Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) threatens global public health. The development of a vaccine is urgently needed for the prevention and control of COVID-19. Here, we report the pilot-scale production of an inactivated SARS-CoV-2 vaccine candidate (BBIBP-CorV) that induces high levels of neutralizing antibodies titers in mice, rats, guinea pigs, rabbits, and nonhuman primates (cynomolgus monkeys and rhesus macaques) to provide protection against SARS-CoV-2. Two-dose immunizations using 2 μg/dose of BBIBP-CorV provided highly efficient protection against SARS-CoV-2 intratracheal challenge in rhesus macaques, without detectable antibody-dependent enhancement of infection. In addition, BBIBP-CorV exhibits efficient productivity and good genetic stability for vaccine manufacture. These results support the further evaluation of BBIBP-CorV in a clinical trial.
Keywords: SARS-CoV-2, inactivated vaccine, BBIBP-CorV
Graphical Abstract
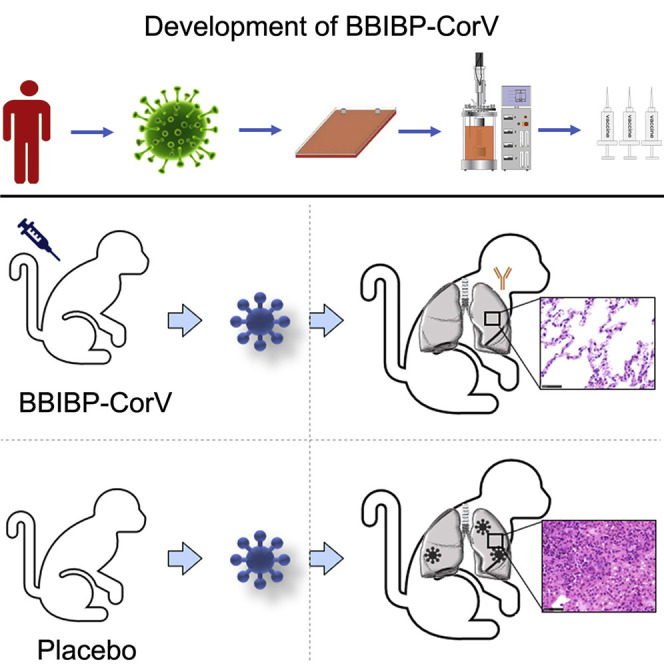
Highlights
-
•
An inactivated SARS-CoV-2 vaccine candidate, BBIBP-CorV, is developed
-
•
BBIBP-CorV induces high levels of neutralizing antibodies titers in animal models
-
•
Two-dose immunization with 2 μg/dose BBIBP-CorV efficiently protects rhesus macaques
-
•
BBIBP-CorV is efficiently produced, genetically stable, and seems to be safe in animals
Wang et al. report the development, characterization, and preclinical evaluation of an inactivated SARS-CoV-2 vaccine candidate for COVID-19 that safely induces high levels of neutralizing antibodies in multiple mammalian species and protective efficacy against SARS-CoV-2 challenge in rhesus macaques.
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has recently emerged throughout the world, resulting in 5.2 million infections and over 337 thousand deaths worldwide as of May 2020 according to the World Health Organization (WHO) report (https://covid19.who.int/) (Chan et al., 2020, Chen et al., 2020, Li et al., 2020, Wang et al., 2020, Zhu et al., 2020). SARS-CoV-2, a member of the Betacoronavirus genus, is closely related to severe acute respiratory syndrome coronavirus (SARS-CoV) and several bat coronaviruses (Lu et al., 2020, Tan et al., 2020, Zhou et al., 2020). Compared to SARS-CoV and Middle East respiratory coronavirus (MERS-CoV), SARS-CoV-2 appears to undergo more rapid transmission (Chan et al., 2020, Chen et al., 2020), leading to the urgent demand for a vaccine. To date, three candidate vaccines (including an inactivated vaccine, an adenovirus-vectored vaccine, and a DNA vaccine) were reported to protect rhesus macaques against SARS-CoV-2 with different efficacy (Gao et al., 2020, Lurie et al., 2020, van Doremalen et al., 2020, Yu et al., 2020a). Inactivated vaccines are widely used for the prevention of emerging infectious diseases (Stern, 2020), and the relatively high speed of the development of this kind of vaccine makes it a promising strategy for COVID-19 vaccine development. It is worthy to note that emerging evidence has shown antibody-dependent enhancement (ADE) in SARS-CoV infection (Wang et al., 2016, Yang et al., 2005), suggesting that particular attention should be paid to the safety evaluation in the development of the vaccine against coronaviruses. Here, we report the study of an inactivated SARS-CoV-2 vaccine candidate (BBIBP-CorV) and show that its potency and safety in preclinical studies warrants further clinical evaluation.
Results
Vaccine Design and Production
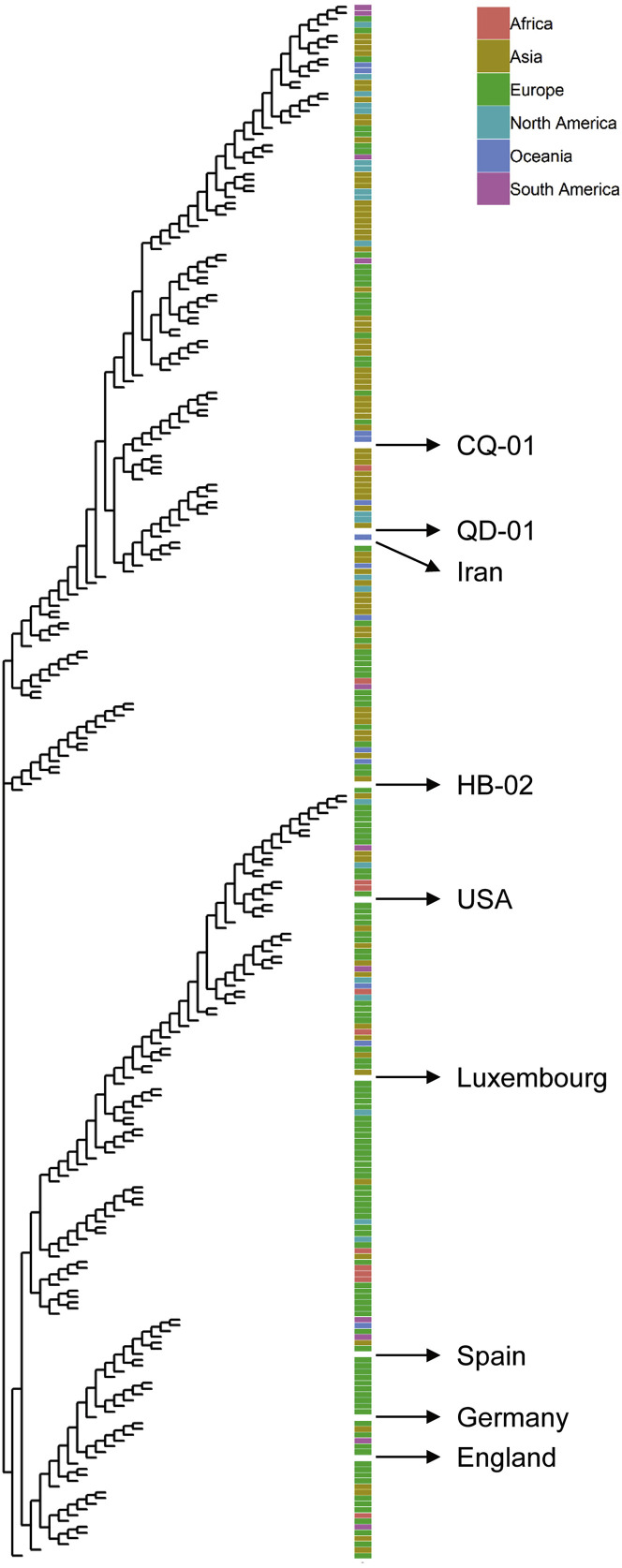
We isolated three SARS-CoV-2 strains from the bronchoalveolar lavage samples or throat swabs of three hospitalized patients from the recent COVID-19 outbreak to develop preclinical in vitro neutralization and challenge models for an inactivated SARS-CoV-2 vaccine candidate (Lu et al., 2020, Zhu et al., 2020). The three strains were 19nCoV-CDC-Tan-HB02 (HB02), 19nCoV-CDC-Tan-Strain03 (CQ01), and 19nCoV-CDC-Tan-Strain04 (QD01), which are scattered on the phylogenetic tree constructed from all available sequences, suggesting coverage of the main SARS-CoV-2 populations (Figure S1 ). Notably, all of these strains were isolated from Vero cells, which have been certified by WHO for vaccine production. Vero cells, but not other cell lines, were infected via the throat swabs of patients to prevent possible mutations during viral culture and isolation.
Figure S1.
SARS-CoV-2 Maximum Likelihood Phylogenetic Tree Related to Figure 1
The SARS-CoV-2 isolates used in this study are indicated with black arrows and labeled. Viral strains were isolated from infected patients who traveled from the indicated continent/area.
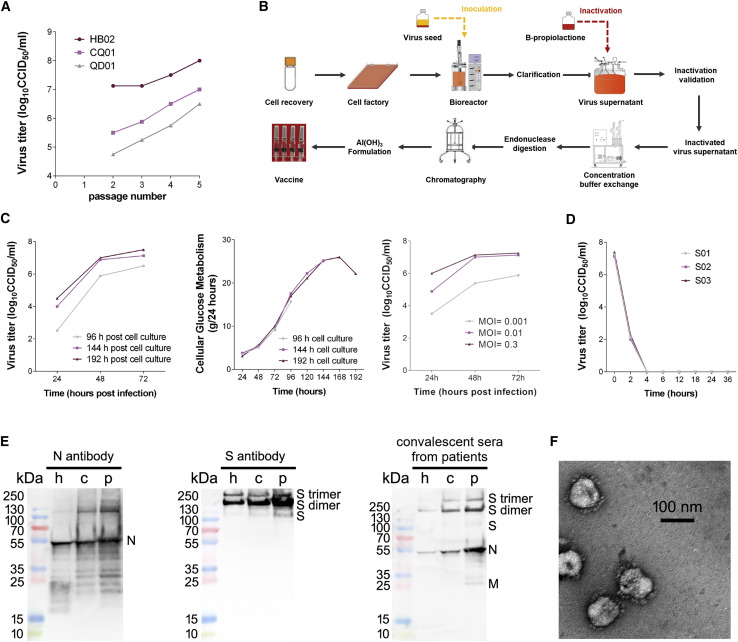
Highly efficient proliferation and high genetic stability are key features for the development of an inactivated vaccine. We first found that the HB02 strain showed the most optimal replication and generated highest virus yields in Vero cells among three viral strains (Figure 1 A). We therefore chose the HB02 strain for the further development of the inactivated SARS-CoV-2 vaccine (BBIBP-CorV). The comparisons on the whole-genome sequences of the HB02 strain and other SARS-CoV-2 strains from domestic and international sources showed that the HB02 strain is homologous to other viral strains and demonstrated that the main protective antigen (the spike protein) has 100% homology, indicating potential broad protection against various SARS-CoV-2 strains (Figures S1 and S2 ).
Figure 1.
Characterization of the SARS-CoV-2 Vaccine Candidate BBIBP-CorV
(A) Viral titers of three strains of different generations.
(B) Flowchart of BBIBP-CorV preparation.
(C) Culture conditions. The left panel shows the effect of cell culture time on BBIBP-CorV stock virus titer, the middle panel shows the growth kinetics of the Vero cells for BBIBP-CorV stock culture, and the right panel shows the effect of inoculation MOI on BBIBP-CorV stock virus titer.
(D) Inactivation kinetics of three batches of virus supernatant.
(E) The protein composition of BBIBP-CorV were evaluated by incubating with antibodies targeting N protein (left panel) and S protein (middle panel) and incubation with convalescent patient sera (right panel). h, harvest; c, concentrated viral solution; p, purified viral solution.
(F) Representative electron micrograph of BBIBP-CorV. Scale bar: 100 nm.
Figure S2.
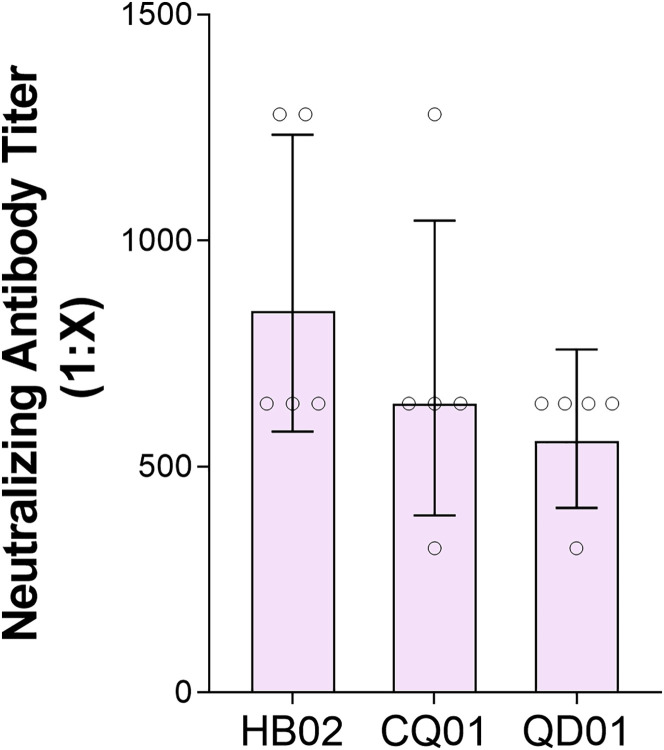
Neutralization of SARS-CoV-2 Strains HB02, CQ01, and QD01 by the Sera of Mice Vaccinated with BBIBP-CorV, Related to Figure 1
Mice were intraperitoneally injected with 8 μg/does of BBIBP-CorV at one time, and the ability of their sera to neutralize three SARS-CoV-2 strains was tested (n = 5) day 14 day after inoculation.
To obtain a viral stock adapted for high productivity, the HB02 strain was purified and passaged in Vero cells to generate the P1 stock. The P1 stock was adaptively cultured, passaged, and expanded on Vero cells. The strain after adaptation for seven generations (BJ-P-0207) was used as the original seed (BJ-P1) for vaccine production. To evaluate the genetic stability, three more passages were performed to obtain the P10 stock. We sequenced the whole genome of the HB02 strain and the P10 stock by deep sequencing analysis, and the results showed that their sequence homology was more than 99.95%. Furthermore, no amino acid variation was found in the full sequence, including the position near the furin cleavage site, in the P10 stock. These results suggest the high genetic stability of the HB02 strain, which is beneficial for further development.
For highly efficient manufacture, we established a strategy for the production of a BBIBP-CorV stock based on a novel carrier in a basket reactor (Figure 1B). Growth kinetic analysis of the P7 stock in Vero cells showed that the stock virus could replicate efficiently and reached a peak titer over 7.0 log10 CCID50 by 48–72 h post-infection (hpi) at multiplicities of infection (MOI) of 0.01–0.3 (Figure 1C). To inactivate virus production, β-propionolactone was thoroughly mixed with the harvested viral solution at a ratio of 1:4,000 at 2°C–8°C. The inactivation of three batches of virus eliminated viral infectivity, validating the good stability, and repeatability of the inactivation process (Figure 1D). Western blot analysis showed that the vaccine stock contained viral structural proteins (protective antigens) (Figure 1E). A negatively stained electron microscopy image visualized oval viral particles with spikes with the diameters of approximately 100 nm (Figure 1F).
Immunogenicity of BBIBP-CorV
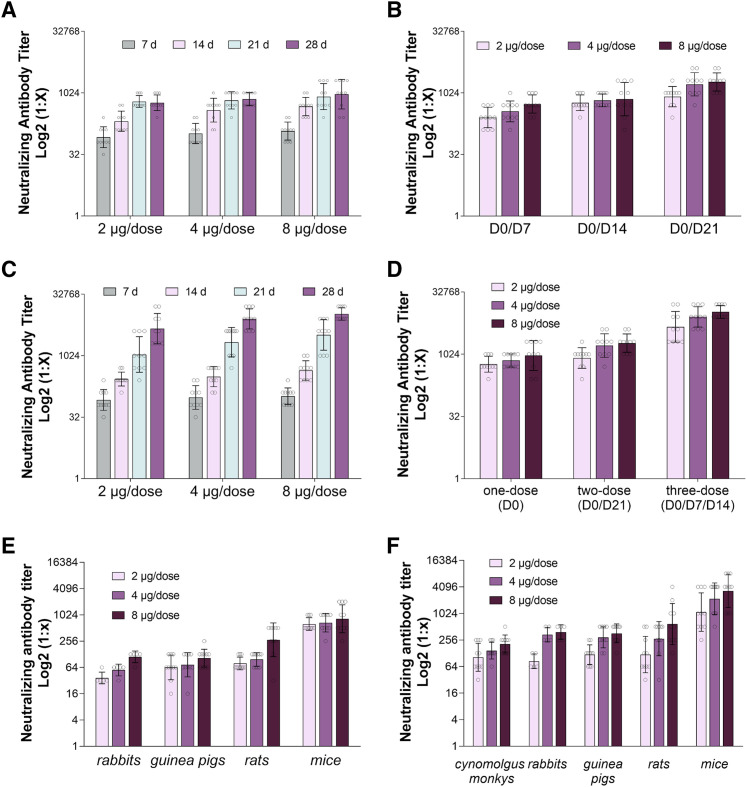
To assess the immunogenicity of BBIBP-CorV, BALB/c mice were injected with different immunization programs and various doses (2, 4, or 8 μg/dose) of vaccine mixed with aluminum hydroxide adjuvant. In the one-dose immunization group, mice were intraperitoneally administered a high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) dose of BBIBP-CorV at day 0 (D0), and the levels of neutralization antibody (NAb) at 7, 14, 21, and 28 days after injection were evaluated. The results showed that the seroconversion rate in the high-, middle-, and low-dose groups reached 100% at 7 days after immunization, and the immunization effect was time dependent (Figure 2 A; Table S1). The NAb levels at 7, 14, and 21 days in the low- and middle-dose groups show significant variation, whereas no significant variation between 21 and 28 days was observed. In the high-dose group, a significant variation only was observed between 7 and 14 days (Figure 2A).
Figure 2.
BBIBP-CorV Immunization Elicits a Neutralizing Antibody Response in Different Animals with Different Doses and Immunization Programs
(A) Mouse neutralization antibody (NAb) levels with one-dose (D0) immunization. Mice were injected intraperitoneally with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine, and the NAb levels at 7 days, 14 days, 21 days, and 28 days after the first immunization were tested by the microtitration method (n = 10).
(B) NAb levels with different immunization interval programs via two-dose immunization. Mice were injected intraperitoneally by using two-time immunization (D0/D7; D0/D14; D0/D21), and the NAb levels at 7 days after the second immunization were tested by the microtitration method (n = 10).
(C) Mouse neutralization antibody levels with three-dose (D0/D7/D14) immunization. Mice were inoculated intraperitoneally with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine at 0, 7, and 14 days, and NAb levels at 7, 14, 21, and 28 days after the first immunization were tested by the microtitration method (n = 10).
(D) Mouse NAb levels with different immunization programs. Mice were injected intraperitoneally with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine by using one-dose (D0), two-dose (D0/D21), and three-dose (D0/D7/D14) immunization programs, respectively, and the NAb levels at 28 days after the first immunization were checked by the microtitration method (n = 10).
(E) Rabbits (n = 5), guinea pigs (n = 10), rats (n = 10), and mice (n = 10) were immunized with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine by one-dose (D0) immunization, and the NAb levels at 21 days after the first immunization were tested by the microtitration method.
(F) Cynomolgus monkeys (n = 10), rabbits (n = 5), guinea pigs (n = 10), rats (n = 10), and mice (n = 10) were immunized with high (8 μg/does), middle (4 μg/dose), and low (2 μg/dose) doses of vaccine by three-dose (D0/D7/D14) immunization, and the NAb levels at 21 days after the first immunization were tested by the microtitration method.
For (A–F), error bars reflect the geometric SD.
In the two-dose immunization group, we tested different immunization programs (D0/D7, D0/D14, and D0/D21 intervals) in which two immunizations at days 0/7, days 0/14, and days 0/21, respectively, were administered. The seropositivity of the high-, medium-, and low-dose groups from all three immunization programs reached 100% at 7 days after the second immunization (Figure 2B; Table S1). The immunogenicity of the two-dose immunization program was significantly higher than that of the one-dose immunization program in the high- and middle-dose groups. Moreover, use of the D0/D21 interval obtained the highest NAb level at 7 days after the second immunization.
We also tested the immunogenicity of a three-dose immunization program, in which the mice were intraperitoneally administered a high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) dose of vaccine at days 0, 7, and 14 (Figure 2C). The NAb levels for all groups were determined at days 7, 14, 21, and 28, and the seroconversion rate in all three groups reached 100% at day 7 after the first immunization (Figure 2C; Table S1). The results showed that the three-dose (D0/D7/D14) immunization program resulted in higher NAb levels than the one-dose (D0) program in all three groups at days 28 (Figures 2A and 2C). Moreover, we analyzed the NAb levels in mice with high, middle, and low doses of vaccine following the one-dose (D0), two-dose (D0/D21), and three-dose (D0/D7/D14) immunization programs and checked the NAb levels at 28 days after the first immunization to maintain the same starting and ending points. The results showed that the immunogenicity of the three-dose (D0/D7/D14) immunization program was higher than that of both the one- and two-dose programs (Figure 2D).
We next measured the immunogenicity of BBIBP-CorV in different animal models: rabbits, guinea pigs, rats, and mice. The animals were immunized with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine by the one-dose (D0) immunization program, and the NAb levels were determined at 21 days after immunization. The results demonstrated that BBIBP-CorV has good immunogenicity, and the seroconversion rate reached 100% at day 21 after immunization in all animal models (Figure 2E; Table S1). In the three-dose (D0/D7/D14) immunization group, cynomolgus monkeys, rabbits, guinea pigs, rats, and mice were immunized with high (8 μg/dose), middle (4 μg/dose), or low (2 μg/dose) doses of vaccine. The seroconversion rate reached 100% at 21 days after immunization in all animal models, and the NAb levels at 21 days after the first immunization showed that the immunogenicity of the three-dose (D0/D7/D14) program with high, middle, and low doses was higher than that of the one-dose (D0) program in rabbit and guinea pig models (Figures 2E and 2F; Table S1).
Protection in a Nonhuman Primate Animal Model
Recent studies have shown that SARS-CoV-2-infected rhesus macaques developed pulmonary infiltrates and histological lesions (Munster et al., 2020, Shan et al., 2020, Yu et al., 2020b). We evaluated the immunogenicity and protective efficacy of BBIBP-CorV in rhesus macaques.
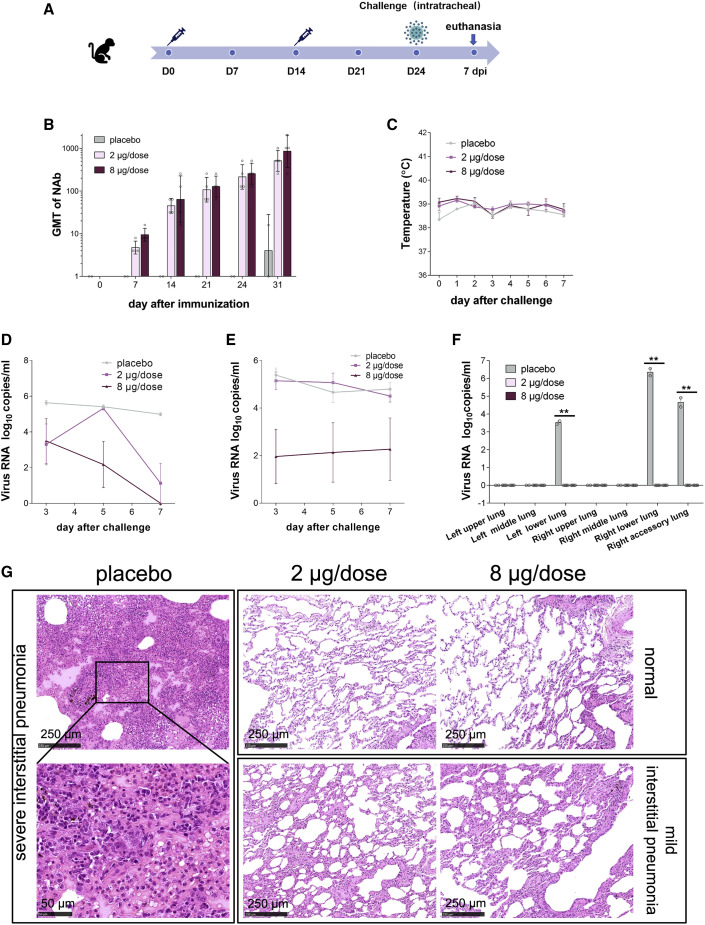
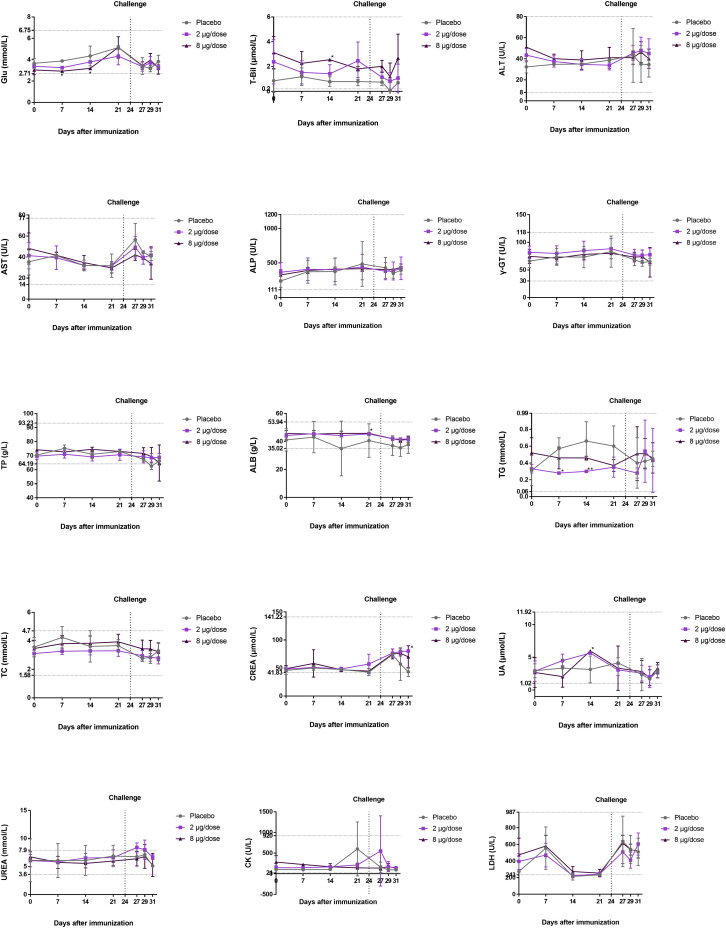
All macaques were immunized twice on days 0 (D0) and 14 (D14). The placebo group was intramuscularly administered physiological saline, and the two experimental groups were intramuscularly injected with low-dose (2 μg/dose) or high-dose (8 μg/dose) BBIBP-CorV (Figure 3 A). Before virus challenge at D24, the geometric mean titer of NAb in the low-dose and high-dose groups reached 215 and 256, respectively (Figure 3B). At D24 (10 days after the second immunization), all macaques were intratracheally challenged with l06 TCID50 of SARS-CoV-2 per monkey under anesthesia. Body temperatures of both the vaccinated groups and placebo group fluctuated within the normal range after virus challenge from 0 to 7 days postinoculation (dpi) (Figure 3C; Figure S4 A). Moreover, the serum biochemical parameters in rhesus macaques after vaccination and challenge with living virus remained constant (Figure S3). Because a recent study showed that SARS-CoV-2 infection does not affect host blood chemistry (Munster et al., 2020), this result suggests that inoculation with BBIBP-CorV does not result in side effects on serum biochemical parameters.
Figure 3.
Immunogenicity and Protective Efficacy of BBIBP-CorV in Nonhuman Primates
(A) Experimental strategy.
(B) Macaques were immunized twice with 2 μg/dose (n = 4) or 8 μg/dose (n = 4) of BBIBP-CorV or placebo (n = 2). The NAb titers were measured. Data are presented as geometric mean with geometric SD.
(C–G) The protective efficacy of BBIBP-CorV against SARS-CoV-2 challenge at 10 days after second immunization was evaluated in macaques. Changes in clinical signs (temperature, C) were recorded. Viral loads in throat (D) and anal (E) swabs obtained from macaques at 3, 5, and 7 days post inoculation. (F) Viral loads in all seven lung lobes collected from all macaques at day 7 post inoculation were determined by real-time PCR. All data are presented as mean ± SEM from four independent experiments for the BBIBP-CorV groups and two independent experiments for the placebo group; error bars reflect the SEM. Data points represent the individual macaques. Asterisks indicate significance: ∗∗p < 0.01. Dotted lines indicate the limit of detection. (G) Histopathological changes in lungs of macaques at day 7 post inoculation. All macaques received vaccination showed normal lung with focal mild interstitial pneumonia in few lobes.
Scale bars are indicated in the panels.
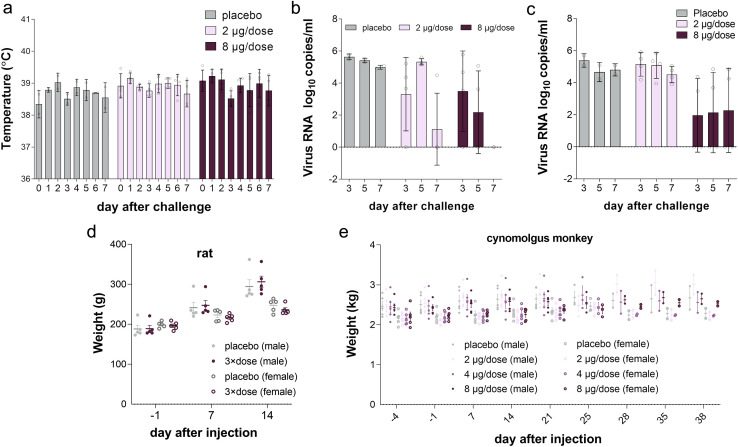
Figure S4.
Individual Data for Temperature, Viral Load, and Body Weight of Animals in Efficacy and Safety Evaluation, Related to Figures 3C–3E
Individual (A) temperature, viral load in (B) throat and (C) anal swab data in nonhuman primate efficacy evaluation. Individual body weight of (D) rats (n = 5) and (E) cynomolgus monkeys (n = 10) in safety evaluation (Related to Figures 4A and 4C).
Figure S3.
Serum Biochemical Parameters in Rhesus Macaques after Vaccination and Challenge with Living Virus, Related to Figure 3
Rhesus macaques were intramuscularly immunized twice on days 0 and 14, and live virus challenge was conducted on day 24 (the dotted line). Blood was collected, and serum biochemical parameters were monitored at different time points. Glu (glucose), T-Bil (total bilirubin), ALT (alanine aminotransferase), AST (aspartate aminotransferase), ALP (alkaline phosphatase), γ-GT (γ-glutamyl transpeptidase), TP (total protein), Alb (albumin), TG (triglycerides), TC (total cholesterol), CREA (creatinine), UA (uric acid), UREA (blood urea), CK (creatine kinase), LDH (lactate dehydrogenase). The data are presented as the mean ± SD. ∗p < 0.05 and ∗∗∗p < 0.001 versus the placebo group (n = 2 in the placebo group, n = 4 in the 2 μg/dose and 8 μg/dose groups). The dotted line on the y axis indicates the normal upper and lower lines of the data.
We next determined the viral load in the throat and anal swabs of macaques by real-time PCR. All placebo macaques showed and maintained a high viral load during the whole evaluation period after virus challenge by both throat and anal swabs (Figures 3D and 3E; Figures S4B and S4C). In contrast, the viral load in the throat swabs of the low-dose group peaked (5.33 log10copies/mL) at 5 dpi and then decreased to 1.12 log10copies/mL at 7 dpi, which was significantly lower than that of the placebo group. In particular, among the four macaques in the low-dose group, three showed a nondetectable viral load at 7 dpi. The throat swabs of all four macaques in the high-dose group were negative for viral load. Moreover, no viral load was detected in the anal swabs of two (out of four) macaques in the high-dose group.
At 7 dpi, all animals were euthanized to determine the viral load in the lung tissue and for pathological examination (Figure 3F and 3G). No macaques in the low-dose and high-dose groups had a detectable viral load in any lung lobe, which was significantly different from the results in the placebo group (Figure 3F). In the placebo group, a high viral load was detected in the left lower lung, right lower lung, and right accessory lung, and the pathological histology analysis results showed severe interstitial pneumonia. To note, only 3 of 7 sections of the lung lobes were detected to have infection in the placebo group, possibly because the virus infection in the lung lobes is dynamically changing. Furthermore, all macaques that received vaccination showed normal lung with focal mild histopathological changes in few lobes (Figure 3G), demonstrating the BBIBP-CorV vaccination could efficiently block the infection of SARS-CoV-2 and COVID-19 disease in monkey. At 7 dpi, the macaques treated with placebo produced low-level NAb with a titer of 1:16, whereas the NAb levels of the vaccinated macaques were highest at 1:2,048 (average 1:860) in the high-dose group and 1:1,024 in the low-dose group (average 1:512) (Figure 3B). Taken together, all these results demonstrated that both low-dose and high-dose BBIBP-CorV conferred highly efficient protection against SARS-CoV-2 in macaques without observed antibody-dependent enhancement of infection.
Safety
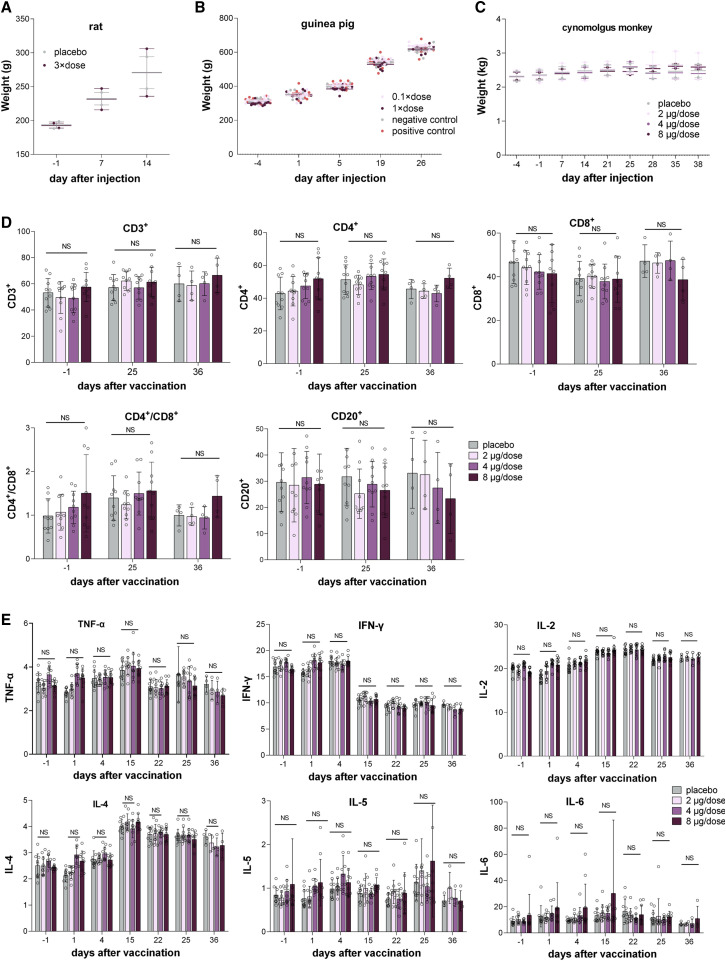
We first performed a single intramuscular injection experiment in Sprague-Dawley rats to evaluate the acute toxicity of BBIBP-CorV. In this study, 20 rats were divided into two groups (n = 10, 5/gender) and intramuscularly injected with 3 × dose (8 μg/dose, 24 μg/rat) of BBIBP-CorV and physiological saline as the control. After inoculation, all rats were continuously observed for 14 days and euthanized at day 15 to assess systematic anatomy and for general observation. No cases of death or impending death or obvious clinical signs were observed in any of the four groups over 14 consecutive days after vaccine inoculation. Moreover, there was no significant difference in weight or feeding state between the experimental groups and control groups (Figure 4 A; Figure S4D). No histopathologic changes were observed after euthanasia. Notably, the maximum tolerated dose (MTD) used for a single intramuscular injection in rats was 24 μg/rat, which is equivalent to 900 times the dose in humans, indicating the potential good safety of BBIBP-CorV in humans.
Figure 4.
Safety Evaluation of BBIBP-CorV in Rats, Guinea Pigs, and Nonhuman Primates
(A) Body weight analysis of rats in the experimental group and control group (n = 5). Male and female rat weight mean are used in this plot. Plot with individual data is presented in Figure S4D.
(B) Body weight analysis of guinea pigs in the experimental groups (0.1 × dose/guinea pig, 1 × dose/guinea pig) and negative control and positive control groups (n = 9).
(C) Cynomolgus monkeys were intramuscularly injected four times on days 1, 8, 15, and 22 with low (2 μg/dose), middle (4 μg/dose), and high (8 μg/dose) doses of BBIBP-CorV or placebo. Body weight analysis of cynomolgus monkeys (n = 10) in all four groups. Male and female cynomolgus monkey weight mean are used in this plot. Plot with individual data is presented in Figure S4E.
(D) Hematological analysis of cynomolgus monkey in all four groups (n = 10). The percentages of the lymphocyte subsets CD3+, CD3+CD4+ (labeled CD4+), CD3+CD8+ (labeled CD8+), CD20+, and CD3+CD4+/CD3+CD8+ (labeled CD4+/CD8+) were monitored at day −1 (1 day before vaccination), day 25 (3 days after the third vaccination), and day 36 (14 days after the fourth vaccination).
(E) The key cytokines TNF-α, IFN-γ, IL-2, IL-4, IL-5, and IL-6 were examined at days −1, 1 (the day for the first vaccination), 4, 15, 22, 25, and 36, respectively.
For (A–E), values are shown as the mean ± SD
Systemic anaphylaxis due to BBIBP-CorV was subsequently evaluated by intramuscular and intravenous injections in guinea pigs. Thirty-six male guinea pigs were divided into 4 groups (9/group), a negative control group (physiological saline), a positive control group (human blood albumin, 20 mg/sensitization, 40 mg/stimulation), a low-dose group (0.1 × dose/sensitization, 0.2 × dose/stimulation), and a high-dose group (1 × dose/sensitization, 2 × dose/stimulation). Sensitization was performed on D1, D3, and D5. The first stimulation (intravenous excitation via the foot) for 3 (out of 9) guinea pigs from each group was performed at D19, and secondary stimulation of the remaining animals of each group (6/9) was performed at D26. The results showed no abnormal reactions during the sensitization period by clinical observation and measurement of the body weights of the guinea pigs (Figure 4B). No allergic reaction symptoms were found in the negative control group or experimental group on D19 or D26. The anaphylaxis of the positive control group was highly positive (1/6 animals were positive, 3/6 animals were strongly positive, and 2/6 animals were extremely positive). In sharp contrast, in the low- and high-dose groups, no allergic reactions at D19 and D26 were found, and the allergic reactions were negative.
The long-term toxicity of BBIBP-CorV was further evaluated in cynomolgus monkeys. Forty cynomolgus monkeys (20/gender) were divided into 4 groups (5/gender/group) and intramuscularly injected with a control solution (physiological saline injection, group 1) or 2, 4, or 8 μg of BBIBP-CorV (groups 2 to 4) in a volume of 0.5 mL. The animals were injected once a week for 3 continuous weeks (four times in total). A total of 3/5 animals of each sex in each group were dissected on D25, and the remaining 2/5 animals of each sex in each group were dissected on D36. The gross anatomy was evaluated, and histopathologic examination was administered. No cases of death or impending death or significant abnormalities in clinical physiological and pathological indicators, lymphocyte subgroup distribution (CD3+, CD3+CD4+, CD3+CD8+, CD20+, CD3+CD4+/CD3+CD8+), cytokines (tumor necrosis factor alpha [TNF-α], interferon [IFN]-γ, interleukin [IL]-2, IL-4, IL-5, and IL-6), c-reactive protein, complement, or body weight were observed in the 2, 4, and 8 μg/dose groups (Figures 4C–4E; Figure S4E). No abnormalities in the gross anatomy of the euthanized animals in each dosed group on D25 and D36 were observed. Granulomatous inflammation was observed in the 2, 4, and 8 μg/dose groups on D25 and remained at the end of the recovery period (D36), with a slight improvement compared to that observed on D25. The animals showed only local irritation characterized by mild to severe granulomatous inflammation due to injection, but this reaction was absent at 2 weeks after injection. The no observed adverse effect level was found to be 8 μg/dose in this trial.
Discussion
The development of vaccines with high immunogenicity and safety is crucial for control of the global COVID-19 pandemic and prevention of further illness and fatalities. Here, we report development of an inactivated SARS-CoV-2 vaccine candidate, BBIBP-CorV, and show that it induced high levels of neutralizing antibody in six mammalian species, including rats, mice, guinea pigs, rabbits, cynomolgus monkeys, and rhesus macaques, protecting them against SARS-CoV-2 infection. Two-dose immunization using 2 μg/dose of BBIBP-CorV conferred highly efficient protection against SARS-CoV-2 in rhesus macaques without observable ADE or immunopathological exacerbation.
Prior to our reporting of this BBIBP-CorV study, three candidate vaccines against SARS-CoV-2 were reported, including an adenovirus-vectored vaccine (ChAdOx1 nCoV-19), a DNA vaccine, and an inactivated vaccine (PiCoVacc) (Gao et al., 2020, Lurie et al., 2020, van Doremalen et al., 2020, Yu et al., 2020a). In the protection studies, ChAdOx1 nCoV-19 and the DNA vaccine were challenged by both lower and upper respiratory tract, whereas PiCoVacc and BBIBP-CorV were intratracheally challenged. Despite the different challenge ways, the pathological changes in lung tissues were observed in all model groups, and the viral RNAs were detected in throat or nasal swabs, indicating that the successful establishment of the animal model for virus challenge. The recombinant adenovirus-vectored vaccine is easily manipulated for genetic modification and capable of inducing potent antigen-specific immune responses; but the neutralizing antibody against the carrier vector virus is still a challenge (Zhang and Zhou, 2016). The DNA vaccine is easy to be produced and is stable for storage with limited recovery or residual toxicity; however, the concerns on their immunogenicity and safety still remain (Gary and Weiner, 2020). For ChAdOx1 nCoV-19, 5 of 6 lung lobes in vaccinated group showed the detectable viral load (van Doremalen et al., 2020); but for BBIBP-CorV, all macaques in the low- and high-dose groups did not show a detectable viral load in any lung lobe at 7 days after inoculation. Nevertheless, both BBIBP-CorV and ChAdOx1 nCoV-19 conferred effective protection and prevented all vaccinated macaques from viral interstitial pneumonia. Compared with the adenovirus-vectored vaccine and the DNA vaccine, the strategy for inactivated vaccine development and production is a conventional and mature technology. In the development of BBIBP-CorV, the HB02 strain generated the highest virus yields in Vero cells among three candidate viral strains and had no amino acid variations within 10 passages, suggesting its good genetic stability. Moreover, we established a strategy for the production of a BBIBP-CorV stock based on a novel carrier in a basket reactor to ensure the highly efficient production. Most importantly, two-dose immunizations using low-dose (2 μg/dose) of BBIBP-CorV provided highly efficient protection against SARS-CoV-2 in rhesus macaques, which might benefit the further clinical usage of the inactivated vaccine with less adverse effects. Of note, the protection effects on the upper respiratory tract are not assessed in this study and warrant further evaluation.
In the absence of an effective antiviral drug against SARS-CoV-2, vaccines with good potency and safety will be needed to effectively establish immunity in population. The development of BBIBP-CorV provides a potential solution to the COVID-19 pandemic, and the pipeline used for BBIBP-CorV pilot-scale production also sheds light on rapid vaccine developing against other coronavirus. Based on the result presented here, a Phase I clinical trial of BBIBP-CorV is currently in progress and a Phase II clinical trial has recently been initiated. These clinical trials have been designed using the same aluminum adjuvant formulation described here, with three different groups of high, medium, and low dose groups to evaluate the appropriate dose for further clinical application.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-N protein rabbit monoclonal Ab | Sino Biological | Cat.# 40143-R019; RRID:AB_2827973 |
| Anti-S protein rabbit polyclonal Ab | Sino Biological | Cat.# 40150-T62-CoV2; RRID: AB_2313773 |
| Human convalescent sera from patients | Wuhan Institute of Biological Products Co., Ltd. | Cat.# XG2020021006; RRID: AB_2313773 |
| Goat anti-rabbit IgG H&L (HRP) | GE Healthcare | Cat.# GE NA934; RRID: AB_772206 |
| Rabbit polyclonal anti-SARS-COV-2 | NIFDC | Cat.# SR20200304; RRID: AB_2313773 |
| FITC labeled Goat Anti-Rabbit IgG | ZSGB-BIO | Cat.# 201130103; RRID: AB_2313773 |
| HRP-labeled goat anti-human IgG (gamma chain) cross-adsorbed secondary antibody | Invitrogen | Cat.# 62-8420; RRID: AB_88136 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2/WH-09/human/2020/CHN | Key Laboratory of Human Disease Comparative Medicine, Chinese Ministry of Health, Beijing Key Laboratory for Animal Models of Emerging and Remerging Infectious Diseases, Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and Comparative Medicine Center, Peking Union Medical College, Beijing, China. | Cat.# MT093631.2 |
| 19nCoV-CDC-Tan-HB02 (HB02) | Beijing Institute of Biological Products Co., Ltd. (BBIBP) | Batch No. W2020(COVID-19-BJ-P7)01 |
| 19nCoV-CDC-Tan-Strain03 (CQ01) | Chinese Center for Disease Control and Prevention (IVDC, China CDC) | IVDC-2020(COVID-19-CT03-P5)01 |
| 19nCoV-CDC-Tan-Strain04 (QD01) | Chinese Center for Disease Control and Prevention (IVDC, China CDC) | IVDC-2020(COVID-19-CT04-P3)01 |
| Biological Samples | ||
| Bovine Serum Albumin | Lanzhou Minhai Biological Engineering Co., Ltd. | Batch No. 20181011 |
| Human Albumin | Chengdu Rongsheng Pharmaceutical Co., Ltd. | Batch No. 201907A099 |
| Typsin | GIBCO | Batch No. 2019271 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| M199 | ToCall (Suzhou) Biotechnology Co.,Ltd | Batch No. 190402 |
| Sodium Bicarbonate | Hebei Huachen Pharmaceutical Co., Ltd. | Batch No. 180914 |
| Sodium Hydroxide | Hunan Erkang Pharmaceutical Co., Ltd. | Batch No. 100920180501 |
| Disodium Hydrogen Phosphate (Dodecahydrate) | Hunan Jiudian Hongyang Pharmaceutical Co., Ltd. | Batch No. TF22190910 |
| Sodium Chloride | Jiangsu Qinfen Pharmaceutical Co., Ltd. | Batch No. 20200115 |
| Sodium Dihydrogen Phosphate (Dihydrate) | Sinopharm Chemical Reagent Co., Ltd. | Batch No. 20180125 |
| Aluminum Adjuvant | Brenntag Biosector A/S | CAS No. 21645-51-2 Batch No. 5443 |
| Critical Commercial Assays | ||
| PrimerScript RT Reagent Kit | TAKARA | Cat.# AJ60658A |
| PowerUp SYBG Green Master Mix Kit | ABI | Cat.# 00799448 |
| RNeasy Mini Kit | QIAGEN | Cat.# 74104 |
| Deposited Data | ||
| All data in this study | This paper | https://data.mendeley.com/datasets/pb9w5vrkxy/1 |
| Experimental Models: Cell Lines | ||
| Vero Cell Lines | WHO (ECACC) | Cat.# 88020401 |
| Experimental Models: Organisms/Strains | ||
| Rhesus macaques | Institute of Laboratory Animal Science, CAMS | SCXK (Beijing) 2019-0014 |
| Balb /C Mouse | Beijing Vital River Laboratory Animal Technology Co., Ltd | SCXK (Beijing) 2019-0010 |
| Guinea pig | Beijing Vital River Laboratory Animal Technology Co., Ltd | SCXK (Beijing) 2016-0011 |
| Wistar Rat | SPF (Beijing) Biotechnology Co., Ltd. | SCXK (Beijing) 2019-0010 |
| Rabbit | Beijing Vital River Laboratory Animal Technology Co., Ltd. | SCXK (Beijing) 2016-0012 |
| cynomolgus monkeys | Guangxi Grandforest Scientific Primate Company | SCXK (Guangxi) 2016-0003 |
| Sprague-Dawley Rat | Beijing Vital River Laboratory Animal Technology Co., Ltd. | SCXK (Beijing) 2016-0006 |
| Oligonucleotides | ||
| RT-PCR Forward Primer: TCGTTTCGGAAGAGACAGGT | This paper | N/A |
| RT-PCR Reverse Primer: GCGCAGTAAGGATGGCTAGT |
This paper | N/A |
| Software and Algorithms | ||
| 7500 Software v2.0.5 | Applied Biosystems | N/A |
Resource Availability
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Zhiyong Lou (louzy@mail.tsinghua.edu.cn).
Materials Availability
The materials used in this study are available upon request.
Data and Code Availability
All data in this study have been deposited to Mendeley Data: http://dx.doi.org/10.17632/pb9w5vrkxy.1.
Experimental Model and Subject Details
Ethics statements
All animals involved in this study were housed and cared for in an Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility. All experimental procedures with mice, rats, rabbits, guinea pigs, and nonhuman primates were conducted according to Chinese animal use guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC). Rhesus macaque studies were performed in an animal biosafety level 3 (ABSL-3) laboratory and approved by the IACUC of the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences with approval number GH20008. All nonhuman primate animals were anesthetized with ketamine hydrochloride (10 mg/kg).
Animal models
Balb /C female mice (12-14 g), female guinea pigs (200-300 g), male chinchilla rabbits (1.5-2.0 kg), were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. Female Wistar rats (175-200 g) were obtained from (Beijing) Biotechnology Co., Ltd. Male cynomolgus monkey (2.8-3-year-old, 2.15-2.97 kg) and female cynomolgus monkey (2.7-3.9-year-old, 1.99- 2.46 kg) were obtained from Guangxi Grandforest Scientific Primate Company. The rhesus macaques: 2.5-3.8 kg, 3-4-year-old. All animals participate in this research are in good health and are not involved in other experimental procedure. All animals were allowed free access to water and diet and provided with a 12 h light/dark cycle (temperature: 18-28°C, humidity: 40%–70%). The mice, guinea pigs, rabbits and rats were bred and maintained in specific pathogen free (SPF) environment at the Laboratory Animal Center of Beijing institute of biological products Co., LTD. All rhesus macaques and cynomolgus monkey were housed and cared in Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) accredited facility. All cynomolgus monkey experiments were approved by the institutional animal ethics committee (IACUC) prior to operation.
Method Details
Virus titration and vaccine preparation
The SARS-CoV-2 virus titer was determined by a microdose cytopathogenic efficiency (CPE) assay. Serial 10-fold dilutions of virus-containing samples were mixed with 3∼5 × 104 Vero cells and then plated in 96-well culture plates. After 4 days of culture in a 5% CO2 incubator at 37°C, cells were checked for the presence of a CPE under a microscope. The virus titer was calculated by the method of Karber (Ramakrishnan, 2016). SARS-CoV-2 isolated from a SARS-CoV-2-infected patient for candidate vaccine preparation was provided by the Chinese Center for Disease Control and Prevention. Viruses were cultured in a 10 L basket bioreactor at the temperature of 36 ± 1°C. The virus solution was harvested 48-72 h after inoculation and then was inactivated with β-propiolactone at a ratio of 1:4000 at 2-8°C for 20-24 h, followed by chromatography purification. The final bulk was prepared by adding 0.45 mg/mL aluminum hydroxide as the adjuvant and dilution buffer containing phosphate.
Validation of the inactivation
Effective inactivation of the virus was validated in a sample from each batch of BBIBP-CorV. Ten milliliters of inactivated SARS-CoV-2 was used to inoculate Vero monolayers in 75 cm2 flasks, and negative control cells were prepared; the cells were then cultured at 36 ± 1°C for 4 days. This was the first passage. Then 20 mL of supernatants from cells in the flask was inoculated onto two additional Vero monolayers in 75 cm2 flasks (10 mL each) and incubated at 36 ± 1°C for 4 days. This was the second passage. Then, 10 mL of supernatants from cells in the flask was inoculated onto further Vero monolayers in 75 cm2 flasks and incubated at 36 ± 1°C for 4 days. This was the third passage. No CPE was observed for three passages.
After consecutively blind passaged for three generations, the supernatant and cells were re-verified by immunofluorescence. The supernatant was inoculated onto Vero cells and cultured in a 37°C incubator. The cells were cultured in a 12-well plate after digestion with trypsin; culture for 48 h then washed three times with potassium-free PBS. The cells were fixed with 4% paraformaldehyde, and primary antibody (rabbit anti-SARS-CoV-2, dilution 1:100) was added overnight at 4°C, then wash three times with PBS. The secondary antibody (FITC-labeled anti-rabbit IgG, dilution 1:100) was added for 40 min at 37°C, then wash three times with PBS. DAPI dye for nuclear staining was add to each sample for 5 min and washed with PBS. Cells were mounted with PBS-glycerol (1:1) solution and examined with a fluorescence microscope.
RT-PCR
Total RNA was extracted from organs with an RNeasy Mini Kit (QIAGEN, USA) and PrimerScript RT Reagent Kit (TaKaRa, Japan). The forward and reverse primers targeting the SARS-CoV-2 nucleocapsid protein (N) gene for RT-PCR were 5′-GGGGAACTTCTCCTGCTAGAAT-3′ and 5′-CAGACATTTTGCTCTCAAGCTG-3′, respectively. RT-PCR was performed under the following reaction conditions: 42°C for 5 min, 95°C for 10 s, and 40 cycles of 95°C for 10 s and 60°C for 30 s.
Vaccine immunogenicity analysis and neutralization assay
Cynomolgus monkeys, rabbits, guinea pigs, rats and mice were randomly divided into three groups (5 rabbits in each group, and for other species, 10 animals in each group) and immunized intraperitoneally and intramuscularly with the trial vaccine at three doses (2 μg/inoculation, 4 μg/inoculation, 8 μg/inoculation). Blood was collected from each model animal before immunization, and the serum was isolated the next day as a control. Each animal was inoculated with 0.5 mL of the test sample (equivalent to 1 human dose).
The serum of the animal to be tested was diluted 1:4 in advance and inactivated in a 56°C water bath for 30 min. Serum was successively diluted 1:4 to the required concentration by a 2-fold series, and an equal volume of challenge virus solution containing 100 CCID50 virus was added. After neutralization in a 37°C incubator for 2 h, a 1.0∼2.5 × 105/mL cell suspension was added to the wells (0.1 mL/well) and cultured in a CO2 incubator at 37°C for 4 days. The Karber method (Ramakrishnan, 2016) by observing the CPE was used to calculate the neutralization endpoint (convert the serum dilution to logarithm), which means that the highest dilution of serum that can protect 50% of cells from infection by challenge with 100 CCID50 virus is the antibody potency of the serum. A neutralization antibody potency < 1:4 is negative, while that ≥ 1:4 is positive.
Safety evaluation
Twenty rats (10/gender) were divided into 4 groups (5/gender/group) and intramuscularly injected with 24 μg/rat of BBIBP-CorV or sodium chloride as a control. Thirty-six male guinea pigs were evenly divided into 4 groups: 1/4 were injected with sodium chloride as a negative control, and 1/4 were injected with human blood albumin as a positive control. The other two groups were injected with different doses and used as the low- and high-dose groups. Sensitization was carried out on D1, D3, and D5 by intramuscular injection. Three guinea pigs from each group were selected for intravenous excitation via the foot, with secondary excitation of the remainder of the guinea pigs in each group performed at D26. Forty cynomolgus monkeys (20/gender) were divided into 4 groups (5/gender/group) and intramuscularly injected with a control solution (sodium chloride injection, group 1) or 2, 4 or 8 μg of BBIBP-CorV (groups 2 to 4) in a volume of 0.5 ml. The animals were injected once a week for 3 continuous weeks (4 times in total). Three of the five animals of each sex in each group were dissected on D25, and the remaining 2/5 animals of each sex in each group were dissected on D36. Collection of data on safety-related parameters (body weight and body temperature) and clinical observation were carried out during and after immunization. Analysis of lymphocyte subset percentages (CD3+, CD3+CD4+, CD3+CD8+, CD20+, CD3+CD4+/CD3+CD8+) and key cytokines (TNF-α, IFN-γ, IL-2, IL-4, IL-5 and IL-6) and biochemical blood tests of the collected blood samples were also carried out. Sixty percent of the monkeys were euthanized at day 25 postimmunization, and the remaining 40% were euthanized at day 36.
Challenge assay in rhesus macaques
Rhesus macaques (3-4 years old) were divided into three groups: 2 in the placebo group, the animals in which were intramuscularly injected with physiological saline; 4 in the low-dose vaccine group, the animals in which were intramuscularly injected with 2 μg/dose vaccine; and 4 in the high-dose vaccine group, the animals in which were intramuscularly injected with 8 μg/dose vaccine. All macaques were immunized at days 0 and 14. A challenge study was conducted 10 days after the second immunization by direct inoculation of l06 TCID50 of SARS-CoV-2 virus through the intratracheal route under anesthesia. The general symptoms of the animals were observed and recorded every day during the experiment, along with the animal’s body temperature before and after challenge. Peripheral blood was collected on days 0, 7, 14 and 21 before immunization; and day 7 postinoculation (dpi), and a neutralizing antibody test and routine blood biochemical test were conducted. Throat and anal swabs were collected 3, 5, and 7 days after challenge and used to determine the viral load. Seven days after challenge, all animals were euthanized, the viral load in the lung tissue was detected, and a pathological examination was conducted.
Phylogenic tree analysis
High-quality genome sequences for SAR-CoV-2 were retrieved from GISAID (https://www.gisaid.org/) on Apr 24th, 2020. After duplicate genomes were removed, 6,659 sequences were clustered into different groups based on the combination of sampling country and date. A representative strain was selected from each group for phylogenetic analysis. For the 266 strains that were retained, multiple sequence alignment and phylogenetic reconstruction were performed by using MAFFT (Katoh et al., 2002) and FastTree (Price et al., 2010), respectively. The inferred maximum likelihood tree was plotted by ggtree (Yu et al., 2017).
Western blotting
Samples containing 30 μg of protein (harvest, concentrated viral solution, purified viral solution) were mixed with loading buffer and then boiled at 100°C for 10 min. The proteins were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane (300 mA, 55 min). The membrane was sealed in phosphate-buffered saline with Tween-20 (PBST) with 5% skim milk at 37°C for 2 h and subsequently incubated overnight with the primary antibodies anti-N protein rabbit monoclonal Ab (Sino Biological) (1:1000 dilution) and anti-S protein rabbit polyclonal Ab (Sino Biological) (1:1000 dilution) and human convalescent sera from patients (1:500 dilution) at 4°C. The membrane was incubated for 1 h at room temperature with the secondary antibodies goat anti-rabbit IgG H&L (HRP) (GE NA934, 1:4000) and HRP-labeled goat anti-human IgG (gamma chain) cross-adsorbed secondary antibody (Invitrogen, 62-8420) (1:1000). Protein bands were visualized using enhanced chemiluminescence (Azure biomolecular imager, USA).
EM sample preparation
Samples were applied to carbon-coated copper grids previously glow-discharged at low air pressure, stained with 2% uranyl acetate, and visualized in a Tecnai T12 electron microscope (FEI) equipped with an LaB6 filament operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a magnification of 23,000 × and a defocus of approximately −1.5 μm by using low-dose procedures.
Serum biochemical evaluation
Blood was collected, and serum biochemical parameters were monitored at different time points. Abbreviations are as following: Glu (glucose), T-Bil (total bilirubin), ALT (alanine aminotransferase), AST (aspartate aminotransferase), ALP (alkaline phosphatase), γ-GT (γ-glutamyl transpeptidase), TP (total protein), Alb (albumin), TG (triglycerides), TC (total cholesterol), CREA (creatinine), UA (uric acid), UREA (blood urea), CK (creatine kinase) and LDH (lactate dehydrogenase). Samples were analyzed with the Mindray’s biochemical reagent kits in a biochemical automatic analyzer (Mindray, BS-360S, Shenzhen, China).
Replications and sample-size
All experiments in this study were performed with replications numbers indicated in Method Details or Figure Legends. All experiments were randomized and no data was excluded from data analysis. The sample size and statistical method were indicated in Method Details or Figure Legends.
Quantification and Statistical Analysis
RT-PCR
For quantification of viral loads by RT-PCR (Figures 3d-3f), A standard curve of CT values to the copy number of viral RNA is generated with serial 10-fold dilutions of recombinant plasmid with a known copy number dilutions of PCR target fragments from 106 to 101 copies per microliter. The viral loads of each sample were converted with Ct value and the standard curve. Statistical analysis was performed by Applied Biosystems 7500 Software v2.0.5.
Flow cytometry
For quantification of lymphocyte percentage by flow cytometry (Figures 4d-4e), values represent the positive cell population percentage. Statistical analyses were performed using a double-tailed analysis with the inspection level at 5% or p ≤ 0.05. The mean number and standard deviations of all data results are calculated in the Provantis system (SAS 9.2 statistic software).
Serum biochemical evaluation
The data were analyzed with SPSS (version 17.0) software. Student’s t test was used to determine the statistical significance of the differences. See Method Details section “Serum biochemical evaluation” for more details.
Acknowledgments
We thank Dr. Taijiao Jiang for his assistance to prepare the phylogenic tree. This work was supported by the National Program on Key Research Project of China (2020YFA0707500, 2016YFD0500301, 2017YFC0840300, and 2020YFC0842100), the National Natural Science Foundation of China (NSFC) (31971126), CAMS Initiative for Innovative Medicine of China (2016-I2M-2-006), National Mega Projects of China for Major Infectious Diseases (2017ZX10304402 and 2016ZX10004001-003), and Beijing Science and Technology Plan (Z201100005420014).
Author Contributions
Conceptualization, X.Y., Y. Zhang, W.T., and H.W. Methodology, C.Q., M.X., C.L., and W.T. Validation, B.H., W.W., L.Z., P.N., Y.Q., Y. Zhao, J.Z., H.L., H.F., S.Y., Z.Z., W. Zhou, W. Zhen, N.L., L.D., W.D., L.B., Y.X., H.G., and J.L. Formal Analysis, B.H., J.L., L.D., H.F., S.Y., and Z.Z. Investigation, all authors. Resources, H.W., C.Q., G.F.G, G.W., W.T., and X.Y. Writing – Original Draft, Z.L. and G.X. Writing – Review & Editing Preparation, Z.L., G.F.G, G.W., and W.T. Visualization Preparation, G.X. Supervision, G.F.G, G.W., W.T., and Y. Zhang. Project Administration, X.Y. Funding Acquisition, W.T. and X.Y.
Declaration of Interests
The authors declare no competing interests.
Published: June 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.06.008.
Supplemental Information
References
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary E.N., Weiner D.B. DNA vaccines: prime time is now. Curr. Opin. Immunol. 2020;65:21–27. doi: 10.1016/j.coi.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Yao Y.-F., Yang X.-L., Zhou Y.-W., Wu J., Gao G., Peng Y., Yang L., Hu W., Xiong J. Infection with Novel Coronavirus (SARS-CoV-2) Causes Pneumonia in the Rhesus Macaques. Research Square. 2020 doi: 10.21203/rs.2.25200/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern P.L. Key steps in vaccine development. Annals of Allergy, Asthma & Immunology. 2020 doi: 10.1016/j.anai.2020.01.025. [DOI] [PubMed] [Google Scholar]
- Tan W.J., Zhao X., Ma X.J., Wang W.L., Niu P.H., Xu W.B., Gao G.F., Wu G. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019−2020. China CDC Weekly. 2020;2:61–62. [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. USA. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020 doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhou D. Adenoviral vector-based strategies against infectious disease and cancer. Hum. Vaccin. Immunother. 2016;12:2064–2074. doi: 10.1080/21645515.2016.1165908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study have been deposited to Mendeley Data: http://dx.doi.org/10.17632/pb9w5vrkxy.1.