Abstract
Copy number variants (CNVs) are important in genome variation and genetic disease, with new mutations arising frequently in the germline and somatic cells. Replication stress caused by aphidicolin and hydroxyurea (HU) is a potent inducer of de novo CNVs in cultured mammalian cells. HU is used extensively for long-term management of sickle cell disease. Here we examined the effects of HU treatment on germline CNVs in vivo in male mice to explore whether replication stress can act as a CNV mutagen in germline mitotic divisions as in cultured cells and whether this would support a concern for increased CNV mutations in offspring of men treated with HU. Several trials of HU administration were performed by oral gavage and subcutaneous pump, with CNVs characterized in C57BL/6 × C3H/HeJ hybrid mouse offspring by microarray and mate-pair sequencing. HU had a short half-life of ~14 min and a narrow dose window over which studies could be performed while maintaining fertility. Tissue histopathology and reticulocyte micronucleus assays verified that doses had a substantial tissue and genetic toxicity. CNVs were readily detected in offspring that originated in both paternal and maternal mouse strains, as de novo and inherited events. However, HU did not increase CNV formation above baseline levels. These results reveal a high rate of CNV mutagenesis in the mouse germline but do not support the hypothesis that HU would increase CNV formation during mammalian spermatogenesis, perhaps due to highly toxic effects on sperm development or experimental variables related to HU pharmacology in mice.
Introduction
Sickle cell disease (SCD) is the most common inherited blood disorder in humans, affecting approximately 100,000 people in the United States and millions of people worldwide [Lopez, et al., 2010]. SCD is most commonly caused by a single inherited point mutation in the β-globin chain of hemoglobin, resulting in expression of HbS in erythrocytes. The disease is characterized by a lifelong hemolytic anemia with a greatly increased risk for acute medical complications and accrual of organ damage [Bonds, 2005]. Chronic treatment problems include increasingly difficult transfusion management and addiction to medications used to treat painful vaso-occlusive crises.
Hydroxyurea (HU) is the most extensively used FDA-approved drug for long-term management of SCD in adults and children and is used to treat some thalassemias and cancers [Algiraigri and Kassam, 2017, Madaan, et al., 2012, Steinberg, et al., 2010, Ware and Aygun, 2009]. Extended HU treatment leads to increased expression of fetal hemoglobin (HbF) and a reduced fraction of sickle hemoglobin (HbS) in erythrocytes, thereby decreasing the rate of painful crises and ameliorating disease severity [Charache, et al., 1995]. The mechanisms of action for HbF induction are not completely understood. Presumably, HU causes suppression of erythroid progenitors and cell stress signaling, which leads to recruitment of erythroid progenitors with increased HbF levels [Agrawal, et al., 2014]. The short-term toxicities of HU are minimal, and a recent report on the long-term effects of HU treatment in a 17.5-year follow-up of adults undergoing HU therapy found no differences in the incidences of stroke, organ dysfunction, infection, or cancer between treated individuals and controls [Steinberg, et al., 2010]. Thus, HU is a valuable and effective therapeutic agent for SCD in adults [Steinberg, et al., 2010, Ware and Aygun, 2009].
HU has both cytotoxic and genotoxic effects leading to concerns about potential effects in treated patients and offspring. Few reproductive studies have been performed for HU despite important potential effects on reproduction and fetal development. Treated males typically have reduced testicular indices, semen quality, sperm counts and serum testosterone levels [Modebe and Ezeh, 1995]. There is also evidence that HU administration is associated with increased risk of leukemia in patients treated for myeloproliferative disorders [Kiladjian, et al., 2006, Mizutani, et al., 2010, Weinfeld, et al., 1994]. Recent studies using alkaline comet and micronucleus assays found that patients treated with HU had a significant increase in DNA damage in leukocytes and erythrocyte precursors [Flanagan, et al., 2010, Friedrisch, et al., 2008]. It remains to be determined whether these genotoxic effects extend to the germline.
We previously showed that HU is a potent inducer of de novo copy number variants (CNVs) in a human cell culture system at low doses (100–300 μM) comparable to the plasma level achieved in treated human patients [Arlt, et al., 2011]. CNVs are generally defined as deletions or duplications ranging in size from tens to millions of base pairs. Tens of thousands of normal polymorphic CNVs have now been described throughout the genome of healthy individuals and represent the major component of human genomic variation [Sudmant, et al., 2015]. In addition, spontaneous or de novo CNVs arise frequently in the germline and are an important and frequent cause of genetic and developmental disorders [Lee and Scherer, 2010, Zhang, et al., 2009]. Despite their importance the risk factors involved in CNV formation remain unknown, but, like all classes of mutation, will likely include exposure to precipitating environmental mutagens and inherited genetic predispositions.
The presumed mechanism of action of HU in CNV induction is its well-described indirect impairment of replication fork progression by direct inhibition of ribonucleotide reductase, an enzyme involved in synthesis of dNTPs. This inference extends from related experimental evidence that replication stress can induce CNVs in mammalian cells [Arlt, et al., 2009, Arlt, et al., 2011, Arlt, et al., 2012, 2014]. Specifically, partial inhibition of replication with the DNA polymerase inhibitor aphidicolin (APH) induces a high frequency of de novo CNVs in cultured mammalian cells with nearly identical features to those induced by HU [Arlt, et al., 2009]. No CNVs induced by HU or APH in vitro contained large stretches of homology indicative of non-allelic homologous recombination (NAHR), which mediates the formation of recurrent CNVs. Rather, junction sequences displayed at most short microhomologies indicative of either end joining or replicative template switching mechanisms, such as MMBIR [Hastings, et al., 2009]. These junction features, as well as the large size of experimentally induced CNVs (median 186 kb) are consistent with descriptions of non-recurrent CNVs, a major class of both normal and disease-associated CNVs in humans [Conrad, et al., 2010, Conrad, et al., 2010]. These combined observations suggest a common mechanism for most CNVs arising in the human germline and those induced by replication stress in cell culture.
Importantly, while recurrent CNV formation by NAHR occurs in meiosis during homologue pairing, constitutional non-recurrent CNVs are much more likely to occur in the mitotic divisions within or leading to formation of the germline, prior to meiosis itself. This inferred mitotic cell origin has important implications for the genetic and environmental factors involved in non-recurrent CNV formation and for their study. For example, the fact that the male germline undergoes mitotic divisions leading to mature sperm throughout adulthood while females complete germline mitotic divisions in a short window during fetal development predicts a male sex bias in risk, including from adult exposures to mutagenic environmental agents. In this context, it is noteworthy that we observed CNV induction in vitro after treatments lasting only one to three cell divisions. Adult men are treated with HU for years, which spans many rounds of mitotic cell divisions leading from prospermatogonia to primary spermatocytes.
In this study, we administered HU to adult male mice and measured CNV formation in their offspring using genomic methods to inform two related questions. First, we sought to test our general hypothesis that replication stress will act as a CNV mutagen in germline mitotic divisions in the same manner as in cultured somatic cells. Second, we sought to determine whether data from a mouse model would support a concern for increased formation of de novo genomic alterations in the sperm and offspring of men treated long-term with HU during their reproductive years.
Materials and Methods
Mouse strains, acquisition and breeding
Animal use was reviewed and approved by the Institutional Animal Care & Use Committee (IACUC) University of Michigan; protocol PRO00006521. Animal husbandry was provided by the staff of the University of Michigan Unit for Laboratory Animal Medicine (ULAM) under the guidance of supervisors who are certified by the American Association for Laboratory Animal Science (AALAS). Veterinary care was provided by ULAM faculty members and veterinary residents. The University of Michigan is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC, Intl) and the animal care and use program conforms to the standards of “The Guide for the Care and Use of Laboratory Animals,” Revised 2011. The University of Michigan has filed an assurance statement on these matters with the Office of Laboratory Animal Welfare (OLAW). All procedures used followed ULAM guidelines.
For HU administration by gavage, male C57BL/6J mice 22 days of age at arrival were purchased from Jackson Laboratories (Bar Harbor, ME). For HU administration by microinfusion pumps, male C57BL/6J mice between 35 and 49 days of age at arrival were purchased from Jackson Laboratories (Bar Harbor, ME) or Charles River Laboratories (Wilmington, MA). Female C3H/HeJ mice for breeding to treated and control males were purchased from Jackson Laboratories (Bar Harbor, ME).
Following the treatment window, each treated C57BL/6J male was bred to two untreated C3H/HeJ females for seven days, after which the females were removed to separate cages. When pups were born, they were counted and visually examined for physical abnormalities. The pups were allowed to grow for 17–21 days and then euthanized. During this time they were monitored for any visible signs of physical or behavioral abnormalities (none noted). Two pups from each litter were arbitrarily chosen for CNV analysis. In addition, any pups that appeared runted, deformed, or sick were also analyzed in the event that a CNV led to the unhealthy phenotype.
HU administration by oral gavage and subcutaneous pumps
For oral administration, the animals were dosed by oral gavage twice daily 9 hours apart for 56 days, starting at 28 days of age. The animals were weighed and arbitrarily assigned into treatment and control groups. Body weights were collected each morning prior to dosing, and daily dose volumes were calculated based on the most recent body weight. Well-being of the mice was evaluated and clinical observations were recorded twice daily at the time of dosing. At the end of the treatment regimen, mice were allowed to recover for one day before being mated.
iPRECIO continuous infusion pumps, model number SMP-300 were used for subcutaneous administration of HU. Pumps were programmed to pump at a rate of 4 μl/hour). Pump implantation was conducted following institutional guidelines for rodent aseptic surgery. The mice were anesthetized in a rodent anesthesia induction chamber with isoflurane (2.5%) and oxygen (1 liter) and placed in an anesthesia mask for surgical prep and surgery. The surgical area was clipped with electric clippers using a #50 surgical blade and cleaned with Nolvasan surgical scrub and sterile saline. The pumps were implanted subcutaneously. The antenna and hose of the pump was tied together with 5–0 PDS and 7mm wound clips were used to close the incision. An application of sterile surgical glue (Indermil) was applied to the incision before placing the animal in a recovery cage. Pumps were refilled twice daily via injection through the pump’s fill port. Mice were weighed prior to each pump refill, with each dose concentration calculated prior to refill based on the previous recorded weight. A second surgery was performed at the end of the 14-day treatment window to remove the pumps. At the end of the treatment regimen, mice were allowed to recover for 35 days before being mated. Untreated control mice did not receive pumps.
Pharmacokinetic assessment of plasma HU levels
Blood samples were taken from each mouse by cardiac puncture after euthanasia with CO2. The blood was shipped on dry ice overnight to the Research Triangle Institute (Research Triangle Park, NC) for quantitation of plasma HU levels using high-performance liquid chromatography tandem mass spectrometry with electrospray ionization.
Histologic assessment of testes and epididymides
Tissues for histology were fixed for 48 hours in 10% neutral buffered formalin and then processed to paraffin by standard methods using an automated tissue processor. Decalcification of bone marrow sections was performed using a formic acid-based decalcifying agent (Immunocal, Decal Chemical Corporation). Tissues were mounted onto glass slides and stained with hematoxylin and eosin on an automated histostainer. Tissues were evaluated by light microscopy using published, standard criteria for lesions of the male reproductive system in rodents [Creasy DM, 2002]. Evaluation was performed by a board-certified veterinary pathologist blinded to the experimental groups (treated and control) at the time of evaluation. Severity grades for testicular and epididymal lesions was based on the following scale: 0: no alteration, 1: minimal (<5% tubules affected), 2: mild (5–25% tubules affected), 3: moderate (26–50% tubules affected), 4: marked (51–75% tubules affected), 5:severe (>75% tubules affected). This scale is similar to standard 5-point grading scales used for the histological evaluation of testicular lesions [Creasy DM, 2002] apart from the use of a more conservative (5–25% as opposed to 10–25%) lower threshold for mild changes in our study.
CNV detection and characterization by microarray
For all genomic analyses, DNA was isolated from tail snips of selected pups using the Blood & Cell Culture DNA Mini Kit (Qiagen). In early studies, this DNA was subjected to CNV detection by Nimblegen mouse MM9 whole genome tiling UX3 microarrays. As a comparative method, a reference DNA was required for which we used a single female C57BL/6 × C3H/HeJ hybrid animal. Array analysis was performed by custom software designed to sensitively detect copy number differences unique to just one pup. We first dropped chrY, which was uninformative given our female reference DNA, and applied a median normalization individually to all other chromosomes per sample to account for DNA content differences in male vs. female animals. All chromosomes thus had an adjusted median probe log reference ratio (LRR) of zero. We next examined the LRR values across all analyzed mouse samples at each probe and tentatively assigned each sample as copy number neutral (within 2.5 standard deviations of the chromosome mean), gain or loss relative to the chromosome. We then applied a second normalization such that the median sample within the majority copy number group was set to an LRR of zero. In this way, a common change in the copy number of interrogated F1 offspring relative to the reference DNA was ignored, while probe-sample pairs at a unique CNV would be substantially deviant from zero.
CNV discovery was executed by three distinct approaches. The first approach was previously described for the VAMP software [Arlt, et al., 2011, Arlt, et al., 2011]. The last two approaches used hidden Markov models (HMMs) with a series of discrete LRR states at 0, 0.1, −0.1, etc. The emission probability that an input LRR value would be observed for a candidate state was calculated from one of two probe standard deviations and the normal distribution. The first standard deviation was for all probes on a chromosome in a given sample, which optimally considers sample array quality. The second standard deviation was for the set of sample LRR values in the majority group determined above for each probe, which optimally considers probe quality. A maximum allowed LRR deviation of 1.2 and a minimum emission probability of 0.0005 ensured that extreme outlier values (i.e. failed probes) did not have an undue influence on the model. The HMMs further invoked a low transition probability of 0.025, given that most of the genome, including CNVs, is comprised of runs of probes in the same copy number state. The Viterbi algorithm was used to solve each resulting HMM to assign an LRR state to each probe in each sample.
For maximal sensitivity, consecutive series of at least 5 probes nominated as CNVs (average LRR deviation of 0.2 or more) for any animal by any algorithm were collapsed into overlapping regions. Array data for all samples were visually examined together at each region in a genome browser to judge CNV call quality and sample specificity. We used the same browser interface to visually scan the genome to ensure that obvious CNVs had not been missed.
CNV detection and characterization by mate-pair sequencing
In later studies we turned to genomic sequencing due to discontinuation of Nimblegen genomic microarrays. Libraries were constructed using tail snip DNA and the Illumina Nextera Mate Pair Library Prep Kit without fragment size selection according to the manufacturer’s instructions. Paired end sequencing (read lengths 52 to 66 nt) was performed at the University of Michigan DNA Sequencing Core on a HiSeq 2500, pooling 4 or 5 samples per lane using standard Illumina indexing. Reads pairs were first processed by NXTrim [O’Connell, et al., 2015] to assign them to appropriate orientation categories, since read pairs from mate-pair libraries sometimes cross the circularization ligation junction and sometimes represent simple genomic fragments. Three samples were excluded from further analysis based on a much lower yield of circularization junctions, indicative a failed library preparation. Resulting reads were mapped to the mm10 reference genome using BWA MEM [Li and Durbin, 2010] with default settings. Fragment-level coverage, i.e. considering the entire length of sequenced DNA fragments assigned as proper pairs by BWA (SAM flag bit 0×2 set), was typically 30 to 40-fold, providing excellent sensitivity for CNV detection. Base-level, i.e. read, coverage was typically 1 to 3-fold, providing sufficient information on SNPs for strain assignment but insufficient coverage for discovery of de novo point mutations.
Two distinct algorithms were used to nominate structural variants in aligned reads, using custom software that follows established approaches and optimizes the detection of CNVs unique to just one sample. To facilitate analysis, samples were grouped into sets of six to eight pups of the same sex where no two animals shared a common parent, so that CNVs that arose in a parent and were inherited by a sib pair would still be detected. The first algorithm follows logic patterns based on the relative spacing and orientation of paired reads [Arlt, et al., 2011, Korbel, et al., 2007], for example that the reads of a pair flanking a novel deletion junction map with a large spacing and a predictable clustering of reads on each side of the junction. We applied this algorithm simultaneously to all non-PCR-duplicated anomalous read pairs (i.e. flag bit 0×2 not set) from all samples in a set with a minimum mapping quality of 5, which permitted detection as well as assessment of the uniqueness of non-reference read-pair patterns. Whereas most non-reference patterns were present in all isogenic animals, patterns unique to one animal were nominated as candidate de novo CNVs if they had at least five supporting read pairs and were <2 Mb, which suppresses mapping artifacts arising from reads in repetitive DNA elements. Notably, no unique inversions were found even though the read pair method is capable of finding them.
For the second algorithm based on fragment depth, we constructed a coverage map of the genome for each set of samples using variable width bins. Each proper read pair was counted such that each covered position was incremented by 1 divided by the fragment length. These position values were summed over all read pairs and samples in a set. Chromosome bins were assigned from left to right, with bases added to each bin until its summed count was equal to 100 times the number of samples. Thus, all bins carry equal and substantial statistical weight. Four samples (three treated, one untreated) showed a notably larger intra-sample variation in bin coverage. These samples were excluded from depth analysis as they yielded many false-positive CNVs, but were included in read pair calling. The typical median bin size was 6 kb, allowing detection of ~25 kb CNVs covering four consecutive bins. Bin sizes are inversely correlated to the mappability of the underlying DNA and the modal copy number in the input samples, i.e. the copy number in typical non-CNV containing animals. We therefore compared all bin sizes to reference span chr1:100000000–200000000, empirically determined to have a stable copy number of 2. A HMM was constructed based on the variance of bin sizes in this region and algebraic expansion to obtain the expected mean and variance for other discrete copy number states. Solution of this HMM by the Viterbi algorithm assigned a modal copy number to all bins genome-wide. This information permitted calculation of expected read counts for a set of discrete copy number change (CNC) states (0, +1, −1, etc.) for each sample, based on the median bin count across all samples after normalizing them to the same scale based on each sample’s total reads. This median count, which represents the modal copy number state, is typically ~100 but deviates from that value when one or more samples have a CNV. For visualization, the CNC of a sample at a bin was calculated as the population modal copy number multiplied by the fractional deviation of the sample’s normalized read count relative to the median count. For CNV detection, a second HMM was constructed and solved for each sample that used the Poisson distribution and the expected read counts to determine emission probabilities for each actual bin count, resulting in an assigned discrete CNC for all bins and samples. CNVs were called as stretches of four or more bins with a CNC different than zero.
The set of all nominated variants from each method was then assembled and all available evidence was visually examined at each candidate genomic locus for all samples, including anomalous read pairings, coverage depth of proper pairs, and the allelic fraction of known C3H/HeJ SNPs [Keane, et al., 2011], where half of sequenced reads were expected to match the C3H/HeJ allele under the null hypothesis of a heterozygous state with a copy number of two. CNVs in only one pup were kept as candidate de novo events. As an internal validation, CNVs were consistently detected by both read pair and depth algorithms except where specific identifiable factors prevented it. For example, it is not possible to detect CNVs that arose by NAHR using read pair orientations. Conversely, the read pair algorithm had higher sensitivity for small events.
Assessment of CNV de novo status, parent of origin, and breakpoint sequences
We first validated that candidate CNVs were real, and not an artifact of the genomic detection method using droplet digital PCR (ddPCR) with the QX-200 ddPCR EvaGreen Supermix (Bio Rad). Test primers were designed within the called CNV. Control primers were designed within a verified copy number 2 region of chromosome 16. ddPCR was performed on DNA from the pup and a control mouse lacking the called CNV. Validated candidate de novo CNVs were unique to F1 pups but might have arisen in either parental germline, or in the mouse colonies that gave rise to the parent animals. Accordingly, we next examined the parents of each pup with a validated CNV using the same ddPCR approach. If the CNV was absent from both of its parents, it was classified as a de novo CNV. Parent of origin of de novo CNVs could be determined in cases where the CNV spanned one or more informative SNPs in the pup. PCR amplification of the followed by Sanger sequencing revealed which allele was affected by the CNV (loss or gain) and thus, in which parent’s germline the CNV arose. CNV breakpoint junctions were amplified using the Expand Long Template PCR System (Roche Applied Science) as previously described [Arlt, et al., 2009]. All products were subjected to standard Sanger sequencing and the resulting sequence was compared to the reference genome (build mm9) to identify the breakpoint junctions.
Detection of Turner 39,X mice
Turner mice with a 39,X karyotype were revealed by mate-pair sequencing read count ratios of sex chromosomes and autosomes consistent with one copy of chrX and no copies of chrY, where expected ratios were established from the cohorts of XX and XY mice. Similar to above, prior knowledge of SNPs in C3H/HeJ relative to C57BL/6 readily established the parental origin of the chrX that was present. For follow-up studies of unsequenced mice, DNA was extracted from tails snips by using the Hot Sodium Hydroxide and Tris (HotSHOT) method [Truett, et al., 2000]. Briefly, 75μl alkaline lysis buffer (25mM NaOH, 0.2mM EDTA, pH of 12) was added to 0.2-cm tail snips. Samples were heated to 95°C for 30 minutes, cooled to 4°C, and followed by an addition of 75μl neutralizing reagent (40mM Tris-HCl). We first used this DNA and sex determining region Y (SRY) primers (Forward: 5’-TTGTCTAGAGAGCATGGAGGGCCATGTCAA-3’; Reverse: 5’-CCACTCCTCTGTGACACTTTAGCCCTCCGA-3’) to identify and discard males with a detectable Y chromosome. PCR was performed on the remaining animals using primers (Forward: 5’-ACACAGGAAAGCCCTTGACA-3’; Reverse: 5’-TGCACCATCAGCATGCTCTT-3’) flanking a C57BL/6 chrX SNP rs13483744 that created a restriction fragment length polymorphism (RFLP) between the two strains. PCR products were cut with Xba I and run on a gel, where XX females showed both cut and uncut products whereas paternally-derived 39,X females showed only uncut products. Animals that had been subjected to mate-pair sequencing where sex chromosome status was known were used to validate the assay, since no additional 39,X pups were found.
Micronucleated reticulocyte assay
For mice treated using gavage, seven dose groups were examined: 0, 50, 55, 60, 65, 70, and 75 mg/kg HU, administered twice daily. Dosing was maintained for 42 days, after which blood was drawn. For mice treated using pumps, two experimental groups were used, untreated (N=5) and 19.5 mg/kg/hr HU (N=4). Blood was drawn from the lateral tail vein with a 21-gauge needle on Day 0 (untreated), Day 1, and Day 3 of treatment and 10 μl analyzed for micronucleated reticulocytes as described in Hayashi et al. [Hayashi, et al., 1990].
Data availability
All microarray and mate-pair sequencing data used for interpreting HU effects have been deposited in NCBI’s Gene Expression Omnibus [Edgar, et al., 2002] under series accession number GSE114754 and Sequence Read Archive [Leinonen, et al., 2011] under study accession number SRP142308, respectively.
Results
Overall experimental strategy
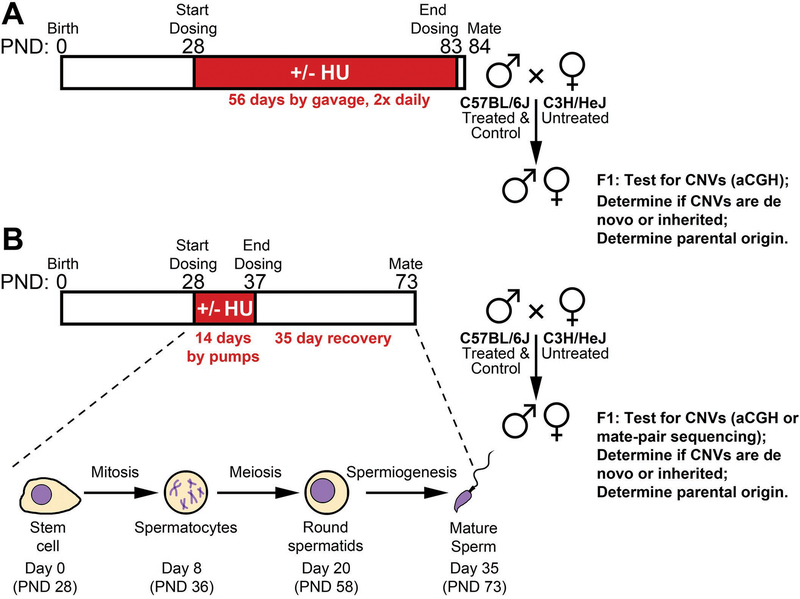
We performed several trials of HU administration with the common design strategy outlined in Fig 1. Throughout, we sought to use the maximally tolerated dose (MTD) of HU to afford the highest possible likelihood of observing an effect of HU on de novo CNV formation, where the dose limiting toxicity was an inability to sire offspring due to reduced spermatogenesis or failure to thrive. Blood plasma drug measurements and reticulocyte micronucleus assays further informed dosing efficacy. HU was administered to male C57BL/6 mice via gavage or subcutaneous microinfusion pumps in the postnatal reproductive period during a time window expected to include mitotic divisions in the germline. Mice treated via gavage received HU for 56 days prior to the initiation of mating. Mice treated via microinfusion pumps received HU for 14 days and allowed to recover for 35 post treatment, during which time cells undergoing mitotic cell divisions during treatment developed into mature sperm [Griswold, 2016]. After the treatment and recovery periods, treated males were mated alongside untreated controls to C3H/HeJ female mice to produce hybrid offspring. CNVs were scored in independent offspring (at most two per sired litter) by whole genomic analysis, either high-density microarray comparative genomic hybridization (aCGH) or low coverage mate-pair sequencing (see Methods). Digital droplet PCR (ddPCR) or aCGH was used to assess the presence of detected CNVs in parents to determine whether CNVs were de novo or inherited. The cross of C57BL/6 males by C3H/HeJ females was chosen to ensure a high number and density of heterozygous polymorphisms for finally determining the parent of origin of observed CNVs.
Figure 1. Overall experimental outline of HU treatment and mating.
Male C57BL/6 mice were treated with HU by either oral gavage (A) or subcutaneous microinfusion pumps (B). In the former, dosing continued up to the time of mating, in the latter, mice were allowed to recover for 35 days during which time spermatogenesis completed from stem cells that were dividing during the treatment period. Common design features included mating of treated C57BL/6 males with C3H/HeJ females to produce F1 offspring heterozygous for a large number of polymorphisms used to assign the parent of origin of CNVs detected by whole genomic analysis, either aCGH or low-coverage mate-pair sequencing.
HU administration and dose finding by gavage
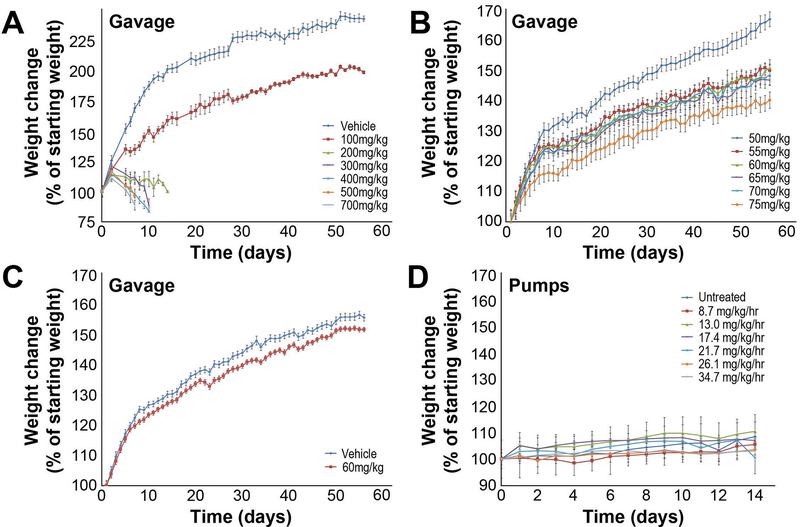
We first administered HU to mice by oral gavage. Based on published reports in mice and rats [Rich and De Kretser, 1977, Wiger, et al., 1995], we estimated that a twice-daily oral dose of 500 mg/kg would approximate the MTD in mice. We performed a dose-range finding study with HU doses of 0, 100, 200, 300, 400, 500, and 700 mg/kg given twice daily to six male mice in each group. For all gavage experiments, dosing began at postnatal day (PND) 28 and continued for 56 days, at which time mice were mated. Strikingly, all mice treated with HU at doses greater than 100 mg/kg showed severe weight loss and distress and were euthanized after 8–12 days of treatment (Fig 2A). Only the 0 and 100 mg/kg group mice completed the 56 day trial. There was a clear effect on weight gain seen at 100 mg/kg but the mice were otherwise healthy. We attempted to breed four males in each group. While plugs were noted, indicating successful mating, none of the 100 mg/kg HU-treated mice sired litters. C57BL/6 mice are thus very sensitive to the effects of HU in a 56 day treatment protocol, even at 100 mg/kg HU, a much lower dose than expected based on published experiments using shorter treatment periods [Wiger, et al., 1995].
Figure 2. Dose range finding studies weight gain.
Weight gain in mice administered with the indicated doses of HU by gavage (A-C) and microinfusion pumps (D). Weight gain is indicated as a percent change in animal weight compared to weight at Day 0. HU treatment began on Day 1 and continued through 56 days (gavage) or 14 days (pumps).
A second dose-range finding study was performed using lower doses of 0, 6, 12, 18, 25, 50, 75, and 100 mg/kg for 56 days. None of these treated mice showed notable ill effects of drug treatment, although weight gain was reduced at the highest doses (Fig 2A). Mice treated with doses of HU up to 50 mg/kg sired normal sized litters while the mice treated with 75 and 100 mg/kg were infertile (Table 1). These results indicated a very narrow range for the MTD between 50 and 75 mg/kg HU.
Table 1.
Summary of histopathology and breeding capacity of HU-treated males.
| Gavage | |||
| HU dose (mg/kg) | Histopathology (testes) # affected/evaluated Severity grade (0 to 5)1 | # of males that sired litters (%)2 | Average Litter Size2 |
| 0 | Normal 5/5 |
3/4 (75%) 23/24 (96%)2 |
6.4 6.12 |
| 6 | n.d. | 4/4 (100%) | 4.7 |
| 12 | n.d. | 4/4 (100%) | 6.2 |
| 18 | n.d. | 4/4 (100%) | 4.6 |
| 25 | Normal 1/1 | 4/4 (100%) | 5.0 |
| 50 | Germ cell depletion/degeneration 1/1 Grade 1 out of 5 |
3/4 (75%) 5/5 (100%) |
6.4 5.8 |
| 55 | n.d. | 4/5 (80%) | 6.7 |
| 60 | Germ cell depletion/degeneration 6/6 Grade 2–4 out of 5 |
3/5 (60%) 31/42 (74%)3 |
6.7 4.93 |
| 65 | n.d. | 4/5 (80%) | 3.3 |
| 70 | n.d. | 3/5 (60%) | 1.7 |
| 75 | Germ cell depletion/degeneration 1/1 Grade 4 out of 5 |
0/4 (0 %) 0/5 (0%) |
- - |
| 100 | Tubular atrophy (complete loss of germ cells) 1/1 Grade 5 of 5 |
0/4 (0 %) | - |
| Pumps | |||
| HU dose (mg/kg/hr) | Histopathology (Testes) # affected/evaluated Severity grade (0 to 5)1 | # of males that sired litters (%)2 | Average Litter Size2 |
| 0 | Normal 4/4 | 4/4 (100%) 18/20 (90%)2 |
5.0 6.42 |
| 8.7 | n.d. | 2/2 (100%) | 5.0 |
| 13.0 | n.d. | 1/2 (50%) 1/4 (25%) |
8.5 7.5 |
| 17.4 | n.d. | 1/3 (33%) 3/4 (75%) 22/38 (58%)3 |
9.0 5.3 5.23 |
| 19.5 | Germ cell depletion/degeneration 5/5 Grade 1–2 out of 5 |
18/47 (38%)3 | 5.93 |
| 21.7 | n.d. | 1/2 (50%) 0/0 (0%) |
4.0 - |
| 26.1 | n.d. | 0/3 (0%) | - |
| 34.7 | n.d. | 1/3 (33%) | 2.0 |
See methods for severity grading descriptions.
Rows with multiple data points are from separate dose range finding studies.
Data from definitive studies.
A final dose-range finding study used 50, 55, 60, 65, 70, and 75 mg/kg HU twice daily for 56 days. There was a dose-dependent effect on weight gain consistent with previous trials (Fig 2B). As in the previous experiment, mice treated with 75 mg/kg HU were infertile, but mice were able to sire litters at all other doses, with 5 out of 5 mice (100%) treated with 50 mg/kg HU siring litters, and 3 out of 5 mice (60%) siring litters at 60 and 70 mg/kg HU. The 70 mg/kg dose also dramatically reduced litter size (Table 1). Based on mouse weights and breeding capacity, a dose of 60 mg/kg HU administered twice daily was chosen for CNV studies. Limitations of this dosing regimen with respect to pharmacokinetics are discussed below.
Histopathology of mice treated with HU by gavage
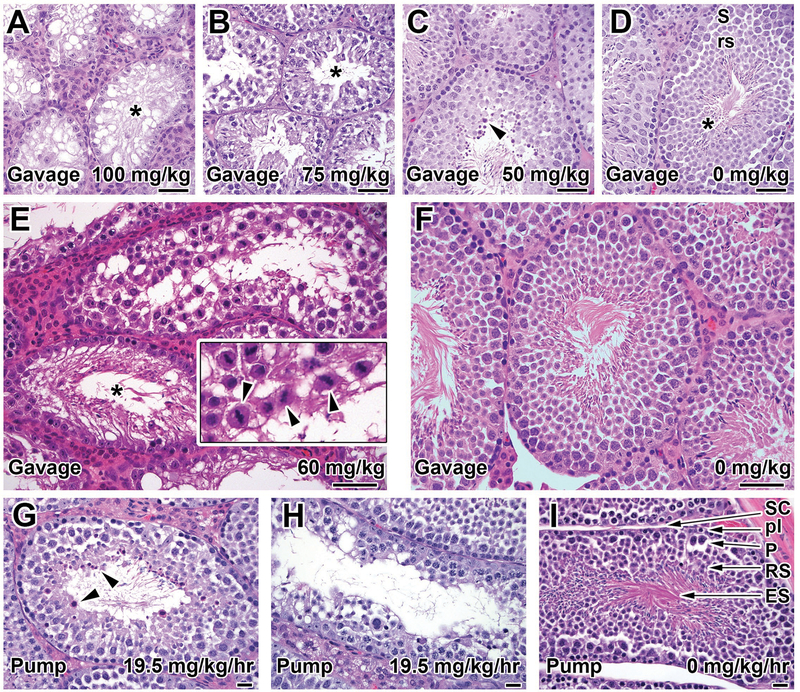
Male reproductive and hematopoietic tissues from mice treated with 0, 25, 50, 75, and 100 mg/kg twice daily in the dose-range finding study were histopathologically assessed (Fig 3A–D). HU-treated mice demonstrated a number of degenerative changes in the testes, with secondary effects in the epididymides, in a dose-dependent manner with little to no functional sperm production at the highest doses. At 100 mg/kg, there was severe atrophy of almost 100% of the testicular seminiferous tubules, with diffuse depletion of germ cells and vacuolation of the remaining Sertoli cells (Fig 3A). The epididymides contained no sperm. At 75 mg/kg, approximately 75–80% of the tubules had partial depletion of germ cells, consisting of greatly depleted round and elongated spermatids (post-meiotic stages) and decreased spermatocytes. The epididymides had markedly decreased sperm and increased exfoliated germ cells. At 50 mg/kg (Fig 3B), there were qualitatively similar effects of germ cell depletion but only 20–25% of the seminiferous tubules were affected. The epididymides contained slightly decreased numbers of sperm. At 25 mg/kg HU the testes were almost histologically normal apart from increased residual bodies and slightly decreased mature spermatids in ~5% of tubules (Fig 3C). The epididymides.at this dose had a normal amount of morphologically normal sperm. Control animals had morphologically normal testes and epididymides (Fig 3D).
Figure 3. Histopathology of testis after HU treatment.
Representative images of seminiferous tubules showing dose-dependent germ cell depletion and degeneration in mice administered the indicated doses of HU by gavage (A-F) and microinfusion pumps (G-I). Germ cell depletion in the gavaged animals ranging from complete loss of germ cells (tubular atrophy) (asterisk) in the high dose animal (A, 100 mg/kg) to decreased spermatids and spermatocytes in a mid-dose animal (B, 50 mg/kg), and minimal changes apart from a few tubules with mildly decreased elongated spermatids and increased residual bodies (arrowheads) in the 25 mg/kg animal (C). Compare to the control animal (D, Stage VIII tubule) with numerous mature elongated spermatids (asterisk), immature round spermatids (rs), and spermatocytes (S). See text for proportion of tubules affected at each dose. Gavage at 60 mg/kg (E) produced occasional tubular atrophy (asterisk) and depleted post-meiotic germ cells (spermatids) and degeneration and sloughing of pre-meiotic stages (spermatocytes) affecting up to 75% of tubules. This included luminal sloughing of late-stage spermatocytes with visible meiotic spindles (arrowheads, inset). Compare the cell density to that in the control animal (F). Animals receiving 19.5 mg/kg/hr HU by peristaltic pump had mild-to-moderate germ cell depletion evidenced by increased residual bodies (arrowheads, G) and decreased spermatocytes and spermatids (H) affecting between 5–25% of tubules. Compare to the cell density in the control animal (I, Stage VIII tubule) showing numerous elongated spermatids (ES), round spermatids (RS), pachytene spermatocytes (P), and preleptotene spermatocytes (pl) supported by Sertoli cells (SC). Hematoxylin and eosin-staining. Bars A-F: 50 μm, G-H: 20 μm.
In contrast to the germline, hematopoietic alterations were confined to the highest dose (100 mg/kg) mouse, which had a mildly increased myeloid (granulocytic):erythroid ratio in the femoral bone marrow and moderately increased extramedullary hematopoiesis in the spleen. The increased extramedullary hematopoiesis in the spleen was predominantly of the erythroid lineage. There were no hematopoietic alterations in other animals and no lymphoid alterations in any animal.
In the definitive study for CNV induction, six mice treated with 60 mg/kg HU and four vehicle-treated controls were analyzed for testicular or epididymal histopathology (Fig 3E–F). Treated mice had significantly smaller absolute and relative (to body weight) testicular weights at necropsy (mean ± S.D. of treated was 0.060g ± 0.016 vs 0.103 ± 0.008 for control, p=0.0018 by unpaired two-tailed t test). This was consistent with the histological findings of decreased spermatogenesis (germ cell depletion and degeneration) in treated animals (Fig 3E). Germ cell depletion/degeneration manifested as severe depletion of round and elongating spermatids and moderate to marked decreases in pachytene spermatocytes. Frequently, there was luminal sloughing of pachytene spermatocytes with visible meiotic spindles (Fig 3E, inset) and absence of post-meiotic germ cell stages (spermatids), suggesting meiotic arrest. Germ cell depletion/degeneration was present in all 6 treated animals and ranged from mild (5–25% tubules affected, 5 out of animals) to marked (51–75% tubules affected, 1 of 6 animals), thus, although spermatogenesis was decreased, it was not entirely abolished at the 60 mg/kg dose., Epididymal changes affected 50% (3/6) of treated animals, consisting of decreased sperm numbers and mildly to moderately increased (above control levels) sloughed intraluminal germ cells. Despite the decrease in sperm number, most of the sperm within the epididymis were morphologically normal, which is consistent with compound effects on earlier (meiotic and/or pre-meiotic) stages of spermatogenesis, as opposed to abnormalities of spermatid (post-meiotic) maturation. This observation is also consistent with the ability of the majority of treated animals to sire offspring (Table 1). Control mice were histologically normal (Fig 3F), apart from minor background histologic findings of occasional tubular atrophy, tubule dilation, and increased residual bodies, each affecting <5% of tubules. These findings are commonly seen at low levels as background findings in normal young mice and were considered unrelated to treatment [Creasy DM, 2002].
CNV formation in mice treated with HU by gavage
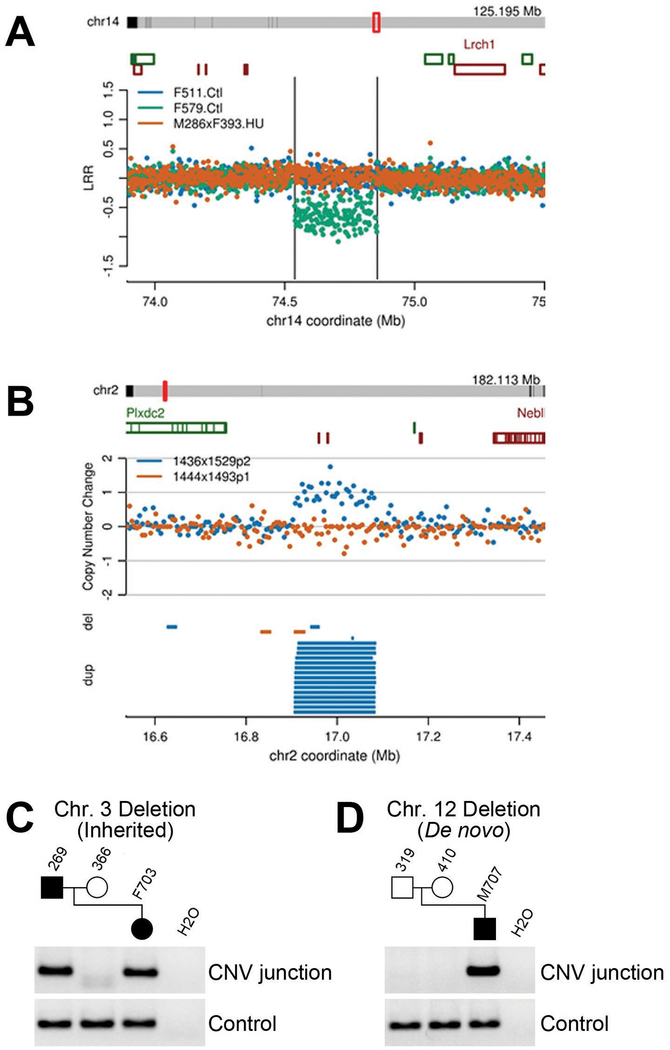
A total of 42 male mice were treated by gavage with 60 mg/kg HU b.i.d. for 56 days and bred to females alongside 24 controls treated with 0 mg/kg HU (water vehicle only). Weight gain data for treated and control animals are shown in Fig 3C. All control males and 34 of 42 (81%) treated males sired litters, with average litter sizes of 6.1 and 4.9 pups, respectively. This modest decrease in fertility was consistent with our targeted MTD. Genomic DNA from 58 pups from HU-treated sires and 22 pups from control sires was analyzed by microarray. We found more than 20 common polymorphic CNV variants segregating in the colony, as well as ten rare CNV variants found in individual pups but inherited from one parent, indicating that they were recent, spontaneous de novo CNV mutations arising during the propagation of these inbred strains (Tables 2 and 3, Supplemental File S1). Five CNVs were confirmed to be de novo, i.e. present in only one pup and absent in both parents. One de novo CNV was maternally-derived in a pup sired by a control male, while the remaining four CNVs were in pups sired by treated males. Three of the latter CNVs were maternally-derived, while a single CNV had an indeterminate parental origin due to a lack of informative heterozygous SNPs. If the latter CNV were of paternal origin, the resulting frequency of de novo, paternally-derived CNVs in treated mice (1/58; 1.72%) and controls (0/22; 0%) would not significant (p=1.00, Fisher’s Exact Test). With these sample sizes, the smallest detectable significant induction of CNVs would have been 11/58 (19.0%), due in part to the low number of control pups tested. In this context, we chose to test more treated pups than control pups because some CNVs must be detected in treated animals if a CNV induction effect were to be possible. Because we observed no such target CNVs with certainty, further testing of controls pups was not indicated or cost efficient.
Table 2:
De novo CNV frequencies
| Delivery | Dose | # pups | De novo CNVs | Maternally derived1 | Paternally derived1 | Unknown parental origin | Maximum frequency of paternally derived CNVs2 | p-value3 |
|---|---|---|---|---|---|---|---|---|
| Gavage | 0 mg/kg | 22 | 1 | 1 | 0 | 0 | 0/22 (0%) | - |
| Gavage | 60 mg/kg | 58 | 4 | 3 | 0 | 1 | 1/58 (1.72%) | 1.00 |
| Pump | 0 mg/kg/hr | 21 | 0 | 0 | 0 | 0 | 0/21 (0%) | - |
| Pump | 17.4 mg/kg/hr | 42 | 4 | 1 | 1 | 2 | 3/42 (7.1%) | 0.299 |
| Combined | NT | 43 | 1 | 1 | 0 | 0 | 0/43 (0%) | - |
| Combined | Treated | 100 | 8 | 4 | 1 | 3 | 4/100 (4.00%) | 0.316 |
Parental origin determined by SNP analysis.
Assumes all CNVs with unknown parental origin are paternal.
Fisher’s exact test, treated compared to untreated in each delivery group.
Table 3:
De novo CNVs
| Delivery | Mouse | HU/Ctrl | CNV coordinates (mm10) | CNV size (kb) | CNV type | Parental origin* |
|---|---|---|---|---|---|---|
| Gavage | F579 | Ctrl | chr14:74138655–74456451 | 317 | Loss | Maternal |
| Gavage | F703 | HU | chr11:73567353–73578788 | 11 | Gain | Unknown |
| Gavage | M707 | HU | chr12:113978427–113993947 | 16 | Loss | Maternal |
| Gavage | M707 | HU | chr6:124510361–124580524 | 70 | Loss | Maternal |
| Pump | 1436x1529p2 | HU | chr16:8982381–9004906 | 22 | Gain | Unknown |
| Pump | 1436x1529p2 | HU | chr2:16905457–17084790 | 178 | Gain | Unknown |
| Pump | 1416x1477p1 | HU | chr2:70453670–70884498 | 427 | Gain/Complex | Paternal |
| Pump | 1442x1475p2 | HU | chr6:16924886–16947509 | 22 | Gain | Maternal |
Parental origin determined by SNP analysis.
The breakpoint junction from one de novo CNV was sequenced: a maternally-derived, 17.7 kb deletion that showed 5 bp of microhomology between the joined ends (Table 4). In addition, junctions from two inherited rare variant CNVs were sequenced. One was a duplication with two base pairs of microhomology and the other was a deletion with one base pair of microhomology. In total, the absence of known paternally derived de novo CNVs did not support the hypothesized induction of germline CNVs by HU. However, lack of HU-induced CNVs was not due to an inability to detect CNVs; CNVs could be and were detected, including 4 of 80 (5%) untreated maternal haplotypes with de novo mutations and ten rare variants arising in the inbred C57/BL6 or C3H colonies, indicating a high rate of this type of mutation in mice.
Table 4.
CNV breakpoint junctions.
| Mouse | Delivery | Treatment | De Novo or Rare | CNV type | Chr | Left breakpoint (mm10) | Right breakpoint (mm10) | # bp homology at junction | Homologous bases |
|---|---|---|---|---|---|---|---|---|---|
| M288xF397 | Gavage | - | Rare | Gain | 6 | 51,318,090 | 51,418,183 | 1 | C |
| M707 | Gavage | HU | De novo | Loss | 12 | 113,977,727 | 113,995,472 | 5 | AACAA |
| F703 | Gavage | - | Rare | Loss | 3 | 82,674,187 | 82,682,462 | 2 | CT |
| 1416x1477 | Pump | HU | De novo | Complex | 2 | 70,881,262 | 70,968,367 | 0 | - |
| 1416x1477 | Pump | HU | De novo | Complex | 2 | 70,453,485 | 70,968,476 | 3 | GGC |
| 1436x1529 | Pump | HU | De novo | Gain | 2 | 16,905,358 | 17,084,979 | 5 | TG(A/T)GG |
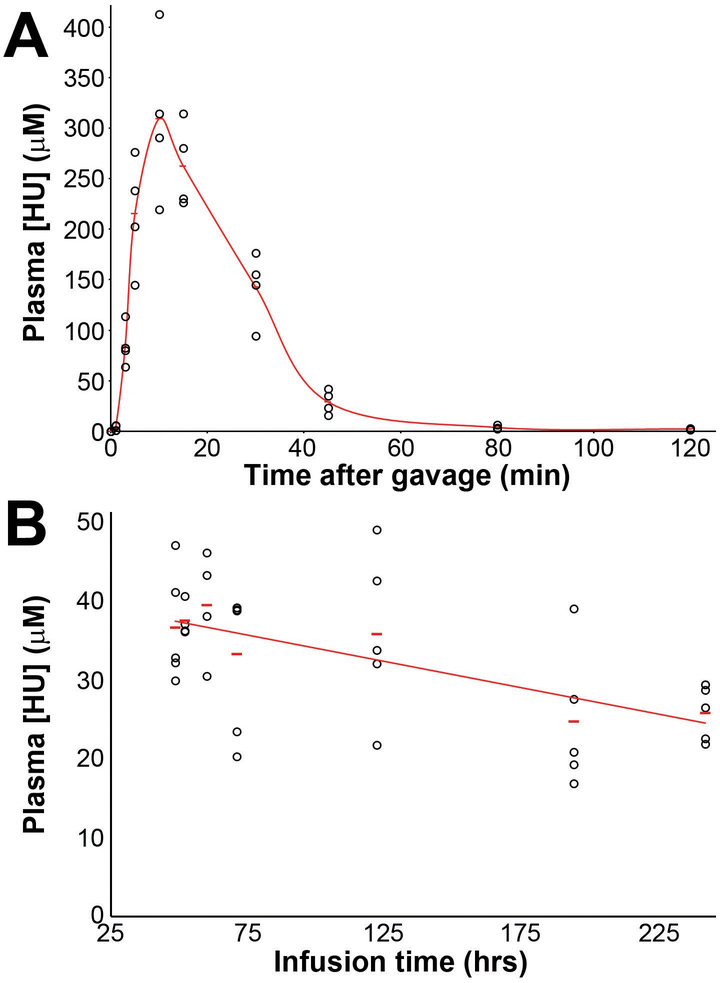
To further understand the nature of HU treatment by gavage, we performed pharmacokinetic analysis of plasma HU levels after treatment with 60 mg/kg HU (Fig 4B). Mice were treated with a single dose of HU and blood samples taken at time points from 48 to 240 hours post-administration. HU levels spiked rapidly in the bloodstream, with a peak concentration of 300 μM within minutes of gavage, and was cleared rapidly, with only trace levels detected at 80 minutes. The elimination rate constant (Ke) was calculated to be −0.049 with half-life of only 13.97 minutes. This resulted in essentially only a 20–30 minute treatment window. We reasoned that the absence of paternally-derived de novo CNVs in the offspring of male mice in the gavage experiment could be the result of this rapid turnover, resulting in long drug-free periods, even with twice-daily dosing, and inefficient perturbation of S-phase replication fork progression considered necessary for CNV formation.
Figure 4. HU pharmacokinetic profiles.
HU levels in plasma were measured by high-performance liquid chromatography tandem mass spectrometry after a single 60 mg/kg dose administered by gavage (A) or a continuous 19.5 mg/kg/hr dose administered by subcutaneous infusion pumps (B). Data are plotted as individual animal measurements, one per animal. In contrast to a high bioavailability and rapid clearance of HU in mice treated orally, HU administered by pumps resulted in a stable continuous HU plasma level for many days.
HU administration and dose finding by subcutaneous peristaltic pumps
To achieve longer windows of persistent replication stress needed for CNV induction, we turned to programmable microinfusion pumps (iPRECIO) as an alternate approach for HU delivery. These battery-operated, surgically-implanted pumps have a refillable reservoir whose contents are released at small time intervals into subcutaneous tissue, thus administering a drug effectively continuously over many days. To test the pumps, mice were treated with 19.5 mg HU/kg/hr and plasma HU concentrations were measured at multiple time points over 10 days (Fig 4B). A nearly constant, steady state concentration of HU was observed in the plasma, ranging from 26 to 40 μM, a marked improvement over oral gavage.
Two 14-day dose range finding studies were conducted using the pumps, again using male fertility as the primary endpoint (Fig 1). The first study used HU doses of 8.7, 13.0, 17.4, 21.7, 26.1, and 34.7 mg/kg/hr. No difference in weight gain in the animals at different doses was noted during the treatment window (Fig 3D). A large drop in the number of mice able to sire litters occurred between 21.7 mg/kg/hr, for which ~40% of treated males sired litters, and 26.1 mg/kg/hr, for which no pups were sired (Table 1). A large drop in litter size occurred between 13.0 and 17.4 mg/kg/hr, with 8 and ~2.5 pups per litter, respectively. The second dose range study examined additional concentrations between 13.0 and 21.7 mg/kg/hr. The greatest decrease in fertility and litter size occurred between 19.5 and 21.7 mg/kg/hr. We therefore chose 19.5 mg/kg/hr for further studies of CNV induction.
Histopathology of mice treated with HU by subcutaneous peristaltic pumps
In the definitive CNV study, five mice treated with 19.5 mg/kg/hr HU by peristaltic pump and four vehicle-treated controls were analyzed histopathologically (Fig 3G–I). Mice treated with 19.5 mg/kg/hr HU showed testicular alterations of germ cell depletion/degeneration that were qualitatively similar to those seen at higher doses in the gavage studies, but were quantitatively much less severe, since they affected only 1–5% (minimal) or 5–25% (mild) of tubules. Affected tubules contained decreased elongated and round spermatids (post-meiotic stages), often with increased luminal or basal residual bodies (Fig 3G). There were also decreased or degenerate pachytene spermatocytes and spermatogonia (Fig 3H). Consistent with the small number of tubules affected, the overall density of sperm in the majority of testicular tubules was normal or only slightly decreased and the epididymal sperm were normal in quantity and morphology. There were no alterations in the testes (Fig 3I) or epididymis of control mice. These observations suggest a reduced toxic effect of HU administered with this technique compared to gavage.
CNV formation in mice treated with HU by subcutaneous peristaltic pumps
In our first experiment using pumps, 47 males were treated at 19.5 mg HU /kg/hr subcutaneously from postnatal day (PND) 28 to 37. These animals were bred to females for seven days beginning 35 days after the end of the treatment period. From our dose-finding studies we expected a fertility rate of ~50% with ~3 pups per litter after HU treatment. In the definitive experiments we obtained a treated fertility rate of 38% with 5.8 pups per litter. Control mice had a fertility rate of 90% with 6.4 pups per litter. Genomic DNAs from 32 pups from HU-treated sires were analyzed for de novo CNV mutations by aCGH. In total, there were three private validated CNVs found in these pups, all of which proved to be rare population variants inherited from one parent.
Given the higher than expected number of treated mice unable to sire litters in this experiment compared to the dose range finding study and the narrow concentration range in which HU has an effect, it was possible that sperm or pups with induced CNVs were being selected against. We therefore repeated the experiment as above, but with a slightly reduced HU dose of 17.4 mg/kg/hr. A total of 42 pups sired by treated mice and 21 pups sired by untreated controls were analyzed for de novo CNVs by mate pair sequencing. Four de novo CNVs were validated in this experiment, all arising in pups sired by HU treated mice (Table 2). One of these CNVs was maternally derived, one was paternally derived, and the remaining two were of indeterminate parental origin because of a lack of informative SNPs. To estimate the largest possible HU effect from these data, we presumptively assigned the indeterminate CNVs to the sires. Thus, potentially 3 of 42 pups from the treatment group had de novo CNVs arising in the paternal germline, as compared to 0 of 21 control pups. This difference is not significant for an HU-induction effect (p=0.299, Fisher’s Exact Test), consistent with findings when HU was administered by oral gavage. Combining the CNV data from this experiment and the gavage experiment, assuming all de novo CNVs of unknown parental origin were paternal, gives a total of 4/100 and 0/43 de novo CNVs in treated pups and controls, respectively (p=0.316, Fisher’s Exact Test).
Breakpoint junctions from two de novo CNVs were sequenced. These breakpoints were again characterized by short stretches of microhomology, characteristic of nonrecurrent CNVs (Table 4). One was a 178 kb duplication with five base pairs of microhomology, the other was a complex duplication/deletion CNV containing two breakpoint junctions, one with three base pairs of microhomology and one with a blunt end.
An incidental finding in this last HU experiment was that two of 42 pups from the initial treatment group had only one copy of chrX and no copies of chrY, as determined from sequencing read counts, and thus had a Turner syndrome 39,X karyotype. SNPs established that the maternal C3H/HeJ chrX was present in both 39,X pups, indicating that a sex chromosome was lost during spermatogenesis in treated males. Published rates of 39,X karyotypes in mice of 0.7% [Russell and Saylors, 1961] and the absence of 39,X karyotypes in the treatment group suggested the possibility of an unexpected induction of germline chromosome segregation defects by HU. However, when we screened additional pups using sex chromosome-targeted PCR, the final tally of 2 of 113 (1.8%) paternally-derived 39,X pups from treated sires and 0 of 141 (0%) from control sires was not significantly different (p=0.197, Fisher’s Exact Test). With these sample sizes, the smallest detectable significant induction of paternally-derived 39,X karyotypes at p<0.05 would have been 4/113 pups (3.5%).
Genotoxicity assessment by micronucleated reticulocytes
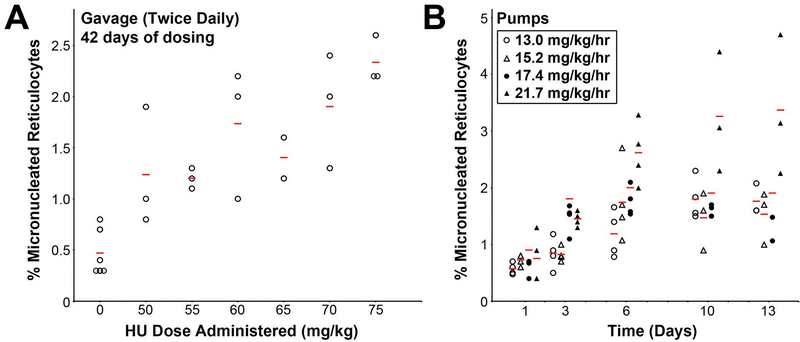
The reproductive deficiencies observed in treated mice verified a tissue effect of HU administration, but we sought further validation of a genotoxic effect. We therefore assessed different doses and methods of HU administration using a peripheral blood reticulocyte micronucleus assay [Hayashi, et al., 1990]. 1000 reticulocytes were evaluated for the presence or absence of micronuclei using fluorescence staining, expressed as a percentage of micronucleated reticulocytes (%MNRET). In the animals treated by gavage for 42 days, untreated controls had a mean baseline %MNRET of 0.47%. At increasing doses of HU, the %MNRET increased, up to 2.33% in mice treated with 75 mg/kg HU (Fig 5). When HU was administered by infusion pumps at a target dose of 13.0 to 21.7 mg/kg/hr, mice had an increase in %MNRET that plateaued by 6–10 day of treatment, with a maximum value of 3.4% at 21.7 mg/kg/hr (Fig 5). These data provide independent confirmation that HU had a substantial impact on DNA damage as administered in our experimental trials.
Figure 5. Micronucleated reticulocytes after HU dosing.
To assess the genotoxic effect of HU, reticulocyte micronuclei were measured after either 42 days of HU administration by gavage at the indicated doses (A), or measured at multiple timepoints after administration by infusion pumps at the indicated doses (B). Data are plotted with individual points for each animal (circles and triangles) and the mean value (bars). In (B), points are offset for each dose for clarity; samples were all collected at either 1, 3, 6, 10 or 13 days. A clear dose- or time-dependent increase in micronuclei was observed in each dosing paradigm.
Discussion
We treated male mice with HU at a MTD determined by reproductive potential and monitored CNV formation genome-wide in offspring. Results support several distinct conclusions with respect to the pharmacologic properties of HU and a high rate of spontaneous de novo CNV formation in mammals as outlined below. Most importantly, within the statistical power of our studies, results fail to support our entry hypothesis that chronic HU administration would increase CNV formation during mammalian spermatogenesis. As discussed, experimental variables related to the short HU half-life in mice and HU administration could have played a substantial role in this result.
Pharmacologic properties of HU in mice
HU has substantial impacts on growth and testicular function [Berthaut, et al., 2008, Modebe and Ezeh, 1995, Wiger, et al., 1995]. Using a different delivery method, there is some evidence that HU has a shorter half-life in the blood of rodents as compared to humans [Iyamu, et al., 2001]. We found each of these effects to be more dramatic than anticipated, which led to experimental and interpretive challenges. Published measurements of blood HU following intravenous administration suggested a half-life in mice ranging from 15 to 19 minutes [Iyamu, et al., 2001]. In contrast, blood HU levels after oral administration are potentially influenced by both absorption and elimination. It was possible that HU would persist longer in the bloodstream after oral administration, due to delayed absorption through the stomach. However, we found that the bioavailability of HU following oral gavage of simple aqueous solutions was very high, with peak plasma levels obtained in minutes. The ensuing elimination half-life was approximately 14 minutes. This led to acute high-dose exposure followed by long drug-free periods following even twice daily oral dosing that are not experienced by patients given the much longer half-life of 3–4 hours in humans [Newman, et al., 1997]. Importantly, more appropriate long-term sustained exposure at levels that perturb but do not completely inhibit DNA replication was readily achieved when HU was administered by surgically implanted subcutaneous microinfusion pumps.
We also found that published HU doses tolerated in rats proved much too high in C57BL/6 mice, especially when treated by oral gavage. Severe growth suppression and toxicity requiring euthanasia were observed at doses above 100mg/kg, even though doses as high as 400 mg/kg/day were tolerated by rats, which maintained some spermatogenesis [Rich and De Kretser, 1977]. We suspect that these half-life and dose-tolerance differences reflect a species- and/or strain-specific difference in HU sensitivity. Most importantly, HU promoted extreme testicular suppression in mice with an extremely narrow dose window regardless of the route of administration. For example, mice progressed from normal sperm production to complete infertility when the gavage dose changed from only 50 to 75 mg/kg. This pattern is potentially highly relevant to our experimental interpretation and for patients, because it reflects an extreme tissue sensitivity of the testis to HU exposure, perhaps realized via apoptosis, which could prevent the long-term replication inhibition in spermatogonial divisions we sought to achieve. Indeed, these highly toxic effects were observed in oral gavage despite the fact that animals were only actually exposed to HU during brief periods of each treatment day. The potential implications of these toxicity mechanisms are discussed further below.
High rate of non-recurrent de novo CNV formation
CNVs and copy number neutral structural variants are now well established as a major form of genetic variation in mammals, and are the predominant form when quantified as the number of affected base pairs. Importantly, the impact of CNVs on human health is large in part because of high rates of de novo CNV formation, which have been estimated in various studies to range from 1.2×10−2 to 3×10−2 [Conrad, et al., 2010, Itsara, et al., 2010]. One consequence of high CNV mutation rates is evident in clinical genetic practice, as many human genomic disorders in patients are caused by CNV mutations absent from either parent [Zhang and Freudenreich, 2007]. Indeed, this study was motivated by the potentially serious implications for offspring if this rate were to increase due to pharmacologic exposure, as possibly suggested by our previous findings that HU at doses comparable to those in treated patients is a potent inducer of CNVs in normal cells in vitro.
Our studies corroborate observations of de novo CNVs in human populations by direct measurement of CNV formation in colonies of inbred mice. In our gavage study, maternally derived de novo CNVs were more frequent than those arising in treated sires, possibly suggesting a higher rate of CNV mutation in C3H/HeJ mice than in C57BL/6. When summed over all experiments and groups (whether treated or not), we found that 8 of 190 (4.2%) pups assessed in this study, four from each administration method, had a de novo CNV. We could also infer a substantial rate of CNV formation from the appearance of additional rare variants in the colony that generated our studied mice. We detected a total of 13 rare variant CNVs in 190 pups (6.8%). While these rare variants did not arise in the gametes giving rise to studied pups, they must have arisen in the approximately ten breeding generations from the frozen embryo stocks giving rise to the C57BL/6 and C3H/HeJ breeding colonies, since only one dam or sire carried them [Taft, et al., 2006]. Still other CNVs were found in pups from a subset of crosses, and therefore inherited from independent parents. Clearly, even inbred mice rapidly acquire large-scale genomic structural alterations that might violate expectations of genetic purity.
Importantly, these collective observations also document that our studied animals did harbor de novo CNVs, and that we were able to detect them with appropriate sensitivity. Thus, we do not believe that the failure to observe an effect of HU on CNV formation arises from technical issues. Nevertheless, we almost certainly did fail to detect any CNVs that were either too small (<~25 kb) or located in excessively repetitive regions, given that our methods are known to be least sensitive for detection of expansions or contractions of genomic repeats.
Most human CNVs, those experimentally induced by aphidicolin and HU in vitro, and the large majority of CNVs detected here, are described as non-recurrent because breakpoint junctions are created by mechanisms independent of long stretches of homology, making them essentially unique in every occurrence. New events of this class were clearly evident in our data. Recurrent CNVs are generated by essentially the same mutational event in multiple independent occurrences by NAHR via long homologous repeats. We did detect one such CNV that segregated within a subset of mice, which appeared to arise via a segmental duplication flanking an intervening duplication. Nevertheless, we had reduced ability to detect such events when there was insufficient unique intervening sequence to reveal a copy number change and so we assume our data underrepresent the rate of spontaneous recurrent CNV formation. This reduced sensitivity was tolerable in the context of our hypothesis that replication inhibition by HU would induce non-recurrent CNVs as seen in cell culture experiments.
Lack of CNV induction by HU
Because of the pharmacologic issues noted above, we ultimately performed several trials of HU administration and in no single experiment, or when considered in aggregate, did we reveal an increased rate of de novo CNV formation in treated animals. Thus, our results support the null hypothesis that HU as administered in these experiments is not a potent germline CNV mutagen, which might provide some reassurance to patients taking the drug. However, a number of limitations of our study must be duly noted. First, for cost reasons our experiments used a relatively small number of total animals in each group. Thus, our study was only powered to reveal large fold differences in CNV formation rates. Such large differences are observed in tissue culture, but the real effect in the mammalian germline might be lesser. For example, our data cannot rule out a 10% increase in CNV formation rates, i.e. approximately one additional CNV in ~190 births, which some might consider a meaningful increase in risk. There are ways that statistical power could be improved in future studies, for example by performing high throughput genomic assessment of individual sperm using rapidly expanding single-cell technologies. Our description of microinfusion pump use and relevant doses will greatly facilitate such an endeavor.
The window in which developing germ cells are dividing mitotically and subjected to the effects of HU-induced replication stress extends over 10 days. This is followed by a long, 35 day period during which sperm mature. Our study design was based on a similar study using the direct-DNA damaging agent benzo[a]pyrene (BaP) [O’Brien, et al., 2016]. BaP is a direct-acting mutagen requiring only a single “hit” and a short exposure. HU, on the other hand, acts indirectly to alter DNA synthesis and must be present throughout the cell cycle and preferably several mitotic divisions for effects on CNV formation. Because HU can delay cell division, it is possible that the cell cycle and sperm maturation timeline were altered such that maximum effects of HU on CNVs were not observed.
Finally, our results may reflect different cellular responses to HU by cells in vitro and those in vivo. There are at least two reasons that CNV detection in the HU-treated mouse germline might occur at a lower fold induction than in tissue culture. First, the basis of our original hypothesis was that HU would act in tissues as a partial mitotic replication inhibitor, allowing the production of cells from stressed but completed S phases. It does have this action in vitro at appropriate doses, but it might not be the predominant effect in tissues even though plasma levels in treated patients are comparable to those that cause replication impairment in tissue culture [Arlt, et al., 2011, Newman, et al., 1997]. The pharmacologic patterns described above suggest that HU has a potent cellular or tissue toxicity that is mediated acutely, not via prolonged inhibition over multiple rounds of replication. This toxicity could be mediated by the sensitive death and removal of cells experiencing impaired replication or by other unknown replication-independent mechanisms. Regardless of the precise mechanism, pre-meiotic spermatogonia and stem cells appear in histopathological analysis to be highly sensitive to HU leading to their elimination before replication-dependent effects can be manifest in gametes. If this toxicity mechanism is not replication-mediated, HU not be an optimal choice for determining the effects of replication impairment on germline CNV formation. Alternatively, programmed sensitivity of sperm progenitors to inhibited replication may serve as a valuable defense mechanism against germline genomic instability. Future studies are needed to probe these important distinctions.
A distinct issue is that 32% and 37% of the CNVs we have observed induced in tissue culture arise in genomic hotspots that strongly correspond to large genes in human fibroblasts and mouse embryonic stem cells, respectively [Wilson, et al., 2015]. Moreover, these genes must be actively transcribed for CNV formation to occur at high frequency. An initial objective of this study was to see if similar transcription-dependent patterns might be observed in the germline. However, we still have incomplete knowledge of the full transcriptional program of cells in the male germline, specifically with respect to the expression of very long (>1 Mb) gene isoforms. If transcription of gene long isoforms is rare in the germline, then the hotspots that contribute to cell culture CNV formation rates might be absent, which would necessitate even higher animal numbers for statistically significant results to be obtained. On the other hand, it is clear that very long gene isoforms are expressed in other tissues, including fibroblasts, lymphocytes and most notably the brain. It will therefore be of considerable interest to explore the impact of HU exposure on somatic CNV formation in tissues that had cells dividing during the time of HU exposure.
Comparison to other germline CNV studies
Only one earlier study examined induced germline CNVs in mice. Adewoye et al. [Adewoye, et al., 2015] found a significant, eight-fold increase in the frequency of de novo CNVs and 2.4-fold increase in de novo indels in offspring of male mice irradiated with 3Gy of X-rays during spermatogenesis. Matings were initiated 8 weeks or 4 days post irradiation, enabling the study of mutation induction at pre- and post-meiotic stages of spermatogenesis, respectively. De novo CNVs were observed in both groups with the highest frequency in offspring of males irradiated during mitotic division stages of spermatogenesis. Among the 14 unique germline CNV mutations found in the offspring of irradiated males, twelve were deletions and two were duplications, with an enrichment for very large deletions (>1Mb) with the loss of up to 80 genes. In this experiment, there was less enrichment for gains, with three deletions and five duplication, including one complex gain. The de novo CNV frequency in controls was 1/93 pups (freq. 0.011), which is lower than the frequency of 0.042 we describe here. This study utilized treated C57BL6/J males crossed to CBA/Ca females, while our study utilized C3H females. Since most of the spontaneous CNVs in our study were maternal in origin, it is possible that spontaneous CNV frequency could differ between mouse stains, thus contributing to this difference.
Supplementary Material
Supplemental File S1. Catalog of all detected CNVs.
This file is a spreadsheet containing information on all CNVs detected in these experiments. CNV genomic coordinates are provided (genome build mm10). “Detection method” indicates whether a CNV was detected by aCGH, anomalous mate pair sequencing fragments (Seq-Pairs), changes in mate pair sequencing read depth (Seq-Depth), or a combination of the above. Log R Ratios (LRR) and Z scores are indicated for CNVs detected by aCGH. “De novo/Common/Rare Variant” indicates whether a CNV was found as a novel CNV in a single pup, not found in either parent (De novo), a common CNV found in multiple pups and parents in multiple crosses (Common), or as a CNV inherited from a parent but not found in other crosses (Rare). HU administration method (Gavage or Pumps) and dose are also indicated. “Notes” contain additional information, such as parent of origin and regions containing segmental duplications.
Supplemental File S2. Comprehensive CNV data images.
This file contains data image panels for all called germline CNVs from the Arlt et al. study of mice treated with hydroxyurea (HU). Panels are organized by experiment, sorted by chromosome/coordinate within each experiment, to match the accompanying Excel table. Each panel shows a chromosome bar at the top, with a red hash/box showing the location of the CNV, followed by an Ensembl gene track. Boxes around plots highlight de novo CNVs. aCGH data use mm9 coordinates and are plotted as the sex-corrected log R Ratio (LRR). A subset of samples are plotted that, when possible, include two pups with and two without the CNV. Vertical lines indicate the span of the CNV region. Mate-pair sequencing data use mm10 coordinates and are plotted first as the copy number change determined from fragment coverage depth, and then as the span of anomalous deletion(del) or duplication (dup) pairs, showing one sample with and one without the CNV.
Figure 6: Detection of CNVs and determination of de novo status.
Examples of de novo CNVs detected by (A) aCGH and (B) mate pair sequencing (see Supplemental File S2 for similar depictions of all CNVs detected in this study). PCR across breakpoint junctions was used to determine whether a CNV in a pup was inherited from its sire (C) or arose de novo (D).
Acknowledgements
We thank Gail Rising for performing iPRECIO pump implantation surgeries and Tsung Tan for help in establishing the use of iPRECIO pump technology for this work. We thank the In Vivo Animal Core (IVAC) of the Unit for Laboratory Animal Medicine at the University of Michigan for necropsy and histopathology preparation. We thank So Hae Park for assistance with mouse handling and drug administration. We also thank Scott Auerbach and Barry Mcintyre for advice on timing and kinetics of HU administration. We thank Manjunath Pai for assistance with calculating HU plasma half-life. This work was supported by grant R01 ES020875 from the National Institutes of Health and University of Michigan M-LEEaD Center grant P30 ES017885.
References
- Lopez C, Saravia C, Gomez A, Hoebeke J and Patarroyo MA. 2010. Mechanisms of genetically-based resistance to malaria. Gene 467:1–12. [DOI] [PubMed] [Google Scholar]
- Bonds DR. 2005. Three decades of innovation in the management of sickle cell disease: the road to understanding the sickle cell disease clinical phenotype. Blood Rev 19:99–110. [DOI] [PubMed] [Google Scholar]
- Algiraigri AH and Kassam A. 2017. Hydroxyurea for hemoglobin E/beta-thalassemia: a systematic review and meta-analysis. Int J Hematol 106:748–756. [DOI] [PubMed] [Google Scholar]
- Madaan K, Kaushik D and Verma T. 2012. Hydroxyurea: a key player in cancer chemotherapy. Expert Rev Anticancer Ther 12:19–29. [DOI] [PubMed] [Google Scholar]
- Steinberg MH, McCarthy WF, Castro O, Ballas SK, Armstrong FD, Smith W, Ataga K, Swerdlow P, Kutlar A, DeCastro L, Waclawiw MA, Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell A and Follow-Up MSHP. 2010. The risks and benefits of long-term use of hydroxyurea in sickle cell anemia: A 17.5 year follow-up. Am J Hematol 85:403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware RE and Aygun B. 2009. Advances in the use of hydroxyurea. Hematology Am Soc Hematol Educ Program 62–9. [DOI] [PubMed] [Google Scholar]
- Charache S, Terrin ML, Moore RD, Dover GJ, Barton FB, Eckert SV, McMahon RP and Bonds DR. 1995. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med 332:1317–22. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Patel RK, Shah V, Nainiwal L and Trivedi B. 2014. Hydroxyurea in sickle cell disease: drug review. Indian J Hematol Blood Transfus 30:91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modebe O and Ezeh UO. 1995. Effect of age on testicular function in adult males with sickle cell anemia. Fertil Steril 63:907–12. [DOI] [PubMed] [Google Scholar]
- Kiladjian JJ, Rain JD, Bernard JF, Briere J, Chomienne C and Fenaux P. 2006. Long-term incidence of hematological evolution in three French prospective studies of hydroxyurea and pipobroman in polycythemia vera and essential thrombocythemia. Semin Thromb Hemost 32:417–21. [DOI] [PubMed] [Google Scholar]
- Mizutani S, Kuroda J, Shimizu D, Horiike S and Taniwaki M. 2010. Emergence of chronic myelogenous leukemia during treatment for essential thrombocythemia. Int J Hematol 91:516–21. [DOI] [PubMed] [Google Scholar]
- Weinfeld A, Swolin B and Westin J. 1994. Acute leukaemia after hydroxyurea therapy in polycythaemia vera and allied disorders: prospective study of efficacy and leukaemogenicity with therapeutic implications. Eur J Haematol 52:134–9. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Howard TA, Mortier N, Avlasevich SL, Smeltzer MP, Wu S, Dertinger SD and Ware RE. 2010. Assessment of genotoxicity associated with hydroxyurea therapy in children with sickle cell anemia. Mutat Res 698:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrisch JR, Pra D, Maluf SW, Bittar CM, Mergener M, Pollo T, Kayser M, da Silva MA, Henriques JA and da Rocha Silla LM. 2008. DNA damage in blood leukocytes of individuals with sickle cell disease treated with hydroxyurea. Mutat Res 649:213–20. [DOI] [PubMed] [Google Scholar]
- Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE and Glover TW. 2011. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci U S A 108:17360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, Konkel MK, Malhotra A, Stutz AM, Shi X, Paolo Casale F, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJ, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HY, Jasmine Mu X, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Genomes Project C, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE and Korbel JO. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C and Scherer SW. 2010. The clinical context of copy number variation in the human genome. Expert Rev Mol Med 12:e8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME and Lupski JR. 2009. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10:451–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST and Glover TW. 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet 84:339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE and Glover TW. 2012. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet 8:e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE and Glover TW. 2014. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ Mol Mutagen 55:103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G and Lupski JR. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ and Hurles ME. 2010. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42:385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Control C, Tyler-Smith C, Carter NP, Lee C, Scherer SW and Hurles ME. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Ozdemir AC, Birkeland SR, Lyons RH Jr., Glover TW and Wilson TE. 2011. Comparison of constitutional and replication stress-induced genome structural variation by SNP array and mate-pair sequencing. Genetics 187:675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J, Schulz-Trieglaff O, Carlson E, Hims MM, Gormley NA and Cox AJ. 2015. NxTrim: optimized trimming of Illumina mate pair reads. Bioinformatics 31:2035–7. [DOI] [PubMed] [Google Scholar]
- Li H and Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M and Snyder M. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318:420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J and Adams DJ. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA and Warman ML. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52, 54. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Morita T, Kodama Y, Sofuni T and Ishidate M Jr. 1990. The micronucleus assay with mouse peripheral blood reticulocytes using acridine orange-coated slides. Mutat Res 245:245–9. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M and Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M and International Nucleotide Sequence Database C. 2011. The sequence read archive. Nucleic Acids Res 39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD. 2016. Spermatogenesis: The Commitment to Meiosis. Physiol Rev 96:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich KA and De Kretser DM. 1977. Effect of differing degrees of destruction of the rat seminiferous epithelium on levels of serum follicle stimulating hormone and androgen binding protein. Endocrinology 101:959–68. [DOI] [PubMed] [Google Scholar]
- Wiger R, Hongslo JK, Evenson DP, De Angelis P, Schwarze PE and Holme JA. 1995. Effects of acetaminophen and hydroxyurea on spermatogenesis and sperm chromatin structure in laboratory mice. Reprod Toxicol 9:21–33. [DOI] [PubMed] [Google Scholar]
- Russell LB and Saylors CL. 1961. Spontaneous and induced abnormal sex-chromosome number in the mouse. Genetics 46:1. [Google Scholar]
- Berthaut I, Guignedoux G, Kirsch-Noir F, de Larouziere V, Ravel C, Bachir D, Galacteros F, Ancel PY, Kunstmann JM, Levy L, Jouannet P, Girot R and Mandelbaum J. 2008. Influence of sickle cell disease and treatment with hydroxyurea on sperm parameters and fertility of human males. Haematologica 93:988–93. [DOI] [PubMed] [Google Scholar]
- Iyamu WE, Lian L and Asakura T. 2001. Pharmacokinetic profile of the anti-sickling hydroxyurea in wild-type and transgenic sickle cell mice. Chemotherapy 47:270–8. [DOI] [PubMed] [Google Scholar]
- Newman EM, Carroll M, Akman SA, Chow W, Coluzzi P, Hamasaki V, Leong LA, Margolin KA, Morgan RJ, Raschko JW, Shibata S, Somlo G, Tetef M, Yen Y, Ahn CW and Doroshow JH. 1997. Pharmacokinetics and toxicity of 120-hour continuous-infusion hydroxyurea in patients with advanced solid tumors. Cancer Chemother Pharmacol 39:254–8. [DOI] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ and Eichler EE. 2010. De novo rates and selection of large copy number variation. Genome Res 20:1469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H and Freudenreich CH. 2007. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell 27:367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RA, Davisson M and Wiles MV. 2006. Know thy mouse. Trends Genet 22:649–53. [DOI] [PubMed] [Google Scholar]
- O’Brien JM, Beal MA, Yauk CL and Marchetti F. 2016. Next generation sequencing of benzo(a)pyrene-induced lacZ mutants identifies a germ cell-specific mutation spectrum. Sci Rep 6:36743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Arlt MF, Park SH, Rajendran S, Paulsen M, Ljungman M and Glover TW. 2015. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res 25:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adewoye AB, Lindsay SJ, Dubrova YE and Hurles ME. 2015. The genome-wide effects of ionizing radiation on mutation induction in the mammalian germline. Nat Commun 6:6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File S1. Catalog of all detected CNVs.
This file is a spreadsheet containing information on all CNVs detected in these experiments. CNV genomic coordinates are provided (genome build mm10). “Detection method” indicates whether a CNV was detected by aCGH, anomalous mate pair sequencing fragments (Seq-Pairs), changes in mate pair sequencing read depth (Seq-Depth), or a combination of the above. Log R Ratios (LRR) and Z scores are indicated for CNVs detected by aCGH. “De novo/Common/Rare Variant” indicates whether a CNV was found as a novel CNV in a single pup, not found in either parent (De novo), a common CNV found in multiple pups and parents in multiple crosses (Common), or as a CNV inherited from a parent but not found in other crosses (Rare). HU administration method (Gavage or Pumps) and dose are also indicated. “Notes” contain additional information, such as parent of origin and regions containing segmental duplications.
Supplemental File S2. Comprehensive CNV data images.
This file contains data image panels for all called germline CNVs from the Arlt et al. study of mice treated with hydroxyurea (HU). Panels are organized by experiment, sorted by chromosome/coordinate within each experiment, to match the accompanying Excel table. Each panel shows a chromosome bar at the top, with a red hash/box showing the location of the CNV, followed by an Ensembl gene track. Boxes around plots highlight de novo CNVs. aCGH data use mm9 coordinates and are plotted as the sex-corrected log R Ratio (LRR). A subset of samples are plotted that, when possible, include two pups with and two without the CNV. Vertical lines indicate the span of the CNV region. Mate-pair sequencing data use mm10 coordinates and are plotted first as the copy number change determined from fragment coverage depth, and then as the span of anomalous deletion(del) or duplication (dup) pairs, showing one sample with and one without the CNV.
Data Availability Statement
All microarray and mate-pair sequencing data used for interpreting HU effects have been deposited in NCBI’s Gene Expression Omnibus [Edgar, et al., 2002] under series accession number GSE114754 and Sequence Read Archive [Leinonen, et al., 2011] under study accession number SRP142308, respectively.