Abstract
Background
The Plasmodium falciparum chloroquine transporter gene (pfcrt) is known to be involved in chloroquine and amodiaquine resistance, and more particularly the mutations on the loci 72 to 76 localized within the second exon. Additionally, new mutations (T93S, H97Y, C101F, F145I, M343L, C350R and G353V) were recently shown to be associated with in vitro reduced susceptibility to piperaquine in Asian or South American P. falciparum strains. However, very few data are available on the prevalence of these mutations and their effect on parasite susceptibility to anti-malarial drugs, and more particularly piperaquine in Africa.
Methods
A molecular investigation of these mutations was performed in 602 African P. falciparum parasites collected between 2017 and 2018 on malaria patients hospitalized in France after a travel in African countries. Associations between genotypes and in vitro susceptibilities to piperaquine and standard antimalarial drugs were assessed.
Results
None of the mutations, previously described as associated with piperaquine resistance, was found in the 602 P. falciparum African isolates. The K76T mutation is associated with resistance to chloroquine (p < 0.0002) and desethylamodiaquine (p < 0.002) in Africa. The K76T mutation is not associated with in vitro reduced susceptibility to piperaquine. The mutation I356T, identified in 54.7% (n = 326) of the African isolates, was significantly associated with reduced susceptibility to quinine (p < 0.02) and increased susceptibility to mefloquine (p < 0.04). The K76T and I356T mutations were significantly associated in West African isolates (p = 0.008).
Conclusion
None of the mutations in pfcrt found to be associated with piperaquine reduced susceptibility in Asia or South America (T93S, H97Y, C101F, F145I, M343L C350R and G353V) were found in the 602 African isolates including the three isolates with reduced susceptibility to piperaquine. The K76T mutation, involved in resistance to chloroquine and amodiaquine, and the I356T mutation were not associated with in vitro reduced susceptibility to piperaquine. Differences in mefloquine susceptibility between I356 and 356T isolates were, while statistically different, minimal. Further analyses are needed with a more important sample size from the same geographic area to confirm the role of the I356T mutation on quinine susceptibility.
Keywords: Malaria, Plasmodium falciparum, Antimalarial drug, Resistance, In vitro, Molecular marker, pfcrt, I356T, Africa
Background
The Plasmodium falciparum chloroquine transporter gene (pfcrt) has been under different drugs pressure during parasites evolution for decades. As a result, acquisition of mutations for adaptation has emerged. These mutations induce an alteration of membrane protein physiochemical properties modifying vacuolar traffic of these drugs in resistant parasites [1]. Quinoline-based compounds like chloroquine, amodiaquine, mefloquine, primaquine, and piperaquine share a similar structure [2]. The common and widely spread mutations on pfcrt gene associated with resistance are localized within its second exon. Precisely, chloroquine resistance was associated with mutations on the loci 72–76 [1, 3]. The K76T mutation was associated with chloroquine resistance and used as molecular marker to survey chloroquine resistance in epidemiological studies [3]. But mutations on other loci were also found to be involved either as amplifying the resistance or as compensatory mutations for the fitness cost [4].
Since 2005, the World Health Organization (WHO) has recommended artemisinin-based combination therapy (ACT) as the first-line treatment against malaria. However, P. falciparum parasites resistant to artemisinin derivatives rapidly emerged in Southeast Asia, and more particularly in western Cambodia, Myanmar, Thailand and Laos [5, 6]. More recently, the emergence of P. falciparum resistance to dihydroartemisinin-piperaquine was observed in Cambodia, where recrudescent infections had rapidly increased [7–9], and then in Vietnam [10, 11]. Duplication of the P. falciparum Plasmepsin 2 gene (pfpm2), encoding a protease involved in haemoglobin degradation, has been found to be associated with reduced in vitro susceptibility to piperaquine in Cambodian P. falciparum parasites and with dihydroartemisinin-piperaquine failures in Cambodia [12, 13]. However, the involvement of pfpm2 in piperaquine resistance seems controversial in Africa [14]. Parasites from patients successfully treated with dihydroartemisinin-piperaquine could carry with two copies of pfpm2 while only a single copy of pfpm2 was detected in isolates collected in imported malaria cases after dihydroartemisinin-piperaquine failures or in P. falciparum isolates with in vitro reduced susceptibility to piperaquine [15–20]. All these data suggest that pfpm2 would not be the only gene that explains the resistance to piperaquine in Africa. Mutations in pfcrt could be involved in piperaquine resistance. The K76T mutation seems to be not associated with in vitro reduced susceptibility to piperaquine [21]. However, several new mutations in pfcrt were found to be associated with piperaquine reduced susceptibility. Piperaquine resistance, selected by in vitro continuous piperaquine pressure on P. falciparum Dd2, was associated with the C101F mutation in pfcrt on the same exon where the loci 72–76 are located [22, 23]. In 2012 in French Guiana, dihydroartemisinin-piperaquine has not been yet officially recommended as first-line treatment against malaria but was used as self-medication by illegal gold miners in rainforest. Many years after the withdrawal of chloroquine as P. falciparum treatment, but still used for the treatment of P. vivax malaria, the prevalence of the mutant haplotype SVMNT (residues 72–76 in PfCRT) remained high (97.5%). In this context, the mutation C350R on pfcrt emerged in 2002 to reach a prevalence of 58% in 2012. This mutation was involved in the decrease of in vitro susceptibility to piperaquine with a restoration of chloroquine susceptibility [23]. In Cambodia, Agrawal and colleagues identified the substitution of phenylalanine by isoleucine on the locus 145 (F145I) associated with a decrease in piperaquine susceptibility [25]. In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of K13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance [26, 27]. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam [28, 29]. Many other mutations were found in pfcrt but few were investigated for their association with anti-malarial drug resistance. The mutation I356T/L for instance is often found both on Asian or South-American parasites [30, 31].
These data suggest that parasite susceptibility to piperaquine is affected by some of these mutations in Southeast Asia and South America. However, very few data are available on the prevalence of these mutations and their effect on parasites susceptibility to piperaquine in Africa. It would, therefore, be essential to study these loci to provide more information on African parasites genotypes and piperaquine susceptibility. To this end, a molecular epidemiologic study of pfcrt genotypes was conducted on 602 African P. falciparum parasites collected from different countries. The association between these mutations and ex vivo susceptibility to piperaquine (PPQ) was assessed. Association with resistance to other common antimalarial drugs, such as chloroquine (CQ), quinine (QN), dihydroartemisinin (DHA), artesunate (AS) monodesethylamodiaquine (DQ), mefloquine (MQ), lumefantrine (LMF) and pyronaridine (PND) was also evaluated.
Methods
Sample collection
A retrospective analysis was performed on 602 African P. falciparum samples collected between January 2017 and October 2018 on malaria patients hospitalized in France after a travel in sub-Saharan African countries (Table 1). The samples were sent from different civilian or military hospitals of the French National Reference Centre for Imported Malaria network (Aix en Provence, Bordeaux, Lyon, Marseille, Montpellier, Nice, Toulon and Toulouse) to the French National Reference Centre for Malaria (IRBA, IHU Méditerranée Infection, Marseille, France).
Table 1.
Number of I256T and K76T mutations per regions and countries
| Regions/countries | Allele I356 or 356T (number of samples) | Allele K76 or 76T (number of samples) | ||||||
|---|---|---|---|---|---|---|---|---|
| I356 | Mixed | 356T | Total | K76 | Mixed | 76T | Total | |
| West Africa | 145 | 17 | 139 | 301 | 197 | 13 | 68 | 278 |
| Benin | 6 | 1 | 6 | 13 | 2 | 1 | 6 | 9 |
| Burkina Faso | 9 | 1 | 10 | 20 | 16 | 1 | 2 | 19 |
| Gambia | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Ghana | 2 | 0 | 4 | 6 | 6 | 0 | 0 | 6 |
| Guinea | 24 | 5 | 19 | 48 | 17 | 5 | 27 | 49 |
| Ivory Coast | 70 | 6 | 65 | 141 | 115 | 3 | 13 | 131 |
| Mali | 4 | 0 | 4 | 8 | 3 | 1 | 3 | 7 |
| Niger | 5 | 0 | 6 | 11 | 8 | 0 | 0 | 8 |
| Nigeria | 6 | 2 | 8 | 16 | 6 | 0 | 7 | 13 |
| Senegal | 7 | 2 | 4 | 13 | 6 | 2 | 5 | 13 |
| Sierra Leone | 1 | 0 | 3 | 4 | 3 | 0 | 3 | 6 |
| Togo | 9 | 0 | 10 | 19 | 15 | 0 | 1 | 16 |
| Cape Verde | 1 | 0 | 0 | 1 | ||||
| Central Africa | 101 | 12 | 116 | 229 | 179 | 10 | 40 | 229 |
| Angola | 0 | 0 | 2 | 2 | 2 | 0 | 0 | 2 |
| Cameroon | 54 | 4 | 65 | 123 | 106 | 4 | 15 | 125 |
| Central African Republic | 14 | 0 | 20 | 34 | 32 | 0 | 0 | 32 |
| Chad | 5 | 0 | 11 | 16 | 10 | 2 | 2 | 14 |
| Congo | 11 | 4 | 8 | 23 | 8 | 2 | 13 | 23 |
| Gabon | 17 | 4 | 10 | 31 | 21 | 2 | 10 | 33 |
| East Africa | 7 | 0 | 3 | 10 | 9 | 0 | 2 | 11 |
| Burundi | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 2 |
| Djibouti | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 2 |
| Ethiopia | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| Mozambique | 1 | 0 | 1 | 2 | 3 | 0 | 0 | 3 |
| Sudan | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| Tanzania | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 2 |
| Indian Ocean | 12 | 0 | 23 | 35 | 31 | 1 | 1 | 33 |
| Comoros | 9 | 0 | 20 | 29 | 26 | 0 | 1 | 27 |
| Madagascar | 3 | 0 | 3 | 6 | 5 | 1 | 0 | 6 |
| Maghreb | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 1 |
| Morocco | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 1 |
| Unknown origin | 10 | 2 | 13 | 25 | 16 | 2 | 8 | 26 |
| Total | 276 | 32 | 294 | 602 | 433 | 26 | 119 | 578 |
Nucleic acid extraction
DNA extraction for each sample was performed and purified using the QIAamp® DNA Mini kit according to the manufacturer’s recommendations (Qiagen, Hilden, Germany).
pfcrt genotyping
Two fragments of the pfcrt gene (PF3D7_0709000) were amplified. The first fragment of 840 nucleotides including the exons 2, 3 and part of exon 4 and covering the positions 72–76, 93, 97, 101, 145, 146, 158 and 159 was amplified by PCR using the primers pairs: 5′-GAT-GGC-TCA-CGT-TTA-GGT-GGA-3′ and 5′-TGT-TAC-AAC-AAT-AAT-AAC-TGC-TCC-G-3′. The second fragment of 339 nucleotides including the exon 10 and covering the positions 343, 350, 353 and 356 was amplified using the primer pairs: 5′-CCA-TAT-AAT-TTT-TCA-TTT-TC-3′ and 5′-GTT-CTC-TTA-CAA-CAT-CAC-3′. The reaction mixture for PCR contained 200 ng of genomic DNA, 0.32 µM of each primer, 1X final of reaction buffer (750 mM Tris-HCl, 200 mM (NH4)2SO4, 0.1% (v/v) Tween 20 and stabilizer, pH 8.8), 2.5 mM MgCl2, 200 µM of dNTP mixture (Euromedex, Souffelweyersheim, France) and 0.2 U of Hot Diamond Taq® polymerase (Eurogentec, Liège, Belgium) in a final volume of 25 µL. For the first fragment, the thermal cycler (Life Eco V 2.04; Bioer, China) was programmed as follows: 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 65 °C of hybridization temperature for 1 min, elongation at 65 °C for 1 min 30 s, and a final 10-min extension step at 65 °C. The amplification programme for the second fragment was 95 °C for 5 min, 40 cycles of 95 °C for 30 s, hybridization temperature 46 °C for 45 s, elongation at 68°C for 1 min, and a final 10 min extension step at 68 °C. To cover the mutations T93S, H97Y and C101F, the purified amplicons were sequenced by the amplification forward primer of the first fragment and the mutations F145I, I146L, F158L and V159S were sequenced using the primer 5′-TTA-GGA-ACG-ACA-CCG-AAG-C-3′. To cover the mutations M343L, C350R and I356T, the amplified second fragment was purified and sequenced using the forward primer. Sequencing was performed on ABI Prism 3100 analyser (Applied Biosystems, Villebon sur Yvette, France) according to the manufacturers’ instructions. Base calling was implemented on Vector NTI 10.3.0 software (Invitrogen, Cergy Pontoise, France). Poor-quality sequences were either re-sequenced or discarded and repeat polymorphisms were retained for analysis if clean individual peaks were observed in the electropherogram.
Drugs and ex vivo assay
The drug susceptibility assays were performed using the HRP2 ELISA-based assay Malaria Ag Celisa kit (ref KM2159, Cellabs PTY LDT, Brookvale, Australia) as previously described [32].
Each batch of plates was validated using the CQ-resistant W2 strain (isolated from Indochina; MRA-157 obtained from MR4, VA, USA) in four independent experiments. The mean 50% inhibitory concentration (IC50) values for all the batches used over 2 years were 495 ± 45 nM for CQ, 401 ± 36 nM for QN, 93 ± 17 nM for DQ, 23.9 ± 3.4 nM for MQ, 57.1 ± 5.6 nM for PPQ, 18.6 ± 3.1 nM for PND, 2.4 ± 0.4 nM for DHA and 1.6 ± 0.4 nM for AS. A comparison of the W2 anti-malarial susceptibility data between the different batches of plates indicated that there was no significant difference in the responses to anti-malarial drugs over the 2 years (0.625 < p < 0.990).
The polymorphic genetic markers msp1 and msp2 and microsatellite markers specific to P. falciparum W2 were genotyped at least once a month to verify W2 clonality as previously described [33–35].
Data management and statistical analysis
IC50 values were calculated with the inhibitory sigmoid Emax model, with estimation of the IC50 through non-linear regression using a standard function of the R software ICEstimator version 1.2 (http://www.antimalarial-icestimator.net/). IC50s estimates values and sequences analysis were collected and analyzed on excel sheets and GraphPad Prism (V7.0a). Samples were grouped by regions for analysis to reflect parasites population structure sharing the same genetic background. The median of the IC50 values was calculated for parasites with identical genotypes. Comparison between groups was implemented with Mann-Whitney test. Proportions were compared by Chi square test or Fisher exact test depending on sample size at significance level of 0.05.
Results
pfcrt mutations
No mutations were detected on the loci 93, 97, 101, 145, 146, 158, 159, 343 and 350 among the 602 P. falciparum samples. Polymorphism on the locus 356 (I356T) was found in 54.7% (n = 326) of samples (mutants and mixed infection). More precisely, 276 isolates (45.8%) carried the wild type (I356) allele, 294 isolates (48.8%) carried the mutant allele (356T) and 32 isolates (5.4%) were mixed with both wild type and mutant allele (Table 1). Among the 578 samples tested for the 72–76 haplotype (24 were not genotyped successfully), the overall proportion of wild allele K76 was 74.9% (n = 433) versus 20.6% (n = 119) for mutant allele 76T and 4.5% (n = 26) for mixed populations (K76 and 76T) (Table 1). Details of I356T and K76T mutations per country are presented in Table 1. The majority of samples were from West African followed by Central African countries. Because other regions had low samples size, only West African and Central African samples were considered for further analysis. No association between K76T and I356T mutations was found for Central African isolates (n = 209, p = 0.1) while for West African isolates, there was a significant association found (n = 248, p = 0.008).
Drug susceptibility
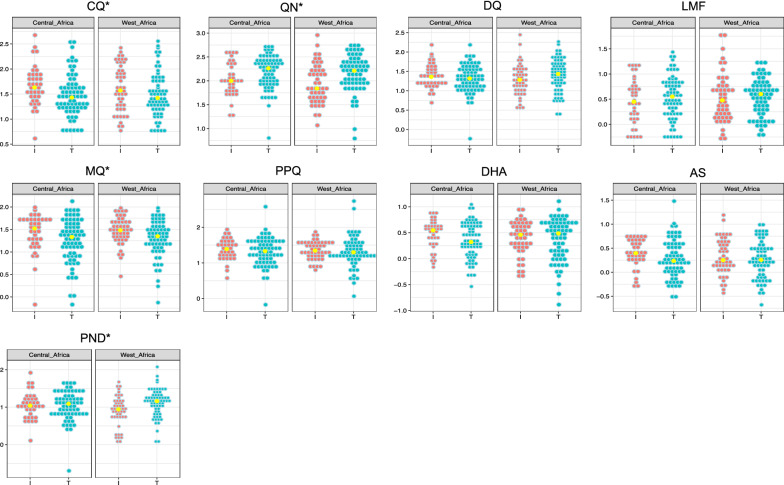
On the 602 samples collected, 296 isolates (117 from West Africa and 113 from Central Africa) were successfully evaluated in vitro. Values of IC50 estimates ranged from 0.7 nM to 544.0 nM for PPQ (mean = 27.9 nM), 4.1 nM to 474.2 nM for CQ (mean = 59.6 nM), 6.2 nM to 906.3 nM for QN (mean = 179.6 nM), 0.5 nM to 61.5 nM for LMF (mean = 5.2 nM), 2.5 nM to 185.1 nM for DQ (mean = 31.2 nM), 0.6 nM to 133.5 nM for MQ (mean = 30.5 nM), 0.2 nM to 119.3 nM for PND (mean = 16.4 nM), 0.2 nM to 18.5 nM for DHA (mean = 3.8 nM) and 0.2 nM to 64.0 nM for AS (mean = 2.6 nM) (Fig. 1). Based on the cut-off values for reduced in vitro susceptibility to CQ (100 nM), QN (800 nM), DQ (80 nM), MQ (30 nM), LMF (150 nM), PPQ (135 nM), (PND 60 nM), DHA and AS (10.5 nM) [36, 37], a proportion of samples had an in vitro reduced susceptibility to CQ (16.3%) and MQ (40.2%).
Fig. 1.
Dot plot of the IC50 values distribution of P. falciparum isolates from West Africa and Central Africa for chloroquine (CQ), quinine (QN), desethylamodiaquine (DQ), lumefantrine (LMF), mefloquine (MQ), piperaquine (PPQ), dihydroartemisinin (DHA), artesunate (AS) and pyronaridine (PND) according to I356T. Each dot represents the log10(IC50) for each isolate, red dots represent wild type I356 isolates (I), blue dots mutant 356T isolates (T) and yellow dots the log10(median IC50) value for each drug. The y-axis represents the log10(IC50) in nM. * Significant differences were found between wild type (I356) and mutant (356T) in West Africa (QN: p = 0.001, MQ: p = 0.01, PND: p = 0.005) and in Central Africa (QN: p = 0.02, MQ: p = 0.04, CQ: p = 0.01)
IC50 values were classified in 2 groups based on their genotype for the locus 356. The average parameters estimates of IC50 values for wild type (I356) and mutant allele (356T) for samples from West African and Central African countries are given in Table 2. There was a significant difference between the alleles I356 or 356T and IC50 median values of QN (p = 0.001) and MQ (p = 0.01) in West African group and QN (p = 0.02), CQ (p = 0.01), and MQ (p = 0.04) in Central Africa group (Table 2).
Table 2.
IC50 average parameters in nM for wild type (I356) and mutant (356T) P. falciparum isolates from West Africa and Central Africa for piperaquine (PPQ), quinine (QN), mefloquine (MQ), chloroquine (CQ), lumefantrine (LMF), desethylamodiaquine (DQ), pyronaridine (PND), dihydroartemisinin (DHA) and artesunate (AS)
| Drugs | West Africa | Central Africa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type I356 | Mutant 356T | p value | Wild type I356 | Mutant 356T | p value | |||||
| Median (no) | Min-Max | Median (no) | Min-Max | Median (no) | Min–Max | Median (no) | Min–Max | |||
| PPQ | 22.2 (49) | 6.8–76.3 | 19.9 (66) | 1.2–544 | 0.32 | 24.5 (36) | 4.1–79 | 21.4 (70) | 0.67–96.3 | 0.4 |
| QN | 69.1 (49) | 11.7–906.3 | 162.6 (68) | 6.2–590.5 | 0.001 | 98.2 (36) | 17.6–365.2 | 183.4 (73) | 28.9–538.4 | 0.02 |
| MQ | 30.4 (49) | 2.9–89.8 | 22.4 (67) | 0.8–100.6 | 0.01 | 33.3 (38) | 0.7–97.9 | 21.5 (72) | 0.62–133.9 | 0.04 |
| CQ | 37.4 (49) | 6.2–283.2 | 26.5 (68) | 5.5–359.5 | 0.1 | 42.8 (39) | 4.1–474.2 | 26.9 (73) | 5.79–325 | 0.01 |
| LMF | 3.0 (49) | 0.5–61.54 | 4.0 (68) | 0.6–18.2 | 0.6 | 2.8 (40) | 0.6–15.5 | 3.6 (73) | 0.55–22.4 | 0.5 |
| DQ | 19.2 (49) | 3.4–169.2 | 27.1 (68) | 2.5–185.1 | 0.07 | 22.8 (40) | 5.1–152.8 | 20.8 (73) | 4.7–152.9 | 0.4 |
| PND | 8.8 (48) | 1.2–50.1 | 14.5 (66) | 0.7–119.3 | 0.005 | 11.1 (38) | 3.9–41 | 12.3 (67) | 0.2–47.9 | 0.8 |
| DHA | 2.9 (49) | 0.4–8.3 | 3.1 (68) | 0.2–12.8 | 0.9 | 3.5 (40) | 0.7–7.7 | 2.1 (73) | 0.47–11.1 | 0.08 |
| AS | 1.8 (49) | 0.4–15.48 | 1.8 (67) | 0.2–9.8 | 0.6 | 2.5 (39 | 0.6–5.7 | 1.7 (71) | 0.29–30.2 | 0.05 |
West Africa: Benin, Burkina Faso, the Gambia, Ghana, Guinea, Ivory Coast, Niger, Mali, Nigeria and Togo
Central Africa: Cameroon, Central African Republic, Chad, Gabon and Republic of Congo
Italic p values refers to significant difference (p < 0.05)
Parasites harbouring the K76T mutation were significantly less susceptible to CQ (p < 0.0002) and DQ (p < 0.002) in Africa (Table 3). PPQ susceptibility was not significantly associated with the K76T mutation.
Table 3.
IC50 average parameters in nM for wild type (K76) and mutant (76T) P. falciparum isolates from West Africa and Central Africa for piperaquine (PPQ), quinine (QN), mefloquine (MQ), chloroquine (CQ), lumefantrine (LMF), desethylamodiaquine (DQ), pyronaridine (PND), dihydroartemisinin (DHA) and artesunate (AS)
| Drugs | West Africa | Central Africa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type K76 (n = 76) | Mutant 76T (n = 27) | p value | Wild type K76 (n = 82) | Mutant 76T (n = 14) | p value | |||||
| Median | Min–Max | Median | Min–Max | Median | Min–Max | Median | Min–Max | |||
| CQ | 23.2 | 5.5–229.6 | 152.6 | 7.9–381 | 4.295E−10 | 28.7 | 5.1–474.4 | 113.1 | 14.2–325 | 0.0002 |
| DQ | 19.3 | 2.5–185.1 | 46.2 | 6.5–169.2 | 0.0002 | 20.1 | 4.7–85.8 | 32.9 | 16.3–152.9 | 0.002 |
| QN | 138.6 | 6.2–906.3 | 163.8 | 36.7–586 | 0.2 | 143.9 | 28.9–538.4 | 180.1 | 17.6–503 | 0.52 |
| LMF | 3.9 | 0.5–31.6 | 3.5 | 0.5–61.5 | 0.3 | 3.1 | 0.5–27.3 | 4.9 | 0.6–14.2 | 0.758 |
| MQ | 25.2 | 0.8–89.8 | 26.2 | 2.9–76.7 | 0.8 | 23.7 | 1.04–86.6 | 40.9 | 0.7–92.3 | 0.11 |
| DHA | 2.9 | 0.2–12.8 | 2.8 | 0.5–8.2 | 0.9 | 2.9 | 0.5–18.5 | 2.9 | 0.7–6.5 | 0.78 |
| PND | 11.8 | 1.1–119.3 | 13.3 | 1.9–50.2 | 0.7 | 11.3 | 0.2–46.6 | 12.3 | 2.7–40.8 | 0.99 |
| AS | 1.8 | 0.3–64 | 2.1 | 0.4–7.3 | 0.8 | 1.9 | 0.3–30.2 | 3.0 | 0.6–5.9 | 0.36 |
| PPQ | 20.2 | 1.2–78.9 | 28.6 | 7.4–544 | 0.08 | 20.6 | 0.7–84.6 | 20.9 | 3.4–67.1 | 0.82 |
West Africa: Benin, Burkina Faso, Gambia, Ghana, Guinea, Ivory Coast, Niger, Mali, Nigeria and Togo
Central Africa: Cameroon, Central African Republic, Chad, Gabon and Republic of Congo
Italic p values refers to significant difference (p < 0.05)
Discussion
Understanding the genetic profile of drug resistance genes in P. falciparum malaria in endemic countries is essential. Knowing the predominant genotype circulating in a country or a region provide to policy makers a valuable information for the treatment regiments to be adopted depending on the predominant resistance genetic marker.
Mutations within the pfcrt haplotype 72–76 are known to confer resistance to chloroquine and other quinoline drugs, like amodiaquine and lumefantrine [1, 3, 38, 39]. As already documented many dozens of times, the present results show that the K76T mutation is associated with in vitro resistance to chloroquine and desethylamodiaquine and is an excellent molecular marker of resistance to chloroquine and amodiaquine for resistance monitoring in Africa. In contrast, the K76T mutation did not appear to be associated with in vitro reduced susceptibility to piperaquine (Table 3), confirming previous results [21, 38].
Previous studies found several new mutations in pfcrt associated with piperaquine reduced susceptibility, like the mutations T93S, H97Y, C101F, F145I, M343L and G353V in Cambodian parasites [22, 23, 25–29] and C350R in isolates from French Guiana [24] and Suriname [30]. However in studies on African P. falciparum samples, there was no report of identification of these mutations [39, 40]. None of these mutations was identified in the present 602 African isolates. These data suggest a very low prevalence of these mutations in African parasites, certainly due to the very low prevalence of P. falciparum parasites resistant to piperaquine in Africa. The present results suggest that susceptible parasites to piperaquine do not carry these mutations before selection pressure. The three isolates with piperaquine IC50 above the threshold of reduced susceptibility (135 nM) carried none of these mutations. Moreover, the absence of mutation on codon 93, 97, 101, 145, 146, 158, 159, 343 and 350 suggest that these mutations are not associated with in vitro resistance to amino-quinoline (chloroquine, amodiaquine) or amino-alcohol compounds (mefloquine) in Africa.
The mutation I356T found in 54.7% of isolates in the present work was already identified. The I356T mutation was previously reported at a rate of 24% in P. falciparum parasites from Malaysia [41]. In a local and global epidemiological study on global spread of mutant PfCRT, the I356T mutation was found in 12.6% of the P. falciparum isolates tested in Africa in 2011–2012 and mainly in the Gambia (78.7%) and the Democratic Republic of Congo (36.5%), and in 70.1% of isolates from Asia and from Thailand (99.2%) and Cambodia (67.7%) [42]. The I356T mutation was detected in 2.4% of isolates, as mixed infections (I356 and 356T alleles) collected before artemether-lumefantrine treatment in Uganda in 2014 [43]. This mutation was not found in P. falciparum positively isolates after artemether-lumefantrine treatment. A different amino acid substitution on the locus 356 (I356L) was also found in isolates from Latin America [1, 30], but this mutation was absent the present 602 African isolates. However, this mutation was not observed in isolates from Suriname [30].
The present study shows that the mutation I356T seems to be involved in quinine and mefloquine susceptibilities in African P. falciparum parasites. The mutation I356T is significantly associated with reduced susceptibility to quinine and increased susceptibility to mefloquine (Table 2). This mutation is also significantly associated with increased susceptibility to chloroquine but only in isolates from Central Africa. Additionally, this mutation is significantly associated with decreased susceptibility to pyronaridine but only in West African parasites. This mutation is not associated with piperaquine susceptibility, arguing against a role for residue 356 in modeling P. falciparum susceptibility to piperaquine. These data confirm the results of Dhingra et al. [42], that showed no difference in the survival of P. falciparum parasites with the FCB (I356) and Dd2 (356T) alleles in the presence of piperaquine. Additionally, the I356T mutation was not associated with ex vivo susceptibility to lumefantrine in the present 602 isolates according to previous data showing that this mutation was not selected in recrudescent P. falciparum parasites after artemether-lumefantrine [43].
Allelic frequencies of K76T and I356T in Central Africa show that there is no association with the two alleles. But on West African isolates, a significant association (p = 0.008) was found.
None of the mutations in PfCRT associated with in vivo piperaquine resistance in Asia or in vitro resistance in South America (T93S, H97Y, C101F, F145I, M343L, C350R and G353V) were found in the three isolates with ex vivo reduced susceptibility to piperaquine. All these isolates carried the I356T mutation and two the K76T mutation. Only one copy of pfpm2 was detected in the three isolates in a previous work [19]. But one of the weaknesses of the study is the low number of samples with reduced susceptibility to piperaquine. Further investigations are required to understand piperaquine resistance in Africa where dihydroartemisinin-piperaquine treatment remains highly effective [44].
Conclusion
None of the mutations in pfcrt found to be associated with piperaquine reduced susceptibility in Asia or South America (T93S, H97Y, C101F, F145I, M343L C350R and G353V) were found in the 602 African isolates including the three isolates with reduced susceptibility to piperaquine. The K76T mutation, involved in resistance to chloroquine and amodiaquine, and the 356T mutation were not associated with in vitro reduced susceptibility to piperaquine. Differences in mefloquine susceptibility between I356 and 356T isolates were, while statistically different, minimal. Further analyses are needed with a more important sample size from the same geographic area to confirm the role of the I356T mutation on quinine susceptibility.
Acknowledgements
The authors thank the patients and the staff of the hospitals of the French National Reference Center for Imported Malaria network.
The members of the French National Reference Centre for Imported Malaria Study Group are as follow: V Augis (Groupe Hospitalier Pellegrin, Bordeaux), P Bastien (Centre Hospitalier Universitaire de Montpellier, Montpellier), F Benoit-Vical (Centre Hospitalier Universitaire de Rangueil, Toulouse), A Berry (Centre Hospitalier Universitaire de Rangueil, Toulouse), P Brouqui (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), P Chauvin (Centre Hospitalier Universitaire de Rangueil, Toulouse), M Cividin (Centre Hospitalier du Pays d’Aix, Aix en Provence), F Courtier (Centre Hospitalier de Valence), P Delaunay (Centre Hospitalier Universitaire de l’Archet, Nice), L Delhaes (Groupe Hospitalier Pellegrin, Bordeaux), M Drancourt (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), N Dubosc (Centre Hospitalier Robert Boulin, Libourne), T Gaillard (Hôpital d’Instruction des Armées Desgenettes, Lyon), A Genin (Centre Hospitalier du Pays d’Aix, Aix en Provence), E Garnotel (Hôpital d’Instruction des Armées Laveran, Marseille), E Javelle (Hôpital d’Instruction des Armées Laveran, Marseille), C L’Ollivier (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), JC Lagier (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), E Ledault (Hôpital d’Instruction des Armées Laveran, Marseille), M Leveque (Centre Hospitalier Universitaire de Montpellier, Montpellier), D Malvy (Groupe Hospitalier Pellegrin, Bordeaux), P Marty (Centre Hospitalier Universitaire de l’Archet, Nice), G Ménard (Hôpital d’Instruction des Armées Saint-Anne, Toulon), E Menu (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), P Millet (Groupe Hospitalier Pellegrin, Bordeaux), P Minodier (Hôpital Nord, Marseille), P Parola (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), S Picot (Hôpital de la Croix Rousse, Lyon), C Pomares-Estran (Centre Hospitalier Universitaire de l’Archet, Nice), S Ranque (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), M-C Receveur (Groupe Hospitalier Pellegrin, Bordeaux), A Robin (Centre Hospitalier du Pays d’Aix, Aix en Provence), E Sappa (Centre Hospitalier du Pays d’Aix, Aix en Provence), H Savini (Hôpital d’Instruction des Armées Laveran, Marseille), J Sevestre (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), F Simon (Hôpital d’Instruction des Armées Laveran, Marseille), Y Sterkers (Centre Hospitalier Universitaire de Montpellier, Montpellier), C Surcouf (Hôpital d’Instruction des Armées Laveran, Marseille), E Varlet (Centre Hospitalier Universitaire de Montpellier, Montpellier), A Wolff (Hôpital d’Instruction des Armées Laveran, Marseille).
Abbreviations
- AS
Artesunate
- CQ
Chloroquine
- DHA
Dihydroartemisinin
- DNA
Deoxyribonucleic acid
- dNTP
Deoxynucleotide triphosphate
- DQ
Monodesethylamodiaquine
- ELISA
Enzyme linked immunosorbent assay
- HRP2
Histidine-rich protein 2
- IC50
50% inhibitory concentration
- LMF
Lumefantrine
- MQ
Mefloquine
- PCR
Polymerase chain reaction
- PfCRT
Plasmodium falciparum chloroquine resistance transporter
- PfK13
Plasmodium falciparum Kelch 13
- Pfpm2
Plasmodium falciparum plasmepsin 2 gene
- PND
Pyronaridine
- PPQ
Piperaquine
- QN
Quinine
- WHO
World Health Organization
Authors’ contributions
FTF, MG, NB and RA carried out the molecular studies. JM and IF carried out the ex vivo evaluation of anti-malarial drug susceptibility. BP, SW and MM conceived and coordinated the study. FTF, HB, MM and BP analysed the data and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the French Institute for Public Health Surveillance (Santé Publique France, grant CNR paludisme) and the Délégation Générale pour l’Armement (Grant No PDH-2-NRBC-4-B-4104. Francis Foguim was supported by the Foundation Méditerranée Infection.
Availability of data and materials
The datasets analysed in this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Informed consent was not required for this study because this work was performed under the statutory auspices of the French national reference centre for imported malaria, and isolates were anonymised by re-coding. Additionally, bio-banking of human clinical samples used for malaria diagnostics and secondary uses for scientific purposes are possible as long as the corresponding patients are informed and have not indicated any objections. This requirement was fulfilled here by giving verbal information to the patients, and no immediate or delayed patient opposition was reported to the hospital clinicians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bruno Pradines, Email: bruno.pradines@gmail.com.
The French National Reference Centre for Imported Malaria Study Group:
V. Augis, P. Bastien, F. Benoit-Vical, A. Berry, P. Brouqui, P. Chauvin, M. Cividin, F. Courtier, P. Delaunay, L. Delhaes, M. Drancourt, N. Dubosc, T. Gaillard, A. Genin, E. Garnotel, E. Javelle, C. L’Ollivier, J. C. Lagier, E. Ledault, M. Leveque, D. Malvy, P. Marty, G. Ménard, E. Menu, P Millet, P Minodier, P. Parola, S Picot, C. Pomares-Estran, S. Ranque, M. C. Receveur, A. Robin, E. Sappa, H. Savini, J. Sevestre, F. Simon, Y. Sterkers, C. Surcouf, E. Varlet, and A. Wolff
References
- 1.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nqoro X, Tobeka N, Aderibigbe BA. Quinoline-based hybrid compounds with antimalarial activity. Molecules. 2017;22:2268. doi: 10.3390/molecules22122268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 4.Gabryszewski SJ, Dhingra SK, Combrinck JM, Lewis IA, Callaghan PS, Hassett MR, et al. Evolution of fitness cost-neutral mutant PfCRT conferring P falciparum 4-aminoquinoline drug resistance is accompanied by altered parasite metabolism and digestive vacuole physiology. PLoS Pathog. 2016;12:e1005976. doi: 10.1371/journal.ppat.1005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin–piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leang R, Taylor WRJ, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 10.Phuc BQ, Rasmussen C, Duong TT, Dong LT, Loi MA, Ménard D, et al. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg Infect Dis. 2017;23:715–717. doi: 10.3201/eid2304.161872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thanh NV, Thuy-Nhien N, Tuyen NT, Tong NT, Nha-Ca NT, Dong LT, et al. Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J. 2017;16:27. doi: 10.1186/s12936-017-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foguim Tsombeng F, Gendrot M, Robert MG, Madamet M, Pradines B. Are k13 and plasmepsin II genes, involved in Plasmodium falciparum artemisinin derivatives and piperaquine resistance in Southeast Asia, reliable to monitor resistance surveillance in Africa? Malar J. 2019;18:285. doi: 10.1186/s12936-019-2916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakolwa MA, Mahende MK, Ishengoma DS, Mandara CI, Ngasala B, Kamugisha E, et al. Efficacy and safety of artemisinin-based combination therapy and molecular markers for artemisinin and piperaquine resistance in Mailand Tanzania. Malar J. 2018;17:369. doi: 10.1186/s12936-018-2524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo G, L’Episcopia M, Menegon M, Souza SS, Dongho BGD, Vullo V, et al. Dihydroartemisinin-piperaquine treatment failure in uncomplicated Plasmodium falciparum malaria case imported from Ethiopia. Infection. 2018;46:867–870. doi: 10.1007/s15010-018-1174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malvy D, Torrentino-Madamet M, L’Ollivier C, Receveur MC, Jeddi F, Delhaes L, et al. Plasmodium falciparum recrudescence two years after treatment of an uncomplicated infection without return to an area where malaria is endemic. Antimicrob Agents Chemother. 2018;62:e01892-17. doi: 10.1128/AAC.01892-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen SA, Ceja FG, Conrad MD, Tumwebase PK, Byaruhanga O, Katairo T, et al. Changing antimalarial drug sensitivities in Uganda. Antimicrob Agents Chemother. 2017;61:e01516–e01517. doi: 10.1128/AAC.01516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert MG, Foguim Tsombeng F, Gendrot M, Mosnier J, Amalvict R, Benoit N, et al. Absence of a high level of duplication of the Plasmepsin II gene in Africa. Antimicrob Agents Chemother. 2018;62:e00374-18. doi: 10.1128/AAC.00374-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robert MG, Foguim Tsombeng F, Gendrot M, Diawara S, Madamet M, Kounta MB, et al. Baseline ex vivo and molecular responses of Plasmodium falciparum isolates to piperaquine before implementation of dihydroartemisinin-piperaquine in Senegal. Antimicrob Agents Chemother. 2019;63:e02445-18. doi: 10.1128/AAC.02445-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascual A, Madamet M, Bertaux L, Amalvict R, Benoit N, Travers D, et al. In vitro piperaquine susceptibility is not associated with the Plasmodium falciparum chloroquine resistance transporter gene. Malar J. 2013;12:431. doi: 10.1186/1475-2875-12-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhingra SK, Redhi D, Combrinck JM, Yeo T, Okombo J, Henrich PP, et al. A variant PfCRT isoform can contribute to Plasmodium falciparum resistance to the first-line partner drug piperaquine. MBio. 2017;8:e00303–e00317. doi: 10.1128/mBio.00303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, et al. Association of a novel mutation in the Plasmodium falciparum chloroquine resistance transporter with decreased piperaquine sensitivity. J Infect Dis. 2017;216:468–476. doi: 10.1093/infdis/jix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kümpornsin K, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun. 2018;9:3314. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhingra SK, Small-Saunders JL, Ménard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect Dis. 2019;19:1168–1169. doi: 10.1016/S1473-3099(19)30543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton WL, Amato R, van der Pluijm RW, Jacobs CG, Quang HH, Thuy-Nhien NT, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chenet SM, Okoth SA, Kelley J, Lucchi N, Huber CS, Vreden S, et al. Molecular profile of malaria drug resistance markers of Plasmodium falciparum in Suriname. Antimicrob Agents Chemother. 2017;61:e02655-16. doi: 10.1128/AAC.02655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gendrot M, Foguim FT, Robert MG, Amalvict R, Mosnier J, Benoit N, et al. The D113N mutation in the RING E3 ubiquitin protein ligase gene is not associated with ex vivo susceptibility to common anti-malarial drugs in African Plasmodium falciparum isolates. Malar J. 2018;17:108. doi: 10.1186/s12936-018-2252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogreau H, Renaud F, Bouchiba H, Durand P, Assi SB, Henry MC, et al. Genetic diversity and structure of African Plasmodium falciparum populations in urban and rural areas. Am J Trop Med Hyg. 2006;74:953–959. [PubMed] [Google Scholar]
- 34.Henry M, Diallo I, Bordes J, Ka S, Pradines B, Diatta B, et al. Urban malaria in Dakar, Senegal: chemosusceptibility and genetic diversity of Plasmodium falciparum isolates. Am J Trop Med Hyg. 2006;75:146–151. [PubMed] [Google Scholar]
- 35.Anderson TJC, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 36.Pascual A, Madamet M, Briolant S, Gaillard T, Amalvict R, Benoit N, et al. Multinormal in vitro distribution of Plasmodium falciparum susceptibility to piperaquine and pyronaridine. Malar J. 2015;14:49. doi: 10.1186/s12936-015-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pradines B, Bertaux L, Pomares C, Delaunay P, Marty P. Reduced in vitro susceptibility to artemisinin derivatives associated with multi-resistance in a traveller returning from South-East Asia. Malar J. 2011;10:268. doi: 10.1186/1475-2875-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol. 2006;6:309–314. doi: 10.1016/j.meegid.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Basco LK. Molecular epidemiology of malaria in Cameroon. XIII. Analysis of pfcrt mutations and in vitro chloroquine resistance. Am J Trop Med Hyg. 2002;67:388–391. doi: 10.4269/ajtmh.2002.67.388. [DOI] [PubMed] [Google Scholar]
- 41.Atroosh WM, Al-Mekhlafi HM, Mahdy MAK, Surin J. The detection of pfcrt and pfmdr1 point mutations as molecular markers of chloroquine drug resistance, Pahang, Malaysia. Malar J. 2012;11:251. doi: 10.1186/1475-2875-11-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhingra SK, Gabryszewski SJ, Small-Saunders JL, Yeo T, Henrich PP, Mok S, et al. Global spread of mutant PfCRT and its pleitropic impact on Plasmodium falciparum multidrug resistance and fitness. MBio. 2019;10:e02731-18. doi: 10.1128/mBio.02731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balikagala B, Mita T, Ikeda M, Sakurai M, Yatsushiro S, Takahashi N, et al. Absence of in vivo selection for k13 mutations after artemether-lumefantrine treatment in Uganda. Malar J. 2017;16:23. doi: 10.1186/s12936-016-1663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West African Network for Clinical Trials of Antimalarial Drugs (WANECAM) Pyronaridine-artesunate or dihydroartemisinin-piperaquine versus current first-line therapies for repeated treatment of uncomplicated malaria: a randomised, multicentre, open-label, longitudinal, controlled, phase 3b/4 trial. Lancet. 2018;391:1378–1390. doi: 10.1016/S0140-6736(18)30291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed in this study are available from the corresponding author on reasonable request.