Abstract
Background
Anthocyanins, which have important biological functions and have a beneficial effect on human health, notably account for pigmentation in purple-fleshed sweet potato tuberous roots. Individual regulatory factors of anthocyanin biosynthesis have been identified; however, the regulatory network of anthocyanin biosynthesis in purple-fleshed sweet potato is unclear.
Results
We functionally determined that IbMYB340 cotransformed with IbbHLH2 in tobacco and strawberry receptacles induced anthocyanin accumulation, and the addition of IbNAC56a or IbNAC56b caused increased pigmentation. Furthermore, we confirmed the interaction of IbMYB340 with IbbHLH2 and IbNAC56a or IbNAC56b via yeast two-hybrid and firefly luciferase complementation assays; these proteins could form a MYB340-bHLH2-NAC56a or MYB340-bHLH2-NAC56b transcriptional complex to regulate anthocyanin biosynthesis by binding to the IbANS promoter rather than the IbUFGT promoter. Furthermore, it was found by a transient expression system in tobacco leaves that IbMYB44 could decrease anthocyanin accumulation. Moreover, the interaction of IbMYB44 with IbMYB340 and IbNAC56a or IbNAC56b was verified. This result suggested that IbMYB44 acts as a repressor of anthocyanin in sweet potato.
Conclusions
The repressor IbMYB44 affected anthocyanin biosynthesis by competitively inhibiting the IbMYB340-IbbHLH2-IbNAC56a or IbMYB340-IbbHLH2-IbNAC56b regulatory complex formation. Overall, the present study proposed a novel regulatory network whereby several vital TFs play key roles in regulating anthocyanin biosynthesis, and it provides strong insight into the potential mechanism underlying anthocyanin biosynthesis in sweet potato tuberous roots with purple color.
Keywords: Sweet potato (Ipomoea batatas), Anthocyanin biosynthesis, IbMYB340, IbMYB44, IbNAC56, Regulatory complex, Repressor
Background
Sweet potato (Ipomoea batatas) is well known for its abundant nutritional value and is the fourth most important crop species in China [1, 2]. The colors of its storage roots mainly include white, yellow, orange and purple. Among the different colored sweet potato types, purple-fleshed sweet potato is widely popular for its high anthocyanin contents in its storage roots. Anthocyanin is now recognized as the most important secondary metabolite in plants and has important biological functions, including disease resistance, UV radiation protection and defense against herbivores and pathogens [3, 4]. Moreover, the addition of anthocyanins in human recipes has a beneficial effect on the prevention of cancer and diabetes and on cardiovascular and neuronal illnesses [5].
The anthocyanin biosynthesis pathway has been extensively documented in different plant species and includes several key structural genes, such as phenylalanine ammonia lyase (PAL), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT) and glutathione S-transferase (GST) [6–8]. Transcription factors, especially the MBW complex, have been verified to be involved in synergistically regulating anthocyanin synthesis [9]. Among them, R2R3-MYBs particularly make a major contribution to anthocyanin biosynthesis [10]. For example, AtMYB75, AtMYB113 and AtMYB114 in Arabidopsis [11]; MdMYB10 and MdMYB110a in apple [12, 13]; PyMYB10 and PyMYB114 in pear [14, 15]; LcMYB5 in litchi [16]; PaMYB10 in apricot [17]; and FaMYB10 in strawberry [18] have been reported to be involved in anthocyanin biosynthesis as activators. In addition to MYB activators, R3-MYB and some R2R3-MYB repressors, including the chrysanthemum CmMYB#7 (an R3-MYB), strawberry FaMYB1 and FaMYB44.1, peach PpMYB18 and potato StMYB44 (R2R3-MYBs) ones, have also been identified as participating in the flavonoid biosynthetic pathway [19–22].
In addition to MYB TFs, other TFs are also involved in anthocyanin biosynthesis. NAC (NAM, ATAF1/2, and CUC2) TFs have also been widely documented to participate in multiple biological processes, such as plant development, disease resistance, and the abiotic stress response [23, 24]. For secondary metabolism, many NAC TFs have been verified to be involved in the phenylpropanoid pathway, thereby regulating lignin biosynthesis [25, 26]; it was reported that ANAC032 acts as a repressor of anthocyanin accumulation in Arabidopsis thaliana during stress conditions [24]. Moreover, ANAC078 and PpBL are characterized as activators of anthocyanin biosynthesis under certain conditions [27, 28].
In purple-flesh sweet potato, individual regulatory factors of anthocyanin biosynthesis have been identified [29, 30]. IbMYB1 can promote the expression of anthocyanin-related genes specifically in purple-fleshed sweet potato [30]. Another factor, IbWD40, was documented to have a positive correlation with anthocyanin contents in different sweet potato cultivars, suggesting that IbWD40 plays a key role in anthocyanin biosynthesis in purple-fleshed sweet potato [29]. However, additional evidence could not be provided due to the slow process brought forth by the complexity of the sweet potato genome, which is hexaploid (2n = 6x = 90) and highly polymorphic [31]. Thus, whether and how other NAC TFs and repressor MYBs participate in anthocyanin biosynthesis are unclear, and the molecular regulatory network of anthocyanin biosynthesis in purple-fleshed tuberous roots has rarely been reported.
In this study, four candidate TFs, IbMYB340, IbMYB44, IbNAC56a and IbNAC56b, were screened by bioinformatics and RT-qPCR analysis in sweet potatoes of different colors, and the gene functions were identified by transient expression assays in tobacco and strawberry receptacles. Further analysis indicated that IbMYB340 contributes greatly to anthocyanin accumulation and could also form a regulatory complex with cofactors IbbHLH2, IbNAC56a and IbNAC56b to regulate anthocyanin biosynthesis by binding to the IbANS promoter, rather than the IbUFGT promoter, whereas IbMYB44 acts as a negative regulator and could interact with IbMYB340 and IbNAC56a or with IbNAC56b to regulate anthocyanin biosynthesis in sweet potato, as reflected by yeast two-hybrid and firefly luciferase complementation assays. Our study reveals a possible underlying mechanism of anthocyanin biosynthesis in purple-fleshed sweet potato, which will provide us with a potential understanding of the regulatory network of sweet potato roots with different colors.
Results
Characterization of IbMYB and IbNAC anthocyanin-related genes in sweet potato
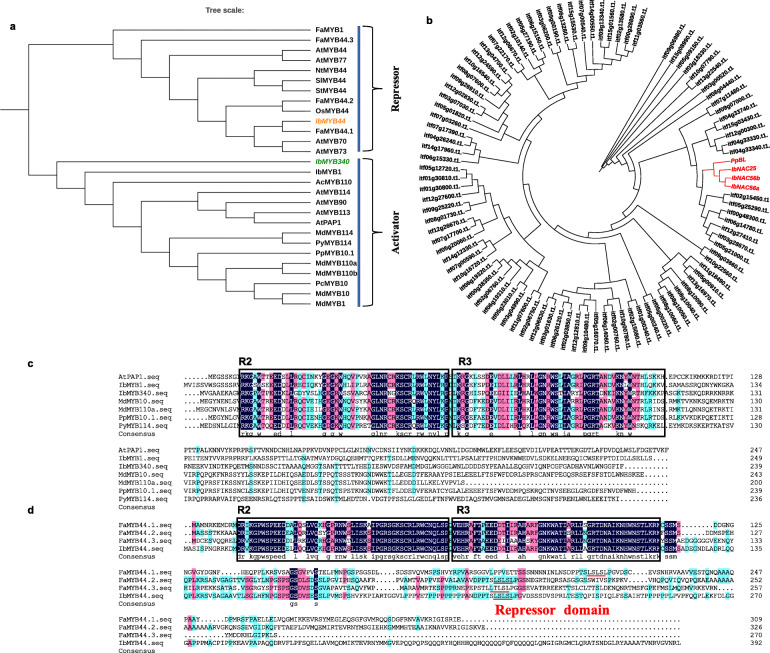
In previous reports, TFs AtMYB75 (AtPAP1) in Arabidopsis thaliana and MdMYB10 (DQ267897) and PyMYB114 (ASY06612.1) in non-model species were found to be R2R3-type MYBs, which may work together with other TFs in regulating anthocyanin biosynthesis [11, 12, 15]. Here, we found that the Itf12g05820.t1 gene was highly homologous with AtPAP1 (AT1G56650) by BLAST alignment against the sweet potato genome database. It was named IbMYB340 after multiple sequence alignment revealed its similarity with other typical R2R3-MYB TFs in other species. In addition, IbMYB44 was identified from FaMYB44.1 by homologous sequence alignment; FaMYB44 is a transcriptional repressor that negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10 [19]. Phylogenetic analysis indicated that IbMYB340 belongs to the activator clade, together with other known MYB transcription factors with the R2R3 domain (Fig. 1a). By contrast, TF IbMYB44, which harbors the transcriptional repressor domain LxLxL (Fig. 1d), was phylogenetically related to FaMYB44.1 (Fig. 1a), which acts as a transcriptional repressor in strawberry [19]. Thus, we speculated that these two MYB TFs may regulate anthocyanin biosynthesis in sweet potato.
Fig. 1.
Phylogenetic analysis and multiple sequence alignments of NACs and MYBs. a Phylogenetic tree derived from the amino acid sequences of R2R3-MYBs in sweet potato and other species. IbMYB340 and IbMYB44 are marked by orange or green letters, respectively. The analysis of evolutionary history inferred using the neighbor-joining method was conducted by MEGA 7. b Phylogenetic analysis of 98 IbNAC genes in sweet potato and other species. In addition, IbNAC25, IbNAC56a, IbNAC56b and PpBL are highlighted with red letters. c-d The protein sequences of R2R3-MYBs in sweet potato and other species were used to perform the sequence alignment. In addition, the R2 and R3 domains are labeled, and the LxLxL negative repressor motif is highlighted with a red underscore
We retrieved sequence information for 98 NAC TFs containing a NAC domain in the N-terminal region from the sweet potato genome database. Amino acid sequences of NACs in sweet potato and peach PpBL (ALK27819.1), which can promote the activity of PpMYB10.1 by acting as a heterodimer with PpNAC1, resulting in anthocyanin accumulation in tobacco leaves [28], were used to construct a phylogenetic tree (Fig. 1b). Three NAC genes, IbNAC56a, IbNAC56b, and IbNAC25, were the candidates according to the phylogenetic analysis results.
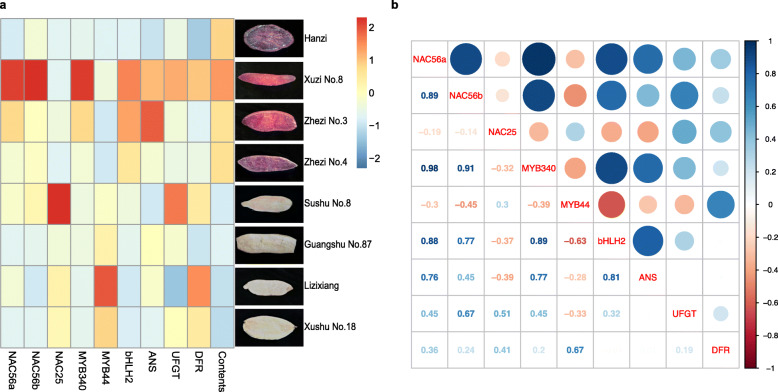
Then, we quantified the transcript abundance of candidate genes in tuberous roots of different sweet potato cultivars, and the sectional drawings for tuberous roots of different cultivars sweet potatoes were shown in Fig. 2a. The IbNAC56a and IbNAC56b expression levels were significantly higher in purple-fleshed sweet potato cultivars ‘Xuzi No. 8’ and ‘Zhezi No. 3’ than in yellow-fleshed sweet potatoes ‘Sushu No. 8’ and ‘Guangshu No. 87’ or in white-fleshed sweet potatoes ‘Lizixiang’ and ‘Xushu No. 18’, whereas the expression pattern of IbNAC25 was the opposite. Therefore, IbNAC56a and IbNAC56b were selected for subsequent experiments (Fig. 2a).
Fig. 2.
a The expression levels of IbNAC56a, IbNAC56b, IbNAC25, IbMYB340, IbMYB44, IbbHLH2, IbANS, IbUFGT and IbDFR, and anthocyanin contents in the tuberous roots of different sweet potato cultivars (‘Hanzi’, ‘Xuzi No. 8’, ‘Zhezi No. 3’, ‘Zhezi No. 4’, ‘Sushu No. 8’, ‘Guangshu No. 87’ and ‘Lizixiang’, ‘Xushu No. 18’). b Correlation analysis among the gene expression levels of IbNAC56a, IbNAC56b, IbNAC25, IbMYB340, IbMYB44, IbbHLH2, IbANS, IbUFGT and IbDFR. Pearson’s correlation coefficients for the data were analyzed using R scripts
Moreover, the transcript abundance of IbMYB340 and IbbHLH2 was significantly higher in ‘Xuzi No. 8’ and ‘Zhezi No. 3’ than in the other cultivars, while the expression of IbMYB44 was maintained at lower levels in purple-fleshed sweet potatoes than in yellow−/white-fleshed sweet potatoes. Furthermore, ‘Lizixiang’ and ‘Xushu No. 18’ presented higher transcript levels of IbMYB44 than did ‘Sushu No. 8’ and ‘Guangshu No. 87’. The expression level of IbANS was also higher in the purple-fleshed cultivars except for ‘Hanzi’, while the yellow−/white-fleshed cultivars showed relatively low gene expression. However, the expression of IbUFGT and IbDFR showed no obvious difference (Fig. 2a).
Further, we analyzed the correlation among anthocyanin biosynthesis-related gene expression levels in the different sweet potato cultivars. As shown in Fig. 2c, the expression of IbNAC56a, IbNAC56b, IbbHLH2 and IbANS displayed noticeably positive correlations with IbMYB340 expression, and the correlation coefficients ranged from 0.77 to 0.98. However, the expression of IbMYB44 had negative correlations with the expression of IbMYB340, IbNAC56a, IbNAC56b, IbbHLH2, IbANS and IbUFGT, suggesting that there might be an opposite effect of IbMYB340 and IbMYB44. In addition, IbNAC25 was negatively correlated with other factors (except IbMYB44, IbDFR and IbUFGT).
Anthocyanin was induced by cotransformingIbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b in tobacco leaves
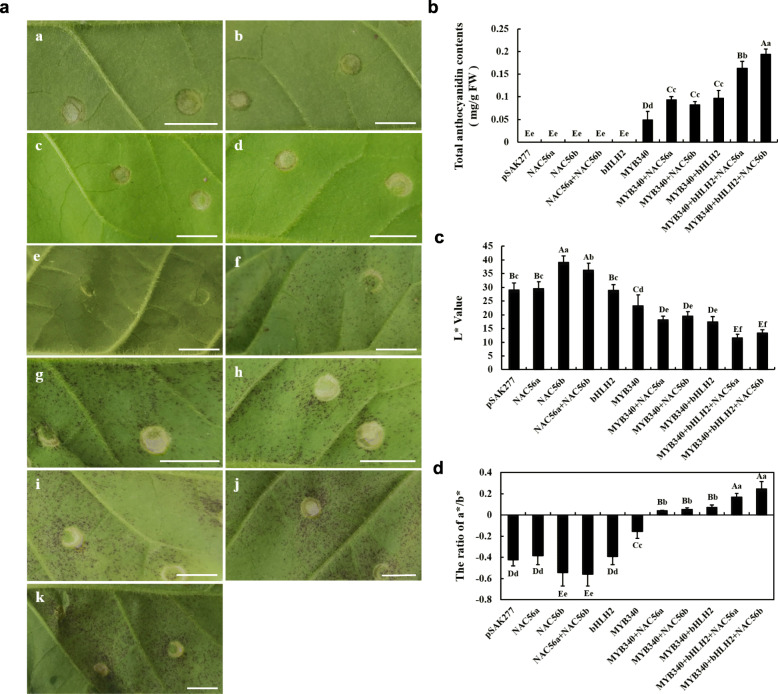
The results of a functional analysis of IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b using a transient expression assay in tobacco leaves are shown in Fig. 3a. No anthocyanin accumulation was detected at the injection areas of tobacco leaves when transformed with IbNAC56a, IbNAC56b or IbbHLH2 alone, while slight pigmentation appeared with IbMYB340 injected alone. However, an enhanced color was visible at injection regions 7 day after transformation with both IbMYB340 and IbNAC56a/IbNAC56b or IbbHLH2. When IbNAC56a or IbNAC56b was cotransformed with IbMYB340 and IbbHLH2, obvious intense pigmentation was detected. The quantification of total anthocyanin contents indicated that anthocyanin biosynthesis was induced by cotransformation of three TFs, which was more than that from two TFs or IbMYB340 alone, whereas tobacco leaves did not show any visible anthocyanin accumulation after transformation with the empty vector or with IbNAC56a or IbNAC56b alone (Fig. 3b). We next analyzed the color of the tobacco leaves. The L* values apparently declined when the pigmentation appeared; by contrast, the a*/b* ratio significantly increased (Fig. 3c-d).
Fig. 3.
Functional analysis of the IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b genes using a transient expression assay in tobacco leaves. a The phenotype of tobacco leaves on the 7th day after infiltration: a, pSAK277; b, IbNAC56a; c, IbNAC56b; d, IbNAC56a + IbNAC56b; e, IbbHLH2; f, IbMYB340; g, IbMYB340 + IbNAC56a; h, IbMYB340 + IbNAC56b; i, IbMYB340 + IbbHLH2; j, IbMYB340 + IbbHLH2 + IbNAC56a; and k, IbMYB340 + IbbHLH2 + IbNAC56b. Scale bar = 1 cm. b Total anthocyanin contents in transformed leaves of tobacco. FW, fresh weight. c-d The color parameter L* and a*/b* ratio values were measured with a Minolta Chroma Meter and were used to indicate phenotypic changes. The values are presented as the means ± SDs (n = 3), and the uppercase and lowercase letters represent significant differences at P < 0.01 or P < 0.05, respectively
Heterologous overexpression of IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b induces anthocyanin biosynthesis in strawberry receptacles
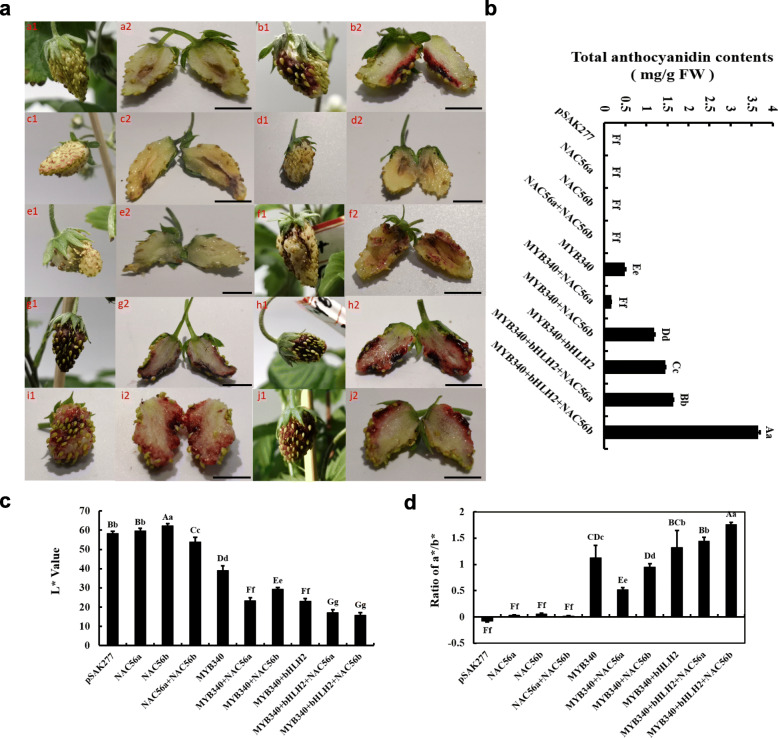
To further confirm the roles of IbMYB340 and IbNAC56a or IbNAC56b in anthocyanin biosynthesis, we cotransformed IbMYB340 and IbNAC56a or IbNAC56b in strawberry receptacles via agroinfiltration, and diploid strawberry (Fragaria vesca) ‘Yellow Wonder’ 5AF7 was used for the transient transformation experiment. The transient expression assays in strawberry receptacles showed that coinfiltration of IbMYB340 with any of the other candidate TFs resulted in apparent pigmentation accumulation 7 day after transformation, while the coinfiltration of three TFs caused deeper red pigmentation than did two TFs or IbMYB340 alone. However, no pigmentation was observed when the empty vector was infiltrated; IbNAC56a or IbNAC56b were infiltrated separately; or IbNAC56a and IbNAC56b were coinfiltrated. In addition, pigmentation was also visible in the injection region of IbMYB340 alone (Fig. 4a). Measurements of the induced anthocyanin are shown in Fig. 4b. The total anthocyanin contents of the three TFs cotransformed were significantly higher than those when the two TFs or one TF alone were cotransformed. The L* and a*/b* values of the injection regions of the strawberry receptacles declined or increased sharply when pigmentation appeared and stayed at significantly relatively low or high levels, respectively (Fig. 4c-d). Heterologous overexpression systems in strawberry receptacles presented phenotypic changes similar to those presented in the tobacco leaves in this study. We observed great anthocyanin pigmentation when IbMYB340 was cotransformed with IbbHLH2 and IbNAC56a or IbNAC56b together.
Fig. 4.
Transient expression of IbMYB340 and IbNACs in strawberry receptacles. a The phenotype of strawberry receptacles: a, pSAK277; b, IbMYB340; c, IbNAC56a; d, IbNAC56b; e, IbNAC56a + IbNAC56b; f, IbMYB340 + IbNAC56a; g, IbMYB340 + IbNAC56b; h, IbMYB340 + IbbHLH2; i, IbMYB340 + IbbHLH2 + IbNAC56a; and j, IbMYB340 + IbbHLH2 + IbNAC56b. Scale bar = 1.5 cm. b Anthocyanin contents in transformed strawberry fruits. FW, fresh weight. c-d The color parameter L* and a*/b* ratio values indicate phenotypic changes. The values are presented as the means ± SDs (n = 3), and the uppercase and lowercase letters represent significant differences at P < 0.01 or P < 0.05, respectively
Expression patterns of anthocyanin-related genes in strawberry receptacles
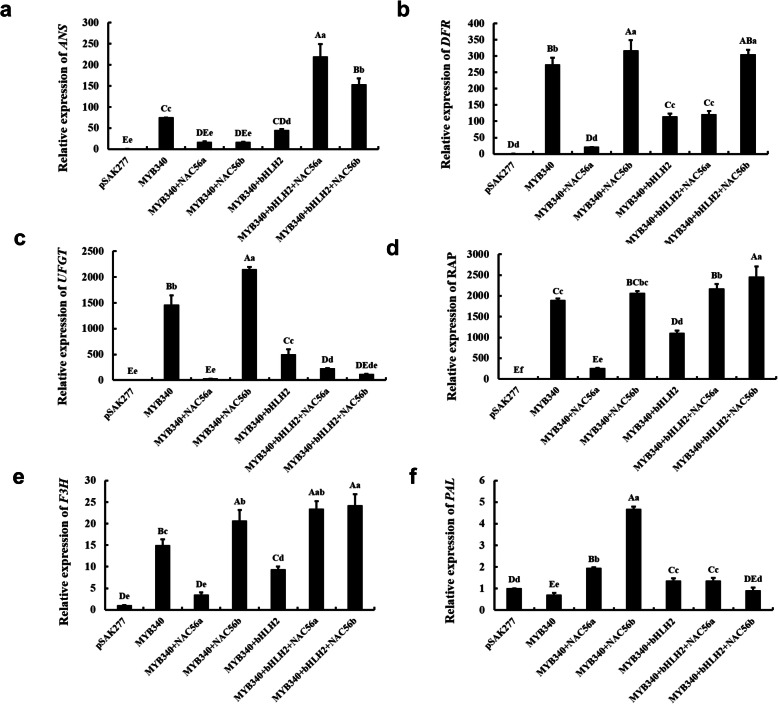
To explore the regulatory models of the TFs IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b, several critical anthocyanin-related genes, FvPAL, FvF3H, FvANS, FvDFR, FvUFGT and FvRAP were analyzed in induced-color strawberry receptaclesby RT-qPCR. FvANS expression was greatly increased when IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b were cotransformed (P < 0.01, Fig. 5a). Interestingly, the transformation of IbMYB340 alone showed a higher FvANS expression level than did cotransformation of IbMYB340 and of IbbHLH2 and IbNAC56a or IbNAC56b (P < 0.01 or P < 0.05, respectively). For FvDFR, transformation of TFs, including IbNAC56b or IbMYB340 alone, resulted in noticeably gene expression levels that were higher than those from the other TF combinations (P < 0.01), while the additional IbbHLH2 with IbMYB340 and IbNAC56a cotransformation caused an obvious increase in FvDFR expression levels (P < 0.01, Fig. 5b). Figure 5d-e illustrates similar changes in the expression levels of FvRAP and FvF3H; these two genes were highly expressed in the cotransformation with different TFs or IbMYB340 alone, except for IbMYB340 + IbNAC56a (P < 0.01). In addition to IbMYB340 + IbNAC56a, IbMYB340 + IbbHLH2 also maintained a relatively low expression level of FvRAP and FvF3H compared with that of other cotransformation types, excluding the empty vector pSAK277 (P < 0.01). From Fig. 5c, we can see that FvUFGT was significantly expressed in response to the cotransformation of IbMYB340 + IbNAC56b or IbMYB340 alone (P < 0.01), whereas other cotransformation types seemed not to activate the transcription of FvUFGT. In addition, it seemed that FvPAL could not be activated by different cotransformation types except for the cotransformation of IbMYB340 and IbNAC56b (Fig. 5f). As mentioned above, almost all the genes involved in the anthocyanin biosynthetic pathway and vacuolar transport were highly expressed in the abovementioned cotransformed strawberries, especially when IbMYB340, IbbHLH2, and IbNAC56a or IbNAC56b were cotransformed. This result suggested that the IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b genes might regulate or coregulate anthocyanin synthesis by forming a regulatory complex.
Fig. 5.
Expression levels of anthocyanin biosynthesis- and vacuolar transport-related genes in induced-color strawberry receptacles from Fig. 4. a-f: FvANS, FvDFR, FvUFGT, FvRAP, FvF3H, and FvPAL. The values are presented as the means ± SDs (n = 3), and the uppercase and lowercase letters represent significant differences at P < 0.01 or P < 0.05, respectively
The regulatory complex MYB340-bHLH2-NAC56 promotes anthocyanin biosynthesis by binding to the IbANS promoter
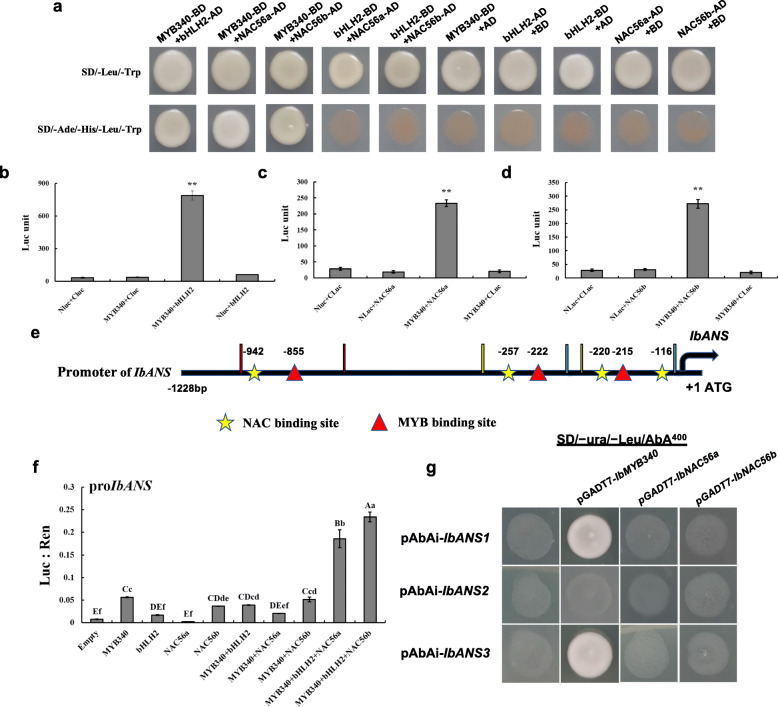
We speculated that the products of the IbMYB340, IbbHLH2, IbNAC56a and IbNAC56b genes might form regulatory complexes to regulate anthocyanin synthesis. As such, the possible interactions of IbMYB340 with IbbHLH2, IbNAC56a and IbNAC56b were studied via Y2H assays. First, we found that cotransformed yeast cells harboring pGADT7 and IbMYB340 failed to grow on SD − Trp/−Leu/−His/−Ade media, indicating that IbMYB340 could not activate downstream gene expression in yeast by itself. As a result, strong growth on SD-Trp-Leu-His-Ade + AbA media was observed when we cotransformed the complete amino acid sequence of IbMYB340 with IbbHLH2, IbNAC56a or IbNAC56b. However, the Y2H assays showed no interaction between IbbHLH2 and IbNAC56a or IbNAC56b (Fig. 6a). These results suggest that IbMYB340 can interact in yeast with IbbHLH2, IbNAC56a and IbNAC56b.
Fig. 6.
Interaction verification of IbMYB340 with IbbHLH2, IbNAC56a or IbNAC56b and coregulatory site evaluation of the IbANS promoter. a Verification of the interaction of IbMYB340 with IbbHLH2, IbNAC56aor IbNAC56b via Y2H assays. b-d Verification of the interaction between IbMYB340 and IbNAC56aor IbNAC56b via firefly luciferase complementation assays. ** indicates a significant difference at the P < 0.01 level. e Schematic of the IbANS promoter. The prediction of the cis-acting elements in the 1228-bp promoter region of IbANS was performed using the PlantCare database [32]. f Validation of the activation effect by cotransformation of IbMYB340, IbbHLH2, and IbNAC56 on the IbANS promoter using a dual-luciferase assay in tobacco leaves. The ratio of Luc to Ren indicates that TFs activate the promoter activity of IbANS. g Y1H assays for IbMYB340, IbNAC56a, and IbNAC56b for the IbANS promoter. The prey vectors pGADT7-IbMYB340/IbNAC56a/IbNAC56b were transformed into Y1H Gold cells harboring pAbAi-IbANS1/2/3 and then tested on SD/−Ura/−Leu/AbA400 plates
Additionally, we further validated the results obtained in yeast via firefly luciferase complementation assays in tobacco. Coexpression of NLuc-IbMYB340 and CLuc-IbbHLH2 or IbNAC56a or IbNAC56b vectors reversed the intense luciferase enzyme activity. In contrast, we detected no apparent luciferase enzyme activity in any of the control groups, including the groups with Nluc-IbMYB340 with CLuc and CLuc-IbbHLH2/−IbNAC56a/−IbNAC56b with NLuc (Fig. 6b-d). Taken together, these results indicated that IbMYB340 can interact with IbNAC56a and IbNAC56b in tobacco leaves. These results were in agreement with the results of the yeast two-hybrid assay.
We next tested the transactivation activity of the candidate TFs with the IbANS promoter via dual-luciferase reporter assays (Fig. 6f). Cotransformation of IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b showed more transactivation on the IbANS promoter than did IbMYB340 cotransformation with IbbHLH2 and IbNAC56a or IbNAC56b. However, it seemed that cotransformation of IbMYB340 and any other TFs or IbMYB340 alone made no difference, while the transformation of IbbHLH2 or IbNAC56b slightly promoted the activity (Fig. 6f). Furthermore, Y1H assays were performed to demonstrate whether the IbANS promoter region is bound directly by IbMYB340 and IbNAC56a or IbNAC56b. Promoter structure analysis revealed multiple cis-regulatory elements, including a MYB motif (T/CAACCA) and a NAC-binding site (CACG) (Fig. 6e). In this assay, we transformed pGADT7-IbMYB340/IbNAC56a/IbNAC56b prey vectors into Y1H Gold cells harboring pAbAi-IbANS1/2/3 bait vectors and tested them on SD/−Ura/−Leu/AbA plates. The transformants coexpressing the prey vectors pGADT7-IbMYB340 and pAbAi-IbANS1/3 were grown on SD/−Ura/−Leu/AbA400 plates, while the pAbAi-IbANS1/2/3 bait vectors could not grow on SD/−Ura/AbA400 plates, suggesting that IbMYB340 was capable of binding to the 1st (− 956 bp to − 755 bp) and 3rd (− 310 bp to − 105 bp) IbANS promoter fragments rather than the 2nd IbANS promoter fragment (Fig. 6g). However, we found no direct association between IbNAC56a or IbNAC56b and the promoter of IbANS, although several NAC-binding sites were located in different individual promoter regions (Fig. 6g). Taken together, these results indicated that the different regulatory complexes (MYB340-bHLH2-NAC56a and MYB340-bHLH2-NAC56b) directly activated the expression of IbANS by binding to the MYB motif element.
IbMYB44 suppresses anthocyanin accumulation by competitively inhibiting the regulatory complex formation of IbMYB340-IbbHLH2-IbNAC56a or IbNAC56b
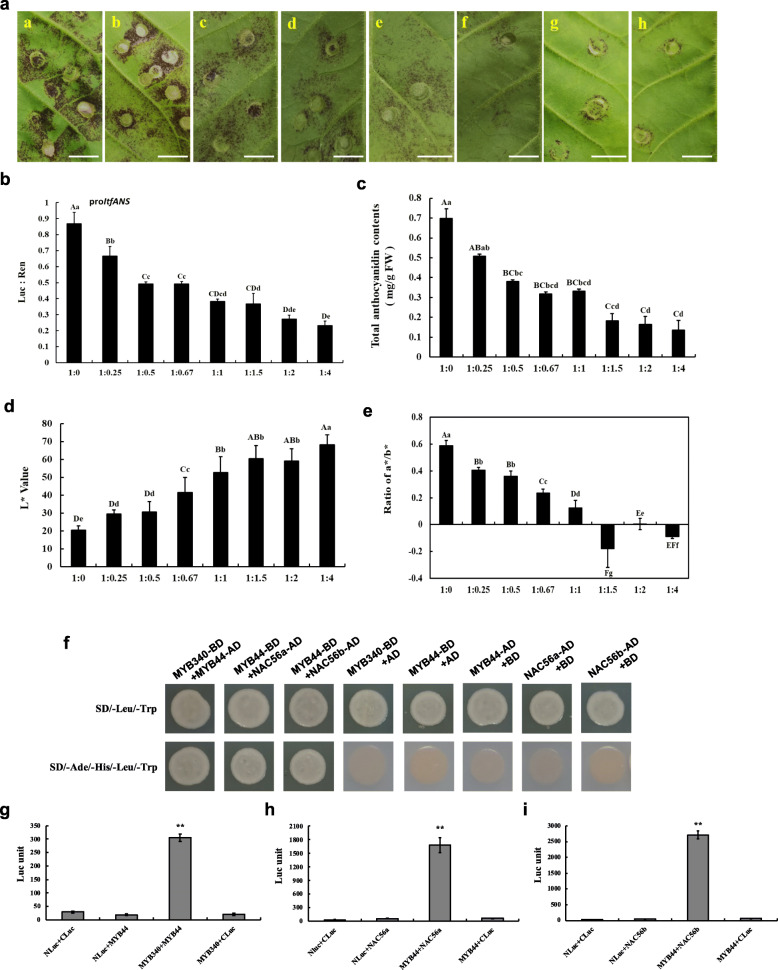
To study the role of IbMYB44 in anthocyanin biosynthesis, we cotransformed IbMYB340 and IbMYB44 at different ratios into the abaxial side of tobacco leaves to test the transcriptional repression effect of IbMYB44. As shown in Fig. 7a, the pigmentation in tobacco leaves gradually diminished with an increasing proportion of IbMYB44. Using a dual-luciferase reporter assay, we then investigated the transactivation activity of the IbANS promoter when IbMYB44 and IbMYB340 were cotransformed in tobacco leaves. The results showed that steadily declining IbANS promoter activity occurred when the IbMYB340:IbMYB44 ratio decreased from 1:0 to 1:4. However, it seemed that there were no changes between several ratios of IbMYB340:IbMYB44: 1:0.5, 1:0.67, 1:1 and 1:1.5 (Fig. 7b). The total anthocyanin contents and a*/b* values declined significantly at different ratios, their changes were consistent with the phenotypes of the tobacco leaves mentioned above, and the L* values apparently increased (Fig. 7c-e). Thus, when cotransformed with IbMYB340, IbMYB44 coulddecrease anthocyanin biosynthesis.
Fig. 7.
Functional analysis of the IbMYB340 and IbMYB44 genes using transient expression assays in tobacco leaves and interaction verification of IbMYB44 with IbMYB340, IbNAC56a or IbNAC56b in vivo. a The phenotypes of tobacco leaves on the 7th day after infiltration; the letters ‘a’ ~ ‘h’ represent the ratio of Agrobacteria harboring the IbMYB340 and IbMYB44 constructs that were coinfiltrated into young tobacco leaves. Scale bar = 1 cm. b Transcriptional effects of coinfiltration of IbMYB340 and IbMYB44 at different ratios on IbANS promoter activity. c Anthocyanin contents in transformed leaves of tobacco. FW, fresh weight. d-e The color parameter L* and a*/b* ratio values were used to indicate phenotypic changes. The values are presented as the means ± SDs (n = 3), and the uppercase and lowercase letters represent significant differences in ratios at P < 0.01 or P < 0.05, respectively. f Verification of the interaction between IbMYB44 and IbMYB340, IbNAC56a,or IbNAC56b via Y2H assays. g Verification of the interaction between IbMYB340 and IbMYB44 via firefly luciferase complementation assays. h-i Verification of the interaction between IbMYB44 and IbNAC56a or IbNAC56b via firefly luciferase complementation assays
We further verified the interaction of IbMYB44 with IbMYB340 and IbNAC56a or IbNAC56b. Y2H analysis showed that the cells cotransformed with IbMYB44 with IbMYB340 and IbNAC56a or IbNAC56b could grow on SD-Trp-Leu-His-Ade + AbA plates (Fig. 7f). Moreover, the marked luciferase enzyme activity was rescued by the control infiltrated with IbMYB340 + IbMYB44, IbMYB44 + IbNAC56a or IbMYB44 + IbNAC56b; by contrast, Nluc-IbMYB340/IbMYB44coinfiltrated with CLuc or CLuc-IbMYB44/IbNAC56a/IbNAC56b coinfiltrated with Nluc did not result in a sufficient level of luciferase enzyme activity (Fig. 7g-i). Overall, IbMYB44 could interact with IbMYB340, IbNAC56a or IbNAC56b, suggesting that IbMYB44 suppressed anthocyanin accumulation probably through competitive inhibition of IbMYB340, IbNAC56a or IbNAC56b.
Discussion
Anthocyanins have gained much attention in recent years and are common flavonoid compounds involved in red or purple pigmentation in plants [33]. To date, it is well known that structural genes participating in the anthocyanin pathway are regulated by the MYB-bHLH-WD40 complex in plants, and R2R3-MYBs particularly contribute greatly to anthocyanin accumulation; TFs have been extensively studied in different plant species, including AtMYB114 in Arabidopsis, PpMYB10.1 in peach, PyMYB114 in pear, MdMYB110a in apple, and IbMYB1 in sweet potato [8, 10, 13, 15, 30]. Moreover, coexpression of PpMYB10.1 and PpbHLH3/33 can promote anthocyanin biosynthesis in peach [9]. In addition, two activators of MYBs, PyMYB114 and PyMYB10, can interact to promote anthocyanin biosynthesis in red pears [15]. In this study, the TF IbMYB340, which was highly homologous to AtPAP1 in Arabidopsis, could induce high levels of anthocyanin pigmentation, especially when cotransformed with other TFs, in tobacco leaves and strawberry receptacles (Figs. 3a, 4a). We further demonstrated that IbMYB340 interacts with IbbHLH2, IbNAC56a or IbNAC56b to regulate anthocyanin synthesis by forming regulatory complexes. However, we detected no interaction between IbbHLH2 and IbNAC56a or IbNAC56b (Fig. 6a). These results revealed that IbMYB340 makes great contributions to anthocyanin accumulation as a transcriptional activator. However, the relationship of IbMYB1 [30] with IbMYB340 and other TFs in regulating anthocyanin biosynthesis in purple-flesh sweet potatoes is unclear, and the synergistic regulation of color formation needs to be studied further.
MYB repressors have recently gradually been shown to participate in regulating the flavonoid biosynthetic pathway. The repressors usually contain an LxLxL negative repressor motif located in the C-terminal region, which is responsible for the repressive effect [22]. As reported recently, the enhanced expression of flesh-specific StMYB44 accounts for the reduced contents of anthocyanin in potato flesh [22]. CmMYB#7, a negative R3-MYB regulator in chrysanthemum, can competitively inhibit CmMYB6 for interaction with CmbHLH2, resulting in a decrease in anthocyanin content [20]. In peach, the PpMYB18 protein can repress anthocyanin accumulation by competitively inhibiting the interaction of MYB activators with bHLHs [21]. Here, the R2R3-MYB IbMYB44 caused the pigmentation to diminish gradually in tobacco in the injection regions (Fig. 7a). This was consistent with the results of a previous study in that the MYB repressors containing the LxLxL motif resulted in a decrease in anthocyanin biosynthesis. Then, we obtained evidence of the interactions between IbMYB44 and IbMYB340 and between IbMYB44 with IbNAC56a or IbNAC56b by Y2H and firefly luciferase complementation assays (Fig. 7f-i). The results indicated that the IbMYB44-IbMYB340, IbMYB44-IbNAC56a or IbMYB44-IbNAC56b complexes might inhibit the formation of the regulatory complexes MYB340-bHLH2-NAC56a or MYB340-bHLH2-NAC56b. Taken together, the results show that IbMYB44 can negatively regulate anthocyanin biosynthesis in sweet potato.
The MBW complex is popularly recognized as a regulator affecting anthocyanin biosynthesis at the transcriptional level. In addition to the MBW complex, numerous TFs, including WRKYs, NACs, ERFs, COP1, SPL9, DELLA proteins and so on, have been shown to corroborate in determining anthocyanin biosynthesis or indirectly affect the activity of the MBW complex [15, 27, 28, 34–37]. For the NAC TF family, it was reported that a NAC TF designated BLOOD (BL) acts as a heterodimer with PpNAC1 to promote the transcriptional activity of PpMYB10.1, leading to anthocyanin accumulation [28]. Moreover, a negative effect of ANAC032 on anthocyanin accumulation was investigated in Arabidopsis in response to stress conditions [24]. In this study, three NAC genes, IbNAC56a, IbNAC56b, and IbNAC25, were identified based on the results of a phylogenetic analysis of 98 IbNACs and PpBL (Fig. 1a). RT-qPCR analysis showed that IbNAC56a/IbNAC56b were significantly expressed in ‘Xuzi No. 8’ and ‘Zhezi No. 3’ with abundant anthocyanin content (Fig. 2a), suggesting that they may function as positive regulators of anthocyanin biosynthesis in sweet potato tuberous roots. More obvious pigmentation was clearly detected when we cotransformed IbNAC56a or IbNAC56b with IbMYB340 and IbbHLH2 in heterologous overexpression systems (Figs. 3a, 4a), suggesting that IbNAC56a or IbNAC56b has an additive effect on the fully functional MYB340-bHLH2 partnership. The regulatory pattern was similar to that reported in peach [28]. In addition, although IbNAC25 is homologous to PpBL, the expression pattern was different from that of IbNAC56a or IbNAC56b in purple-flesh sweet potatoes, and the correlation analysis also showed that IbNAC25 was negatively correlated with other factors except IbMYB44, IbDFR and IbUFGT (Fig. 2b). As such, we speculated that IbNAC25 might negatively regulate anthocyanin biosynthesis; however, the function of this gene needs to be studied further.
As reported, the storage roots of ‘Yamakawamurasaki’, a purple-fleshed sweet potato cultivar, had a markedly higher expression level of IbANS than the white-fleshed cultivar ‘Yubeibai’, while IbDFR or IbUFGT showed no obvious differences [29]. In this study, we found similar results: the yellow−/white-fleshed cultivars showed lower IbANS expression than did the purple-fleshed cultivars, while the expression levels of IbUFGT and IbDFR had no obvious correlation in these cultivars (Fig. 2a). In this study, cotransformation of IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b induced apparent anthocyanin accumulation in tobacco leaves and strawberry receptacles (Figs. 3a, 4a). In addition, RT-qPCR analysis showed that the expression of FvANS was highly upregulated with the cotransformation of the above mentioned TFs (Fig. 5a). The Y1H assay illustrated that IbMYB340 could directly bind to the IbANS promoter, and the IbANS promoter activity was the strongest when IbMYB340, IbbHLH2 and IbNAC56a or IbNAC56b were cotransformed, as reflected by a dual-luciferase reporter assay (Fig. 6b-c), suggesting that the IbANS gene may play a vital role in the regulatory network of anthocyanin biosynthesis in various sweet potato cultivars.
Conclusions
In summary, using Y2H, firefly luciferase complementation, and dual-luciferase reporter assays, we investigated whether different regulatory complexes (MYB340-bHLH2-NAC56a or MYB340-bHLH2-NAC56b) can be formed to promote anthocyanin biosynthesis by binding to the IbANS promoter and upregulating the expression of other anthocyanin-related genes. In addition, our findings illustrated that IbMYB44 acts as a repressor of anthocyanin biosynthesis in sweet potato by interacting with IbMYB340 and IbNAC56a or IbNAC56b gene function or by indirectly affecting the MYB340-bHLH2-NAC56a or MYB340-bHLH2-NAC56b complex, which can enhance IbANS promoter activity. These results can help us develop a stronger understanding of the possible underlying mechanism of anthocyanin biosynthesis in purple-flesh sweet potato, although additional scientific research needs to be done to give a comprehensive and profound interpretation.
Methods
Plant materials
The storage roots of ‘Hanzi’, ‘Xuzi No. 8’, ‘Zhezi No. 3’, ‘Zhezi No. 4’, ‘Sushu No. 8’, ‘Guangshu No. 87’, ‘Lizixiang’, and ‘Xushu No. 18’ sweet potato were collected at 120 days after budding in the field at the National Sweet Potato Improvement Center (Xuzhou, Jiangsu Province, China) on 10 October 2018.
The seeds of tobacco (Nicotiana tabacum ‘NC89’ and Nicotiana benthamiana) are preserved and presented in School of Food and Biological Engineering, Hefei University of Technology (Hefei, Anhui Province, China), and the plants were grown in the glasshouse at 24 °C under artificial irradiance (daylight, 16 h). Four-week-old Nicotiana tabacum ‘NC89’ leaves were used for the transient transformation experiment and for the dual-luciferase reporter system assay, and tobacco (Nicotiana benthamiana) plants with six leaves were used for firefly luciferase complementation assays.
The seeds of ‘Yellow Wonder’ 5AF7 (YW5AF7) was provided by Prof. Jun Wu (Nanjing Agricultural University, China), which is a diploid strawberry (F. vesca) cultivar with yellow-white colored fruit. It was grown in the glasshouse under 12 h of light, while the temperature was maintained at 25 °C during the daytime and at 20 °C during the night. Receptacles 2 weeks after anthesis were used for transient expression assays.
RNA extraction and qPCR analysis
The total RNA of 0.8 g of sweet potato tuberous roots and 0.5 g of strawberry samples was extracted using a Plant Total RNA Isolation Kit (Foregene). First-strand cDNA was synthesized from total RNA using Prime Script™ RT Master Mix (Takara). qRT-PCR was conducted using SYBR® Premix Ex Taq™ II (Takara) in a total reaction volume of 20 μl consisting of 150 ng of template cDNA, each primer at 0.2 μM and 10 μl of SYBR® Premix Ex Taq™ II, and the amplification program was as follows: 1 cycle of 95 °C for 10 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. A strawberry gene, FvTubulin (gene11892), and a sweet potato gene, IbTubulin (Itf04g29110), were used as internal controls. The primers used for RT-qPCR are listed in additional file 1: Table S1.
Transient assays in tobacco and strawberry
For the transient expression analysis, PCR amplification was conducted using Phanta® Super-Fidelity DNA Polymerase (Vazyme), and the full-length coding sequences of IbMYB340 (Itf12g05820.t1), IbbHLH2 (Itf14g18730.t2), IbNAC56a (Itf02g15460.t1) and IbNAC56b (Itf01g19290.t1) were inserted into a pSAK277 vector under the control of the 35S promoter with EcoRI and XbaI. The primer sequences used for expression vector construction are listed in Additional file 1: Table S2. The recombinant vectors were individually transformed into GV3101 strains of Agrobacterium tumefaciens using the chemical method. The Agrobacterium cells harboring the recombinant pSAK277 vectors was resuspended in an injection solution (OD600 = 1.0) and incubated at 25 °C under 60 rpm at 4–5 h before injection.
For the functional assay of IbMYB340, IbbHLH2 and IbNAC56, Agrobacterium cultures containing the abovementioned TFs were mixed equally. In addition, Agrobacterium cells harboring IbMYB340 and IbMYB44 were mixed at different ratios to test the repressing effect of IbMYB44. The mixed Agrobacterium cells were injected into young N. tabacum leaves and strawberry receptacles according to the methods described by Zhou et al. [21]. The plants were incubated in darkness for 24 h and then moved to a greenhouse with artificial irradiance (16 h daylength). Photos were taken 7 d after injection, anthocyanins were collected for quantification, and total RNA was collected for extraction where necessary.
Extraction and quantification of anthocyanins in tobacco and strawberry
Anthocyanins were extracted and quantified according to the methods described by Yao et al. [15]. In brief, we immersed 0.2 g of tobacco leaf tissue or 0.2 g of strawberry receptacle tissue around the injection regions, which had been fully groundin liquid nitrogen, in 1 ml of cold methanol consisting of 0.1% HCl at 4 °C for 24 h. The supernatant was collected after centrifugation of the mixture at 12000 g for 15 min. The anthocyanin levels were estimated from the methanolic extracts according to the eq. A = (A530 - A620) – 0.1 (A650 - A620), and the absorbance was measured using a Multiskan Spectrum device (Thermo Scientific Multiskan GO 1510, Finland).
Yeast one-hybrid assay
A yeast one-hybrid (Y1H) assay was performed using a Matchmaker® Gold Yeast One-Hybrid System. Briefly, three amplifications of IbANS promoter fragments (− 956 bp to − 755 bp, − 396 bp to − 224 bp and − 310 bp to − 105 bp) were cloned into a pAbAi vector with Hind III and Xho I, and the complete sequences of IbMYB340, IbNAC56a and IbNAC56b were cloned into a pGADT7 vector with EcoR I and Xho I. Then, we transformed prey vectors into Y1H Gold cells harboring the pAbAi-bait and tested them on SD/−Ura/−Leu/AbA plates. The primer sequences used for vector construction are listed in Additional file 1: Table S2.
Dual-luciferase reporter assay of tobacco leaves
To construct the dual-luciferase reporter vector, the 1.9 kb upstream promoter region of IbANS (Itf13g04110.t1) (from the ATG start codon) was amplified from the genomic DNA of tuberous roots from the sweet potato cultivar ‘Xuzi No. 8’ and inserted into a pGreen II 0800-LUC binary vector. Moreover, Agrobacterium transformation and injection preparation were the same as the methods described for the transient transformation assay in tobacco leaves and strawberries.
Agrobacterium cells harboring the pGreen II 0800-LUC recombinant vector, pSAK277 vector, IbMYB340, IbbHLH2, and IbNAC56 were mixed at a 1:3:3:3 ratio. The mixture of Agrobacterium cells was injected into young N. tabacum leaves that were 2 weeks old. At 48–72 h after infiltration, the LUC and Ren activity were measured with an E1910 Dual-Luciferase® Reporter Assay System (Promega).
Yeast two-hybrid assay
A yeast two-hybrid (Y2H) assay was performed using a Matchmaker® Gold Yeast Two-Hybrid System. The full-length coding sequences of IbMYB340, IbbHLH2 and IbMYB44 were amplified and cloned into a pGBKT7 vector with NdeI and PstI; moreover, the complete sequences of IbbHLH2, IbNAC56a, IbNAC56b and IbMYB44 were cloned into pGADT7 with EcoRI and XhoI. Then, we cotransformed the integrated vectors into Y2H Gold cells with the LiCl-PEG method according to the manufacturer’s instructions and tested the protein-protein interactions on SD/−Leu/−Trp/−His/−Ade + AbA plates.
Firefly luciferase complementation assay
The firefly luciferase complementation assay was performed according to the methods reported by Chen et al. [38]. Full-length coding sequences of IbMYB340 and IbMYB44 without stop codons were cloned into a pCambia1300-NLuc binary vector, and the complete sequences of IbNAC56 and IbMYB44 were cloned into a pCambia1300-CLuc binary vector. Agrobacterial transformation and injection preparation were performed as previously described for the transient transformation assay. Then, the activity of firefly luciferase was measured with a Steady-Glo® Luciferase Assay System (Promega) during 48–72 h.
Statistical analysis
All samples were replicated at least three times independently, and all the data are represented as the means ±SDs. Statistical analyses were performed using Student’s t-test embedded in Excel 2013 software. Pearson’s correlation coefficients (R values) and the constructed heatmap were analyzed using R scripts. Significance is indicated by * (P < 0.05) or ** (P < 0.01) or by different letters.
Supplementary information
Additional file 1: Figure S1. Amino acid sequence alignment of IbNACs and the NAC transcription factor PpBL (ALK27819.1) in peach (Prunus persica). Table S1. List of primers used for RT-qPCR. Table S2. The list of primers used for developing the constructs. Table S3. Protein sequences subjected to phylogenetic analysis and multiple sequence alignments.
Acknowledgments
We thank Dr. Andrew C. Allan, Dr. Lin-Wang Kui and Dr. Richard Espley for the dual vector pGreen II 0800-LUC in The New Zealand Institute for Plant & Food Research Limited, Auckland, New Zealand.
Abbreviations
- TF
Transcription factor
- PAL
Phenylalanine ammonia lyase
- F3H
Flavanone 3-hydroxylase
- DFR
Dihydroflavonol 4-reductase
- ANS
Anthocyanidin synthase
- UFGT
UDP-glucose flavonoid 3-O-glucosyltransferase
- GST
Glutathione S-transferase
- Y2H
Yeast two-hybrid
- Y1H
Yeast one-hybrid
- MBW
MYB-bHLH-WD40
Authors’ contributions
Z.Z.W., G.F.Y., K.D.H. D.L.Z. and H.Z. conceived and designed the experiments; Z.Z.W., G.F.Y. performed the experiments; J.T., Z.Q.H. and D.L.Z. provided the experiment materials; Z.H., Y.H.L., J.T., P.J., and H.L.Y. analyzed the data; Z.Z.W., G.F.Y. wrote the paper; K.D.H., G.F.Y., Z.Q.H. and H.Z. interpreted the data and revised the manuscript. All authors have read and approved the final version of this manuscript.
Funding
This work was supported by the National Key R&D Program of China (2019YFD1001303, 2019YFD1001300), the National Natural Science Foundation of China (31901993, 31970312, 31970200, 31670278), the Natural Science Foundations of Anhui Province (1908085MC72), the Key Research and Development Program of Anhui Province (201904a06020031), the Fundamental Research Funds for the Central Universities (JZ2018HGTB0241). The funders had no role in the experimental design, data collection and analysis or writing the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zeng-Zheng Wei, Kang-Di Hu and Dong-Lan Zhao contributed equally to this work.
Contributor Information
Zeng-Zheng Wei, Email: 649000983@qq.com.
Kang-Di Hu, Email: kangdihu@hfut.edu.cn.
Dong-Lan Zhao, Email: 271047544@qq.com.
Jun Tang, Email: tangjun5886@163.com.
Zhong-Qin Huang, Email: 395383266@qq.com.
Peng Jin, Email: 82769551@qq.com.
Yan-Hong Li, Email: 1326769208@qq.com.
Zhuo Han, Email: kanahan80@163.com.
Lan-Ying Hu, Email: 1659421583@qq.com.
Gai-Fang Yao, Email: yaogaifang@hfut.edu.cn.
Hua Zhang, Email: hzhanglab@hfut.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02451-y.
References
- 1.Munoz-Rodriguez P, Carruthers T, Wood JRI, Williams BRM, Weitemier K, Kronmiller B, Ellis D, Anglin NL, Longway L, Harris SA, Rausher MD, Kelly S, Liston A, Scotland RW. Reconciling conflicting phylogenies in the origin of sweet potato and dispersal to polynesia. Curr Biol. 2018;28(8):1246–1256. doi: 10.1016/j.cub.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 2.Ghasemzadeh A, Talei D, Jaafar HZE, Juraimi AS, Mohamed MTM, Puteh A, Halim MRA. Plant-growth regulators alter phytochemical constituents and pharmaceutical quality in sweet potato (Ipomoea batatas L.). BMC Complem Altern M. 2016;16:152. [DOI] [PMC free article] [PubMed]
- 3.Kong JM, Chia LS, Goh NK, Chia TF, Brouillard R. Analysis and biological activities of anthocyanins. Phytochemistry. 2003;64(5):923–933. doi: 10.1016/S0031-9422(03)00438-2. [DOI] [PubMed] [Google Scholar]
- 4.Veeriah S, Kautenburger T, Habermann N, Sauer J, Dietrich H, Will F, Pool-Zobel BL. Apple flavonoids inhibit growth of HT29 human colon cancer cells and modulate expression of genes involved in the biotransformation of xenobiotics. Mol Carcinog. 2006;45(3):164–174. doi: 10.1002/mc.20158. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Ling W, Du Z, Chen Y, Li D, Deng S, Liu Z, Yang L. Effects of anthocyanins on cardiometabolic health: a systematic review and meta-analysis of randomized controlled trials. Adv Nutr. 2017;8(5):684–693. doi: 10.3945/an.116.014852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12(12):2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 9.Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015;167(4):1448–1470. doi: 10.1104/pp.114.256172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahim MA, Busatto N, Trainotti L. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 2014;240(5):913–929. doi: 10.1007/s00425-014-2078-2. [DOI] [PubMed] [Google Scholar]
- 11.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49(3):414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chagne D, Kui LW, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, Mcghie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013;161(1):225–239. doi: 10.1104/pp.112.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng SQ, Wang YL, Yang S, Xu YT, Chen XS. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 2010;232(1):245–255. doi: 10.1007/s00425-010-1170-5. [DOI] [PubMed] [Google Scholar]
- 15.Yao GF, Ming ML, Allan AC, Gu C, Li LT, Wu X, Wang RZ, Chang YJ, Qi KJ, Zhang SL, Wu J. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017;92(3):437–451. doi: 10.1111/tpj.13666. [DOI] [PubMed] [Google Scholar]
- 16.Lai B, Du LN, Hu B, Wang D, Huang XM, Zhao JT, Wang HC, Hu GB. Characterization of a novel litchi R2R3-MYB transcription factor that involves in anthocyanin biosynthesis and tissue acidification. BMC Plant Biol. 2019;19:62. [DOI] [PMC free article] [PubMed]
- 17.Xi WP, Feng J, Liu Y, Zhang SK, Zhao GH. The R2R3-MYB transcription factor PaMYB10 is involved in anthocyanin biosynthesis in apricots and determines red blushed skin. BMC Plant Biol. 2019;19:287. doi: 10.1186/s12870-019-1898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodriguez-Franco A, Caballero JL, Schwab W, Munoz-Blanco J, Blanco-Portales R. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria ananassa fruits. J Exp Bot. 2014;65(2):401–417. doi: 10.1093/jxb/ert377. [DOI] [PubMed] [Google Scholar]
- 19.Wei LZ, Mao WW, Jia MR, Xing SN, Ali U, Zhao YY, Chen MT, Cao ML, Dai ZR, Zhang K, Dou ZC, Jia WS, Li BB. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot. 2018;69(20):4805–20. doi: 10.1093/jxb/ery249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, Chen K. CmMYB# 7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019; 70(12):3111-23. [DOI] [PMC free article] [PubMed]
- 21.Zhou H, Lin-Wang K, Wang F, Espley RV, Ren F, Zhao J, Ogutu C, He H, Jiang Q, Allan AC. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019;221(4):1919–1934. doi: 10.1111/nph.15486. [DOI] [PubMed] [Google Scholar]
- 22.Liu YH, Lin-Wang K, Espley RV, Wang L, Li YM, Liu Z, Zhou P, Zeng LH, Zhang XJ, Zhang JL, Allan AC. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J Exp Bot. 2019;70(15):3809–3824. doi: 10.1093/jxb/erz194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Seo PJ, Lee HJ, Park CM. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012;70(5):831–844. doi: 10.1111/j.1365-313X.2012.04932.x. [DOI] [PubMed] [Google Scholar]
- 24.Mahmood K, Xu ZH, El-Kereamy A, Casaretto JA, Rothstein SJ. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front Plant Sci. 2016;7:1548. [DOI] [PMC free article] [PubMed]
- 25.Zhong RQ, Lee CH, Zhou JL, Mccarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20(10):2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis vascular-related NAC-domain 6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22(10):3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009;50(12):2210–2222. doi: 10.1093/pcp/pcp159. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Kui LW, Wang HL, Gu C, Dare AP, Espley RV, He HP, Allan AC, Han YP. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82(1):105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
- 29.Dong W, Niu LL, Gu JT, Gao F. Isolation of a WD40-repeat gene regulating anthocyanin biosynthesis in storage roots of purple-fleshed sweet potato. Acta Physiol Plant. 2014;36(5):1123–1132. doi: 10.1007/s11738-014-1487-y. [DOI] [Google Scholar]
- 30.Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 2007;143(3):1252–1268. doi: 10.1104/pp.106.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Moeinzadeh MH, Kuhl H, Helmuth J, Xiao P, Haas S, Liu GL, Zheng JL, Sun Z, Fan WJ, Deng GF, Wang HX, Hu FH, Zhao SS, Fernie AR, Boerno S, Timmermann B, Zhang P, Vingron M. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat Plants. 2017;3(9):696–703. doi: 10.1038/s41477-017-0002-z. [DOI] [PubMed] [Google Scholar]
- 32.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van De Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesschaeve I, Noble AC. Polyphenols: factors influencing their sensory properties and their effects on food and beverage preferences. Am J Clin Nutr. 2005;81(1):330s–335s. doi: 10.1093/ajcn/81.1.330S. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Wu J, Hu KD, Wei SW, Sun HY, Hu LY, Han Z, Yao GF, Zhang H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic Res. 2020;37(7):1–12. doi: 10.1038/s41438-020-0254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hulskamp M, Hoecker U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013;74(4):638–651. doi: 10.1111/tpj.12153. [DOI] [PubMed] [Google Scholar]
- 36.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23(4):1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Tan HJ, Ma ZX, Huang JR. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol Plant. 2016;9(5):711–721. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Chen HM, Zou Y, Shang YL, Lin HQ, Wang YJ, Cai R, Tang XY, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146(2):368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Amino acid sequence alignment of IbNACs and the NAC transcription factor PpBL (ALK27819.1) in peach (Prunus persica). Table S1. List of primers used for RT-qPCR. Table S2. The list of primers used for developing the constructs. Table S3. Protein sequences subjected to phylogenetic analysis and multiple sequence alignments.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.