Abstract
Background
The aim of this study was to investigate the association among biofilm formation, virulence gene expression, and antibiotic resistance in P. mirabilis isolates collected from diarrhetic animals (n = 176) in northeast China between September 2014 and October 2016.
Results
Approximately 92.05% of the isolates were biofilm producers, whereas 7.95% of the isolates were non-producers. The prevalence of virulence genes in the biofilm producer group was significantly higher than that in the non-producer group. Biofilm production was significantly associated with the expression of ureC, zapA, rsmA, hmpA, mrpA, atfA, and pmfA (P < 0.05). The results of drug susceptibility tests revealed that approximately 76.7% of the isolates were multidrug-resistant (MDR) and extensively drug-resistant (XDR). Biofilm production was significantly associated with resistance to doxycycline, tetracycline, sulfamethoxazole, kanamycin, and cephalothin (P < 0.05). Although the pathogenicity of the biofilm producers was stronger than that of the non-producers, the biofilm-forming ability of the isolates was not significantly associated with morbidity and mortality in mice (P > 0.05).
Conclusion
Our findings suggested that a high level of multidrug resistance in P. mirabilis isolates obtained from diarrhetic animals in northeast China. The results of this study indicated that the positive rates of the genes expressed by biofilm-producing P. mirabilis isolates were significantly higher than those expressed by non-producing isolates.
Keywords: Proteus mirabilis, Animal, Diarrhea, Biofilm, Virulence gene, Antibiotic resistance, Pathogenicity
Background
Proteus mirabilis is a motile gram-negative bacillus belonging to the family Enterobacteriaceae. It is an opportunistic pathogen of great importance that is found in water and soil as well as in the intestinal tracts of mammals. It has been recognized as a leading cause of urinary tract infections [1] and the primary infectious factor in patients with indwelling urinary catheters [2]. P. mirabilis can cause food poisoning, respiratory and wound infections, bacteremia, and other infections [3–6]. In the past decade, diseases associating with P. mirabilis infection have also been reported in birds with reproductive failure [7], weaned infant rhesus monkeys (Macaca mulatta) and ferrets (Mustela putorius furo) with diarrhea [8], and dogs with chronic otitis externa [9]. Although P. mirabilis, as an opportunistic pathogen capable of causing serious infections, should not be neglected.
Various virulence factors contribute to the pathogenicity of P. mirabilis. These factors include the presence of fimbriae and specific outer membrane proteins; flagella-based and swarming motility; urease activity;environmental iron binding;cell invasiveness,in addition to lipopolysaccharides and hemolysins,most of which are involved in the ability of bacteria to adhere, colonize, and invade tissues, thereby promoting pathogenicity [10, 11]. Virulence gene expression is not the only factor responsible for the pathogenicity of P. mirabilis; biofilm formation exacerbates the complexity of P. mirabilis infection [12], as biofilms are recognized as the ultimate cause of persistent and destructive infections and inflammatory processes [13]. A biofilm is an assemblage of microbial cells that adhere to specific surfaces and neighboring cells, and it is covered with an extracellular matrix [14, 15]. Biofilms inadvertently contribute to bacterial survival, thereby enabling better adaptation to the conditions of the external environment and more effective use of nutrition [16]. P. mirabilis has been found to produce biofilms on a wide range of surfaces, including polystyrene, silicone, latex, glass, and various biological surfaces [17, 18]. In recent years, studies have reported a correlation between biofilm formation and various virulence factors in P. mirabilis isolates from humans [19, 20]. For instance, catheter encrustation has been reported to be brought about by the activity of urease-producing biofilms [19, 21]. Jansen et al. discovered that mannose-resistant Proteus-like fimbriae produced by P. mirabilis, which infects the urinary tract, can induce biofilm formation, with the fimbriae aiding in the aggregation of the bacteria [20]. However, most reports of P. mirabilis describe urinary tract infections in humans, and there are few reports on P. mirabilis isolated from diarrhetic animals. In addition, the association between biofilm formation and various virulence factors in P. mirabilis isolates from diarrhetic animals is still unknown.
Over the past two decades, due to the identification of multiple multidrug-resistant (MDR) and extensively drug-resistant (XDR) P. mirabilis isolates, the treatment of P. mirabilis infections has become increasingly difficult [22–25]. For instance, the production of biofilms by P. mirabilis exacerbates the complexity of bacterial resistance, prolongs the treatment time, and further aggravates the infection. In essence, biofilms protect organisms from the host immune system and antimicrobial agents [26]. Furthermore, a previous study has demonstrated that certain antibiotics can induce biofilm formation [27]. However, it is unclear whether antibiotics are linked to biofilm formation by P. mirabilis isolates obtained from diarrhetic animals.
Here, we investigate biofilm formation, antimicrobial susceptibility, and virulence gene expression in P. mirabilis isolates recovered from feces of various diarrhetic animals in northeast China and discuss the association among the pathogenicity, drug resistance, and virulence of P. mirabilis from the perspective of biofilm formation.
Results
Prevalence of P. mirabilis isolates
The prevalence rates of P. mirabilis in the diarrheal samples are listed in Table 1. Overall, 28.66% (176/614) of diarrheal specimens were positive for P. mirabilis. The positive isolation rate of P. mirabilis in different species was less than 40%. The positive rates of P. mirabilis isolated from specimens was 32.76% for dog, followed by 28.7% for mink, 23.26% for cattle, and 22.5% for fowl.
Table 1.
Prevalence of P. mirabilis in collected diarrheal samples
| Host | Number of samples | Number of positive samples (%) |
|---|---|---|
| Dog | 232 | 76 (32.76) |
| Mink | 216 | 62 (28.7) |
| Cattle | 86 | 20 (23.26) |
| Fowl | 80 | 18 (22.5) |
| Total | 614 | 176 (28.66) |
Biofilm formation
Of the P. mirabilis isolates (n = 176) tested, 162 (92.05%) were biofilm producers and 14 (7.95%) were non-producers. Of the biofilm producers (n = 162), 78 (48.15%) were moderate biofilm producers, whereas 62 (38.27%) and 22 (13.58%) were strong and weak biofilm producers, respectively.
Virulence gene expression
The prevalence rates of ureC, zapA, rsmA, hmpA, mrpA, atfA, pmfA, FliL, and ucaA in P. mirabilis are listed in Table 2. Of the P. mirabilis isolates, the most prevalent gene was ureC, which was identified in 90.91% of the isolates, followed by zapA, rsmA, hmpA, mrpA, atfA, pmfA, FliL, and ucaA, which were detected in 85.8, 81.25, 70.45, 65.91, 64.77, 60.23, 56.82, and 32.95% of the isolates, respectively. The positive rates of genes identified in the biofilm-producing isolates were significantly higher than those in the non-producing isolates. Furthermore, except for hmpA,eight genes tested in the moderate biofilm producing isolates were higher than those in the strong and weak biofilm producing isolates. All nine genes tested in this study showed the lowest prevalence rates among the non-producers. Biofilm production was significantly associated with the expression of ureC, zapA, rsmA, hmpA, mrpA, atfA, and pmfA (P < 0.05).
Table 2.
Prevalence of ureC, zapA, rsmA, hmpA, mrpA, atfA, pmfA, FliL and ucaA genes in P. mirabilis isolates
| Virulence gene | P. mirabilis | ||||
|---|---|---|---|---|---|
| High biofilm producer n = 62 |
Moderate biofilm producer n = 78 |
Weak biofilm producer n = 22 |
Non biofilm producer n = 14 |
P value | |
| ureC | 58(93.54%) | 76 (97.43%) | 16 (72.72%) | 10 (71.43%) | P < 0.0001 |
| zapA | 52 (83.87%) | 72 (92.31%) | 15 (68.18%) | 12 (85.71%) | 0.037 |
| rsmA | 50 (80.64%) | 70 (89.74%) | 14 (63.64%) | 9 (64.29%) | 0.013 |
| hmpA | 48 (77.42%) | 60 (76.92%) | 10 (45.45%) | 6 (42.85%) | 0.002 |
| mrpA | 41 (66.13%) | 65 (83.33%) | 8 (36.36%) | 2 (14.29%) | P < 0.0001 |
| atfA | 40 (64.52%) | 62 (79.49%) | 10 (45.45%) | 2 (14.29%) | P < 0.0001 |
| pmfA | 38 (61.29%) | 58 (74.36%) | 8 (36.36%) | 2 (14.29%) | P < 0.0001 |
| FliL | 33 (53.23%) | 50 (64.1%) | 10 (45.45%) | 7 (50%) | 0.329 |
| ucaA | 14 (22.58%) | 32 (41.03%) | 7 (31.82%) | 5 (35.71%) | 0.146 |
Antimicrobial susceptibility testing
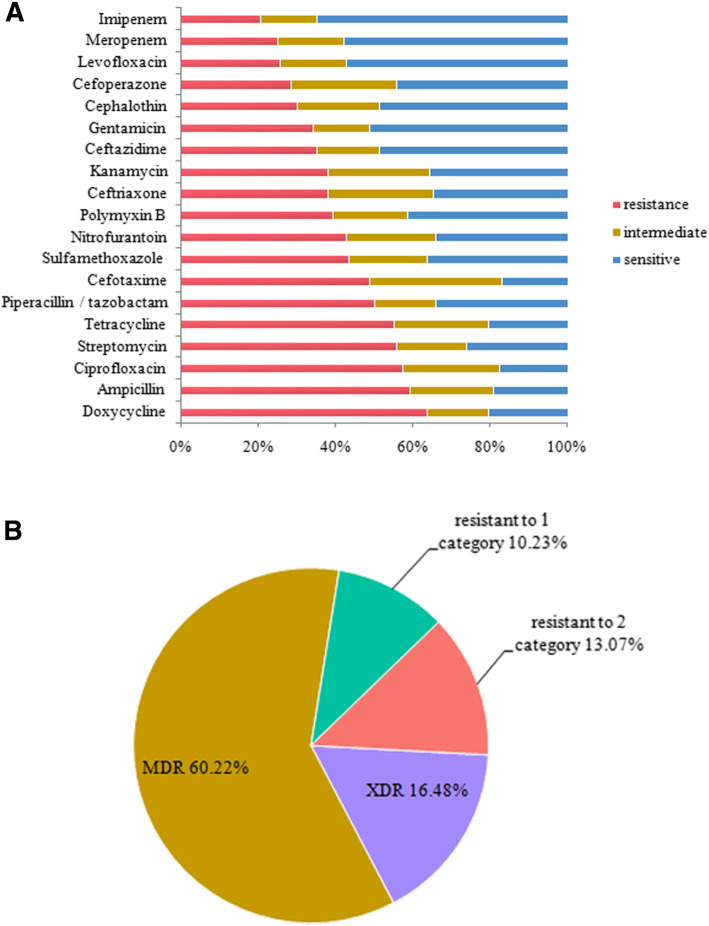
The antimicrobial resistance patterns of the P. mirabilis isolates are shown in Fig. 1. Variable degrees of resistance of these isolates to all antibiotics tested were observed. The resistance to doxycycline was the highest (112, 63.64%), followed by ampicillin (104, 59.09%), ciprofloxacin (101, 57.39%), streptomycin (98, 55.68%), tetracycline (97, 55.12%), piperacillin/tazobactam (88, 50%), cefotaxime (86, 48.87%), sulfamethoxazole (76, 43.19%), nitrofurantoin (75, 42.61%), polymyxin B (69, 39.2%), ceftriaxone (67, 38.07%), kanamycin (67, 38.07%), ceftazidime (62, 35.23%), gentamicin (60, 34.09%), cephalothin (53, 30.12%), cefoperazone (50, 28.41%), levofloxacin (45, 25.57%), meropenem (44, 25%), and imipenem (36, 20.45%). Of the sensitive strains, meropenem (57.96%) and imipenem (64.78%) showed the strongest antimicrobial effect on P. mirabilis (Fig. 1a).
Fig. 1.
Antibiotic resistance phenotypes of P. mirabilis isolates examined in this study. a Resistance rates of all isolates to 19 antibiotics. b Approximately 76.7% of the isolates exhibited multidrug or extensive drug resistance
The MDR patterns of the P. mirabilis isolates are shown in Fig. 1b. No isolate was sensitive to all the antibiotics. Of the P. mirabilis isolates, 18 and 23 isolates were resistant to only one or two of the 19 antibiotics tested, respectively, and 106 isolates were MDR, whereas 29 isolates were XDR. Thus, approximately 76.7% of the strains exhibited MDR or XDR.
The antibiotic susceptibility patterns of the biofilm producing and non-producing P. mirabilis isolates are shown in Table 3. Both biofilm producers and non-producers were highly resistant to doxycycline and moderately resistant to cefotaxime. Of the 19 antibiotics, doxycycline, ampicillin, tetracycline, cefotaxime, and kanamycin were found to be non-susceptible to non-producers. A sensitivity of 62.95% was observed for the biofilm-producing isolates to imipenem, whereas a sensitivity of 85.71% was noticed for the non-producing isolates to meropenem and imipenem. Isolates showing sensitivity to doxycycline, ampicillin, ciprofloxacin, streptomycin, tetracycline, piperacillin/tazobactam, cefotaxime, nitrofurantoin, sulfamethoxazole, and ceftriaxone produced more biofilm than strains showing resistance to these antibiotics. For other antibiotics, such as polymyxin B, ceftazidime, kanamycin, gentamicin, cefoperazone, cephalothin, meropenem, levofloxacin, and imipenem, we observed opposite findings. Biofilm production was significantly associated with resistance to doxycycline, tetracycline, sulfamethoxazole, kanamycin, and cephalothin (P < 0.05).
Table 3.
Antibiotic resistance pattern of the biofilm producing and non-producing P. mirabilis isolates
| Biofilm producer (n = 162) | Non biofilm producer (n = 14) | ||||||
|---|---|---|---|---|---|---|---|
| Antibiotic | resistance | intermediate | sensitive | resistance | intermediate | sensitive | P value |
| Doxycycline | 98 (60.49%) | 28 (17.28%) | 36 (22.23%) | 14 (100%) | 0 (0%) | 0 (0%) | 0.015 |
| Ampicillin | 92 (56.79%) | 36 (22.23%) | 34 (20.98%) | 12 (85.71%) | 2 (14.29%) | 0 (0%) | 0.078 |
| Ciprofloxacin | 90 (55.56%) | 44 (27.16%) | 28 (17.28%) | 11 (78.57%) | 0 (0%) | 3 (21.43%) | 0.084 |
| Streptomycin | 88 (54.32%) | 30 (18.52%) | 44 (27.16%) | 10 (71.42%) | 2 (14.29%) | 2 (14.29%) | 0.445 |
| Tetracycline | 85 (52.47%) | 41 (25.3%) | 36 (22.23%) | 12 (85.71%) | 2 (14.29%) | 0 (0%) | 0.034 |
| Piperacillin/tazobactam | 84 (51.85%) | 26 (16.05%) | 52 (32.1%) | 4 (28.58%) | 2 (14.29%) | 8 (57.13%) | 0.15 |
| Cefotaxime | 78 (48.15%) | 54 (33.33%) | 30 (18.52%) | 8 (57.13%) | 6 (42.87%) | 0 (0%) | 0.255 |
| Nitrofurantoin | 67 (41.36%) | 39 (24.07%) | 56 (34.57%) | 8 (57.13%) | 2 (14.29%) | 4 (28.58%) | 0.492 |
| sulfamethoxazole | 64 (39.5%) | 36 (22.23%) | 62 (38.27%) | 12 (85.71%) | 0 (0%) | 2 (14.29%) | 0.004 |
| Ceftriaxone | 62 (38.27%) | 45 (27.78%) | 55 (33.95%) | 5 (35.7%) | 3 (21.43%) | 6 (42.87%) | 0.776 |
| Polymyxin B | 61 (37.66%) | 32 (19.75%) | 69 (42.59%) | 8 (57.13%) | 2 (14.29%) | 4 (28.58%) | 0.357 |
| Ceftazidime | 58 (35.8%) | 28 (17.28%) | 76 (46.92%) | 4 (28.58%) | 0 (0%) | 10 (71.42%) | 0.131 |
| Kanamycin | 57 (35.19%) | 42 (25.93%) | 63 (38.88%) | 10 (71.42%) | 4 (28.58%) | 0 (0%) | 0.005 |
| Gentamicin | 52 (32.1%) | 24 (14.82%) | 86 (53.08%) | 8 (57.13%) | 2 (14.29%) | 4 (28.58%) | 0.143 |
| Cefoperazone | 48 (29.63%) | 46 (28.4%) | 68 (41.97%) | 2 (14.29%) | 2 (14.29%) | 10 (71.42%) | 0.104 |
| Cephalothin | 45 (27.78%) | 37 (22.84%) | 80 (49.38%) | 8 (57.13%) | 0 (0%) | 6 (42.87%) | 0.030 |
| Meropenem | 42 (25.93%) | 30 (18.52%) | 90 (55.55%) | 2 (14.29%) | 0 (0%) | 12 (85.71%) | 0.061 |
| Levofloxacin | 39 (24.07%) | 30 (18.52%) | 93 (57.41%) | 6 (42.87%) | 0 (0%) | 8 (57.13%) | 0.117 |
| Imipenem | 36 (22.23%) | 24 (14.82%) | 102 (62.95%) | 0 (0%) | 2 (14.29%) | 12 (85.71%) | 0.114 |
Pathogenicity test in mice
The morbidity and mortality of mice that received 32 different isolates of pathogenic bacteria is demonstrated in Table 4. Mice in the negative control group were obviously asymptomatic and in good overall health. Within 12 h of challenge, different symptoms of variable degrees were observed in mice of the experimental groups. These symptoms included gloomy spirit, inactivity, and loss of appetite. In the biofilm-producing group, some mice died within 12 h of challenge. Mice in both the biofilm producing and non-producing groups showed severe clinical symptoms, such as diarrhea, abrosia, subdued behavior, hunched appearance, and absence of grooming within 24 h of challenge. Eleven mice (45.83%) in the biofilm producing and non-producing groups died within 48 h of challenge. After 72 h of challenge, the symptoms began to gradually disappear, and the health of most mice returned to normal. Only two mice (25%) in the non-producing group died within 72 h of challenge. The biofilm-forming ability of P. mirabilis was not significantly associated with morbidity and mortality in mice (P > 0.05).
Table 4.
Pathogenicity to mice in 32 biofilm producing and non-producing P. mirabilis isolates from animal with diarrhea
| Pathogenicity | P. mirabilis | ||
|---|---|---|---|
| Biofilm producer (n = 24) |
Non biofilm producer (n = 8) |
P value | |
| morbidity | 17 (70.83%) | 5 (62.5%) | 0.660 |
| mortality | 11 (45.83%) | 2 (25%) | 0.299 |
Discussion
Biofilm formation by P. mirabilis has recently become an issue of increasing concern. In a previous study, P. mirabilis isolates recovered from urine samples showed a higher degree of biofilm production than those isolated from different catheter segments [28]. In this study, the prevalence of P. mirabilis was high, which is in agreement with the results of studies on urine from catheterized patients [12, 28]. Taken collectively, these findings indicate that further studies on P. mirabilis biofilm formation are needed to better understand the disease process and to develop new preventive and therapeutic options.
As a target gene, ureC, was used to positively identify P. mirabilis as described previously [29]. Ali et al. reported that 96.66% of human P. mirabilis isolates (n = 30) recovered from the urinary tract expressed ureC [30]. In this study, 90.91% of the isolates were positive for ureC. However, its prevalence was relatively low when compared to the results of an earlier study [29]. Our findings revealed that testing only ureC increased the likelihood of obtaining P. mirabilis negative results. Therefore, we used a PCR method based on 16S rRNA expression to detect P. mirabilis. The results of our study showed that the prevalence rates of zapA, rsmA, hmpA, mrpA, atfA, pmfA, FliL, and ucaA were relatively high. Furthermore, the overall prevalence of P. mirabilis was much higher in our study than that in previous studies [31–36].
The prevalence rates of eight genes expressed by moderate biofilm producers were significantly higher than those expressed by strong and weak biofilm producers. These results show that moderate biofilm producers are highly virulent. This is also the first study reporting that the biofilm-forming ability of P. mirabilis is significantly associated with the expression of ureC, zapA, rsmA, hmpA, mrpA, atfA, and pmfA (P < 0.05). Biofilm formation is associated with the adhesion and aggregation of bacteria [20], and rsmA, mrpA, and atfA have been reported to be involved in bacterial adhesion and aggregation [31–33], which is consistent with the results of this study. Biofilm formation was also associated with ureC expression, as previously reported [19, 21]. However, the association between zapA and hmpA, and biofilm formation needs further study.
The results of antimicrobial susceptibility tests revealed high resistance rates of the P. mirabilis isolates to several antibiotics such as doxycycline and ampicillin. The resistance rates ranged from 30 to 50%. The P. mirabilis isolates that were resistant to antibiotics were similar to those previously isolated from chicken products in Hong Kong [37]. However, the resistance rates of P. mirabilis to streptomycin, sulfamethoxazole, kanamycin, ampicillin, ciprofloxacin, cephalothin, gentamicin, cefotaxime, and ceftazidime were significantly higher than those reported in an earlier study, which isolated P. mirabilis from dogs [38]. Our results on the resistance rates of P. mirabilis to nitrofurantoin, tetracycline, and polymyxin B were much lower than those reported in a human study [39], although the resistant rates to other antibiotics, except nitrofurantoin, tetracycline, and polymyxin B, were generally high [39, 40]. In a previous study, P. mirabilis isolates from dogs were found to be highly sensitive to ciprofloxacin and gentamicin [41]. In this study, meropenem and imipenem were the most effective antibiotics against P. mirabilis, which is consistent with the results of an earlier study [42]. Meropenem and imipenem are classified as carbapenems, and our results showed that carbapenems were highly effective against P. mirabilis. We also found that approximately 76.7% of the isolates were MDR or XDR, which is very high compared to the rate reported in a previous study [43]. This finding reveals that resistance to P. mirabilis was steadily increasing. It is also important to mention that the high number of MDR P. mirabilis isolates obtained from companion animals may pose a potential threat to human health. Further studies are needed to define the mechanism of resistance, which may improve the treatment of P. mirabilis infections in the future.
In this study, we found that the resistance to several antibiotics, including piperacillin/tazobactam, ceftazidime, cefoperazone, ceftriaxone, meropenem, and imipenem, was significantly higher among biofilm producers than non-producers, indicating that biofilm producers were more resistant to antibiotics than non-producers. Presently, few studies have described an association between biofilm formation and drug resistance of P. mirabilis, although similar studies have been performed for other pathogens such as uropathogenic E. coli [44, 45], coagulase-negative staphylococci [46], and Haemophilus parasuis [47]. Except for piperacillin/tazobactam, ceftazidime, cefoperazone, ceftriaxone, meropenem, and imipenem, the antibiotic resistance rates of non-producers were higher than those of producers. This finding is consistent with an earlier study that reported non-MDR Acinetobacter baumannii isolates to participate in robust biofilm formation [27]. Different resistance mechanisms are likely to be responsible for the differences in antibiotic resistance and biofilm formation in various bacteria. We also found that the biofilm-forming ability of the P. mirabilis isolates was significantly associated with the resistance to doxycycline, tetracycline, sulfamethoxazole, kanamycin, and cephalothin (P < 0.05). β-lactamase has been reported to decrease the ability of E. coli to form biofilms by inhibiting peptidoglycans, which are required for the assembly of surface molecules on the biofilm. Under sub-inhibitory concentrations of tetracycline and ampicillin, the overexpression of the TetA(C) pump, which contributes to the osmotic stress response and induces capsular colanic acid production, promoted the formation of mature biofilms [48]. In addition, there was an association between the resistance to aminoglycoside and extracellular DNA in Pseudomonas aeruginosa [49]. Thus, we hypothesize that antibiotic resistance is associated with the composition of P. mirabilis biofilms, thereby affecting biofilm formation.
Although the pathogenicity of the biofilm producers was stronger than that of the non-producers, the biofilm-forming ability of the isolates was not significantly associated with morbidity and mortality in mice (P > 0.05). The high pathogenicity of the biofilm producers may have been due to the fact that the biofilm-forming ability of P. mirabilis associated with the expression of ureC, zapA, rsmA, hmpA, mrpA, atfA, and pmfA (P < 0.05). Furthermore, it has been reported that ureC and zapA expressed by P. mirabilis associate with diarrhea in goats [50]. Gabidullin et al. found that diarrhea caused by P. mirabilis was related to enterotoxins [51]. Thus, the mechanism responsible for P. mirabilis-induced diarrhea needs further study.
Conclusions
In conclusion, our study revealed a high level of multidrug resistance in P. mirabilis isolates recovered from diarrhetic animals in northeast China. Therefore, the use of antimicrobial agents in animals needs to be controlled so as to minimize the emergence and eventual spread of resistant pathogens, which is warranted in order to protect human health. The results of this study indicated that the positive rates of the genes expressed by biofilm-producing P. mirabilis isolates were significantly higher than those expressed by non-producing isolates. Furthermore, this is the first study to report that the biofilm-forming ability of P. mirabilis isolates from diarrhetic animals is significantly associated with the expression of ureC, zapA, rsmA, hmpA, mrpA, atfA, and pmfA and the resistance to doxycycline, tetracycline, sulfamethoxazole, kanamycin, and cephalothin. Further studies on biofilm formation by P. mirabilis are necessary to better understand the disease process and to develop effective treatments for mammals with antibiotic resistant P. mirabilis.
Methods
Sample collection
Six hundred and fourteen fecal swabs were collected randomly from different farms in northeast China between September 2014 and October 2016 and transferred to the laboratory in ice-filled containers. Fecal swabs were collected from different animals (i.e., dog, mink, cattle, and fowl) with diarrhea.
P. mirabilis screening
P. mirabilis was isolated as described previously [37]. To grow the bacteria, the fecal swabs were incubated with 1 mL of Luria–Bertani broth for 6 h at 37 °C, and a 30 μL aliquot of the broth was added to buffered peptone water (Aobox, Beijing, China). The resultant mixture was incubated for 24 h at 37 °C to enrich the culture, and then streaked onto xylose lysine deoxycholate agar plates (Hopebiol, Qingdao, China). All plates were incubated at 37 °C for a minimum of 16 h. One isolate from each sample was selected for further characterization.
Identification of isolates
To identify P. mirabilis isolates, standard biochemical tests were used as described previously [52]. The isolates were gram-negative bacilli that were positive for glucose, methyl red, and urease but negative for maltose, sucrose, mannitol, indole, and gelatin liquefaction. The Voges–Proskauer test yielded negative results for the isolates. Urease C (ureC) and mannose-resistant Proteus-like fimbriae (mrpA) were amplified by the polymerase chain reaction (PCR) to identify P. mirabilis from the presumptive isolates [36]. Positive results were randomly selected for sequencing. When the results were negative for ureC and mrpA expression, a segment of the 16S rRNA gene was amplified. The 16S rRNA segment was sequenced [53]. The sequences were compared with ureC, mrpA, and 16S rRNA in GenBank using BLAST.
Quantification of biofilm production
Biofilm production was assessed using a microtiter plate assay as described previously [54]. P. mirabilis isolates were grown in tryptic soy broth (Hopebiol) supplemented with 0.25% glucose overnight at 37 °C. Subsequently, the cultures were diluted to 1 × 106 CFU mL− 1 in fresh tryptic soy broth supplemented with 0.25% glucose, and 200 μL of the diluted culture was transferred into each well of a sterile flat-bottom 96-well polystyrene microtiter plate. After incubation for 24 h at 37 °C without shaking, the wells were gently washed three times with 200 μL of distilled water. The biofilms were fixed with 200 μL of 99% methanol for 15 min. The supernatant was removed, and the plate was air-dried. The biofilms were then stained with 200 μL of 1% crystal violet for 10 min. The excess dye was removed by washing the plate under running tap water. Finally, the bound crystal violet was released by adding 200 μL of 33% acetic acid. The optical density (OD) of each well was measured at 590 nm using a microtiter plate reader. To determine the background OD, control experiments were performed with uninoculated broth. This assay was repeated three independent times. The biofilm formation capacity of each isolate was analyzed using the method of Khoramian et al. [55].
Expression of virulence genes
P. mirabilis virulence genes were detected by PCR. These genes were ureC, extracellular metalloprotease (zapA), swarming behavior (rsmA), hemolysin (hpmA), mrpA, ambient-temperature fimbriae (atfA), P. mirabilis fimbriae (pmfA), flagellar basal body protein (FliL), and uroepithelial cell adhesin fimbriae (ucaA) (Table 5) [10]. The base sequences and predicted sizes of the PCR products for the specific oligonucleotide primers used in this study are listed in Table 5. Subsequently, total genomic DNA was isolated from stationary-phase broth cultures that were grown overnight in Luria–Bertani broth with the TIANamp Bacterial DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. All PCR products were analyzed by agarose gel electrophoresis, and the results were recorded with a gel documentation system. All amplifications were repeated three independent times in parallel with a negative control (distilled water served as the PCR template).
Table 5.
Primers used in the PCRs carried out in this study
| Target gene | Primer | Nucleotide Sequence (5′-3′) | Amplicon (bp) | AT* | Reference |
|---|---|---|---|---|---|
| 16S rRNA | 27F | AGAGTTTGATCCTGGCTCAG | 1463 | 49 | Leite et al. (2015) [53] |
| 1492R | GGTTACCTTGTTACGACTT | ||||
| ureC | ureC-F | GTTATTCGTGATGGTATGGG | 317 | 52 | Stankowska et al. (2008) [36] |
| ureC-R | ATAAAGGTGGTTACGCCAGA | ||||
| zapA | zapA-F | ACCGCAGGAAAACATATAGCCC | 540 | 53 | Stankowska et al. (2008) [36] |
| zapA-R | GCGACTATCTTCCGCATAATCA | ||||
| rsmA | rsmA-F | TAGCGAGTGTTGACGAGTGG | 562 | 49 | Shi et al. (2016) [31] |
| rsmA-R | AGCGAGGTGAAGAACGAGAA | ||||
| hpmA | hpmA-F | CCAGTGAATTAACGGCAGGT | 654 | 49 | Shi et al. (2016) [31] |
| hpmA-R | CGTGCCCAGTAATGGCTAAT | ||||
| mrpA | MRP-F | ACACCTGCCCATATGGAAGATACTGGTACA | 550 | 40 | Barbour et al. (2012) [32] |
| MRP-R | AAGTGATGAAGCTTAGTGATGGTGATGGTG | ||||
| ATGAGAGTAAGTCACC | |||||
| FliL | FliL-F | CTCTGCTCGTGGTGGTGTCG | 770 | 40 | Barbour et al. (2012) [32] |
| FliL-R | GCGTCGTCACCTGATGTGTC | ||||
| ucaA | ucaA-F | GTAAAGTTGTTGCGCAAAC | 560 | 50 | Sosa et al. (2006) [35] |
| ucaA-L | TTGAGCCACTGTGGATACA | ||||
| pmfA | pmfA-F | CAAATTAATCTAGAACCACTC | 618 | 54 | Zunino et al. (2003) [34] |
| pmfA-R | ATTATAGAGGATCCCTTGAAGGTA | ||||
| atfA | atfA-F | CATAATTTCTAGACCTGCCCTAGCA | 382 | 50 | Zunino et al. (2000) [33] |
| atfA-R | CTGCTTGGATCCGTAATTTTTAACG |
AT* anneling temperature
Antimicrobial resistance test
The antimicrobial resistance of the isolates to 19 antibiotics, including doxycycline, ampicillin, ciprofloxacin, streptomycin, tetracycline, piperacillin/tazobactam, cefotaxime, nitrofurantoin, sulfamethoxazole, ceftriaxone, polymyxin B, ceftazidime, kanamycin, gentamicin, cefoperazon, cephalothin, meropenem, levofloxacin, and imipenem (BIO-KONT, Wenzhou, China), was determined by the Kirby–Bauer disc diffusion method [56]. In brief, 0.5 McFarland P. mirabilis inoculum was spread onto Mueller–Hinton agar plates (Hopebiol), and the antibiotic discs were dispensed on the agar. The plates were incubated at 37 °C for 18 h. Escherichia coli (E. coli) ATCC 25922 was used as the control microorganism. The inhibitory zone around each disc was measured, and the results were interpreted according to the guidelines provided by the manufacturers and the Clinical and Laboratory Standards Institute [57]. The results were interpreted as resistant, intermediate, and susceptible. MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories. XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories [58]. Non-MDR was defined as resistance to none or up to two antimicrobial categories [27].
Pathogenicity test in mice
The pathogenicity test was performed as described previously [59]. In brief, healthy, 6-week-old, female BALB/c mice (n = 99) (Changsheng Biotechnology Co., Ltd., Liaoning, China) were used to investigate the pathogenicity of P. mirabilis. The mice were randomly divided into thirty-three groups (n = 3/per group) as follows: one control group and thirty-two experimental groups, which tested 32 P. mirabilis isolates for their pathogenicity to produce biofilm, to express various virulence genes, and to resist different drugs. Each mouse received a 0.2-mL intraperitoneal injection of a bacterial suspension at a concentration of 1 × 108 CFU mL− 1, and the control mice were challenged with vehicle alone. All the mice were fed normally and observed daily for the activity level and water intake for up to 14 days, and deaths were recorded,then the mice alive were euthanized by intraperitoneal injection of 40 mg/kg b.w. of sodium thiopental [60].
Statistical analysis
Data were analyzed with SPSS 18.0 Software (SPSS Statistics, Inc., Chicago, IL, USA). The Chi-square test was adopted for analysis. P-values < 0.05 were considered statistically significant.
Acknowledgments
The authors thank Dr. Songfeng Zheng (Missouri State University, USA) for his help with statistical analysis.
Abbreviations
- P. mirabilis
Proteus mirabilis
- PCR
Polymerase chain reaction
- MDR
Multidrug-resistant
- XDR
Extensively drug-resistant
- BPW
Buffered peptone water
- OD
Optical density
- E. coli
Escherichia coli
- AT
Annealing temperature
Authors’ contributions
All authors participated in study design and planning. YDS and SSW conceived and designed the study as well as acquired and extracted the data. YDS performed data analysis. YDS, SSW, LLZ, Y P, and QQX drafted the manuscript. YDS, SSW, HHL, CWW, HYC, JWG, and HBW interpreted the data and revised the manuscript. All authors have read and approved this version of the manuscript for publication.
Funding
This work was supported by the “Academic Backbone” Project of Northeast Agricultural University (17XG10), the National Natural Science Foundation of China (31672532), and the SIPT Program (201910224139). The role of the funding bodies was to pay for the consumables used in this study. They had no role in the design of the study; the collection, analysis, and interpretation of data and in the writing of the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in National Center for Biotechnology Information (NCBI) repository, under these GenBank accession numbers MT294143-MT294148 and MT276297-MT276312.
Ethics approval and consent to participate
This study was performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China. The protocol was reviewed and approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (2014-SRM-24). The specimens were collected from animals for laboratory analyses only, and unnecessary pain and suffering of the animals was avoided. The animal owners provided written consent for the collection of specimens. Animal experiments were carried out according to animal welfare standards.
Consent for publication
Not applicable.
Competing interests
The authors have no financial or personal relationships with other individuals or organizations that could have inappropriately influenced or biased the contents of this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen CY, Chen YH, Lu PL, Lin WR, Chen TC, Lin CY. Proteus mirabilis urinary tract infection and bacteremia: risk factors, clinical presentation, and outcomes. J Microbiol Immunol Infect. 2012;45:228–236. doi: 10.1016/j.jmii.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Liu MC, Lin SB, Chien HF, Wang WB, Yuan YH, Hsueh PR, et al. 10′(Z), 13′(E)-heptadecadienylhydroquinone inhibits swarming and virulence factors and increases polymyxin B susceptibility in Proteus mirabilis. PLoS One. 2012;7:e45563. doi: 10.1371/journal.pone.0045563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mobley HL, Belas R. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 1995;3:280–284. doi: 10.1016/s0966-842x(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Zhang S, Yu J, Zhang H, Yuan Z, Sun Y, et al. An outbreak of Proteus mirabilis food poisoning associated with eating stewed pork balls in brown sauce, Beijing. Food Control. 2010;21:302–305. [Google Scholar]
- 5.Wang JT, Chen PC, Chang SC, Shiau YR, Wang HY, Lai JF, et al. Antimicrobial susceptibilities of Proteus mirabilis: a longitudinal nationwide study from the Taiwan surveillance of antimicrobial resistance (TSAR) program. BMC Infect Dis. 2014;14:486. doi: 10.1186/1471-2334-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habibi M, Karam MRA, Bouzari S. Construction and evaluation of the immune protection of a recombinant divalent protein composed of the MrpA from MR/P fimbriae and flagellin of Proteus mirabilis strain against urinary tract infection. Microb Pathog. 2018;117:348–55. [DOI] [PubMed]
- 7.Cabassi C, Taddei S, Predari G, Galvani G, Ghidini F, Schiano E, et al. Bacteriologic findings in ostrich (Struthio camelus) eggs from farms with reproductive failures. Avian Dis. 2004;48:716–722. doi: 10.1637/7142. [DOI] [PubMed] [Google Scholar]
- 8.Yu W, He Z, Huang F. Multidrug-resistant Proteus mirabilis isolated from newly weaned infant rhesus monkeys and ferrets. Jundishapur J Microbiol. 2015;8:e16822. 10.5812/jjm.8(5)2015.16822. [DOI] [PMC free article] [PubMed]
- 9.Malayeri HZ, Jamshidi S, Salehi TZ. Identification and antimicrobial susceptibility patterns of bacteria causing otitis externa in dogs. Vet Res Commun. 2010;34:435–444. doi: 10.1007/s11259-010-9417-y. [DOI] [PubMed] [Google Scholar]
- 10.Rózalski A, Sidorczyk Z, Kotełko K. Potential virulence factors of Proteus bacilli. Microbiol Mol Biol Rev. 1997;61:65–89. doi: 10.1128/mmbr.61.1.65-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legnani-Fajardo C, Zunino P, Piccini C, Allen A, Maskell D. Defined mutants ofProteus mirabilislacking flagella cause ascending urinary tract infection in mice. Microb Pathog. 1996;21:395–405. doi: 10.1006/mpat.1996.0070. [DOI] [PubMed] [Google Scholar]
- 12.Hola V, Peroutkova T, Ruzicka F. Virulence factors in Proteus bacteria from biofilm communities of catheter-associated urinary tract infections. FEMS Immunol Med Microbiol. 2012;65:343–349. doi: 10.1111/j.1574-695X.2012.00976.x. [DOI] [PubMed] [Google Scholar]
- 13.Fusco A, Coretti L, Savio V, Buommino E, Lembo F, Donnarumma G. Biofilm formation and Immunomodulatory activity of Proteus mirabilis clinically isolated strains. Int J Mol Sci. 2017;18:414. doi: 10.3390/ijms18020414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwiecinska-Pirog J, Skowron K, Bartczak W, Gospodarek-Komkowska E. The ciprofloxacin impact on biofilm formation by Proteus mirabilis and P. vulgaris strains. Jundishapur J Microbiol. 2016;9:e32656. 10.5812/jjm.32656. [DOI] [PMC free article] [PubMed]
- 16.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 17.McLean RJ, Lawrence JR, Korber DR, Caldwell DE. Proteus mirabilis biofilm protection against struvite crystal dissolution and its implications in struvite urolithiasis. J Urol. 1991;146:1138–1142. doi: 10.1016/s0022-5347(17)38026-6. [DOI] [PubMed] [Google Scholar]
- 18.Rocha SP, Elias WP, Cianciarullo AM, Menezes MA, Nara JM, Piazza RM, et al. Aggregative adherence of uropathogenic Proteus mirabilis to cultured epithelial cells. FEMS Immunol Med Microbiol. 2007;51:319–326. doi: 10.1111/j.1574-695X.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- 19.Stickler D, Ganderton L, King J, Nettleton J, Winters C. Proteus mirabilis biofilms and the encrustation of urethral catheters. Urol Res. 1993;21:407–411. doi: 10.1007/BF00300077. [DOI] [PubMed] [Google Scholar]
- 20.Jansen AM, Lockatell V, Johnson DE, Mobley HL. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect Immun. 2004;72:7294–7305. doi: 10.1128/IAI.72.12.7294-7305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjbar-Omid M, Arzanlou M, Amani M, Shokri Al-Hashem SK, Amir Mozafari N, Peeri DH. Allicin from garlic inhibits the biofilm formation and urease activity of Proteus mirabilis in vitro. FEMS Microbiol Lett. 2015;362:fnv049. doi: 10.1093/femsle/fnv049. [DOI] [PubMed] [Google Scholar]
- 22.Rahal K, Wang F, Schindler J, Rowe B, Cookson B, Huovinen P, et al. Reports on surveillance of antimicrobial resistance in individual countries. Clin Infect Dis. 1997;24:S169–SS75. doi: 10.1093/clinids/24.supplement_1.s169. [DOI] [PubMed] [Google Scholar]
- 23.Alabi OS, Mendonça N, Adeleke OE, da Silva GJ. Molecular screening of antibiotic-resistant determinants among multidrug-resistant clinical isolates of Proteus mirabilis from SouthWest Nigeria. Afr Health Sci. 2017;17:356–365. doi: 10.4314/ahs.v17i2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bie L, Wu H, Wang XH, Wang M, Xu H. Identification and characterization of new members of the SXT/R391 family of integrative and conjugative elements (ICEs) in Proteus mirabilis. Int J Antimicrob Agents. 2017;50:242–246. doi: 10.1016/j.ijantimicag.2017.01.045. [DOI] [PubMed] [Google Scholar]
- 25.Bameri Z, Karam MRA, Habibi M, Ehsani P, Bouzari S. Determination immunogenic property of truncated MrpH. FliC as a vaccine candidate against urinary tract infections caused by Proteus mirabilis. Microb Pathog. 2018;114:99–106. doi: 10.1016/j.micpath.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen SM, Shirtliff ME. Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence. 2011;2:460–465. doi: 10.4161/viru.2.5.17783. [DOI] [PubMed] [Google Scholar]
- 27.Qi L, Li H, Zhang C, Liang B, Li J, Wang L, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483. [DOI] [PMC free article] [PubMed]
- 28.Kwiecinska-Piróg J, Bogiel T, Skowron K, Wieckowska E, Gospodarek E. Proteus mirabilis biofilm-qualitative and quantitative colorimetric methods-based evaluation. Braz J Microbiol. 2014;45:1423–1431. doi: 10.1590/s1517-83822014000400037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Niu Z, Yin K, Liu P, Chen L. Quick identification and quantification of Proteus mirabilis by polymerase chain reaction (PCR) assays. Ann Microbiol. 2013;63:683–689. [Google Scholar]
- 30.Ali HH, Yousif MG. Detection of some virulence factors genes of Proteus mirablis that isolated from urinary tract infection. Int J. 2015;3:156–163. [Google Scholar]
- 31.Shi X, Lin Y, Qiu Y, Li Y, Jiang M, Chen Q, et al. Comparative screening of digestion tract toxic genes in Proteus mirabilis. PLoS One. 2016;11:e0151873. doi: 10.1371/journal.pone.0151873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbour EK, Hajj ZG, Hamadeh S, Shaib HA, Farran MT, Araj G, et al. Comparison of phenotypic and virulence genes characteristics in human and chicken isolates of Proteus mirabilis. Pathogens and global health. 2012;106:352–357. doi: 10.1179/2047773212Y.0000000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zunino P, Geymonat L, Allen AG, Legnani-Fajardo C, Maskell DJ. Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol Med Microbiol. 2000;29:137–143. doi: 10.1111/j.1574-695X.2000.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 34.Zunino P, Sosa V, Allen AG, Preston A, Schlapp G, Maskell DJ. Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology. 2003;149:3231–3237. doi: 10.1099/mic.0.26534-0. [DOI] [PubMed] [Google Scholar]
- 35.Sosa V, Schlapp G, Zunino P. Proteus mirabilis isolates of different origins do not show correlation with virulence attributes and can colonize the urinary tract of mice. Microbiology. 2006;152:2149–2157. doi: 10.1099/mic.0.28846-0. [DOI] [PubMed] [Google Scholar]
- 36.Stankowska D, Kwinkowski M, Kaca W. Quantification of Proteus mirabilis virulence factors and modulation by acylated homoserine lactones. J Microbiol Immunol Infect. 2008;41:243–253. [PubMed] [Google Scholar]
- 37.Wong MHY, Wan HY, Chen S. Characterization of multidrug-resistant Proteus mirabilis isolated from chicken carcasses. Foodborne Pathog Dis. 2013;10:177–181. doi: 10.1089/fpd.2012.1303. [DOI] [PubMed] [Google Scholar]
- 38.Harada K, Niina A, Shimizu T, Mukai Y, Kuwajima K, Miyamoto T, et al. Phenotypic and molecular characterization of antimicrobial resistance in Proteus mirabilis isolates from dogs. J Med Microbiol. 2014;63:1561–1567. doi: 10.1099/jmm.0.081539-0. [DOI] [PubMed] [Google Scholar]
- 39.Adamus-Bialek W, Zajac E, Parniewski P, Kaca W. Comparison of antibiotic resistance patterns in collections of Escherichia coli and Proteus mirabilis uropathogenic strains. Mol Biol Rep. 2013;40:3429–3435. doi: 10.1007/s11033-012-2420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanayama A, Kobayashi I, Shibuya K. Distribution and antimicrobial susceptibility profile of extended-spectrum β-lactamase-producing Proteus mirabilis strains recently isolated in Japan. Int J Antimicrob Agents. 2015;45:113–118. doi: 10.1016/j.ijantimicag.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen K, Pedersen K, Jensen H, Finster K, Jensen VF, Heuer OE. Occurrence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J Antimicrob Chemother. 2007;60:775–781. doi: 10.1093/jac/dkm269. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Xu Y, Wang Z, Lin X. Antimicrobial resistance and genotype analysis of extended-Spectrum-β-lactamase-producing Proteus mirabilis. Open J Clin Diagnostics. 2014;4:57. [Google Scholar]
- 43.Okonko I, Nkang A, Fajobi E, Mejeha O, Udeze A, Motayo B, et al. Incidence of multi-drug resistant (MDR) organisms in some poultry feeds sold in Calabar Metropolis, Nigeria. Br J Pharmacol Toxicol. 2010;1:15–28.
- 44.Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrob Resist Infect Control. 2016;5:5. doi: 10.1186/s13756-016-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5:11. doi: 10.1186/s13756-016-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrestha LB, Bhattarai NR, Khanal B. Antibiotic resistance and biofilm formation among coagulase-negative staphylococci isolated from clinical samples at a tertiary care hospital of eastern Nepal. Antimicrob Resist Infect Control. 2017;6:89. doi: 10.1186/s13756-017-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Xu C, Shen H, Li J, Guo L, Cao G, et al. Biofilm formation in Haemophilus parasuis: relationship with antibiotic resistance, serotype and genetic typing. Res Vet Sci. 2014;97:171–175. doi: 10.1016/j.rvsc.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 48.May T, Ito A, Okabe S. Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob Agents Chemother. 2009;53:4628–4639. doi: 10.1128/AAC.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 50.Yue L. Identification of Proteus mirabilis from goat and detection of its major virulence genes Southwest University for nationalities. 2016. [Google Scholar]
- 51.Gabidullin Z, Zhukova S, Ezepchuk I, Bondarenko V. The detection of a choleriform thermolabile enterotoxin in clinical strains of Proteus isolated in different infections. Zh Mikrobiol Epidemiol Immunobiol. 1989;12:14–6. [PubMed]
- 52.Songer J, Post K. Veterinary Microbiology: bacterial and fungal agents of animal disease. St Louis: Elsevier Saunders, St Louis; 2005. p. 687. [Google Scholar]
- 53.Leite A, Miguel M, Peixoto R, Ruas-Madiedo P, Paschoalin V, Mayo B, et al. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J Dairy Sci. 2015;98:3622–3632. doi: 10.3168/jds.2014-9265. [DOI] [PubMed] [Google Scholar]
- 54.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 55.Khoramian B, Jabalameli F, Niasari-Naslaji A, Taherikalani M, Emaneini M. Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections. Microb Pathog. 2015;88:73–77. doi: 10.1016/j.micpath.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Bauer A, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493. [PubMed] [Google Scholar]
- 57.Jean B. Patel, Franklin R. Cockerill III, et al. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI Document M100-S25, Clinical and Laboratory Standards Institute. 2015;35(3).
- 58.Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 59.Osman KM, Samir A, Abo-Shama UH, Mohamed EH, Orabi A, Zolnikov T. Determination of virulence and antibiotic resistance pattern of biofilm producing Listeria species isolated from retail raw milk. BMC Microbiol. 2016;16:263. doi: 10.1186/s12866-016-0880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jing X, Gu S, Liu D, Zhao L, Xia S, He X, Chen H, Ge J. Lactobacillus brevis 23017 Relieves Mercury Toxicity in the Colon by Modulation of Oxidative Stress and Inflammation Through the Interplay of MAPK and NF-κB Signaling Cascades. Front Microbiol. 2018;9:2425. doi: 10.3389/fmicb.2018.02425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in National Center for Biotechnology Information (NCBI) repository, under these GenBank accession numbers MT294143-MT294148 and MT276297-MT276312.