Significance
Current technologies for the semisynthesis of modified proteins are optimized for in vitro reactions and are not easily transferable to more biologically relevant systems such as live cell culture. Split inteins have been a leading tool for protein chemists ever since their discovery, but as of yet have not been used broadly in live cells. Using the shortest naturally occurring split intein, VidaL, we report examples of traceless protein modification in live cells. We expect that the robust splicing activity and synthetic accessibility of VidaL will find broad utility within chemical and cellular biology.
Keywords: protein semisynthesis, intein splicing, protein engineering, chemical biology
Abstract
Split inteins are privileged molecular scaffolds for the chemical modification of proteins. Though efficient for in vitro applications, these polypeptide ligases have not been utilized for the semisynthesis of proteins in live cells. Here, we biochemically and structurally characterize the naturally split intein VidaL. We show that this split intein, which features the shortest known N-terminal fragment, supports rapid and efficient protein trans-splicing under a range of conditions, enabling semisynthesis of modified proteins both in vitro and in mammalian cells. The utility of this protein engineering system is illustrated through the traceless assembly of multidomain proteins whose biophysical properties render them incompatible with a single expression system, as well as by the semisynthesis of dual posttranslationally modified histone proteins in live cells. We also exploit the domain swapping function of VidaL to effect simultaneous modification and translocation of the nuclear protein HP1α in live cells. Collectively, our studies highlight the VidaL system as a tool for the precise chemical modification of cellular proteins with spatial and temporal control.
Protein splicing is an autoprocessing event whereby an intervening protein, known as an intein, excises itself from a precursor polypeptide, resulting in the ligation of its flanking (extein) sequences (1). Inteins are known to exist in two forms: contiguous versions, which are transcribed and translated as a single polypeptide along with their exteins, and split inteins that are fragmented between two different genes and are transcribed and translated separately. In the latter case, the N-terminal intein fragment (IntN) must associate with the C-terminal fragment (IntC) before they can undergo protein trans-splicing (PTS) to generate the spliced product (ExtN-ExtC) (2–4). PTS can be used to ligate chemically defined polypeptide fragments with temporal control, which enables investigations into protein structure and function, including the effects of posttranslational modifications (PTMs) (5–9).
While intein-based technologies are widely used in chemical biology (1), there exists a yet unsolved disconnect between experiments performed in vitro and those that are possible in vivo or in live cells. An ideal method would involve the delivery of a minimal synthetic fragment to undergo a rapid and bio-orthogonal splicing reaction in living cells. Initial progress toward this goal has been made over the past two decades (10–12); however, the need for a more robust platform persists. In particular, protein semisynthesis has not yet been achieved in live cells, a limitation linked to the size of the IntN and IntC fragments in canonically split intein systems that has constrained PTS to the tagging of protein termini in cells.
Atypically split inteins, which feature much shorter IntN fragments compared to their canonically split counterparts, hold great promise for studying challenging protein substrates (13–16). In principle, the small size of the IntN fragment expands the range of N-extein cargoes that can be appended through synthetic means, opening new possibilities for protein semisynthesis, including potential live-cell applications. Here, we report the characterization of the atypically split intein VidaL and demonstrate the utility of this ligase system for protein semisynthesis in multiple contexts. Informed by a crystal structure, we investigate the molecular basis of VidaL fragment association and splicing, gaining insights that we exploit in the generation of challenging protein substrates in vitro as well as the semisynthesis of posttranslationally modified proteins in live cells. This work represents an important advance in our ability to chemically modify cellular proteins with spatial and temporal control.
Results
VidaL Displays Robust Activity under a Wide Array of Splicing Conditions.
The majority of known split inteins are fractured nearer to their C termini, resulting in IntN and IntC fragments, which are ∼100 and 35 amino acids (aa) in length, respectively (17). The discovery of atypically split inteins, for which the size of the fragments is reversed, has opened new possibilities for protein semisynthesis (Fig. 1A). A recent report described an atypical split intein, named VidaL, with a remarkably short IntN fragment of just 16 aa (15). This system employs a serine residue as the nucleophile at the C-terminal splice junction (as opposed to the more common cysteine nucleophile), an attractive feature since this residue is always retained in the final spliced product. These properties of the VidaL system, in principle, make it well suited to protein semisynthesis. As a prelude to such applications, we undertook an investigation of the molecular recognition and protein splicing properties of this split intein system.
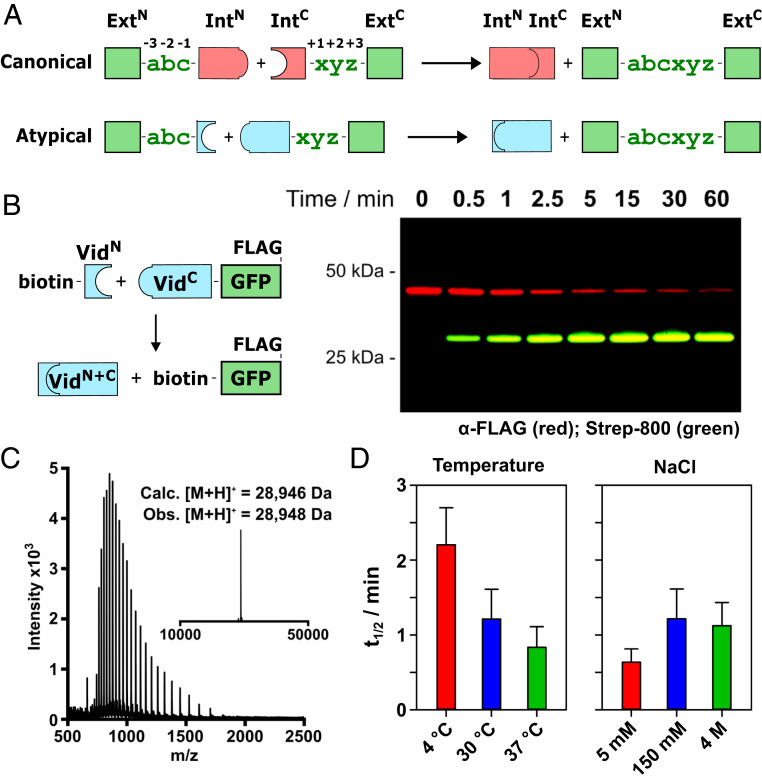
Fig. 1.
In vitro characterization of VidaL. (A) Schematic for protein trans-splicing reactions with canonically split (Top; in red) and atypically split (Bottom; in blue) inteins. Three N- and C-extein residues on either side of the splice junction are shown in green. (B) Schematic for in vitro PTS with VidaL to generate labeled GFP (Left) and a representative Western blot of the reaction (Right). FLAG-tagged starting material (in red) decreases over time, and spliced product (dually tagged) increases. (C) ESI mass spectrum (deconvoluted inset) for biotin-GFP-FLAG spliced product. (D) PTS half-lives across a range of temperatures (Left) and NaCl concentrations (Right); reaction as in B. Conditions: 2 μM VidC-GFP-FLAG, 4 μM biotin-VidN, splicing buffer (pH 7.2). Error bar indicates SD (n = 3).
The protein trans-splicing rate of VidaL was determined in vitro using model proteins with native N- and C-extein residues at the splice junction (Fig. 1B). Specifically, VidaLC was fused to GFP-FLAG via a native extein spacer comprising Ser-Gly-Lys (VidC-GFP-FLAG), while VidaLN was fused via a Glu-Ser-Gly native extein spacer to either HA-tagged MBP (HA-MBP-VidN) or biotin (biotin-VidN; SI Appendix, Fig. S1). Upon mixing the N- and C-intein constructs, we observed rapid spliced-product generation with a reaction half-life of ∼1 min (Fig. 1 B and C and SI Appendix, Fig. S2), placing VidaL among the fastest-splicing split inteins (18, 19). Importantly, and despite the extended C-intein fragment, the rate of premature C-terminal cleavage was exceptionally slow, with <2% observed over the course of 8 h (SI Appendix, Fig. S3). This is in stark contrast to artificially split inteins with short N-terminal fragments, which suffer from elevated levels of this undesirable side-reaction (20).
VidaL was discovered from metagenomic data obtained from Lake Vida in Antarctica, at low temperatures in water with exceptionally high salinity (∼4 M NaCl) (21). We therefore determined the splicing efficiency of VidaL over a range of NaCl concentrations and temperatures. The rates of PTS between VidC-GFP-FLAG and biotin-VidN were measured at 4, 30, and 37 °C (Fig. 1D and SI Appendix, Fig. S4). While we initially suspected that VidaL might be more active in conditions closer to the temperature of its parent organism, we found little difference in the splicing rate across the temperatures tested. Furthermore, we were encouraged by the robust splicing activity at 37 °C as a prerequisite for the use of VidaL in live cells. The electrostatic contribution to split-intein folding can be probed by comparing the splicing rate and binding affinity of the fragments over a range of salt concentrations. Similar to our observations at different temperatures, we found the VidaL splicing rate to be largely unaffected by low and high salt concentrations (5 mM and 4 M NaCl; Fig. 1D and SI Appendix, Fig. S4), further highlighting the robust splicing activity of VidaL. Fluorescence anisotropy was used to measure the binding affinity of the VidaL fragments at the three salt concentrations (SI Appendix, Fig. S5). To enable binding studies, splicing was inactivated through Asn123Ala and Ser+1Ala mutations. The resultant 10-fold increase in Kd despite a near 1,000-fold increase in NaCl concentration (38 ± 12 nM at 5 mM NaCl; 416 ± 86 nM at 4 M NaCl) supports an association and folding mechanism with little contribution from electrostatic interactions. This finding is similar to a recent report for another family of atypical inteins with significantly longer N-intein fragments (16), but stands in contrast to the well-studied “capture and collapse” mechanism of the DnaE family of canonically split inteins (4).
Structural Characterization of VidaL Informs Extein Dependence.
To gain additional insight into the molecular basis of VidaL association and splicing, we generated a fused construct with minimal exteins and Cys1Ala- and Asn123Ala-inactivating mutations for crystallization studies. This construct, along with a selenomethionine-containing variant, was expressed and purified (SI Appendix, Fig. S6), and diffraction-quality crystals were obtained for each. A 1.51-Å–resolution dataset was obtained from a SeMet crystal, and single-wavelength anomalous dispersion phasing was used to solve the structure (SI Appendix, Table S1). This model (Rwork = 0.169; Rfree = 0.190; PDB ID code 6VGW) was used as a search model for molecular replacement in the 1.65-Å–resolution native dataset (Rwork = 0.163; Rfree = 0.209; PDB ID code 6VGV). The all-atom rmsd between the SeMet and native structure was 0.1 Å.
The structure of VidaL revealed a Hedgehog INTein (HINT) fold, conserved between both canonically and atypically split inteins (Fig. 2A) (22). The 16-residue VidaLN region is threaded through VidaLC, completing the β-sheet and placing extein residues in close proximity. Examination of the structure revealed that, as suggested by the biochemical studies, the binding interaction between the intein fragments is largely driven by hydrophobic interactions (Fig. 2B) (23). The structure also provides a possible explanation for the low levels of premature C-terminal cleavage associated with VidaLC–extein fusions; the composite β-sheet within the complex would be disrupted in the isolated C-intein fragment, perfectly priming VidaL for splicing at its evolved split site.
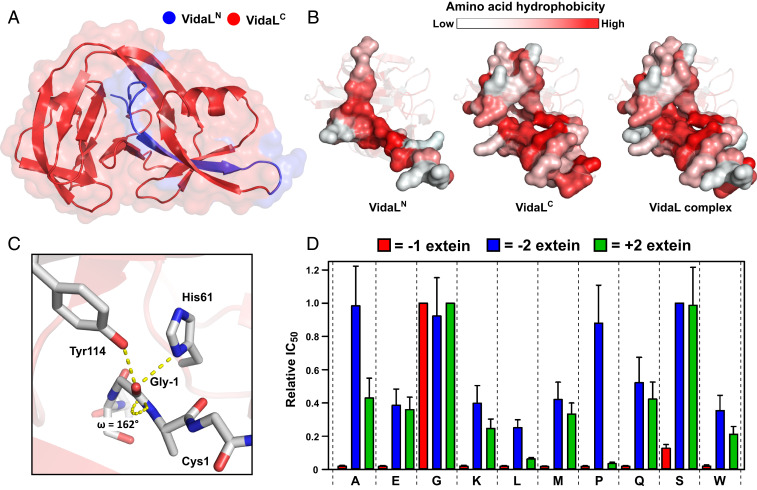
Fig. 2.
X-ray crystal structure of VidaL informs extein dependence. (A) A 1.65-Å–resolution crystal structure of an engineered fused version of VidaL bearing inactivating mutations. The regions corresponding to the N- and C-intein fragments in the split version are colored blue and red, respectively. (B) Space-filling models displaying the interaction between VidaLN and VidaLC with residues colored by hydrophobicity (23). The binding is largely driven by hydrophobic (red) interactions. (C) View of the N-extein of VidaL, displaying H-bonding interactions between Gly-1, His61, and Tyr114 resulting in −1 amide bond distortion (ω = 162°). (D) Extein dependence of VidaL PTS explored using an E. coli-based antibiotic selection assay. Histogram displaying the relative IC50 values of kanamycin for each of the extein mutants tested at the −1 (red), −2 (blue), and +2 (green) extein residues (mean ± SD, n = 3). The value for the wild-type residue at each position is normalized to 1.
The crystal structure also guided our efforts to understand the extein dependence of VidaL. The presence of a Gly-1 residue suggested that either a side chain is not tolerated at this position or the backbone adopts dihedral angles outside of α-helical or β-sheet secondary structure space. Examination of the crystal structure revealed that, while Gly-1 adopts α-helical ϕ and ψ backbone dihedral angles (SI Appendix, Fig. S7), the side chains of His61 and Tyr114 are in position to form hydrogen bonds with the carbonyl oxygen of Gly-1, likely contributing to the observed distortion of the scissile amide bond (ω dihedral angle of 162°) and priming the intein for splicing (Fig. 2C). Attempts to mutate His61 and Tyr114 to liberate space for a different −1 residue were unsuccessful, resulting in either C-terminal cleavage or no splicing activity (SI Appendix, Fig. S8). Given that these two residues are necessary for splicing, we reasoned that the −1 Gly residue would exhibit the least mutational flexibility. Before beginning a systematic extein analysis, we designed constructs varying the −3 and +3 extein residues to determine the scope of our investigation. We observed no loss in splicing rate when mutating Glu-3 or Lys+3 (SI Appendix, Figs. S9 and S10), allowing us to focus our studies on the −2, −1, and +2 extein residues.
We used an assay that selects for intein splicing based on reconstitution of the kanamycin resistance gene in Escherichia coli cultures to systematically test the −2, −1, and +2 extein tolerance of VidaL (24). At each of the three sites, we mutated the wild-type residue to nine other residues (Ala, Glu, Gly, Lys, Leu, Met, Pro, Gln, Ser, Trp), transformed E. coli with plasmids encoding each of the split intein fusions, and compared their survival over a gradient of kanamycin concentrations (Fig. 2D). The results confirmed that mutations to Gly-1 are not tolerated, while mutations to Ser-2 were largely acceptable, with mutations to Gly+2 also moderately tolerated.
Finally, we asked whether the insights into extein dependence gained in vitro were amenable to PTS carried out in isolated nuclei. To this end, we performed PTS reactions between VidaLN peptides varying the −3, −2, and −1 N-extein residues (ESG, RSG, PRG, APR) and VidaLC fused to histone H3. We observed comparable levels of spliced product generation for all but the APR extein construct, consistent with a Gly-1 requirement, and flexibility at the −2 and −3 extein residues (SI Appendix, Figs. S11 and S12). Importantly, understanding the extein dependence of VidaL allows us to tailor our semisyntheses with a minimal extein requirement of a Gly-Ser motif, which is commonly found in the proteome.
Traceless In Vitro Assembly of macroH2A Mononucleosomes.
Next, we set out to use VidaL in the traceless assembly of proteins, exploiting the ubiquitous nature of the minimal Gly-Ser splice junction needed for PTS. We initially focused on systems where an insoluble domain is fused to a soluble domain. These proteins present a significant impediment to biochemical manipulation because of the difficulty inherent in their purification and reconstitution. Insoluble proteins require specific isolation and refolding protocols, which are often incompatible with soluble sequences. We therefore envisioned a protocol where the insoluble and soluble portions are purified separately, followed by a traceless reconstitution step.
An unusual histone variant, macroH2A (25), features this domain architecture, with a linker-histone mimic sequence tethering the folded histone and macro domains (26). macroH2A is proposed to play key roles in the DNA damage response and transcriptional repression (27), but in vitro biochemical studies are lacking due to challenges involved in obtaining homogeneously modified nucleosomal substrates. Recombinant expression in E. coli results in an insoluble histone domain and a soluble macro domain. We posited that the short N-intein fragment of VidaL could be fused to the insoluble histone portion, and VidaLC fused to the soluble macro domain, allowing separate expression and purification and subsequent reconstitution. Accordingly, VidaLN was fused to the C terminus of the histone domain of macroH2A1.1 (macroH2A1.11–119-VidN-H6), and this was expressed and refolded in the presence of wild-type H2B to generate macroH2A1.11–119-VidN-H6-H2B dimers (Fig. 3A). Separately, a soluble H2A macro domain VidaLC fusion (VidC-macroH2A1.1120–372) was expressed and purified (SI Appendix, Fig. S13). Combining these two constructs led to the efficient generation of the desired macroH2A1.1-H2B dimers with an isolated yield of 45% (Fig. 3A and SI Appendix, Fig. S13). These were then combined with wild-type H3–H4 tetramers and successfully assembled into mononucleosomes on the Widom 601 DNA template, resulting in semisynthetic and entirely native macroH2A1.1-containing mononucleosomes (Fig. 3B).
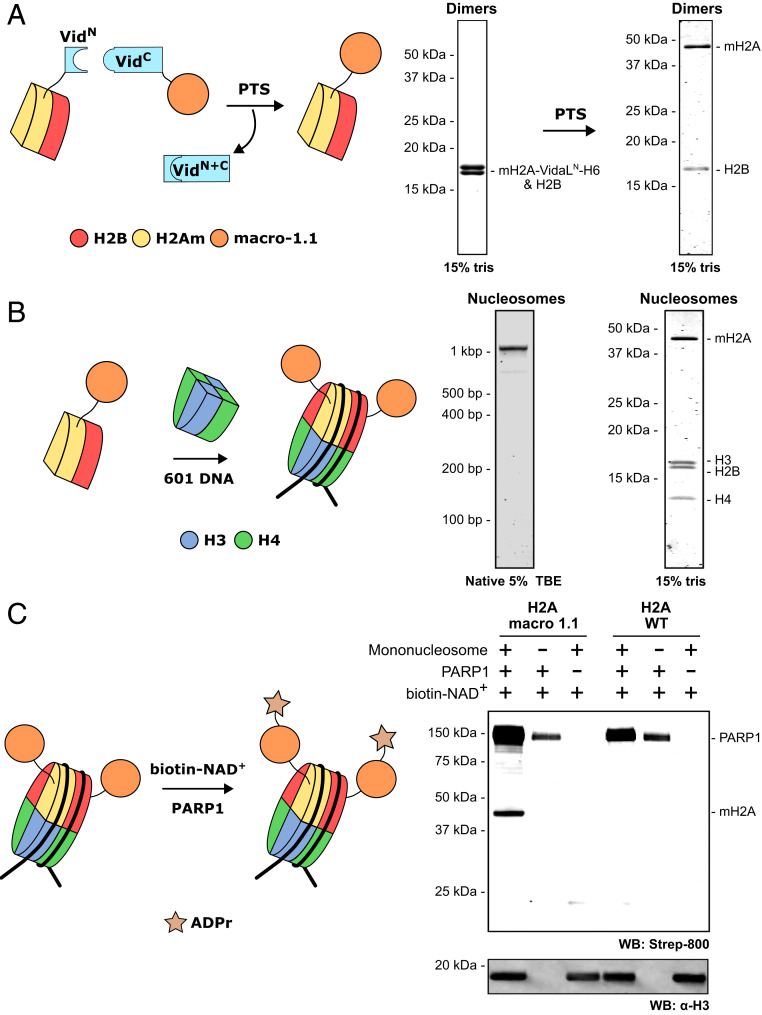
Fig. 3.
Traceless semisynthesis of macroH2A1.1-containing mononucleosomes. (A) Schematic for on-dimer splicing to generate macroH2A1.1-H2B dimers (Left). PTS between assembled macroH2A1.11–119-VidN-H6-H2B dimers and VidC-macroH2A1.1120–372 generates macroH2A1.1-H2B dimers (Right). Histones are visualized on a 15% Tris gel after Coomassie staining. (B) Schematic for macroH2A1.1 mononucleosome assembly with wild-type H3–H4 tetramers and 601 DNA (Left). Homogenous mononucleosomes are observed on a native 5% TBE gel by DNA staining (Center). The four component histones are observed on a denaturing 15% Tris gel after Coomassie staining (Right). (C) Schematic for in vitro ADP ribosylation of macroH2A1.1 mononucleosomes by PARP1, using biotinylated NAD+ to detect modification (Left). (Right) Western blot with biotin detection displaying ADP ribosylation of macro H2A only in the presence of PARP1 and biotin-NAD+. ADP ribosylation of canonical histones is not observed when using wild-type mononucleosomes. H3 serves as a loading control for mononucleosome-containing lanes.
With these substrates in hand, we sought to probe whether nucleosomal macroH2A1.1 is ADP-ribosylated by poly-ADP ribose polymerase 1 (PARP1), a key component of the DNA damage response (28). PARP1 is rapidly recruited to sites of DNA damage, where it installs poly-ADP ribose (PAR) chains on target proteins, in turn recruiting DNA repair proteins to the damage sites. Recently, histone parylation factor 1 (HPF1) was identified as a PARP1-interacting protein, which alters PARP1 substrate specificity to direct PARylation to nucleosomal serine residues (29, 30). Furthermore, the PARylation of core histones has been shown to be almost undetectable in the absence of HPF1. We employed macroH2A1.1 and wild-type mononucleosomes (SI Appendix, Fig. S14) in an in vitro ADP ribosylation assay with PARP1, using biotinylated NAD+ as a cofactor for ease of detection. Robust ADP ribosylation of macroH2A1.1 was observed in our assay (Fig. 3C), with no modification of core histones observed. Interestingly, this reaction occurs in an HPF1-independent manner, in contrast to canonical histones.
Live-Cell Semisynthesis of Dually Modified Histones.
To fully realize the capabilities of VidaL in semisynthesis, we looked to generate posttranslationally modified proteins in live cells. The short length of VidaLN should allow facile cell delivery of selected cargoes when compared to canonically split inteins. Moreover, there are no limitations on the number or chemical composition of selected PTMs that can be installed. It is worth noting that current genetic code expansion methods for installing PTMs in live mammalian cells are largely restricted to one modification per protein, with limited chemotypes (31, 32).
Histones are ideal targets for this method, as their function is regulated through a plethora of PTM chemotypes located on their N-terminal “tail” regions (33). The ability to access chemically modified histone substrates in their native context would represent a significant advance in exploring chromatin biology at the molecular level. Our goal was to install dually modified H3 tails bearing monomethylation of H3 lysine 4 (H3K4me1) and acetylation of H3 lysine 27 (H3K27ac) onto the chromatin of live cells. While it is well established that these modifications characterize active enhancer regions, the molecular mechanism that establishes this epigenetic signature remains unknown (34).
We began experiments in live cells by working at a splice junction between residues 12 and 13 of the H3 tail and delivering the single H3K4me1 modification (Fig. 4A). Upon splicing, a single Ser insertion is present in the H3 tail. Initial studies confirmed that this insertion does not impact cell viability (SI Appendix, Fig. S15). Next, synthetic peptides bearing H3K4me1 or H3K4me0 fused to VidaLN (biotin-H31–12-VidN; SI Appendix, Fig. S16) were delivered via electroporation to HEK 293T cells expressing FLAG-tagged VidaLC fused to truncated H3 and GFP (FLAG-VidC-H313–135-GFP). After 16 h, the cells were harvested and fractionated in the presence of iodoacetamide (critical to prevent ex vivo splicing when using electroporation or cell-penetrating peptides), and, following biotin-IP, Western blotting was used to probe PTS and installation of H3K4me1. We observed robust PTS only when electroporation was performed, confirming the installation of H3K4me1 in a delivery-dependent manner using modification-specific antibodies (Fig. 4B). To confirm that the reactions were occurring inside of the live cells, and not ex vivo, we also visualized the reactions through immunofluorescence and confocal microscopy using fluorescent streptavidin detection of the installed biotin (Fig. 4C).
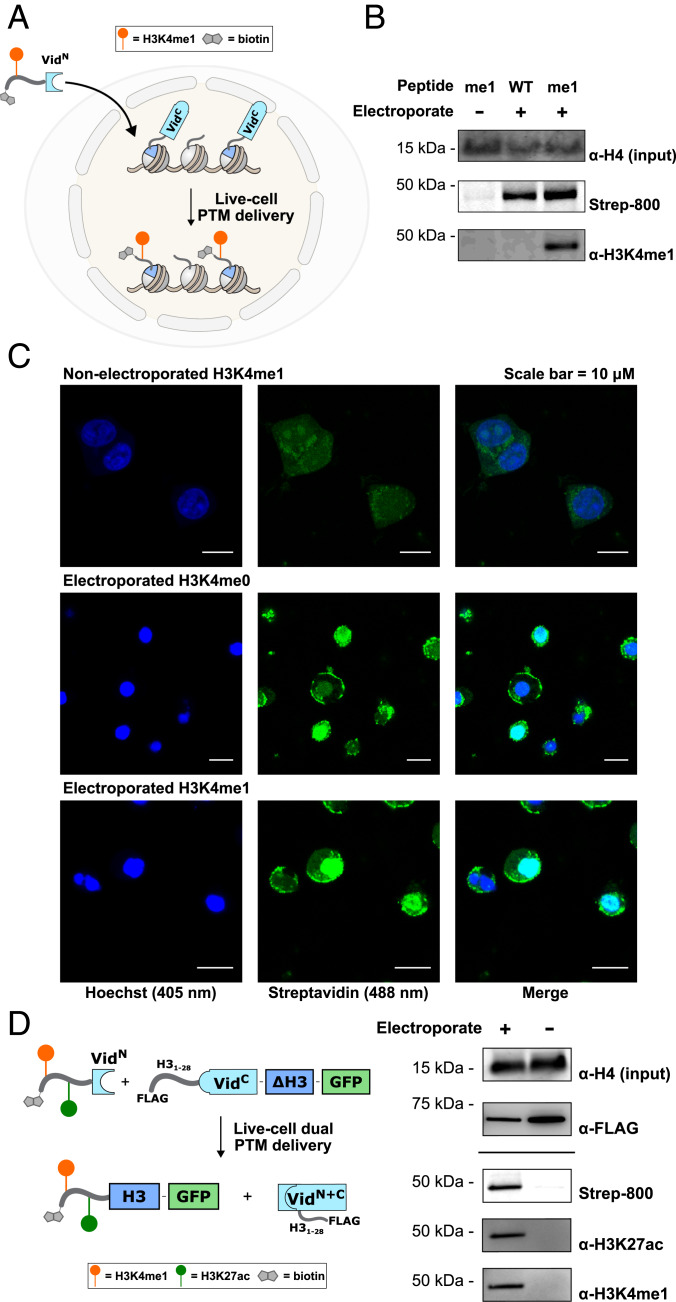
Fig. 4.
In-cell semisynthesis of dually modified histone tails. (A) Schematic for the in-cell semisynthesis of modified histones. Delivery of a biotinylated H3 tail peptide-VidaLN fusion to a truncated H3 protein fused to VidaLC results in chemically modified chromatin after PTS. (B) Semisynthetic H3K4me1-containing chromatin is observed by Western blotting by streptavidin detection of biotin, and by modification-specific antibodies, only when the modified tail is delivered by electroporation to HEK 293T cells. Anti-H4 serves as an input control. (C) Immunofluorescence microscopy images of nonelectroporated HEK 293T cells (Top), cells electroporated with an unmodified H3 tail (Center), and cells electroporated with the H3K4me1 tail (Bottom). Fluorescent nuclear biotin signal is only observed in electroporated cells. (D) Schematic (Left) for the live cell semisynthesis of H3K4me1-H3K27ac modified H3. Western blot (Right) displaying spliced product (biotin detection) and PTM installation by modification-specific antibodies only upon electroporation. Anti-H4 and anti-FLAG serve as input controls.
Next, we extended this approach to the semisynthesis of a dually modified H3 tail (H3K4me1-H3K27ac). For this, we truncated H3 between Gly-33 and -34. A peptide bearing the dual modifications [biotin-H31–33(H3K4me1-H3K27ac)-VidN; SI Appendix, Fig. S16] was delivered via electroporation to cells expressing the corresponding VidaLC construct (FLAG-H31–28-VidC-H334–135-GFP). Note that the C-intein was embedded within the histone tail in order to improve expression (35). After a biotin IP, we observed the presence of both PTMs by Western blotting (Fig. 4D) and confirmed the presence of the unique splice junction and the H3K27ac PTM by LCMS/MS (SI Appendix, Fig. S17). This result represents a semisynthesis of a dually modified protein in cells and highlights the utility of VidaL for accessing substrates with multiple different PTM chemotypes.
Simultaneous Protein Modification and Control of Subcellular Localization in Live Cells.
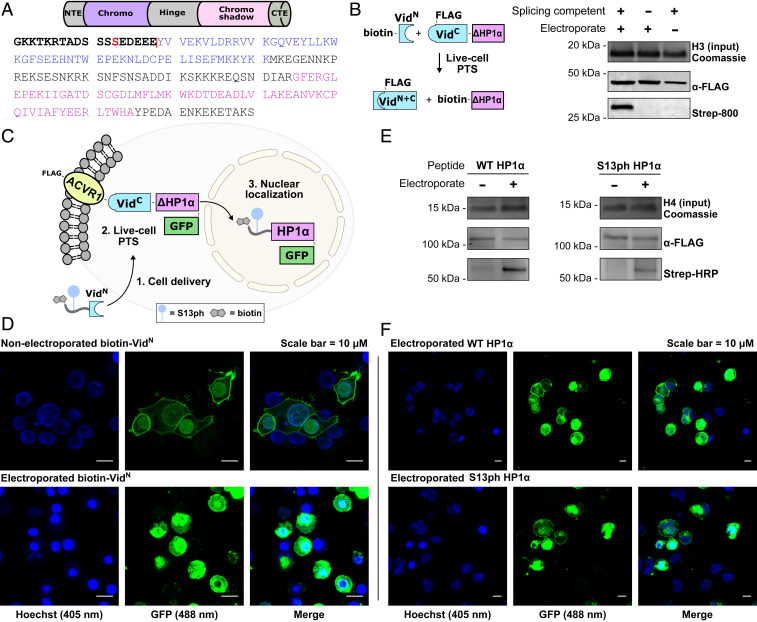
Protein trans-splicing affords us the ability to switch the functionality of a protein, rather than simply ligating polypeptide fragments, as residues fused to the N terminus of IntC will be lost in the reaction and switched with whatever is being spliced on. To illustrate this capability, we chose to simultaneously trigger the nuclear localization and alter the modification state of a protein using PTS. Our protein target was heterochromatin protein 1α (HP1α), a key epigenetic protein involved in the establishment and maintenance of silenced genomic regions (36). A serine patch in the N-terminal extension (NTE) of HP1α is phosphorylated (Fig. 5A), which has implications on the genomic localization and biological function of the protein (37).
Fig. 5.
Semisynthesis and nuclear translocation of HP1α. (A) Annotated HP1α domains displaying the N-terminal extension (NTE) in bold, S13 phosphorylation in red, the splice junction as a red line, the chromo domain in purple, and the chromo-shadow domain in pink. (B, Left) Schematic for in-cell HP1α tagging with VidaL. (B, Right) Western blot displaying spliced product observed by streptavidin detection of the installed biotin only when electroporated with splicing-competent biotin-VidN. No product is observed in cells electroporated with an inactive (biotin-C1A-VidN) delivery construct. H3 staining serves as a loading control. (C) Schematic for the simultaneous semisynthesis and nuclear localization of HP1α using VidaL. HP1α-GFP is fused to the cell membrane by tethering to ACVR1. Upon PTS, the semisynthetic HP1α translocates to the nucleus. (D) Confocal microscopy images for the membrane-bound HP1α-GFP fusion (Top; green) and the spliced product observed in the nucleus after electroporation with biotin-VidN (Bottom; green). Nuclei are stained with Hoechst stain (blue). (E) In-cell semisynthesis of HP1α installing the wild-type (Left) and S13ph tail (Right). Spliced product is observed by streptavidin detection by Western blot when cells are electroporated, with no product observed in the absence of electroporation. FLAG-tagged, membrane-bound starting material is observed by anti-FLAG antibody, and H4 staining serves as a loading control in each case. (F) Confocal microscopy images showing spliced product observed in the nucleus after electroporation with wild-type HP1α NTE VidN (Top; green) and S13ph HP1α NTE VidN (Bottom; green). Nuclei are stained with Hoechst stain (blue).
We first validated HP1α as a semisynthesis target using a splice junction between residues 18 and 19. The corresponding VidaLC construct (FLAG-VidC-HP1α19–190) was expressed in HEK 293T cells, and spliced product was observed upon delivery of the test peptide (biotin-VidN), with no product generated using a splicing-incompetent VidaLN construct (biotin-C1A-VidN; SI Appendix, Fig. S18) or in the absence of electroporation (Fig. 5B).
Next, the HP1α construct was fused to the single-pass transmembrane Activin A receptor (ACVR1) (38), and a C-terminal GFP tag was added to aid with visualization by microscopy (FLAG-ACVR1-VidC-HP1α19–190-GFP; Fig. 5C). Upon expression of the construct, GFP signal was enriched on the membrane of the cells (Fig. 5D). By contrast, delivery of the biotin–VidaLN construct led to a redistribution of the GFP signal to the nucleus, consistent with splicing-dependent liberation of biotin-HP1α19–190-GFP from the cell membrane, leading to nuclear translocation. Finally, a delivery peptide bearing the HP1α N-terminal extension phosphorylated at S13 (S13ph) fused to VidaLN [biotin-HP1α-NTE(S13ph)-VidN] was synthesized, along with the wild-type peptide (SI Appendix, Fig. S19). Each were delivered by electroporation to cells expressing the FLAG-ACVR1-VidC-HP1α19–190-GFP construct, and spliced product in a streptavidin Western blot was observed in each case (Fig. 5E). The reactions were also monitored by live-cell confocal microscopy and displayed strong fluorescent nuclear signal of spliced product with each of the delivered peptides (Fig. 5F), confirming semisynthesis of the HP1α proteoforms.
Discussion
Protein site-directed mutagenesis is a pervasive tool for investigating the mechanisms of cellular processes. However, many questions cannot be easily addressed using standard genetic manipulations, including the role of PTMs in regulating protein function. While several powerful approaches are now available for accessing chemically modified proteins in the test tube (39), manipulating the covalent structure of proteins with the same level of precision is much more challenging in a cellular context, and represents an important frontier in protein science (40). In this study, we performed an extensive structural and biochemical characterization of the atypically split intein, VidaL, work that successfully guided the application of this system for protein semisynthesis both in vitro and in live cells. Indeed, while previous work from our laboratory (12) and others have made strides toward the same goal, the technology described here expands these approaches to perform complete and traceless protein semisynthesis in live cells, rather than being restricted to isolated nuclei, or the tagging of C-termini in cells. Importantly, the current system allows access to the N-terminal regions of proteins, which is of particular significance to the study of posttranslational modifications in chromatin biology.
Key to the utility of VidaL is that it contains the shortest known natural N-intein fragment of just 16 aa. This offers particular utility for N-terminal protein modification given the almost undetectable levels of C-terminal cleavage observed with VidaL. This is in stark contrast to the elevated levels of premature C-terminal cleavage observed when moving the split site of natural inteins closer to the N terminus (15). Hydrophobically driven binding between VidN and VidC at the evolutionally optimized split site results in key catalytic residues being positioned to affect PTS only upon intein fragment association.
Our biochemical studies reveal that VidaL supports rapid protein splicing under a range of conditions and requires a minimal Gly-Ser extein motif for PTS, making it an attractive alternative to existing split intein systems that typically leave behind a cysteine-containing sequence scar at the splice junction (17). Indeed, we note that ∼50% (279,073 of 560,537) of proteins in the UniProt database (41) contain at least one Gly-Ser motif that would allow for completely traceless semisynthesis. As a demonstration of this technology, we semisynthesized H3K4me1- and H3K4me1-H3K27ac–modified histone H3 within the chromatin of live cells. Additionally, we exploited the unique splicing mechanism of inteins to combine protein modification with control of subcellular localization into a single step. The ability to generate site-specifically modified proteins in cell culture with no restriction on the number or composition of the modifications represents a unique advantage of intein-mediated protein semisynthesis. Combinatorial PTM landscapes can be established and evaluated in cells, allowing interrogation of complex epigenetic processes (33). Moreover, the traceless installation of small molecule fluorophores, affinity handles, or other bioorthogonal moieties presents advantages over existing genetic code expansion and protein-mediated labeling technologies (42). This is particularly important for situations where fusion proteins (e.g., GFP, HaloTag) are perturbative to the selected biological setting. Small proteins such as histones (15–20 kDa) are a good example of this, where such fusions more than double the size of the target protein.
The current study relied on electroporation to deliver the synthetic intein-fusion into cultured cells. While we found this to be a reliable method when using HEK 293T cells, we recognize that alternative delivery modalities may be preferable in other contexts. Thus, the VidaL-mediated PTS method will benefit from ongoing efforts to improve cellular delivery of peptides and proteins using various transduction vehicles (43). Indeed, the small size of VidaLN will likely be advantageous in this regard. The technology described herein can also be extended through combination with conditionally splicing intein designs, enabling the triggering of PTS at selected time points or a chosen cellular state (44). Moreover, the combination of the approach described here with dCas9-guided localization could be exploited to enable the precise chemical modification of selected genomic regions or even single loci—an ongoing challenge in genome engineering (45).
Overall, we envision that the VidaL intein will find broad utility for protein engineering. In particular, the ability to affect the precise chemical modification of cellular proteins with spatial and temporal control represents an important advance, opening up new possibilities for studying biochemical processes at the systems level.
Methods
A detailed description of all methods, equipment, and materials used in this study is provided in the SI Appendix. A brief description is provided here.
Split-Kanamycin Resistance Assay.
Assays coupling intein trans-splicing activity to kanamycin resistance in E. coli were performed as previously described (8, 24), sampling selected −2, −1, and +2 extein residues (A, E, G, K, L, M, P, Q, S, and W).
X-Ray Crystallography.
Full-length VidaL, with C-A and N-A mutations, was expressed and purified as described in SI Appendix, Methods. Crystals for the native and SeMet-substituted protein were obtained within 1 wk from a buffer containing 0.1 M sodium acetate (pH 4.6) with 2 M ammonium sulfate. Full experimental details are available in SI Appendix, Methods, and data collection and refinement statistics for the structures are displayed in SI Appendix, Table S1.
In Vitro Protein trans-Splicing.
VidaLN and VidaLC constructs (8 and 4 μM, respectively) in splicing buffer (100 mM phosphate, 150 mM NaCl, 1 mM EDTA, 10% vol/vol glycerol, pH 7.2) were treated with 1 mM TCEP for 10 min at 30 °C. The constructs were then mixed (4 and 2 μM final concentration, respectively), and aliquots were removed at selected time points, quenched by addition of 4× SDS loading buffer, and boiled for 5 min. Where appropriate, the temperature and concentration of NaCl in the assay was adjusted.
MacroH2A1.1 Semisynthesis.
macroH2A1–119-VidN-H6 and H2B dimers in refolding buffer (4 μM final concentration) were mixed in a 1:1 volume ratio with VidC-macroH2A1.1120–372 in purification buffer (3 μM final concentration) in a total volume of 300 μL. The splicing reaction was left to proceed at room temperature for 3 h. The reaction was injected onto an S200 Increase 10/300 GL gel filtration column preequilibrated with refolding buffer. Fractions containing macroH2A1.1-H2B dimers (as identified by SDS/PAGE gel electrophoresis) were concentrated to 8 μM, diluted to 50% vol/vol glycerol, and stored at −20 °C.
Protein trans-Splicing in Isolated Nuclei.
Protein trans-splicing reactions in isolated nuclei were performed as previously described (12). At selected time points, aliquots were removed from the reaction mixture and immediately quenched by addition of 80 mM iodoacetamide and flash-frozen in liquid N2. After thawing the frozen nuclei, chromatin was isolated as described in SI Appendix, Methods, and samples were resolved by SDS/PAGE (12% Bis-Tris gel, 150 V, 60 min) and analyzed by Western blotting.
In-Cell Delivery of VidaLN by Electroporation.
HEK 293T cells were transfected with the appropriate VidaLC construct using Lipofectamine 2000 following the manufacturer’s instructions. After 26 h, the cells were trypsinized and resuspended in cold DMEM (10 mL per 10-cm plate). The cells were pelleted by centrifugation at 1,300 rpm for 5 min, resuspended, washed with cold PBS (2 × 10 mL), and isolated by centrifugation. Finally, the cells were resuspended in PBS at a density of 1 × 107 cells per mL. Each electroporation cuvette was filled with 400 μL of cells, with 6 μM of the appropriate VidaLN peptide. Electroporation was performed on a BTX-830 electroporator (Harvard Biosciences) using 5 × 0.1 msec pulses at 415 V with a 1.1-s delay time. The cells were replated into poly-lysine–coated plates or microscopy dishes containing DMEM at 37 °C with no antibiotics. After 16 h, the cells were harvested in PBS supplemented with iodoacetamide (80 mM final concentration) and recovered by centrifugation at 1,200 × g for 5 min at 4 °C.
Data Availability.
All relevant data are provided in the manuscript and SI Appendix. X-ray crystallographic coordinates and structure factors have been deposited in the PDB under the accession codes 6VGV (VidaL) and 6VGW (VidaL SeMet variant).
Supplementary Material
Acknowledgments
The authors thank Dr. Phil Jeffrey from the Princeton Macromolecular Crystallography Facility, Saw Kyin from the Princeton Proteomics and Mass Spectrometry Core, Dr. Gary Laevsky from the Princeton Confocal Microscopy Core, and Dr. Jenna Gaska for technical assistance. We thank Dr. Robert Thompson and other members of the Muir laboratory for valuable discussions. A.J.B. is a Damon Runyon Fellow of the Damon Runyon Cancer Research Foundation (DRG-2283-17). This work was supported by the U.S. National Institutes of Health (NIH Grant R37-GM086868).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: X-ray crystallographic coordinates and structure factors have been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/) under the accession codes 6VGV (VidaL) and 6VGW (VidaL SeMet variant).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003613117/-/DCSupplemental.
References
- 1.Shah N. H., Muir T. W., Inteins: Nature’s gift to protein chemists. Chem. Sci. 5, 446–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu H., Hu Z., Liu X. Q., Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 95, 9226–9231 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwai H., Züger S., Jin J., Tam P. H., Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Shah N. H., Eryilmaz E., Cowburn D., Muir T. W., Naturally split inteins assemble through a “capture and collapse” mechanism. J. Am. Chem. Soc. 135, 18673–18681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamazaki T. et al., Segmental isotope labeling for protein NMR using peptide splicing. J. Am. Chem. Soc. 120, 5591–5592 (1998). [Google Scholar]
- 6.Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J., Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vila-Perelló M. et al., Streamlined expressed protein ligation using split inteins. J. Am. Chem. Soc. 135, 286–292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens A. J. et al., A promiscuous split intein with expanded protein engineering applications. Proc. Natl. Acad. Sci. U.S.A. 114, 8538–8543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhagawati M. et al., A mesophilic cysteine-less split intein for protein trans-splicing applications under oxidizing conditions. Proc. Natl. Acad. Sci. U.S.A. 116, 22164–22172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giriat I., Muir T. W., Protein semi-synthesis in living cells. J. Am. Chem. Soc. 125, 7180–7181 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Borra R., Dong D., Elnagar A. Y., Woldemariam G. A., Camarero J. A., In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. J. Am. Chem. Soc. 134, 6344–6353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David Y., Vila-Perelló M., Verma S., Muir T. W., Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 7, 394–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel I. V., Volkmann G., Pietrokovski S., Mootz H. D., An atypical naturally split intein engineered for highly efficient protein labeling. Angew. Chem. Int. Ed. Engl. 53, 1306–1310 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Bachmann A. L., Mootz H. D., An unprecedented combination of serine and cysteine nucleophiles in a split intein with an atypical split site. J. Biol. Chem. 290, 28792–28804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neugebauer M., Böcker J. K., Matern J. C., Pietrokovski S., Mootz H. D., Development of a screening system for inteins active in protein splicing based on intein insertion into the LacZα-peptide. Biol. Chem. 398, 57–67 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Stevens A. J., Sekar G., Gramespacher J. A., Cowburn D., Muir T. W., An atypical mechanism of split intein molecular recognition and folding. J. Am. Chem. Soc. 140, 11791–11799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranko A. S., Wlodawer A., Iwaï H., Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 27, 263–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah N. H., Dann G. P., Vila-Perelló M., Liu Z., Muir T. W., Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvajal-Vallejos P., Pallissé R., Mootz H. D., Schmidt S. R., Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J. Biol. Chem. 287, 28686–28696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedel K. et al., A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine. Chem. Sci. 10, 239–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doran P. T., Fritsen C. H., McKay C. P., Priscu J. C., Adams E. E., Formation and character of an ancient 19-m ice cover and underlying trapped brine in an “ice-sealed” east Antarctic lake. Proc. Natl. Acad. Sci. U.S.A. 100, 26–31 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eryilmaz E., Shah N. H., Muir T. W., Cowburn D., Structural and dynamical features of inteins and implications on protein splicing. J. Biol. Chem. 289, 14506–14511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg D., Schwarz E., Komaromy M., Wall R., Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 (1984). [DOI] [PubMed] [Google Scholar]
- 24.Lockless S. W., Muir T. W., Traceless protein splicing utilizing evolved split inteins. Proc. Natl. Acad. Sci. U.S.A. 106, 10999–11004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pehrson J. R., Fried V. A., MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Chakravarthy S. et al., Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25, 7616–7624 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Z., Bernstein E., Histone variant macroH2A: From chromatin deposition to molecular function. Essays Biochem. 63, 59–74 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ray Chaudhuri A., Nussenzweig A., The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs-Seymour I., Fontana P., Rack J. G. M., Ahel I., HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell 62, 432–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonfiglio J. J. et al., Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin J. W., Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y., Addy P. S., Mukherjee R., Chatterjee A., Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem. Sci. 8, 7211–7217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Karnuta J. M., Scacheri P. C., Enhancers: Bridging the gap between gene control and human disease. Hum. Mol. Genet. 27, R219–R227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gramespacher J. A., Stevens A. J., Thompson R. E., Muir T. W., Improved protein splicing using embedded split inteins. Protein Sci. 27, 614–619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eissenberg J. C., Elgin S. C., HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 30, 103–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiragami-Hamada K. et al., N-terminal phosphorylation of HP1alpha promotes its chromatin binding. Mol. Cell. Biol. 31, 1186–1200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petschnigg J. et al., The mammalian-membrane two-hybrid assay (MaMTH) for probing membrane-protein interactions in human cells. Nat. Methods 11, 585–592 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Thompson R. E., Muir T. W., Chemoenzymatic semisynthesis of proteins. Chem. Rev. 120, 3051–3126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David Y., Muir T. W., Emerging chemistry strategies for engineering native chromatin. J. Am. Chem. Soc. 139, 9090–9096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider A. F. L., Hackenberger C. P. R., Fluorescent labelling in living cells. Curr. Opin. Biotechnol. 48, 61–68 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Herce H. D. et al., Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 9, 762–771 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Gramespacher J. A., Burton A. J., Guerra L. F., Muir T. W., Proximity induced splicing utilizing caged split inteins. J. Am. Chem. Soc. 141, 13708–13712 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liszczak G. P. et al., Genomic targeting of epigenetic probes using a chemically tailored Cas9 system. Proc. Natl. Acad. Sci. U.S.A. 114, 681–686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are provided in the manuscript and SI Appendix. X-ray crystallographic coordinates and structure factors have been deposited in the PDB under the accession codes 6VGV (VidaL) and 6VGW (VidaL SeMet variant).