Significance
Most of the thousands of proteins that comprise a human cell have specific subcellular localization patterns essential for their function. “Proximity labeling” (PL) is a method for mapping the localization of endogenous cellular proteins on a proteome-wide scale. To improve the specificity and versatility of PL, we developed split-TurboID, a promiscuous biotinylating enzyme split into two inactive fragments. The fragments are coexpressed in cells and brought together by a drug, protein–protein interaction, or organelle contact to reconstitute TurboID enzymatic activity. We used split-TurboID to map the protein composition of endoplasmic reticulum–mitochondria contact sites, which are essential for mitochondrial fission, lipid biosynthesis, and calcium signaling. For conditional or higher-specificity PL, split-TurboID may be a valuable tool for biological discovery.
Keywords: proximity labeling, ER–mitochondria contacts, split-TurboID
Abstract
Proximity labeling catalyzed by promiscuous enzymes, such as TurboID, have enabled the proteomic analysis of subcellular regions difficult or impossible to access by conventional fractionation-based approaches. Yet some cellular regions, such as organelle contact sites, remain out of reach for current PL methods. To address this limitation, we split the enzyme TurboID into two inactive fragments that recombine when driven together by a protein–protein interaction or membrane–membrane apposition. At endoplasmic reticulum–mitochondria contact sites, reconstituted TurboID catalyzed spatially restricted biotinylation, enabling the enrichment and identification of >100 endogenous proteins, including many not previously linked to endoplasmic reticulum–mitochondria contacts. We validated eight candidates by biochemical fractionation and overexpression imaging. Overall, split-TurboID is a versatile tool for conditional and spatially specific proximity labeling in cells.
Proximity labeling (PL) has been shown to be a valuable tool for studying protein localization and interactions in living cells (1–3). In PL, a promiscuous enzyme such as APEX (4, 5), BioID (6), or TurboID (7) is genetically targeted to an organelle or protein complex of interest. Addition of a biotin-derived small-molecule substrate then initiates biotinylation of endogenous proteins within a few nanometers of the promiscuous enzyme, via a diffusible radical intermediate in the case of APEX, or an activated biotin adenylate intermediate in the case of BioID and TurboID. After cell lysis, biotinylated proteins are harvested using streptavidin beads and identified by mass spectrometry.
PL has been applied in many cell types and species to map the proteome composition of organelles, including mitochondria (5, 8–10), synapses (11, 12), stress granules (13), and primary cilia (14). However, to increase the versatility of PL, new enzyme variants are needed. In particular, split enzymes could enable greater spatial specificity in the targeting of biotinylation activity, as well as PL activity that is conditional on a specific input, such as drug, calcium, or cell–cell contact. For example, contact sites between mitochondria and the endoplasmic reticulum (ER) mediate diverse biology, from lipid biosynthesis and Ca+2 signaling to regulation of mitochondrial fission (15). There is great interest in probing the proteomic composition of ER–mitochondria contacts. However, direct fusion of a PL enzyme to one of the known ER–mitochondria contact resident proteins (e.g., Drp1 or Mff) would generate PL activity outside of ER–mitochondria contacts as well, because these proteins also reside in other subcellular locations (16, 17). On the other hand, use of a split PL enzyme, with one fragment targeted to the mitochondria and the other targeted to the ER, would restrict biotinylation activity to ER–mitochondria contact sites specifically.
Split forms of APEX (18) and BioID (19–21) have previously been reported. However, split-APEX (developed by us) has not been used for proteomics, and the requirement for exogenous H2O2 and heme addition limits its utility in vivo. Split-BioID was first reported by De Munter et al. (19), followed by more active versions from Schopp et al. (20) and Kwak et al. (21). All are derived from the parental enzyme BioID, which requires 18 to 24 h of biotin labeling. We show below that the Schopp et al. (20) split-BioID does not produce detectable activity, while the Kwak et al. (21) split-BioID requires 16+ h of labeling to generate sufficient signal.
Hence we sought to develop an improved, more active split PL enzyme by starting from TurboID. In contrast to APEX, TurboID does not require any cofactors or cooxidants; just biotin addition initiates labeling in cells or animals. TurboID is also >100-fold faster than BioID, requiring only 1 to 10 min of labeling time (7). We performed a screen of 14 different TurboID split sites to identify optimal fragments for high-affinity and low-affinity reconstitution. We converged upon TurboID split at L73/G74, which gave rapamycin-dependent reconstitution when fused to FRB and FKBP in multiple subcellular organelles. We then used this split-TurboID to perform proteomic mapping of ER–mitochondria contact sites in mammalian cells. The resulting proteome of 101 proteins is highly specific and identifies many new ER–mitochondria contact site candidates, eight of which we validated by biochemical fractionation or overexpression imaging.
Results
Development of a Split Promiscuous Biotin Ligase with High Activity.
We started with TurboID, for the reasons given above, and sought to design split protein fragments with no detectable activity on their own, but high reconstituted activity. Given the diversity of ways in which split proteins are used, we envisioned engineering both a low-affinity fragment pair, whose reconstitution could be driven by a protein–protein or membrane–membrane association (Fig. 1A), and a high-affinity pair that would spontaneously reconstitute upon cocompartmentalization of fragments. Previously, we developed split enzymes [split-APEX (18) and split-HRP (22)] by manually selecting cut sites in exposed loops, guided by protein crystal structures. Here, we utilized a recently developed computational algorithm for predicting optimal protein split sites (23). SPELL (split protein reassembly by ligand or light) calculates the energy profile of each candidate fragment relative to that of the full-length protein, and combines this information with solvent accessibility, sequence conservation, and geometric constraints to evaluate potential split sites, aiming for fragment pairs that give high reconstitution efficiency and minimal spontaneous assembly (23). Because crystal structures for TurboID and BioID are not available, we applied the SPELL algorithm to wild-type Escherichia coli biotin ligase [BirA; PDB ID 1HXD (24) and PDB ID 2EWN (25)], from which both enzymes are derived.
Fig. 1.
Engineering split-TurboID. (A) Schematic of split-TurboID reconstitution using the chemically inducible FRB-FKBP dimerization system. Upon rapamycin treatment, two inactive fragments of TurboID reconstitute to form an active enzyme capable of generating biotin-5′-AMP for promiscuous proximity-dependent labeling. N-terminal fragments [Tb(N)] were fused to FKBP and V5. C-terminal fragments [Tb(C)] were fused to HA, HaloTag, and FRB. The HaloTag was used for initial screening as previous studies have shown that it can improve fragment stability (18). (B) Split sites tested. Ten split sites were tested in the first round. In the second round, four additional sites around 73/74 were tested. Split sites are indicated as red lines along the TurboID protein sequence. The α-helices are shown in blue and the β-sheets are shown in purple. (C) Results of split site screen. Split-BioID (split at E256/G257) (20) and Contact-ID (split at G78/G79) (21) are shown for comparison. Each fragment pair was tested in HEK293T cells with 24 h biotin incubation in the presence or absence of rapamycin. At right, cells expressing full-length (FL) TurboID were incubated with biotin for 30 min. FL BioID was incubated with biotin for 24 h. Cell lysates were analyzed by streptavidin blotting as in D, and quantification was performed by dividing the streptavidin sum intensity by the anti-V5 intensity. Values were normalized to that of FL TurboID. (D) Streptavidin blot comparing our best split-TurboIDs to FL TurboID and BioID, and the previously described split-BioID and Contact-ID (20, 21). Labeling conditions were the same as in C. For each construct pair, lanes are shown with both fragments present (B), N-terminal fragment only (N), or C-terminal fragment only (C). Anti-V5 and anti-HA blotting show expression levels of N-terminal fragments (V5-tagged), C-terminal fragments (HA-tagged), and full-length enzymes (V5-tagged). Dashed lines indicate separate blots performed at the same time and developed simultaneously. Asterisks indicate ligase self-biotinylation. Full blots are shown in SI Appendix, Fig. S1A. (E) N- and C-terminal fragments (blue and purple, respectively) of split-TurboID (73/74), depicted on a structure of E. coli biotin ligase (PDB ID 2EWN), from which TurboID was evolved (7). Biotin-AMP in the active site is shown in yellow. The low-affinity split-TurboID cut site is shown in red, the high-affinity split-TurboID cut site is shown in blue, and the previous split-BioID cut site is shown in black (20).
SPELL identified 10 potential split sites, all of which are in exposed loops. We rejected some of them based on prior experimental data: for example, cut site 62/63 was predicted by SPELL, but our previously developed miniTurbo is truncated at amino acid 64 and retains high activity (7). We selected five of the SPELL-predicted cut sites for experimental testing (Fig. 1B). In addition, we included in our screen five more cut sites used in previous split-BioIDs (20, 21). Each fragment pair was cloned as fusions to FKBP and FRB, proteins whose association can be induced by the small-molecule rapamycin (Fig. 1A). The constructs were expressed in the cytosol of HEK293T cells and incubated with biotin for 24 h in the presence or absence of rapamycin. Cell lysates were run on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with streptavidin to evaluate the extent of promiscuous biotinylation. Fig. 1 C and D and SI Appendix, Fig. S1A show that split-TurboIDs cut at 73/74, 78/79, and 98/99 give high reconstituted activity. Cut site 78/79 is the most active, in both the presence and absence of rapamycin, suggesting that the split fragments have high affinity for one another. The SPELL-predicted cut site 73/74 gave the greatest rapamycin-dependent activity, suggesting that it is a low-affinity, or conditional, split-TurboID.
We performed a secondary screen around the cut site 73/74 to further optimize low-affinity split-TurboID. Neighboring cut sites were tested (SI Appendix, Fig. S1B), in addition to pairing of fragments with overlapping or gapped ends (SI Appendix, Fig. S1C). None of these were better than the original 73/74 pair, so we selected this as our optimal low-affinity split-TurboID (referred to simply as “split-TurboID” henceforth).
In a side-by-side comparison to previous split-BioIDs (Fig. 1 C and D), both our high-affinity and low-affinity split-TurboIDs were far more active. The Kwak et al. (21) split-BioID (also termed Contact-ID) showed rapamycin-dependent reconstitution with activity ∼12-fold lower than that of split-TurboID. This is consistent with the reported difference in catalytic activities of the parent enzymes TurboID and BioID (7). Interestingly, when the Contact-ID cut site (78/79) is used to split TurboID, this yields our best high-affinity pair (78/79). The discrepancy between the rapamycin-dependence of Contact-ID and the rapamycin-independence of high-affinity split-TurboID is likely explained by their different regimes of activity; Contact-ID labeling may not be detectable in the omit-rapamycin condition because the intrinsic activity is so low.
In our hands, the previously reported split-BioID from Schopp et al. (20) did not give any detectable signal over background after 24 h of biotin incubation. Interestingly, TurboID split at the same position (256/257) did show some labeling (Fig. 1C), but this activity was also observed with the N-terminal fragment alone (SI Appendix, Fig. S1A), suggesting that this cut site may not yield a true protein complementation system. Notably, we found that the activity of split-TurboID is even greater than that of full-length BioID (Fig. 1D; side-by-side comparison using 24 h of biotin incubation), suggesting that split-TurboID’s activity level should be adequate for a wide range of applications.
By referencing the protein structure of E. coli biotin ligase (PDB ID 2EWN) (25), from which TurboID was evolved, we see that the split-TurboID site (L73/G74) separates the protein into two globular domains (Fig. 1E). It is intriguing that just by moving the cut site five residues away (to 78/79), we produce a split-TurboID system that is high-affinity/rapamycin-independent rather than low-affinity/rapamycin-dependent.
Further Characterization of Split-TurboID.
To further characterize split-TurboID, we confirmed by confocal fluorescence imaging that the constructs catalyze biotinylation in a biotin- and rapamycin-dependent manner (Fig. 2A). Reconstituted split-TurboID is not as active as full-length TurboID, but gave detectable biotinylation after just 30 min of biotin incubation (Fig. 2B). To probe the kinetics of reconstitution, we compared rapamycin preincubation to rapamycin coaddition with biotin. There was no difference in biotinylation activity (SI Appendix, Fig. S2A), suggesting that split-TurboID becomes active and begins catalyzing biotinylation immediately upon rapamycin addition.
Fig. 2.
Characterization of low-affinity split-TurboID. (A) Confocal fluorescence imaging of low-affinity split-TurboID (split site L73/G74). HEK293T cells were transiently transfected and incubated with 50 μM biotin and 100 nM rapamycin for 1 h, then fixed and stained with anti-V5 to detect the N-terminal fragment [Tb(N)], anti-HA to detect the C-terminal fragment [Tb(C)], and neutravidin-647 to detect biotinylated proteins. (Scale bars, 20 μm.) (B) Split-TurboID time course. HEK293T cells transiently transfected with split-TurboID constructs were treated with 50 μM biotin and 100 nM rapamycin for the indicated times, and whole-cell lysates were analyzed by streptavidin blotting. FL TurboID (10 min) and FL BioID (1 h and 18 h) were included for comparison. (C) Design of constructs used to target split-TurboID fragments to various cellular compartments. Full descriptions of constructs are available in SI Appendix, Table S1 (MTS, mitochondrial targeting sequence; SS, signal sequence). (D) Confocal fluorescence imaging of split-TurboID targeted to various cellular compartments. HEK293T cells were labeled and imaged as in A. Fluorescence intensities are not normalized across cellular compartments. Zoomed images of the boxed regions are shown. (Scale bars, 10 μm.)
We also generated constructs fusing split-TurboID with various localization sequences to target the fragments to different subcellular compartments (cytosol, nucleus, mitochondrial matrix, and ER lumen) (Fig. 2C). Confocal fluorescence imaging of cells expressing these constructs labeled with rapamycin and biotin for 1 h shows compartment-specific targeting and rapamycin-dependent biotinylation in all compartments tested (Fig. 2D and SI Appendix, Fig. S2 B and C).
Using Split-TurboID for Proximity Labeling at ER–Mitochondria Contacts.
ER–mitochondria contacts are important in a variety of biological processes, including Ca+2 signaling, lipid metabolism, nutrient signaling, and mitochondrial fission (15, 26, 27). There is tremendous interest in understanding the molecular composition of these contacts. Biochemical purification of mitochondria-associated membranes (MAMs) has frequently been used to study ER–mitochondria contacts (15), but MAMs encompass much more than just mitochondria-associated ER microsomes; they also include contaminants from the plasma membrane, Golgi, peroxisomes, and nuclear membrane (28). To provide a more specific alternative, we recently applied APEX PL to produce separate proteomic maps of the ER membrane (ERM) and outer mitochondrial membrane (OMM), and then intersected the datasets to identify candidate ER–mitochondria contact residents (9). This resulted in the discovery of an ER–mitochondria tethering protein (SYNJ2BP), but many of the hits were merely dual-localized ER and mitochondria proteins.
We sought to use split-TurboID reconstitution across ER–mitochondria contacts in order to map this compartment directly, with much greater specificity than both MAM purifications and separate APEX tagging plus dataset intersection. To target split-TurboID to the ERM and OMM, we fused the fragments to the transmembrane domains of ERM-resident protein Cb5 and OMM-resident protein Tom20, respectively. We also included FKBP and FRB domains to enable rapamycin-induced heterodimerization (Fig. 3 A and B). In U2OS cells, we could observe some biotinylation activity in the absence of rapamycin, but it was substantially increased upon rapamycin addition (Fig. 3C). Thus, it appears that close apposition of mitochondrial and ER membranes is sufficient to mediate some split-TurboID reconstitution, but rapamycin addition further enhances the reconstitution.
Fig. 3.
Reconstitution of split-TurboID at ER–mitochondria contact sites. (A) Schematic of split-TurboID reconstitution across ER–mitochondria contacts in the presence or absence of rapamycin for inducing dimerization. (B) Design of constructs targeting split-TurboID fragments to the OMM and ERM. (C) Confocal fluorescence imaging of split-TurboID activity at ER–mitochondria contacts. Constructs were introduced into U2OS cells using lentivirus. Two days after transduction, cells were incubated with 50 μM biotin and 100 nM rapamycin for 1 h, then fixed and stained with anti-V5 to detect the N-terminal fragment [Tb(N)], anti-HA to detect the C-terminal fragment [Tb(C)], and neutravidin-647 to detect biotinylated proteins. Zoomed images of the boxed regions are shown. (Scale bars, 20 μm.) (D) Localization of split-TurboID in HEK293T cells stably expressing constructs from B. Cells were fixed and stained with anti-V5 to detect the OMM-targeted N-terminal fragment [Tb(N)] or with anti-HA to detect the ERM-targeted C-terminal fragment [Tb(C)]. Tom20 and mCherry-KDEL were used as mitochondrial and ER markers, respectively. (Scale bars, 10 μm.) Colocalization of V5 with Tom20 and HA with mCherry-KDEL are shown on the right. Quantitation from five fields of view per condition. (E) Enrichment of known ER–mitochondria contact proteins by split-TurboID-catalyzed PL. HEK293T cells stably expressing OMM/ERM-targeted split-TurboID constructs were treated with 50 μM biotin and 100 nM rapamycin for 4 h. HEK293T cells stably expressing NES-, OMM-, or ERM-targeted FL-TurboID were treated with 50 μM biotin for 1 min. Biotinylated proteins were enriched from lysates using streptavidin beads, eluted, and analyzed by blotting with streptavidin and antibodies against FACL4 and Mff. Asterisks indicate ligase self-biotinylation. (F) Enrichment of biotinylated proteins for proteomics. Samples were generated as in E. Biotinylated proteins were enriched from lysates using streptavidin beads, eluted, and analyzed by silver stain.
Split-TurboID Enables Proteomic Mapping of ER–Mitochondria Contacts.
We designed our proteomic experiment to probe ER–mitochondria contact sites in HEK293T, both in the absence of rapamycin addition (when split-TurboID reconstitution is mediated only by native ERM and OMM proximity) and in the presence of rapamycin (which enhances split-TurboID reconstitution at ER–mitochondria contacts). We generated stable HEK293T cells expressing the split-TurboID constructs, or the reference constructs TurboID-NES (nuclear export sequence; full-length TurboID in the cytosol), TurboID-OMM (full-length TurboID on the OMM facing cytosol), and ERM-TurboID (full-length TurboID on the ERM facing cytosol). Imaging showed correct localization of all constructs to respective mitochondria and ER organelles (Fig. 3D and SI Appendix, Fig. S4). For split-TurboID, biotinylation activity was again observed in the absence of rapamycin, but was substantially increased upon rapamycin addition (SI Appendix, Fig. S3A).
Due to their differences in activity levels, the split-TurboID samples were treated with biotin (with or without rapamycin) for 4 h, while the full-length TurboID samples were labeled for only 1 min. Under these conditions, we observed comparable levels of biotinylated proteins in our split-TurboID and full-length TurboID samples both before and after streptavidin bead enrichment (Fig. 3F and SI Appendix, Fig. S3 D and E). We also verified that these labeling conditions did not perturb organelle morphology or artificially increase ER–mitochondria contacts in our stable cells (SI Appendix, Fig. S3 B and C). To test if OMM/ERM-targeted split-TurboID could preferentially enrich known ER–mitochondria contact site proteins, we performed Western blot analysis of the streptavidin-enriched material. Fig. 3E shows greater enrichment of the known ER–mitochondria contact proteins FACL4 and Mff (15, 17, 29) in split-TurboID samples than in TurboID-NES samples.
Next, we performed mass spectrometry on our streptavidin-enriched samples. After on-bead digestion of the protein samples, we labeled the released peptides with isotopically distinct tandem mass tag (TMT) labels, enabling us to quantify the relative abundance of each protein across samples (Fig. 4A). Input levels per sample were normalized prior to analysis, and we found that replicate samples were highly correlated (SI Appendix, Fig. S5 A–D). We analyzed our ERM and OMM datasets obtained from full-length TurboID using a ratiometric approach, as previously described (30), and found our datasets to be highly specific both when compared to previously published datasets (7, 9) and when performing gene ontology (GO) term enrichment analysis (SI Appendix, Figs. S5 E–P and S6).
Fig. 4.
Proteomic mapping of ER–mitochondria contacts in HEK293T cells. (A) Experimental design and labeling conditions for MS-based proteomics. Cells stably expressing the indicated constructs were labeled with 50 μM biotin and 100 nM rapamycin. Split-TurboID (ERM/OMM) samples were labeled for 4 h and FL-TurboID samples were labeled for 1 min. Cells were then lysed, and biotinylated proteins were enriched using streptavidin beads, digested to peptides, and conjugated to TMT labels. All samples were then combined and analyzed by LC-MS/MS. (B) Filtering scheme for mass spectrometric data. +R and −R refer to rapamycin, and +B and −B refer to biotin. For each dataset, proteins were first ranked by the extent of biotinylation (ratiometric analysis referencing omit biotin controls, filter 1). Next, proteins were ranked by relative proximity to ER–mitochondria contacts versus cytosol (ratiometric analysis referencing TurboID-NES, filter 2). (C) Scatterplot showing log2(128C/130C) (filter 1) versus log2(128C/126C) (filter 2) for each protein in replicate 1 of split-TurboID cells treated with rapamycin and biotin. Known ERM and OMM proteins (as annotated by GOCC) are labeled blue and red, respectively; ERM and OMM dual-annotated proteins are colored purple, and all other proteins are shown in black. Cutoffs used to filter the mass spectrometric data and obtain the filtered proteome are shown by dashed lines. Zoom of boxed region is shown in the upper left.
For the analysis of our split-TurboID proteomic datasets, we began with 2,496 detected proteins with two or greater unique peptides. We then took the replicates of each experimental condition (plus rapamycin or minus rapamycin) and applied two sequential filtering steps. First, the data were filtered by the extent of biotinylation by split-TurboID, where we established a cutoff at a 10% false-discovery rate (FDR) for detection of mitochondrial matrix false-positive proteins, referencing the “omit biotin” negative control samples. Second, the data were filtered further by the extent of biotinylation by split-TurboID at ER–mitochondria contacts relative to proteins biotinylated by TurboID in the cytosol, where we established a cutoff at a 10% FDR for detection of cytosolic false-positive proteins (Fig. 4B and SI Appendix, Fig. S7). Applying both filters enriched for proteins with prior ERM and OMM annotation, as well as proteins that have been previously associated with ER–mitochondria contacts, including DNM1L, BCAP31, MAVS, and AKAP1 (16, 29, 31, 32) (Fig. 4C). After filtering, we reached proteome lists of 67 proteins (+rapamycin list) and 63 proteins (omit-rapamycin list), 29 proteins of which were found in both lists (Figs. 4B and 5A and Dataset S1).
Fig. 5.
Analysis of ER–mitochondria contact proteome. (A) Venn diagram comparing proteome lists obtained using split-TurboID. Proteins that were previously annotated ERM, OMM, both, or neither were labeled blue, red, purple, or black, respectively. Proteins previously enriched in MAMs are highlighted in yellow. (B) Specificity analysis for proteomic datasets generated using split-TurboID compared to the entire human proteome and to previously published datasets. Bar graph shows the percentage of each proteome with prior OMM annotation only, ERM annotation only, or both OMM and ERM annotation. Each bar is labeled with the size of the proteome. Hung et al. used APEX (9), Cho et al. used APEX and MAM fractionation (36), Kwak et al. used Contact-ID (21), and Wang et al. used MAM purification (35). (C) Markov clustering of split-TurboID proteome using protein–protein interaction scores from the STRING database (33). Gray lines denote protein–protein interactions. Nodes are colored based on whether the corresponding protein was found in the +rapamycin proteome, omit-rapamycin proteome, or both proteomes. Node size is correlated with the relative enrichment of each protein at ER–mitochondria contacts versus cytosol [log2(128C/126C) or log2(129C/126C)]. Each cluster is labeled with associated GO terms (org., organization).
GO term enrichment analysis of each list largely recovered ER and mitochondria membrane-associated terms, suggesting high specificity (SI Appendix, Fig. S9 A and B). We calculated the fraction of proteins in each dataset with prior ERM or OMM annotation, and arrived at 44% for our combined dataset (proteins present in either +rapamycin or omit-rapamycin lists), and 55% for our intersected dataset (proteins present in both lists), both much greater than the equivalent percentages for the entire human proteome (6%) or our pre-filter proteome (11%). We next performed Markov clustering of our proteomic datasets, using known protein–protein interactions from the STRING database (33). Fig. 5C shows the resulting network and GO terms associated with each cluster. The GO terms mitochondrial organization, mitochondrial fission, sterol metabolism, and calcium ion transport are consistent with known roles of ER–mitochondria contacts (15, 27, 34).
We compared our split-TurboID proteomes to previous ER–mitochondria contact proteomes obtained by other methods. The four comparison datasets were 1) A MAM preparation from human tissue (35); 2) a study combining MAM isolation and PL (36); 3) our previous study using APEX labeling on the OMM and ERM separately, followed by dataset intersection (9); and 4) the Contact-ID–generated ER–mitochondria proteome (21). Fig. 5B shows that specificity, as measured by the fraction of proteins with prior OMM or ERM annotation, is similar for all of the PL-based studies, but much poorer for the MAM dataset, which contains contaminants from many other organelles. To quantify sensitivity, we compiled a list of 20 human proteins with prior literature evidence of ER–mitochondria contact localization (Dataset S2). We detected only two of these proteins, similar to other PL studies, while the MAM dataset recovered much more (SI Appendix, Fig. S9C). Overall, MAM purifications are more sensitive, but at the expense of specificity. PL has the opposite characteristics: high specificity, but poorer recovery, particularly for dual-localized proteins, which are known to be removed by the ratiometric filtering process. For example, a protein dual-localized to ER–mitochondria contacts and the cytosol would be removed in the second ratiometric filtering step using cytosolic TurboID-NES as a reference.
Validation of ER–Mitochondria Contact “Orphans”.
Of the 29 proteins detected in both +rapamycin and omit-rapamycin datasets, 12 have previously been detected in MAMs or localized to ER–mitochondria contacts in the literature (Fig. 5A and SI Appendix, Fig. S8). The remainder are “orphans,” or proteins with no prior literature connection to ER–mitochondria contacts. To determine if these are bona fide ER–mitochondria contact residents or false positives, we selected six proteins for which high-quality commercial antibodies exist, and blotted for their presence in purified MAMs. All six were found to be enriched in MAMs, as well as in other compartments consistent with their literature annotation (e.g., MAM + ER for LBR [a lamin B receptor]; MAM + mitochondria for EXD2) (Fig. 6A).
Fig. 6.
Validation of proteomic hits in MAMs and by imaging. (A) Western blotting of candidate ER–mitochondria contact proteins ABCD3, EXD2, OCIAD1, NEK4, LBR, and MLF2 in MAM fractions. HEK293T cells were collected and subjected to subcellular fractionation to obtain cytosol, mitochondria (mito), ER, and MAM fractions. GRP75, IP3R1, PDI, and VDAC1 are known MAM proteins and were included as positive controls. Negative controls are COX4 (IMM), β-tubulin (cytosol), and PCNA (nucleus). (B) Overexpression imaging assay. Candidate ER–mitochondria contact proteins FUNDC2 and MTFR1 (each V5-tagged) were overexpressed in HeLa cells. V5-tagged SYNJ2BP (9) was included as a positive control. MitoTracker stains mitochondria and anti-CANX antibody stains the ER. Zoomed images of the boxed regions are shown. (Scale bars, 20 μm.) (C) Quantification of ER–mitochondria overlap (colocalization of CANX and MitoTracker signals) in B and SI Appendix, Fig. S9F. Five fields of view were analyzed per condition.
In addition to MAM blotting, we selected a subset of orphans for functional analysis. Previously, we showed that overexpression of the ER–mitochondria tether SYNJ2BP in HeLa cells causes a dramatic increase in the extent of overlap between ER and mitochondria by imaging (9). We V5-tagged the orphan proteins FUNDC2, LBR, MTFR1, OCIAD1, and USP30 and overexpressed them in HeLa cells, which were then stained for mitochondria and ER markers. Confocal imaging shows that FUNDC2 and MTFR1, along with the positive control SYNJ2BP, cause a significant increase in colocalization of ER and mitochondria, compared to untransfected control HeLa cells (Fig. 6 B and C). The other three proteins tested did not give as substantial a phenotype (SI Appendix, Fig. S9F). Our data suggest that FUNDC2 and MTFR1 may have tethering functions at ER–mitochondria contacts that are up-regulated in this gain-of-function assay.
Cell–Cell Contact-Dependent Reconstitution of Split-TurboID.
In addition to permitting PL with greater spatial specificity, split-TurboID could potentially enable conditional PL dependent upon specific inputs or signaling events. For example, in neuroscience, immunology, and cancer biology, there is great interest in characterizing the transcriptomes and proteomes of cell subpopulations that have made contact with specific “sender” cells (for example, neurons downstream of specific presynaptic inputs, or immune cells that come into contact with a tumor cell). If we could drive intracellular split-TurboID reconstitution, specifically in “receiver” cell populations that come into contact with defined “sender” cells, then this may enable PL-based proteomic analysis of functionally relevant cellular subpopulations.
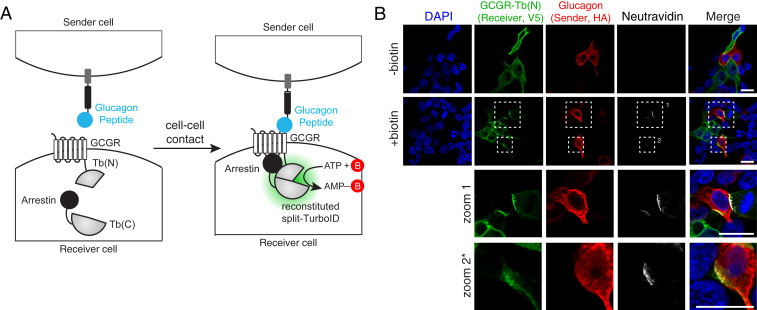
To test this, we designed a synthetic signaling network that utilizes the transcellular interaction between glucagon peptide and its receptor (GCGR), employed in the transsynaptic tracing tool trans-Tango (37). In our design, “receiver” cells express the glucagon receptor (GCGR) and arrestin, each fused to a split-TurboID fragment. Upon cell–cell contact with “sender” cells expressing surface glucagon peptide, GCGR is activated, resulting in arrestin recruitment and reconstitution of split-TurboID (Fig. 7A). Confocal fluorescence imaging of receiver and sender cell co-cultures after biotin treatment for 1 h showed highly specific localization of biotinylated proteins, specifically at sender:receiver cell–cell contacts (red:green overlap sites in Fig. 7B).
Fig. 7.
Cell–cell contact dependent reconstitution of intracellular split-TurboID. (A) Schematic of cell–cell contact-dependent split-TurboID reconstitution. (B) Confocal fluorescence imaging of “sender” cells expressing cell surface targeted glucagon peptide co-cultured with “receiver” cells expressing split-TurboID fragments. Zoomed images of the boxed regions are shown. (Scale bars, 20 μm.) *The contrast has been increased in the zoom 2 row.
Discussion
We have engineered both high-affinity and low-affinity forms of split-TurboID, which are simpler to use and much more active than previously reported split enzymes for PL (18–21). We show that reconstitution is fast and effective in multiple cellular subcompartments (Fig. 2D). Low-affinity split-TurboID reconstitution can be driven by a drug, protein–protein interaction, organelle–organelle contact, or cell–cell contact.
We used split-TurboID for proteomic mapping of ER–mitochondria contact sites and identified 67 proteins in our +rapamycin dataset and 63 proteins in our omit-rapamycin dataset, which were reproducibly enriched over controls. While these lists contain many proteins that were previously detected in MAMs, there are more than 70 “orphans” with no prior literature connection to ER–mitochondria contacts. One interesting example is MIGA1, or mitoguardin 1, identified in both our +rapamycin and omit-rapamycin datasets. MIGA1 has been previously shown to function downstream of the mitofusins and regulate mitochondrial fusion (38). A prior study showed that MIGA1 localizes to the OMM but not specifically to ER–mitochondria contact sites (38). Interestingly, MIGA2, which is highly related to MIGA1, was recently shown to bind VAPA and VAPB and link ER–mitochondria contacts to lipid droplets to promote adipocyte differentiation (39).
We validated eight split-TurboID–identified ER–mitochondria contact orphans, using biochemical fractionation and overexpression imaging. These proteins span a range of functions and their assignment to ER–mitochondria contacts has interesting biological implications. For example, ABCD3 is a known peroxisomal membrane protein that has been shown to be a transporter of branched-chain fatty acids into the peroxisome (40, 41). Our finding that ABCD3 localizes to ER–mitochondria contacts is consistent with previous studies that suggest peroxisome biogenesis may occur at ER–mitochondria contacts (42, 43). EXD2 is an endonuclease that has been shown to localize to both mitochondria and the nucleus (44, 45); its localization to ER–mitochondria contacts by our study suggests a possible link between mitochondrial function and nuclear nucleic acid maintenance, which has also been suggested in previous work (45). LBR is a lamin B receptor that has been localized to the nuclear envelope and is involved in nuclear envelope disassembly during mitosis (46). Our finding that LBR localizes to ER–mitochondria contacts (Fig. 6A), along with our analysis showing that a number of proteins involved in lamin binding, nuclear envelope organization, and mitosis may be enriched at ER–mitochondria contact sites (Fig. 5C), suggest that these processes may be regulated in part by ER–mitochondria contacts. MTFR1 and FUNDC2 are the two split-TurboID–identified orphans that caused an increase in ER–mitochondria overlap upon overexpression (Fig. 6 B and C). While MTFR1 has been shown to regulate mitochondrial dynamics (47), and FUNDC1, related to FUNDC2, has been shown to mediate hypoxia-mediated mitophagy (48), MTFR1 and FUNDC2 have not been previously localized to ER–mitochondria contacts or shown to have organelle tethering activity.
Comparing our +rapamycin and omit-rapamycin datasets, we observed somewhat more proteins with prior ER, mitochondria, and MAM annotations in the +rapamycin condition. Because in our construct design, FRB and FKBP also function like linkers that extend the reach of the split-TurboID fragments, we hypothesize that the +rapamycin proteome may be probing closer ER–mitochondria contacts, while the omit-rapamycin proteome may also be capturing proteins present at wider contacts, as shown in Fig. 3A. Perhaps MAM preparations in previous studies are biased toward closer ER–mitochondria contacts if these are more likely to survive the fractionation process, while wider contacts may be underrepresented. Thus, while there is less prior annotation information for proteins in the omit-rapamycin proteome, these proteins may still be bona fide ER–mitochondria contact proteins.
In addition to generating ER–mitochondria contact candidates, our proteomic experiment also produced OMM and ERM proteomes, via the full-length TurboID constructs we targeted to OMM and ERM, respectively. Using these datasets, we could categorize our ER–mitochondria contact hits as additionally localized to the OMM and/or ERM, or not (SI Appendix, Fig. S9D; each protein is classified as being a resident of “contacts only,” “contacts and OMM,” “contacts and ERM,” or “contacts, OMM, and ERM”). The categorizations based on our proteomic data are largely consistent with prior literature, and with our fractionation blotting and imaging data as well (Fig. 6 and SI Appendix, Fig. S9 E and F). For several proteins, such as OCIAD1 and NEK4, our proteomic data newly assign them to mitochondrial and ER compartments of the cell.
When comparing our proteome with that obtained via Contact-ID (21), we find that both proteomes detected a similar number of proteins (101 proteins using split-TurboID and 115 proteins using Contact-ID). Of the eight proteins we validated, four (FUNDC2, EXD2, OCIAD1, and LBR) were also detected using Contact-ID (SI Appendix, Fig. S8). Conversely, FKBP8, which the authors of Contact-ID show facilitates ER–mitochondria contact formation and local calcium transport (21), was also present in our proteome. While both proteomes have high specificity, measured by the fraction of proteins with prior OMM or ERM annotation, we find that the proteome generated from Contact-ID is more biased toward ERM proteins, whereas we observe more balance between OMM and ERM proteins in our split-TurboID proteome (Fig. 5B). This difference may possibly arise from the longer labeling period used for Contact-ID (16 h vs. 4 h for split-TurboID).
Overall, we have developed a split-TurboID tool that can be conditionally reconstituted for spatially specific PL in cells. Especially when combined with functional assays and screens, split-TurboID–based PL can be a powerful tool for biological discovery around organelle contact sites or macromolecular complexes. Split-TurboID may also improve signal-to-noise for challenging targeting applications, such as dCas9-directed PL of specific genomic loci (49, 50), or dCas13-directed PL of specific cellular RNAs (51).
Methods
Methods related to cloning, split-site pair selection, Western blots, confocal fluorescence imaging, proteomic sample preparation and analysis, additional data analysis, and subcellular fractionation are detailed in SI Appendix.
Mammalian Cell Culture, Transfection, and Stable Line Generation.
HEK293T cells from ATCC were cultured as a monolayer in DMEM with 4.5 g/L glucose and l-glutamine supplemented with 10% (vol/vol) fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37 °C under 5% CO2. For confocal imaging experiments, glass coverslips were coated with 50 μg/mL fibronectin in DPBS (Dulbecco's phosphate-buffered saline) for at least 20 min at room temperature before plating; cells were grown on glass coverslips in 24-well plates with 500 μL growth medium. For Western blot experiments, cells were grown in six-well plates with 2 mL growth medium.
For transient expression, cells were transfected at ∼50% confluency using 5 μL/mL Lipofectamine2000 and 250 ng/mL plasmid in serum-free media. To generate lentiviruses, HEK293T cells were cultured in T25 flasks and transfected at ∼60% confluency with 2,500 ng of the lentiviral vector containing the gene of interest and lentiviral packaging plasmids pVSVG (250 ng) and Δ8.9 (2,250 ng) with 30 μL polyethyleneimine (PEI) (1 mg/mL in water, pH 7.3) in serum-free media. After 48 h, the cell medium containing the lentivirus was harvested and filtered using a 0.45-μm filter.
For generation of stable cell lines, HEK293T cells were infected with crude lentivirus at ∼50% confluence, followed by selection with 8 μg/mL blasticidin (at least 4 d) and 250 μg/mL hygromycin (at least 7 d) in growth medium before use in experiments.
Biotin Labeling with TurboID and Split-TurboID.
For biotin labeling of transiently transfected cells, biotin and rapamycin were added 18 h following transfection. Biotin (10 mM stock in DMSO) was diluted in complete media and added directly to cells to a final concentration of 50 μM. For full-length TurboID, cells were treated with 50 μM biotin and incubated at 37 °C for 1–30 min; for split-TurboID, cells were treated with 50 μM biotin ± 100 nM rapamycin and incubated at 37 °C for 30 min–24 h (as indicated). In general, for split-TurboID labeling, 1 h is a good starting point, but optimization of labeling times may be necessary depending on the application. For both imaging and Western blot experiments, labeling was stopped after the indicated time periods by transferring cells on plates to ice and washing gently with cold DPBS.
Sample Preparation for ER–Mitochondria Proteomics.
For each cell sample shown in Fig. 4A, HEK293T cells were cultured in T150 flasks. All cells used in proteomics experiments were stably expressing the indicated TurboID or split-TurboID constructs. Split-TurboID samples were labeled with 50 μM biotin and 100 nM rapamycin for 4 h, and full-length TurboID samples were labeled with 50 μM biotin for 1 min. Labeling was stopped by placing cells on ice and washing gently with cold DPBS twice. Cells were then detached from the well or flask by pipetting a stream of cold DPBS directly onto cells. The cells were collected and pelleted by centrifugation at 2,500 rpm for 3 min at 4 °C. The supernatant was removed, and the pellet was lysed by resuspension in RIPA lysis buffer and incubation for 10 min at 4 °C. Lysates were clarified by centrifugation at 12,000 rpm for 10 min at 4 °C.
For enrichment of biotinylated proteins, 300 μL streptavidin-coated magnetic beads (Pierce) were washed twice with RIPA lysis buffer and incubated with clarified lysates with rotation at 4 °C overnight. The beads were then washed twice with 1 mL of RIPA lysis buffer, once with 1 mL 1M KCl, once with 1 mL 0.1M Na2CO3, once with 1 mL 2M urea in 10 mM Tris⋅HCl (pH 8.0), and twice with 1 mL RIPA lysis buffer. The beads were subsequently washed with 1 mL digestion buffer (75 mM NaCl, 1 mM EDTA, 50 mM Tris⋅HCl, pH 8.0) twice. The beads were then resuspended in 80 μL digestion buffer, transferred to a new Eppendorf tube, frozen on dry ice, and shipped for further processing and preparation for LC-MS/MS analysis.
For each proteomic sample, 0.2% of the lysate was removed prior to enrichment to verify construct expression and confirm successful biotinylation, as shown in SI Appendix, Fig. S3D. After enrichment, 5% of beads were removed and biotinylated proteins were eluted by boiling the beads in 80 μL 3× protein loading buffer supplemented with 20 mM DTT and 2 mM biotin. The eluted proteins were separated by SDS-PAGE gel and analyzed by Western blotting (1.25%) and silver stain (3.75%) to verify successful enrichment of biotinylated proteins (Fig. 3F and SI Appendix, Fig. S3E).
Generation of ER–Mitochondria Proteome Lists.
Unprocessed mass spectrometry data for split-TurboID targeted to ERM/OMM (both +R+B and −R+B) are shown in Dataset S1, Tabs A–C. These data were filtered in three steps to generate our proteome lists, as follows. First, to identify proteins biotinylated by TurboID over nonspecific binders, we generated a true positive (TP) list of literature-validated ER–mitochondria contact proteins (8, 16, 31, 52–73; Dataset S2, Tab A) and a false positive list (FP1) of mitochondrial matrix proteins, which should not be biotinylated by split-TurboID localized to ER–mitochondria contacts, as determined by Gene Ontology Cellular Component (GOCC) (Dataset S2, Tab B; GO:0005759, but no additional annotations for inner mitochondrial membrane [IMM], OMM, intermembrane space [IMS], or membrane). TMT log2 ratios for split-TurboID (+R+B) over split-TurboID (+R−B) and for split-TurboID (−R+B) over split-TurboID (−R−B) were ranked from highest to lowest. For each possible TMT log2 ratio cutoff, the false discovery rate (FDR), defined as the fraction of detected FP1 proteins detected above the cutoff, was calculated, and the cutoff was determined by setting the FDR < 0.1.
Second, we identified proteins preferentially biotinylated by ER–mitochondria contact-localized split-TurboID over cytosolic TurboID. To do this, we generated another false positive list (FP2) of cytosol-resident proteins that should not be enriched at ER–mitochondria contacts, as determined by GOCC (Dataset S2, Tab C; GO:0005829, but no annotations for membrane). TMT log2 ratios for split-TurboID (+R+B) over TurboID-NES and for split-TurboID (−R+B) over TurboID-NES were ranked from highest to lowest. For each possible TMT log2 ratio cutoff, the false discovery rate (FDR), defined as the fraction of detected FP2 proteins detected above each cutoff, was calculated. The cutoff was determined by setting the FDR < 0.1.
Third, after applying both cutoff steps to each experimental replicate, we intersected the resulting lists to obtain our final +rapamycin and omit-rapamycin proteome lists, which are shown in Dataset S1, Tabs A and B. FKBP1 and mammalian target of rapamycin (mTOR) were removed from the lists as our split-TurboID fusions themselves generate these peptides (because they incorporate FKBP and FRB domains). An additional list intersecting split-TurboID (+R+B) and split-TurboID (−R+B) lists is also shown in Dataset S1, Tab C.
Data Availability.
Proteomics data and all log2 ratio values associated with each protein detected (with two or greater unique peptides) are available in Dataset S1.
Supplementary Material
Acknowledgments
This work was supported by the NIH Grant R01-DK121409 (to A.Y.T. and S.A.C.); Stanford Wu Tsai Neurosciences Institute Big Ideas Initiative (A.Y.T.); Korea Health Technology R&D Project Grant KHIDI HI16C1501 (to I.-K.L.); National Research Foundation of Korea (NRF) Grants 2017R1A2B3006406 (to I.-K.L.) and 2019R1A2C3008463 (to H.-W.R.); and Organelle Network Research Center Grant NRF-2017R1A5A1015366 (to H.-W.R.). K.F.C. was supported by NIH Training Grant 2T32CA009302-41 and the Blavatnik Graduate Fellowship. T.C.B. was supported by Dow Graduate Research and Lester Wolfe Fellowships. A.Y.T. is an investigator of the Chan Zuckerberg Biohub.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919528117/-/DCSupplemental.
References
- 1.Kim D. I., Roux K. J., Filling the void: Proximity-based labeling of proteins in living cells. Trends Cell Biol. 26, 804–817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C. L., Perrimon N., Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip. Rev. Dev. Biol. 6, e272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li P., Li J., Wang L., Di L. J., Proximity labeling of interacting proteins: Application of BioID as a discovery tool. Proteomics 17, 1700002 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Lam S. S. et al., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee H. W. et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branon T. C. et al., Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung V. et al., Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung V. et al., Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife 6, e24463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han S. et al., Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells. Cell Chem. Biol. 24, 404–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh K. H. et al., Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166, 1295–1307.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uezu A. et al., Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markmiller S. et al., Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mick D. U. et al., Proteomics of primary cilia by proximity labeling. Dev. Cell 35, 497–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csordás G., Weaver D., Hajnóczky G., Endoplasmic reticulum-mitochondrial contactology: Structure and signaling functions. Trends Cell Biol. 28, 523–540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman J. R. et al., ER tubules mark sites of mitochondrial division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandre-Babbe S., van der Bliek A. M., The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y. et al., Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Munter S. et al., Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 591, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Schopp I. M. et al., Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8, 15690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak C. et al., Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl. Acad. Sci. U.S.A. 117, 12109–12120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martell J. D. et al., A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dagliyan O. et al., Computational design of chemogenetic and optogenetic split proteins. Nat. Commun. 9, 4042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver L. H., Kwon K., Beckett D., Matthews B. W., Corepressor-induced organization and assembly of the biotin repressor: A model for allosteric activation of a transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 98, 6045–6050 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood Z. A., Weaver L. H., Brown P. H., Beckett D., Matthews B. W., Co-repressor induced order and biotin repressor dimerization: A case for divergent followed by convergent evolution. J. Mol. Biol. 357, 509–523 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Rieusset J., The role of endoplasmic reticulum-mitochondria contact sites in the control of glucose homeostasis: An update. Cell Death Dis. 9, 388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowland A. A., Voeltz G. K., Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 13, 607–625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poston C. N., Krishnan S. C., Bazemore-Walker C. R., In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteomics 79, 219–230 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Horner S. M., Liu H. M., Park H. S., Briley J., Gale M. Jr., Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 108, 14590–14595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung V. et al., Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwasawa R., Mahul-Mellier A. L., Datler C., Pazarentzos E., Grimm S., Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 30, 556–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrill R. A., Strack S., Mitochondria: A kinase anchoring protein 1, a signaling platform for mitochondrial form and function. Int. J. Biochem. Cell Biol. 48, 92–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D. et al., STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrera-Cruz M. S., Simmen T., Of yeast, mice and men: MAMs come in two flavors. Biol. Direct 12, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Wen Y., Dong J., Cao C., Yuan S., Systematic in-depth proteomic analysis of mitochondria-associated endoplasmic reticulum membranes in mouse and human testes. Proteomics 18, e1700478 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Cho I. T. et al., Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J. Biol. Chem. 292, 16382–16392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talay M. et al., Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y. et al., Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol. Cell 61, 111–124 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Freyre C. A. C., Rauher P. C., Ejsing C. S., Klemm R. W., MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell 76, 811–825.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Imanaka T., Aihara K., Suzuki Y., Yokota S., Osumi T., The 70-kDa peroxisomal membrane protein (PMP70), an ATP-binding cassette transporter. Cell Biochem. Biophys. 32, 131–138 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Kashiwayama Y. et al., Role of Pex19p in the targeting of PMP70 to peroxisome. Biochim. Biophys. Acta 1746, 116–128 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Sugiura A., McLelland G. L., Fon E. A., McBride H. M., A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiura A., Mattie S., Prudent J., McBride H. M., Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Broderick R. et al., EXD2 promotes homologous recombination by facilitating DNA end resection. Nat. Cell Biol. 18, 271–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hensen F., Moretton A., van Esveld S., Farge G., Spelbrink J. N., The mitochondrial outer-membrane location of the EXD2 exonuclease contradicts its direct role in nuclear DNA repair. Sci. Rep. 8, 5368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.English A. R., Voeltz G. K., Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5, a013227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monticone M. et al., The nuclear genes Mtfr1 and Dufd1 regulate mitochondrial dynamic and cellular respiration. J. Cell. Physiol. 225, 767–776 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Wu W. et al., FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 35, 1368–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers S. A. et al., Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat. Methods 15, 437–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X. D. et al., C-BERST: Defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat. Methods 15, 433–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han S., et al. , RNA-protein interaction mapping via MS2 or Cas13-based APEX targeting. bioRxiv:10.1101/2020.02.27.968297 (28 February 2020). [DOI] [PMC free article] [PubMed]
- 52.Lee K. S. et al., Altered ER-mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc. Natl. Acad. Sci. U.S.A. 115, E8844–E8853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmen T. et al., PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirabayashi Y. et al., ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Schwarz T. L., The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De vos K. J. et al., VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21, 1299–1311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoica R. et al., ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5, 3996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wortmann S. B. et al., Mutations in the phospholipid remodeling gene SERAC1 impair mitochondrial function and intracellular cholesterol trafficking and cause dystonia and deafness. Nat. Genet. 44, 797–802 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Hayashi T., Su T. P., Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Sugo M. et al., Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 37, e98899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vecchione A. et al., MITOSTATIN, a putative tumor suppressor on chromosome 12q24.1, is downregulated in human bladder and breast cancer. Oncogene 28, 257–269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerqua C. et al., Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 11, 854–860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez-Suaga P. et al., The ER-mitochondria tethering complex VAPB-PTPIP51 regulates autophagy. Curr. Biol. 27, 371–385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu W. H., Wolfgang W., Forte M., Subcellular localization of human voltage-dependent anion channel isoforms. J. Biol. Chem. 270, 13998–14006 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Hamasaki M. et al., Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Raturi A., Simmen T., Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 1833, 213–224 (2013). [DOI] [PubMed] [Google Scholar]
- 67.de Brito O. M., Scorrano L., An intimate liaison: Spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 29, 2715–2723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Area-Gomez E. et al., Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106–4123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Missiroli S. et al., Mitochondria-associated membranes (MAMs) and inflammation. Cell Death Dis. 9, 329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szabadkai G. et al., Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–911 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korobova F., Ramabhadran V., Higgs H. N., An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poston C. N., Duong E., Cao Y., Bazemore-Walker C. R., Proteomic analysis of lipid raft-enriched membranes isolated from internal organelles. Biochem. Biophys. Res. Commun. 415, 355–360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schon E. A., Area-Gomez E., Mitochondria-associated ER membranes in Alzheimer disease. Mol. Cell. Neurosci. 55, 26–36 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data and all log2 ratio values associated with each protein detected (with two or greater unique peptides) are available in Dataset S1.