, 100%, table 3), where ethnicity was known, were white of European ancestry is consistent with previous reports.9,10 Of the heterozygotes with race and ethnicity data available, 96% (47/49, table 2) were of white ancestry, and overall, single or biallelic expansions were detected in 7.1% and 3.6%, respectively, of the total white population surveyed in this study. The high rate of heterozygosity (6.8%) in our cohort is notable but is similar to 1 previous study, which calculated an allele frequency of 4% in control populations of 69 and 133 individuals based on haplotype in next-generation sequencing data sets,10 although another study found a frequency of 0.7% in a cohort of 304 healthy controls using RP-PCR.9 Although our methods cannot accurately size larger repeats, standard PCR analysis indicated that under the conditions of our AAGGG RP-PCR assay, we were able to detect small expansions up to 60 repeats above wild type (∼650 bp, figure 1 and figure e-1, links.lww.com/NXG/A266), and therefore, we suspect that our high rate of detection may be due, in part, to the detection of confounding mildly expanded alleles below the estimated 400 repeat margin of pathogenicity9 (figure 1 and figure e-1, links.lww.com/NXG/A266). It is interesting to note that small AAGGG expansions have not previously been reported in patients or controls,9,10 and because all subjects tested presented with some form of cerebellar ataxia, we cannot exclude the possibility that expanded AAGGG repeats may contribute to the development of cerebellar ataxia in some heterozygous individuals. We also cannot rule out a contribution of false-positive detection of other small polymorphic nonpathogenic non-AAGGG repeats9 in some heterozygous individuals. We did confirm all biallelic subjects with RP-PCR and standard PCR in at least 2 experiments each across 2 separate laboratories and further determined that none of these individuals harbored the most common nonpathogenic expanded repeat, AAAAG9 (data not shown).
In addition, the pathogenic (ATTTC) repeat in the DAB1 gene, causative for SCA37, was not observed in our large undiagnosed ataxia cohort of 596 individuals of mostly white, non-Hispanic ancestry, consistent with the observation of a founder effect in the Iberian Peninsula11,13 and suggesting that although this disorder should be considered in that population, it is likely extremely rare in the United States. Although no pathogenic ATTTC insertions were found, it is possible that extremely large repeats of ATTTT flanking a pathogenic ATTTC insertion might prevent amplification of products from the RP-PCR, so false negatives cannot be ruled out in this study, although this has not been commonly observed in published reports.11,12
Consistent with previous reports, our study identified biallelic RFC1 expansions in a high percentage of patients with CANVAS (n = 8, 73%) and cerebellar ataxia with neuropathy9 (n = 14, 22%). Although we do not have electrophysiologic data on all subjects, of the biallelic patients identified, all appeared to have a large fiber neuropathy (data not shown), which would be an important focus for further clinical investigation. In addition, we also observed biallelic expansion in a notable percentage of patients with pure cerebellar ataxia (n = 4, 2.1%) and generalized spinocerebellar ataxia (n = 3, 1.0%), a frequency on par with the majority of rare ataxic disorders identifiable by clinical sequencing.2 Based on this study, the presence of neuropathy confers a 20.1% absolute benefit to testing (95% CI 9.7%–30.6%) over pure cerebellar ataxia alone, and the presence of both neuropathy and vestibulopathy further increases this to 70.6% (95% CI 44.2%–97.0%), with biallelic RFC1 expansions anticipated in 1 of every 5.0 (95% CI 3.3–10.3) and 1 of every 1.4 (95% CI 1.0–2.3) patients tested, respectively. Taken together, these results suggest that RFC1 expansion testing is high yield in cases of CANVAS and cerebellar ataxia with neuropathy but should also be considered in the genetic workup of patients with undiagnosed pure cerebellar and spinocerebellar ataxia.
Acknowledgment
The authors thank all the patients and their families for their participation in this study.
Glossary
- CANVAS
cerebellar ataxia, neuropathy, and vestibular areflexia syndrome
- DAB1
Disabled Adaptor Protein 1
- RFC1
replication factor C subunit 1
- SCA37
spinocerebellar ataxia type 37
Appendix. Authors
Footnotes
Editorial, page e436
Study funding
This work was supported by the National Institute for Neurological Disorders and Stroke [R01NS082094 to BLF]. BLF acknowledges support through donations to the University of California by the Rochester Ataxia Foundation. PJL was supported by the Vincent Chiodo Foundation. CMG and VK acknowledge support from the National Ataxia Foundation and the Brigham Research Institute.
Disclosure
All authors declare that there is no conflict of interest. Go to Neurology.org/NG for full disclosures.
Publication history
This manuscript was previously posted on bioRxiv: doi: 10.1101/790006. Received by Neurology: Genetics November 7, 2019. Accepted in final form March 15, 2020.
References
- 1.Sun M, Johnson AK, Nelakuditi V, et al. . Targeted exome analysis identifies the genetic basis of disease in over 50% of patients with a wide range of ataxia-related phenotypes. Genet Med 2019;21:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogel BL, Lee H, Deignan JL, et al. . Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol 2014;71:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rexach J, Lee H, Martinez-Agosto JA, Nemeth AH, Fogel BL. Clinical application of next-generation sequencing to the practice of neurology. Lancet Neurol 2019;18:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngo KJ, Rexach JE, Lee H, et al. . A diagnostic ceiling for exome sequencing in cerebellar ataxia and related neurological disorders. Hum Mutat 2020;41:487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulson H. Repeat expansion diseases. Handb Clin Neurol 2018;147:105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundwiler A, Shakkottai VG. Autosomal-dominant cerebellar ataxias. Handb Clin Neurol 2018;147:173–185. [DOI] [PubMed] [Google Scholar]
- 7.Fogel BL. Autosomal-recessive cerebellar ataxias. Handb Clin Neurol 2018;147:187–209. [DOI] [PubMed] [Google Scholar]
- 8.Wallace SE, Bird TD. Molecular genetic testing for hereditary ataxia: what every neurologist should know. Neurol Clin Pract 2018;8:27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortese A, Simone R, Sullivan R, et al. . Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet 2019;51:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafehi H, Szmulewicz DJ, Bennett MF, et al. . Bioinformatics-based identification of expanded repeats: a non-reference intronic pentamer expansion in RFC1 causes CANVAS. Am J Hum Genet 2019;105:151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seixas AI, Loureiro JR, Costa C, et al. . A pentanucleotide ATTTC repeat insertion in the non-coding region of DAB1, mapping to SCA37, causes spinocerebellar ataxia. Am J Hum Genet 2017;101:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loureiro JR, Oliveira CL, Sequeiros J, Silveira I. A repeat-primed PCR assay for pentanucleotide repeat alleles in spinocerebellar ataxia type 37. J Hum Genet 2018;63:981–987. [DOI] [PubMed] [Google Scholar]
- 13.Corral-Juan M, Serrano-Munuera C, Rabano A, et al. . Clinical, genetic and neuropathological characterization of spinocerebellar ataxia type 37. Brain 2018;141:1981–1997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.