Abstract
Background:
Wolff-Parkinson-White (WPW) syndrome is a relatively common arrhythmia affecting ~1-3/1000 individuals. Mutations in PRKAG2 have been described in rare patients in association with cardiomyopathy. However, the genetic basis of WPW in individuals with a structurally normal heart remains poorly understood. Sudden death due to atrial fibrillation (AF) can also occur in these individuals. Several studies have indicated that despite ablation of an accessory pathway, the risk of AF remains high in patients compared to general population.
Methods:
We applied exome sequencing in 305 subjects, including 65 trios, 80 singletons, and 6 multiple affected families. We used de novo analysis, candidate gene approach, and burden testing to explore the genetic contributions to WPW.
Results:
A heterozygous deleterious variant in PRKAG2 was identified in one subject, accounting for 0.6% (1/151) of the genetic basis of WPW in this study. Another individual with WPW and left ventricular hypertrophy carried a known pathogenic variant in MYH7. We found rare de novo variants in genes associated with arrhythmia and cardiomyopathy (ANK2, NEBL, PITX2, and PRDM16) in this cohort. There was an increased burden of rare deleterious variants (MAF <= 0.005) with CADD score ≥25 in genes linked to AF in cases compared to controls (P value=0.0023).
Conclusions:
Our findings show an increased burden of rare deleterious variants in genes linked to AF in WPW syndrome, suggesting that genetic factors that determine the development of accessory pathways may be linked to an increased susceptibility of atrial muscle to AF in a subset of patients.
Keywords: Wolff-Parkinson-White (WPW) syndrome, atrial fibrillation, exome sequencing, ANK2
INTRODUCTION
Wolff-Parkinson-White (WPW) syndrome is a common cause of paroxysmal supraventricular tachycardia (SVT) with a reported prevalence between 0.1%-0.3% in the general population (Davidoff et al., 1981; Packard et al., 1954) . It is characterized by the presence of an accessory pathway between the atria and the ventricles which bypasses the atrioventricular node and causes premature ventricular excitation. Mostly occurring as a sporadic disease, rare reports of familial WPW syndrome have been described (Chia et al., 1982; Gollob et al., 2001a; Harnischfeger 1959; Vidaillet et al., 1987). In some families, WPW is associated with cardiomyopathy, in particular, hypertrophic cardiomyopathy (HCM). Aberrant accessory connections in WPW are hypothesized to occur during embryonic development due to either abnormal growth of myocardial tissue bridges that span the atrioventricular (AV) valves, or alternatively, due to disruptions of the AV valves that result in myocardial tissue connections between the atria and ventricles. While gain of function mutation in PRKAG2 (encoding the gamma-2 regulatory subunit of adenosine monophosphate activated protein kinase) is the most well characterized genetic cause of familial WPW syndrome in association with cardiomyopathy, with glycogen-engorged cardiac myocytes causing disruption of the annulus fibrosus (Ahmad et al., 2005; Arad et al., 2003; Gollob et al., 2001a; Gollob et al., 2001b; Wolf et al., 2008), no causation has yet been established for the majority of individuals with isolated or familial WPW syndrome with apparent structurally normal hearts. WPW pattern is a recognized feature in some autosomal recessive lysosomal storage diseases, such as Pompe (MIM: 232300), X-linked Danon disease (MIM: 300257), MELAS (mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes), and tuberous sclerosis in association with cardiac rhabdomyomas (Miyake et al., 2011). Despite exponential advances in understanding the mechanism of the WPW pattern in these rare genetic disorders, the molecular and genetic underpinnings responsible for WPW syndrome in the vast majority of individuals remains unknown.
Mostly occurring as a sporadic condition in the majority of families, the inheritance pattern of WPW is complex, with likely a continuum ranging from monogenic causes in some families to oligogenic/polygenic etiology with environmental interactions in others. There is significant evidence to suggest that isolated WPW can be inherited as a Mendelian trait, most likely as an autosomal dominant trait with incomplete penetrance (Chia et al., 1982; Ehtisham and Watkins 2005; Mc and Freed 1955). Family studies have previously suggested that at least 3% of the affected individuals have a symptomatic first degree relative (Vidaillet et al., 1987). Few studies have attempted to uncover risk alleles other than PRKAG2 in sporadic WPW families. A deleterious variant in MYH6 (c.5653G>A; p.Glu1885Lys) was reported by Bowles et al. in a large pedigree with multiple affected individuals with WPW syndrome (Bowles et al., 2015). Previously linked to atrial septal defect, dilated, and HCM (MIM: 614089; MIM: 613252; MIM: 613251), this was the first report in which MYH6 was implicated in WPW in a single family. Likewise, a rare variant in MYH7 was described in an individual with HCM and WPW syndrome (Bobkowski et al., 2007). These reports underscore the need for large systematic studies to address the genetic susceptibility in individuals with WPW, for both prognostication and genetic counseling for at-risk families.
The risk of sudden cardiac death (SCD) in WPW, albeit small, is pertinent with reported incidence of approximately 0.25-0.39% annually (Novella et al., 2014; Obeyesekere et al., 2012). The primary mechanism of SCD in patients with WPW is the rapid conduction of AF down an accessory pathway causing life-threatening ventricular fibrillation (VF). It has been shown that patients with WPW syndrome who are most susceptible to VF have a history of atrial fibrillation (AF) (Klein et al., 1979). Indeed, several studies indicate that despite ablation, the risk of AF remains significant in patients with WPW syndrome (Bunch et al., 2015b). Long-term follow up studies have shown that individuals with high susceptibility to AF tend to be younger, are more likely to have inducible arrhythmia, and have a short antegrade effective refractory period of accessory pathway (APERP) of ≤250 ms (Santinelli et al., 2009a; Santinelli et al., 2009b). The genetic determinants that plausibly could contribute to this inherent risk in patients with WPW remain unknown.
We hypothesized that rare variant analysis using de novo and candidate gene approach would identify monogenic causes of WPW in a subset of families. We employed exome sequencing (ES) in 151 families to understand the genetic underpinnings of WPW and to interrogate risk alleles related to cardiac arrhythmias in this cohort.
PATIENTS AND METHODS
2.1. Patients
The study was performed in accordance with the institutional guidelines for human research, with approval by the Institutional Review Board of Baylor College of Medicine. Written informed consents were obtained from all subjects. For the purposes of this study, WPW was defined as a short PR interval with evidence of early ventricular activation, specifically a delta wave on ECG, and was confirmed in all 151 individuals based on standard 12-lead electrocardiography (ECG). Individuals with known genetic diagnoses of mitochondrial disease, MELAS, tuberous sclerosis, and Danon disease were excluded from this study. We recruited 305 subjects through Texas Children’s Hospital in Houston between the years 2008 and 2015. In all, there were 151 affected probands, 148 parents, 5 siblings, and 1 grandparent (Supporting Information Table S1). Of the parents, 4 were affected, one in each individual family; the remaining self-reported to be unaffected. The probands included 89 males and 62 females. The median age at the time of recruitment was 14 years. Within the cohort, there were 65 complete case-parent trios, 80 singleton affected individuals, and 6 families with more than one affected family member (Supporting Information Figure S1A). There were two families with two affected children (WPW232, WPW409), three families with an affected parent (WPW026, WPW173, WPW239), and one family with mother and both children affected (WPW209) (Supporting Information Figure S2).
Of the 151 families, 121 (~80%) had apparently isolated WPW without any evidence of cardiomyopathy or other systemic disease. WPW in association with left ventricular (LV) noncompaction cardiomyopathy (LVNC) was observed in 4/151 (2.6%) subjects, dilated cardiomyopathy and HCM in one subject each (0.7%), and Ebstein anomaly in 7 (4.6%) individuals (Supporting Information Table S2 and Figure S1B). These diagnoses were confirmed by experienced cardiologists at Texas Children’s Hospital. The phenotypes of other individuals are summarized in Supplementary Table S1. Of the enrolled subjects, 63.6% (194/305) were of European descent, 25.2% (77/305) were of Hispanic origin, and 9.8% (30/305) of African American ancestry (Supporting Information Table S1; Figure S1C). The remaining 4 subjects included two of Asian origin, one of mixed Hispanic/European descent, and one of African American/European ancestry.
2.2. Exome sequencing and annotation
Saliva samples were obtained for ES using OrageneTM DNA Self-Collection kits (OGR500, OGR-575). DNA was extracted using the Oragene PrepIT DNA extraction kit (PrepIT-L2P). Exome sequencing was performed at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) initiative (Supporting Information Methods). Variants were filtered by their observed frequencies in databases such as dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), the 1000 Genomes Project (http://www.1000genomes.org/), the NHLBI Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/), and Exome Aggregation Consortium (ExAC) database (N=60,706 individuals)-(http://exac.broadinstitute.org/), and Atherosclerosis
Risk in Communities Study (ARIC) (N=10,940 individuals) database in order to filter common polymorphisms and high frequency, probably benign variants. Variants were classified into exonic, intronic, or intragenic, their potential functional effects (whether synonymous, missense, frameshift, or nonsense variants), and their frequencies in these populations as well as in the BHCMG dataset (n= 6,677). We focused on rare variant alleles with a global minor allele frequency (MAF) of ≤ 0.005 based on NCBI dbSNP, ESP, 1000 Genomes Project, ExAC, for frameshift, stopgain, canonical splice site, non-frameshifting indels, and nonsynonymous variants. Algorithms for bioinformatic prediction of potential functional effects of variants, such as Polyphen2 (Adzhubei et al., 2010), SIFT (Kumar et al., 2009), Mutation Taster (Schwarz et al., 2014), and CADD (Combined Annotation Dependent Depletion) (Kircher et al., 2014), along with Phylop conservation scores were incorporated as part of the annotation process, to prioritize the likely damaging effects of candidate variants.
Ancestry-matched controls were chosen from ARIC (n=10,940 individuals) database (Supporting Information Figure S3). For both cases and controls, ES was performed at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine. Using 1 μg of DNA, an Illumina paired-end pre-capture library was constructed according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D) with modifications as described in the BCM-HGSC Illumina Barcoded Paired-End Capture Library Preparation protocol. This procedure was followed by pooling of pre-capture libraries into 4-plex library pools and their hybridization in solution to the HGSC-designed Core capture reagent (52 Mb, NimbleGen) or 6-plex library pools using the custom VCRome 2.1 capture reagent (42 Mb, NimbleGen) according to the manufacturer’s protocol (NimbleGen SeqCap EZ Exome Library SR User’s Guide) with minor revisions. The sequencing run was performed in paired-end mode using the Illumina HiSeq 2000 platform, with sequencing-by-synthesis reactions extended for 101 cycles from each end and an additional 7 cycles for the index read. With a sequencing yield of 11 Gb, the sample achieved 92% of the targeted exome bases covered with an average depth of coverage of 20X or greater. Data produced were aligned and mapped to the human genome reference sequence (hg19) using the Mercury in-house bioinformatics pipeline (Reid et al., 2014). Variants were called using the ATLAS variant calling method and the Sequence Alignment/Map (SAMtools) suites and annotated with an in-house-developed Cassandra annotation pipeline that uses Annotation of Genetic Variants (ANNOVAR) and additional tools and databases.
2.3. Candidate gene analysis
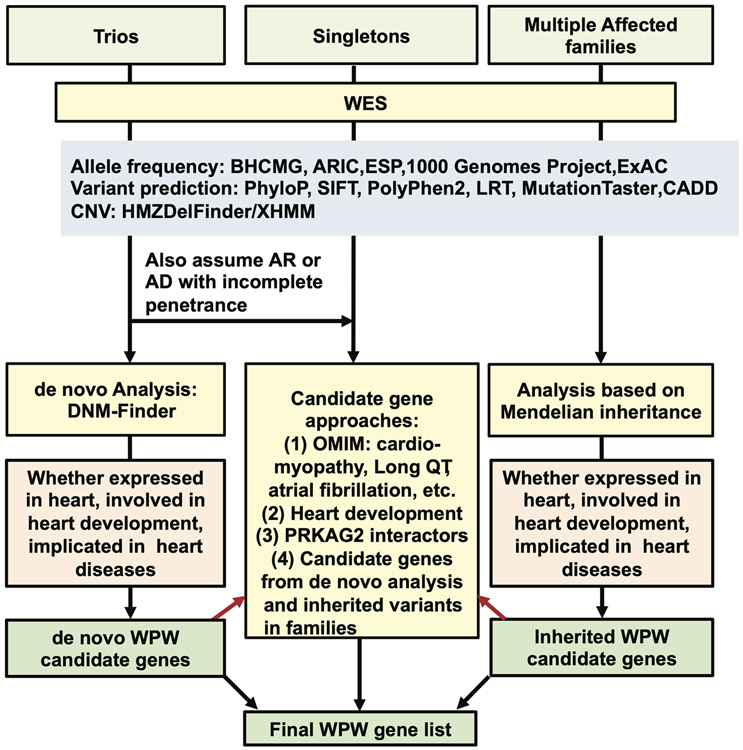
We prioritized candidate genes based on: (1) association with inherited arrhythmia and/or cardiomyopathy; (2) relationship to cardiovascular malformations including the cardiac conduction system (CCS) in Online Mendelian Inheritance in Man (MIM); (3) involvement in cardiac patterning in animal models (Christoffels and Moorman 2009; Sylva et al., 2014) (4); interaction with PRKAG2; and (5) potential contributors from the de novo variant, enrichment analysis, and family studies (Figure 1). Variants in the final candidate genes (Supporting Information Table S3) in 151 affected individuals were further prioritized with reference to lowest MAF in ESP, 1000 Genomes Project, and ExAC databases; pathogenicity scores; phenotypes in OMIM and Mouse Genome Informatics (MGI); and reported alleles in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). In addition, Human Gene Mutation Database (HGMD, Qiagen) was consulted to assess described variants in the literature. Since there were numerous variants of unknown significance (VUS) in TTN, we only analyzed loss-of-function alleles in this analysis. Variants were submitted to the National Center for Biotechnology Information ClinVar database under accession numbers SCV000678334 - SCV000678418.
Figure 1: Schematic of methodology applied for prioritization of candidate genes.
The candidate variants were classified into four categories, Y0-Y3. The highest scoring variants were designated as Y1, representative of the ClinVar alleles known to affect function (designated as pathogenic in the database) in cardiac-specific genes. Y2 variants were predicted deleterious/likely deleterious alleles (based on SIFT, Polyphen2, LRT, Mutation Taster, CADD Phred score), with relative rare allele frequency (based on ExAC, BHCMG internal database, ARIC, ESP6500, and Thousand Genomes). Y3 variants were the least deleterious alleles, with relatively higher allele frequency, or being observed in some controls. Y0 variants were incidental findings, either a loss-of-function (LoF) allele or reported ClinVar pathogenic alleles unrelated to the phenotype (Supporting Information Tables S4 and S5).
2.4. Burden Analysis
First, single nucleotide variants (SNVs), were retrieved from unfiltered vcf files of the BHCMG (n = 6,677) and ARIC (n=10,940) databases for further analysis. To minimize the influence of differences between the two sequence capture designs, HGSC-designed Core and Vcrome 2.1 designs, on the results of the SNV detection method, we identified the intersection of the capture designs and excluded SNVs located outside the regions of overlap. Then, the retrieved SNV variants were reannotated using ANNOVAR against the hg19 RefSeq transcript reference set. Second, SNVs annotated as stopgain, stoploss, nonsynonymous changes were included in further analysis. We performed variant prioritization as follows: If a variant had a variant read number (vR) greater or equal to 5, it was retained. Second, if an SNV had frequency less than 0.5% in the 1000 Genomes Project (1000GP) phase 3 data and the ESP6500 version of Exome Variant Server data and its CADD score was greater or equal to 25, it was included in further analysis. Third, filtered rare and deleterious variants were used for burden analysis of genes associated with AF (N=66) (Supporting Information Table S6) in the WPW European cohort (n=83 affecteds and n=106 unaffecteds) and in the ARIC controls (n=3,064) of European ancestry (Supporting Information Figure S4). The genes associated with AF were selected from peer-reviewed articles (Hayashi et al., 2017; Low et al., 2017a; Lubitz et al., 2016; Perez-Serra et al., 2017). The alleles for rare and deleterious variants in AF genes were counted for any individual sample and then the total resulting average allele count in each group of patients was compared to the other group of patients using Mann-Whitney U test. After this, 10,000 permutations were applied by shuffling the allele counts between each group of patients. For each permutation, a Mann-Whitney U test P-value was calculated (10,000 P-values). Then, the observed P-value was examined in terms of its location of the distribution of 10,000 P-values.
RESULTS
Among all 305 samples, 78,097 high-quality distinct coding variants were identified of which 9,011 (11.53%) were predicted loss-of-function (LoF) SNVs, including canonical splice site disrupting variants, stop-gain variants, frameshift indels, and 69,087 (88.47%) were nonsynonymous SNVs.
3.1. PRKAG2 and MYH7-related WPW syndrome
A predicted deleterious PRKAG2 variant (NM_016203.3:c.359G>A: variant reads/total reads (vR/tR) = 20/29: p.(Arg120His)) with CADD score of 34 was observed in WPW423, an individual with WPW and Ebstein anomaly of the tricuspid valve (Table 1; Supporting Information Tables S3 and S4), accounting for 0.67% (1/151) of the genetic burden in this WPW cohort. WPW385 with WPW and newly developed concentric LVH at age 13 years was found to carry a variant, known to affect function in MYH7, (NM_000257.3:c.2389G>A:54/103:p.(Ala797Thr)), and classified as pathogenic based on the American College of Medical Genetics and Genomics guidelines for classification of variant pathogenicity (Richards et al., 2015), and previously reported in several individuals with HCM (Kassem et al., 2013; Laredo et al., 2006; Van Driest et al., 2004).
Table 1.
Rare variants observed in the WPW cohort
| ID | Genes | Variants | Mutation _type |
Parent al studies |
AR IC |
Tho u |
gnomA D |
ExA C |
esp650 0 |
CAD D_ph red |
pLI | Inhe rita nce |
Ethn icity |
Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| De novo variants in AF genes | ||||||||||||||
| WPW046 | ANK2 | NM_020977.3:c.5437A>G:p.(Lys1813Glu) | Missense | DN | 0 | 0 | NA | 0 | 0 | 32 | 1 | AD | C | WPW, SVT |
| WPW035 | NEBL | NM_006393.2:c.2473C>T:p.(His825Tyr) | Missense | DN | 0 | 0 | NA | 0 | 0 | 24.3 | 0 | n/a | C | WPW, SVT |
| WPW098 | PITX2 | NM_000325.5:c.194C>T:p.(Pro65Leu) | Missense | DN | 0 | 0 | 5.09E-06 | 0 | 0 | 26.5 | 0.98 | AD | C | WPW |
| WPW192 | PRDM16 | NM_022114.3:c.2666C>T:p.(Pro889Leu) | Missense | DN | 1 | 0 | 2.00E-04 | 2.00E-04 | 5.00E-04 | 32 | 1 | n/a | C | WPW, SVT |
| De novo variants in novel genes | ||||||||||||||
| WPW425 | FNIP1 | NM_133372.2:c.2753_2756del:p.(Lys918Argfs*9) | frameshift_deletion | DN | 0 | 0 | NA | 0 | 0 | NA | 1 | n/a | C | WPW, speech delay |
| WPW006 | RHBDF1 | NM_022450.3:c.1384G>A:p.(Gly462Arg) | Missense | DN | 0 | 0 | 4.28E-06 | 0 | 0 | 29.4 | 0 | n/a | C | WPW |
| WPW247 | RHBDF1 | NM_022450.3:c.47A>C:p.(Lys16Thr) | Missense | DN | 0 | 0 | NA | 0 | 0 | 24.5 | 0 | n/a | C | WPW, ADHD |
| WPW282 | WWP1 | NM_007013.3:c.1879A>T:p.(Met627Leu) | Missense | DN | 0 | 0 | NA | 0 | 0 | 27.7 | 1 | AA | WPW, SVT | |
| Rare variants in known WPW genes | ||||||||||||||
| WPW423 | PRKAG2 | NM_016203.3:c.359G>A:p.(Arg120His) | Missense | N/A | 0 | 0 | 0.0001 | 0.00008261 | 0 | 34 | 1 | AD/AR | H | WPW and Ebstein anomaly |
| WPW238 | MYH6 | NM_002471.3:c.4430G>T:p.(Arg1477Leu) | Missense | N/A | 0 | 0 | 4.07E-06 | 0 | 0 | 35 | 0 | AD | AA | WPW |
| WPW385 | MYH7 | NM_000257.3:c.2389G>A:p.(Ala797Thr) | Missense | Inherited | 0 | 0 | 2.03E-05 | 3.30E-05 | 7.70E-05 | 20.5 | 0 | AD | C | WPW, SVT, new diagnosis of LVH |
| WPW381 | MYH7 | NM_000257.3:c.728G>A:p.(Arg243His) | Missense | Inherited | 0 | 0 | 8.12E-06 | 8.24E-06 | 0 | 34 | 0 | AD | C | WPW and LV non compaction cardiomyopathy |
| High CADD score (≥30) variants observed in WPW cohort in cardiac arrhythmia and cardiomyopathy genes | ||||||||||||||
| WPW052 | ACTN2 | NM_001103.3:c.2075T>A:p.(Ile692Asn) | Missense | Inherited | 0 | 0 | NA | 0 | 0 | 33 | 1 | AD | C | WPW, SVT |
| WPW108 | AKAP9 | NM_147185.2:c.11273G>A:p.(Arg3758Gln) | Missense | N/A | 0 | 5.00E-04 | 1.00E-04 | 1.00E-04 | 7.70E-05 | 34 | 0 | AD | C | WPW |
| WPW075 | ANK2 | NM_020977.3:c.742G>A:p.(Val248Met) | Missense | N/A | 0 | 0 | 1.22E-05 | 1.65E-05 | 0 | 33 | 1 | AD | C | WPW, SVT, cardiomegaly |
| WPW006 | KCNQ1 | NM_181798.1:c.1240G>A:p.(Val414Ile) | Missense | Inherited | 0 | 0 | 1.69E-05 | 3.87E-05 | 0 | 32 | 0 | AD/AR | C | WPW |
| WPW079 | KCNQ1 | NM_181798.1:c.343G>A:p.(Asp115Asn) | Missense | Inherited | 0 | 0 | 4.09E-06 | 8.67E-06 | 0 | 32 | 0 | AD/AR | AA | WPW, SVT, and LQTS; has a sister with LQTS |
| WPW238 | KCNQ1 | NM_181798.1:c.808C>T:p.(Arg270Trp) | Missense | N/A | 0 | 0 | 2.00E-04 | 2.00E-04 | 4.00E-04 | 33 | 0 | AD/AR | AA | WPW |
| WPW126 | LAMA4 | NM_001105207.2:c.2377C>T:p.(Arg793Cys) | Missense | Inherited | 7 | 0 | 7.32E-05 | 8.27E-05 | 5.00E-04 | 33 | 0 | n/a | AA | WPW, LV non compaction cardiomyopathy, and VSD |
| WPW416 | MYBPC3 | NM_000256.3:c.1219G>C:p.(Gly407Arg) | Missense | N/A | 0 | 0 | 9.68E-06 | 0 | 0 | 33 | 0 | AD | H | WPW, SVT |
| WPW077 | PRDM16 | NM_022114.3:c.2855C>T:p.(Thr952Met) | Missense | Inherited | 0 | 0 | 2.49E-05 | 9.05E-06 | 0 | 33 | 1 | H | WPW, SVT, ASD, VSD, hypothyroidism | |
| WPW101 | TTN | NM_133432.3:c.40459C>T:p.(Arg13487*) | stopgain | Inherited | 0 | 0 | 4.08E-06 | 0 | 0 | 62 | 0 | AD/AR | C | WPW, SVT |
| WPW416 | TTN | NM_133432.3:c.65863C>T:p.(Arg21955*) | stopgain | N/A | 0 | 0 | NA | 0 | 0 | 67 | 0 | AD/AR | H | WPW, SVT |
| Other rare variants in WPW cohort in cardiac arrhythmia and cardiomyopathy genes | ||||||||||||||
| WPW075 | ACTC1 | NM_005159.4:c.944T>A:p.(Met315Lys) | Missense | N/A | 0 | 0 | NA | 0 | 0 | 29 | 0.74 | AD | C | WPW, SVT, cardiomegaly |
| WPW306 | ACTC1 | NM_005159.4:c.524_525insC:p.(Ala176Cysfs*14) | frameshift_insertion | Inherited | 0 | 0 | NA | 0 | 0 | NA | 0.74 | AD | C | WPW, SVT, and subaortic stenosis |
| WPW279 | ANK2 | NM_020977.3:c.1427A>G:p.(Gln476Arg) | Missense | Inherited | 0 | 0 | NA | 0 | 0 | 28.2 | 1 | AD | C | WPW, SVT |
| WPW079 | CACNA1C | NM_001129842.1:c.1485C>A:p.(His495Gln) | nonsynonymous_SNV/exonic_splicing | Inherited | 2 | 0 | 2.84E-05 | 4.14E-05 | 8.30E-05 | 14.54 | 1 | AD | AA | WPW, SVT, and LQTS; has a sister with LQTS |
| WPW199 | JUP | NM_021991.3:c.773A>G:p.(Glu258Gly) | Missense | N/A | 0 | 0 | NA | 0 | 0 | 29.1 | 0 | AD/AR | AA | WPW |
| WPW209 | LAMA4 | NM_001105207.2:c.1399G>T:p.(Val467Phe) | Missense | Inherited | 0 | 0 | 4.06E-06 | 0 | 0 | 29.5 | 0 | n/a | H | WPW, SVT, and Ebstein anomaly |
| WPW312 | SCN5A | NM_001099405.1:c.1705C>G:p.(Arg569Gly) | Missense | N/A | 0 | 0 | NA | 0 | 0 | 25 | 0.91 | AD/AR | C | WPW |
| WPW159 | SCN5A | NM_001099405.1:c.1567C>T:p.(Arg523Cys) | Missense | Inherited | 0 | 0 | 8.47E-06 | 0 | 0 | 24.5 | 0.91 | AD/AR | H/C | WPW |
DN: de novo
SVT: supraventricular tachycardia
Blue font indicates variants designated to be pathogenic/likely pathogenic in ClinVar
3.2. De novo variants in cardiac arrhythmia susceptibility genes
In the de novo analysis of trios, we identified an average of 2.73 variants per trio by our in-house developed tool, DNM-Finder (https://github.com/BCM-Lupskilab/DNM-Finder) (Eldomery et al., 2017). Out of 65 trios, 41 families had 0-2 de novo variants (Supporting Information Figure S5A). The 178 de novo variants were identified in 65 trios in total, of which 89% were annotated as nonsynonymous, 10% as truncating (frameshift or stopgain), and 1% annotated as canonical splicing variants (Supporting Information Table S7). All genes with de novo nonsynonymous, truncating, and canonical splicing variants in trio families are displayed in a circos plot (Supporting Information Figure S5C). We found de novo variants, confirmed as de novo by Sanger sequencing of trios, in known arrhythmia and cardiomyopathy genes such as ANK2 (NM_020977.3:c.5437A>G:25/110:p.(Lys1813Glu)) in WPW046, PITX2 (NM_000325.5:c.194C>T:24/47:p.(Pro65Leu)) in WPW098, NEBL (NM_006393.2:c.2473C>T:68/149:p.(His825Tyr)) in WPW035, and PRDM16 (NM_022114.3:c.2666C>T:107/240:p.(Pro889Leu)) in WPW192 (Table 1; Supporting Information Table S7 and Figure S6). All subjects with these de novo variants were diagnosed with isolated WPW without cardiomyopathy. Sanger sequencing also confirmed de novo variants in additional genes shown in table 1 (Supporting Information Figure S6).
3.3. Burden analysis
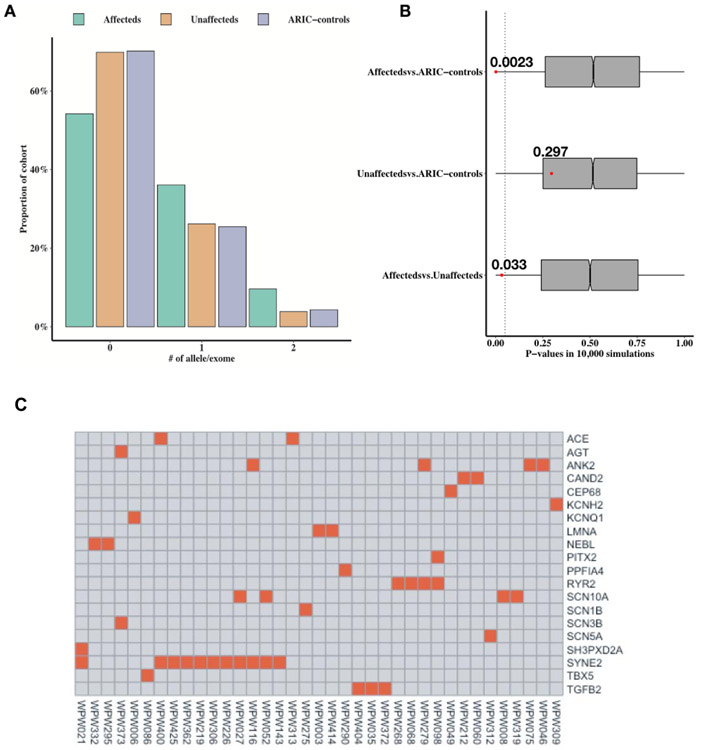
After confirming de novo variants in multiple genes linked to AF in our cohort by Sanger sequencing (ANK2, PITX2, and NEBL), we performed a burden analysis using rare deleterious variants (MAF <= 0.005) with CADD score ≥25 in ~60 known genes associated with AF (Supporting Information Table S6) (PMID: 27589061, 27861186, 28169950, 28416822) (Hayashi et al., 2017; Low et al., 2017a; Lubitz et al., 2016; Perez-Serra et al., 2017). We found that the average allele count of AF genes with CADD score ≥25 in European cases [n=83] was significantly greater (0.277) as compared to unaffected ARIC controls [n=3,064] (average allele count=0.18) (Permutation test one-tailed P-value =0.0023). The average allele count in cases was also greater than the unaffected first-degree relatives [n=106] (Permutation test one-tailed P-value =0.033, 10,000 simulations) (Figure 2A and 2B). Some of these genes included ANK2, KCNQ1, KCNH2, LMNA, PITX2, RYR2, SCN3B, SCN5A, SCN10A, and SYNE2 (Figure 2C). There was no significant increase in rare and deleterious variant burden in AF genes in unaffected first-degree relatives [n=106] (average allele count=0.207) compared to ARIC unaffected controls [n=3,064] (average allele count=0.18) (Permutation test one-tailed P-value =0.297).
Figure 2. Burden analysis of rare deleterious variants in AF genes in WPW affecteds.
(A) An increased proportion of WPW European cases have 1 or 2 rare (MAF<=0.005) and deleterious (CADD>=25) alleles in AF genes (N=66). The column plot shows the proportion of exomes in WPW European affecteds (n=83, depicted in green), WPW European unaffected first degree relatives (n=106, depicted in brown) and ARIC European unaffected controls (ARIC-controls) (n=3,137, depicted in purple) (y-axis) in terms of the number of rare (MAF <=0.005) and deleterious (CADD score >=25) alleles in AF genes carried in their personal genomes (x-axis). (B) The average rare and deleterious allele count in AF genes was compared between affecteds vs. ARIC-controls (top), unaffected first degree relatives vs. ARIC-controls (middle) and affecteds vs. unaffected first degree relatives (bottom) using Mann-Whitney U test. 10,000 permutations were applied by shuffling the allele counts between each pair of group of patients. Gray boxplots shows the distribution of Mann-Whitney U test P-values regarding each permutation. The observed P-value was examined in terms of its location the distribution of 10,000 P-values. This analysis revealed that the average allele rare and deleterious count of AF genes in affecteds was significantly greater (0.277) as compared to ARIC-controls (average allele count=0.18) (Permutation test one-tailed P-value =0.0023) and unaffected first-degree relatives (average allele count=0.207) (Permutation test one-tailed P-value =0.033). There was not a significant increase in rare and deleterious variant burden in AF genes in unaffected relatives [n=106] (average allele count=0.207) compared to ARIC- controls [N=3,064] (average allele count=0.18) (Permutation test one-tailed P-value =0.297). (C) The heatplot shows the AF genes, in which an affected carries a rare variant. Each row corresponds to a gene and each column corresponds to a patient. Red rectangles show that there is a rare and deleterious variant in the AF gene with its HUGO gene symbol presented in the corresponding row in the individual with ID presented in the corresponding column.
We then repeated this analysis using genes unrelated to cardiac arrhythmia and employed an unselected set of neurodevelopmental disorder genes (NDD; n=93) with rare deleterious alleles (Supporting Information Table S8). Our analysis revealed no significant difference in average count of rare (MAF<=0.005) and deleterious (CADD≥25) variant burden in this set of NDD genes. In cases [n=83], there was no significant difference (average allele count=0.163) compared to ARIC controls [n=3,064] (average allele count=0.128) (Permutation test one-tailed P-value =0.104). There was also no significant difference in average allele count in cases compared to the unaffected first-degree relatives [n=106] (average allele count=0.15) (Permutation test one-tailed P-value =0.256, 10,000 simulations) (Supporting Information Figure S8). These data suggest that there is an increased burden of rare deleterious alleles in genes related to AF distinctively in WPW cases, compared to ARIC control, as well as the unaffected first-degree relatives.
3.4. Identification of rare de novo and inherited truncating variants in FNIP1, a negative regulator of AMPK
Disease causing variants in PRKAG2 are known to activate AMPK to cause WPW related cardiomyopathy. In our study, two subjects were ascertained with LoF alleles in FNIP1 (Folliculin-Interacting Protein 1), a negative regulator of AMPK (Siggs et al., 2016); WPW425 had a de novo frameshift variant (NM_133372.2:c.2753_2756del:56/130:p.(Lys918Argfs*9)) and WPW073 had a splicing variant (NC_000005.9(NM_133372.2):c.455+1G>A:9/33) in FNIP1 (Supporting Information Figure S9 and Table S4). Parental samples were unavailable for further studies in WPW073. Neither variant was found in the external databases including ExAC and gnomAD. Both presented with isolated WPW with structurally normal hearts without cardiomyopathy.
3.5. High CADD score variants in cardiac arrhythmia and cardiomyopathy genes
In this cohort, 25 subjects (25/151; 16.5%) were found to have highly deleterious variants in 21 genes, assessed by CADD score of 30 and over (top 0.1% of deleterious variants in the human genome) in genes linked to cardiac arrhythmia and cardiomyopathy (Table 1; Supporting Information Table S9). Aside from PRKAG2 and PRDM16 discussed above, some of the other genes in this category included ACTN2 (Cardiomyopathy, dilated, 1AA [MIM: 612158]), AKAP9 (Long QT syndrome-11 [MIM: 611820]), ANK2 (Long QT syndrome-4 [MIM: 600919], KCNQ1 (Long QT Syndrome, Type 1[MIM: 192500]), LAMA4 (Cardiomyopathy, dilated, 1JJ [MIM: 615235]), MYBPC3 (Cardiomyopathy, familial hypertrophic, 4 [MIM: 115197]), MYH6 (Atrial septal defect 3 [MIM: 614089]), and MYH7 (Cardiomyopathy, dilated, 1S [MIM: 613426]). Likely pathogenic truncating heterozygous variants in TTN with CADD scores of 62 and 67 respectively were observed in two subjects (WPW101 and WPW416) with a structurally normal heart. The TTN variants, (NM_133432.3:c.40459C>T:45/100:p.(Arg13487*) and NM_133432.3:c.65863C>T:89/174:p.(Arg21955*)) were not observed in any of the large control datasets described in the study. WPW126 with WPW and dilated LVNC cardiomyopathy was found to have a LAMA4 rare variant with CADD score of 33 (NM_001105207.2:c.2377C>T:27/56:p.(Arg793Cys)). This gene is known to be associated with dilated cardiomyopathy. In another family, a non-synonymous variant in this gene (NM_001105207.2:c.1399G>T:33/82:p.(Val467Phe)) with CADD score of 29.5 was observed in a multiplex family (WPW209) with an affected mother and proband, both with WPW and Ebstein anomaly. The carrier sibling was diagnosed with supraventricular tachycardia (SVT) (Supporting Information Figure S2).
DISCUSSION
To date, very few genes have been implicated as causative in WPW syndrome. Despite the challenges of predicted incomplete penetrance and undetermined inheritance pattern in the majority of individuals with WPW, this study provides a genetic landscape of rare genetic determinants in a subset of families with WPW syndrome. We used a gene-based burden test for the expected genetic heterogeneity of this disorder. Using very stringent allele filtering thresholds, we identified a number of risk allele variants in our cohort. This approach not only revealed rare de novo variants in genes associated with AF/cardiomyopathy in WPW syndrome (including ANK2, NEBL, PITX2, PRDM16), but also successfully identified genes that were previously linked to WPW in sporadic and familial cases, including PRKAG2 and MYH7. In our study, we found both de novo and rare inherited variants in ANK2 in multiple individuals with WPW syndrome. ANK2 encodes ankyrin-B and plays critical roles in anchoring and stabilizing multiple ion channels in the cardiomyocyte membrane. Arrhythmias related to ANK2 are now known to cause “ankyrin-B syndrome”(Mohler et al., 2003; Mohler et al., 2004). Evidence supporting causality in WPW also comes from a previous report showing ANK2 variant (NM_020977.3:n.2037C>T:p.(Ser646Phe)) segregating with long QT syndrome (LQTS), dilated cardiomyopathy, congenital heart malformation, and WPW syndrome without cardiomyopathy in a group of individuals in the First Nations Population (Swayne et al., 2017). Our data strongly indicates expanding the spectrum of Ankyrin B syndrome to include WPW syndrome.
Importantly, our study identified an increased burden of rare deleterious alleles in genes associated with AF in WPW syndrome compared to controls (P =0.0023). It is recognized that the incidence of AF in WPW is much higher than the general population, estimated to be between 15- 20% in the absence of any clinical evidence of structural heart disease (Fukatani et al., 1990; Haissaguerre et al., 1992; Pietersen et al., 1992; Sharma et al., 1985). While ablation of accessory pathway has been shown to abolish the conduction through the aberrant pathway and reduce the recurrence of AF, many studies have proven that despite ablation, the risk of AF remains high in adult individuals with WPW compared to a control population (Borregaard et al., 2015; Bunch et al., 2015a). Several lines of evidence suggest that there is an underlying atrial muscle vulnerability in patients with the WPW syndrome (Bunch et al., 2015a). Despite intense efforts to unravel the mechanism of AF in WPW syndrome (Centurion 2011) and risk stratification of individuals with WPW syndrome, determining which WPW patients are at highest risk for life-threatening arrhythmia remains unclear.
Sudden cardiac death or adverse events such as life-threatening AF or VF are rare in children with WPW syndrome and were not observed in our pediatric cohort during the length of the study. Without long-term follow up data, it is indeed challenging to determine if the subset of children carrying high deleterious alleles in AF genes would be at an increased risk of AF during adulthood. During electrophysiology (EP) studies, pacing protocols to induce AF are frequently performed by electrophysiologists to identify those children in whom an ablation is indicated. While most children have inducible non-sustained AF (lasting < 30 seconds), only a small minority have sustained AF induced. It has never been elucidated, why some children are easier to induce and sustain than others and raises questions as to whether these children with inducible sustained AF have an underlying genetic predisposition to arrhythmia. Long-term follow up studies have shown that individuals with high susceptibility to AF tend to be younger and are more likely to have inducible arrhythmia (Santinelli et al., 2009). The pediatric subject with a de novo variant in ANK2 (WPW046) in our study had inducible sustained AF during EP study. Whether individuals with high-risk alleles would later develop AF despite ablation in adulthood remains to be determined.
PRKAG2 is the most well characterized gene contributing to a familial form of WPW syndrome where missense variants are known to activate AMP-activated protein kinase (AMPK) and cause HCM with glycogen accumulation and ventricular preexcitation (Arad et al., 2005). In our study, we found only one individual with isolated WPW with a likely deleterious variant in this gene, accounting for <1% of the molecular diagnostic yield. This individual was found to have WPW and Ebstein anomaly. No evidence of HCM was seen in this child. Remarkably, we found truncating variants in FNIP1 with a pLI score of 1, encoding PRKAG2 interacting protein, in two subjects with isolated WPW. FNIP1 negatively regulates γ2-containing AMPK complexes and has an essential role in B-cell development (Baba et al., 2012) (Park et al., 2012). Increased mitochondrial biogenesis in muscle-targeted Fnip1 knockout mice is also reported (Hasumi et al., 2015). Homozygous Fnip1 mice show basal activation of γ2-containing AMPK complexes in the heart with reduced AMP responsivity, and have cardiomyopathy with left ventricular hypertrophy, and abnormal QRS complex (Siggs et al., 2016). These mice present with cardiac glycogen accumulation, which phenocopies the presentation of the mice carrying PRKAG2 variants (Arad et al., 2003; Blair et al., 2001; Patel et al., 2003). Our data combined with the animal studies support FNIP1 as a novel WPW candidate gene. It remains to be seen if disruption of annulus fibrosus due to glycogen accumulation is a similar mechanism of WPW in FNIP1 LoF variants.
From our de novo analysis, we also identified an individual (WPW282) with WPW and structurally normal heart with a de novo missense variant in WWP1, encoding a ubiquitin ligase. The variant, c.1879A>T:p.(Met627Leu) has a CADD score of 27 and is not present in gnomAD. While this gene has not yet been implicated in cardiac phenotype in humans, animal data indicate that overexpression of Wwp1 in mice causes lethal ventricular arrhythmias due to decrease in Cx43 protein in the heart muscle (Basheer et al., 2015). The mice also exhibit mild to moderate left ventricular hypertrophy. We also ascertained rare de novo variants in human rhomboid family-1 (RHBDF1) in two subjects (WPW006 and WPW247). In WPW247, the low variant to total read ratio (32/217) of c.47A>C:p.(Lys16Thr) was confirmed as mosaicism by droplet digital PCR (Supporting Information Figure S7). Though intriguing, the functional relevance of variants in these two genes in WPW remains to be elucidated.
Both PITX2 and NEBL have been identified as risk alleles for AF in genome-wide association studies (GWAS) (Low et al., 2017b) (Yang et al., 2013). We now describe de novo coding variants in both PITX2 and NEBL genes in isolated WPW with normal hearts. NEBL is known to cause dilated cardiomyopathy and endocardial fibroelastosis (Purevjav et al., 2010). PITX2, encoding a transcription factor, regulates the development of left atrium and cardiac conduction system. Variants in this dosage sensitive gene in humans cause Axenfeld-Rieger syndrome, type 1 (MIM: 180500). In addition, variants in this gene have also been implicated in congenital heart disease. Studies have shown that Pitx2 loss of function also predisposes to atrial arrhythmogenesis (Chinchilla et al., 2011; Wang et al., 2010). Our data suggest that rare variants in PITX2 and NEBL may be present in rare individuals with isolated WPW.
CONCLUSIONS
In summary, our study identifies significant rare de novo and inherited genetic variants in the pediatric cohort of WPW and provides a framework for future work for ascertaining high-risk affected individuals. This study provides a landscape of genetic determinants in a large WPW cohort, including 151 affected subjects of all major ethnicities, studied by ES and rare variant family-based genomics approach. Despite its strengths, there are some limitations of our study. Although the vast majority of subjects had isolated WPW without any evidence of cardiomyopathy or other systemic disease, a subset of patients had WPW in association with LVNC, dilated cardiomyopathy, HCM, or extracardiac findings. While de novo and candidate gene analyses were employed for all ethnicities in the study including, Europeans, Hispanics, African Americans, and Asian subjects, the burden testing was undertaken only in the European subjects. Another limitation of the study was related to the young age group of the study population. The median age of patients at the time of study recruitment was only 14 years. Since our cohort was largely pediatric, we were unable to address the correlation of the identified genetic risk alleles to AF for long-term risks in adulthood. Question also remains whether pediatric subjects with WPW carrying rare high CADD score variants in AF genes would also be at an increased risk for developing cardiomyopathy in the future, due to the known allelic heterogeneity of some of these genes within the group. Further studies are required to relate molecular findings in WPW syndrome with long-term outcomes in patients.
Supplementary Material
ACKNOWLEDGEMENTS
We are deeply grateful to all our patients who participated in this study. We are grateful to Chad Shaw for providing assistance in statistical analysis. Support for this work was provided by the National Institutes of Health (National Human Genome Research Institute–National Heart, Lung, and Blood Institute) grant UM1 HG006542 to the Baylor Hopkins Center for Mendelian Genomics, the National Institute of Neurological Disorders and Stroke grant R01 NS058529 and R35NS105078 to J.R.L., the National Human Genome Research Institute U54 HG003273 to RAG. W-L.C. was supported by Cancer Prevention Research Institute of Texas (CPRIT) training Program RP140102. J.E.P was supported by K08 HG008986 from the National Human Genome Research Institute. C.Y.M is supported by 17SDG33410183 from the American Heart Association Scientist Development Grant and K23 HL1386932 from the National Heart Lung and Blood Institute. I.S.P. was supported by 2T32NS043124-16 through the National Institute of Health.
Footnotes
CONFLICT OF INTEREST
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), formerly the Baylor Miraca Genetics Laboratories (BMGL), which performs clinical exome sequencing. J.R.L. serves on the Scientific Advisory Board of BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. J.W.B. contributed to this work while employed by BCM and is currently a full-time employee of Illumina, Inc. Other authors have no disclosures relevant to the manuscript.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, …Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nat Methods, 7(4), 248–9. doi:nmeth0410-248 [pii] 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F, Arad M, Musi N, He H, Wolf C, Branco D, …Seidman JG (2005). Increased alpha2 subunit-associated AMPK activity and PRKAG2 cardiomyopathy. Circulation, 112(20), 3140–8. doi:CIRCULATIONAHA.105.550806 [pii] 10.1161/CIRCULATIONAHA.105.550806 [DOI] [PubMed] [Google Scholar]
- Arad M, Maron BJ, Gorham JM, Johnson WH Jr., Saul JP, Perez-Atayde AR, …Seidman JG (2005). Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med, 352(4), 362–72. doi:352/4/362 [pii] 10.1056/NEJMoa033349 [DOI] [PubMed] [Google Scholar]
- Arad M, Moskowitz IP, Patel VV, Ahmad F, Perez-Atayde AR, Sawyer DB, …Seidman JG (2003). Transgenic mice overexpressing mutant PRKAG2 define the cause of Wolff-Parkinson-White syndrome in glycogen storage cardiomyopathy. Circulation, 107(22), 2850–6. doi: 10.1161/01.CIR.0000075270.13497.2B 01.CIR.0000075270.13497.2B [pii] [DOI] [PubMed] [Google Scholar]
- Baba M, Keller JR, Sun HW, Resch W, Kuchen S, Suh HC, …Casellas R (2012). The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dube syndrome is required for murine B-cell development. Blood, 120(6), 1254–61. doi:blood-2012-02-410407 [pii] 10.1182/blood-2012-02-410407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer WA, Harris BS, Mentrup HL, Abreha M, Thames EL, Lea JB, …Matesic LE (2015). Cardiomyocyte-specific overexpression of the ubiquitin ligase Wwp1 contributes to reduction in Connexin 43 and arrhythmogenesis. J Mol Cell Cardiol, 88, 1–13. doi:S0022-2828(15)30057-2 [pii] 10.1016/j.yjmcc.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair E, Redwood C, Ashrafian H, Oliveira M, Broxholme J, Kerr B, …Watkins H (2001). Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet, 10(11), 1215–20. [DOI] [PubMed] [Google Scholar]
- Bobkowski W, Sobieszczanska M, Turska-Kmiec A, Nowak A, Jagielski J, Gonerska M, …Siwinska A (2007). Mutation of the MYH7 gene in a child with hypertrophic cardiomyopathy and Wolff-Parkinson-White syndrome. J Appl Genet, 48(2), 185–8. doi:393 [pii] 10.1007/BF03194677 [DOI] [PubMed] [Google Scholar]
- Borregaard R, Lukac P, Gerdes C, Moller D, Mortensen PT, Pedersen L, …Jensen HK (2015). Radiofrequency ablation of accessory pathways in patients with the Wolff-Parkinson-White syndrome: the long-term mortality and risk of atrial fibrillation. Europace, 17(1), 117–22. doi:euu176 [pii] 10.1093/europace/euu176 [DOI] [PubMed] [Google Scholar]
- Bowles NE, Jou CJ, Arrington CB, Kennedy BJ, Earl A, Matsunami N, …Gruber PJ (2015). Exome analysis of a family with Wolff-Parkinson-White syndrome identifies a novel disease locus. Am J Med Genet A, 167A(12), 2975–84 doi: 10.1002/ajmg.a.37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TJ, May HT, Bair TL, Anderson JL, Crandall BG, Cutler MJ, …Day JD (2015a). Long-Term Natural History of Adult Wolff-Parkinson-White Syndrome Patients Treated With and Without Catheter Ablation. Circ Arrhythm Electrophysiol, 8(6), 1465–71. doi:CIRCEP.115.003013 [pii] 10.1161/CIRCEP.115.003013 [DOI] [PubMed] [Google Scholar]
- Bunch TJ, May HT, Bair TL, Jacobs V, Crandall BG, Cutler M, …Day JD (2015b). Five-year outcomes of catheter ablation in patients with atrial fibrillation and left ventricular systolic dysfunction. J Cardiovasc Electrophysiol, 26(4), 363–370. doi: 10.1111/jce.12602 [DOI] [PubMed] [Google Scholar]
- Centurion OA (2011). Atrial Fibrillation in the Wolff-Parkinson-White Syndrome. J Atr Fibrillation, 4(1), 287. doi: 10.4022/jafib.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia BL, Yew FC, Chay SO, & Tan AT (1982). Familial Wolff-Parkinson-White syndrome. J Electrocardiol, 15(2), 195–8. [DOI] [PubMed] [Google Scholar]
- Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpon E, …Franco D (2011). PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet, 4(3), 269–79. doi:CIRCGENETICS.110.958116 [pii] 10.1161/CIRCGENETICS.110.958116 [DOI] [PubMed] [Google Scholar]
- Christoffels VM, & Moorman AF (2009). Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others? Circ Arrhythm Electrophysiol, 2(2), 195–207. doi:2/2/195 [pii] 10.1161/CIRCEP.108.829341 [DOI] [PubMed] [Google Scholar]
- Davidoff R, Schamroth CL, & Myburgh DP (1981). The Wolff-Parkinson-White pattern in health aircrew. Aviat Space Environ Med, 52(9), 554–8. [PubMed] [Google Scholar]
- Ehtisham J, & Watkins H (2005). Is Wolff-Parkinson-White syndrome a genetic disease? J Cardiovasc Electrophysiol, 16(11), 1258–62. doi:JCE50139 [pii] 10.1111/j.1540-8167.2005.50139.x [DOI] [PubMed] [Google Scholar]
- Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, …Lupski JR (2017). Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med, 9(1), 26. doi: 10.1186/s13073-017-0412-6 10.1186/s13073-017-0412-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatani M, Tanigawa M, Mori M, Konoe A, Kadena M, Shimizu A, & Hashiba K (1990). Prediction of a fatal atrial fibrillation in patients with asymptomatic Wolff-Parkinson-White pattern. Jpn Circ J, 54(10), 1331–9. [DOI] [PubMed] [Google Scholar]
- Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, …Roberts R (2001a). Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med, 344(24), 1823–31. doi: 10.1056/NEJM200106143442403 [DOI] [PubMed] [Google Scholar]
- Gollob MH, Seger JJ, Gollob TN, Tapscott T, Gonzales O, Bachinski L, & Roberts R (2001b). Novel PRKAG2 mutation responsible for the genetic syndrome of ventricular preexcitation and conduction system disease with childhood onset and absence of cardiac hypertrophy. Circulation, 104(25), 3030–3. [DOI] [PubMed] [Google Scholar]
- Haissaguerre M, Fischer B, Labbe T, Lemetayer P, Montserrat P, d'Ivernois C, …Warin JF (1992). Frequency of recurrent atrial fibrillation after catheter ablation of overt accessory pathways. Am J Cardiol, 69(5), 493–7. [DOI] [PubMed] [Google Scholar]
- Harnischfeger WW (1959). Hereditary occurrence of the pre-excitation (Wolff-Parkinson-White) syndrome with re-entry mechanism and concealed conduction. Circulation, 19(1), 28–40. [DOI] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, …Schmidt LS (2015). Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci U S A, 112(13), E1624–31. doi:1419502112 [pii] 10.1073/pnas.1419502112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Tada H, & Yamagishi M (2017). The genetics of atrial fibrillation. Curr Opin Cardiol, 32(1), 10–16. doi: 10.1097/HCO.0000000000000356 [DOI] [PubMed] [Google Scholar]
- Kassem H, Azer RS, Saber-Ayad M, Moharem-Elgamal S, Magdy G, Elguindy A, …Yacoub MH (2013). Early results of sarcomeric gene screening from the Egyptian National BA-HCM Program. J Cardiovasc Transl Res, 6(1), 65–80. doi: 10.1007/s12265-012-9425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, & Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet, 46(3), 310–5. doi:ng.2892 [pii] 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein GJ, Bashore TM, Sellers TD, Pritchett EL, Smith WM, & Gallagher JJ (1979). Ventricular fibrillation in the Wolff-Parkinson-White syndrome. N Engl J Med, 301(20), 1080–5. doi: 10.1056/NEJM197911153012003 [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, & Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc, 4(7), 1073–81. doi:nprot.2009.86 [pii] 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Laredo R, Monserrat L, Hermida-Prieto M, Fernandez X, Rodriguez I, Cazon L, …Castro-Beiras A (2006). [Beta-myosin heavy-chain gene mutations in patients with hypertrophic cardiomyopathy]. Rev Esp Cardiol, 59(10), 1008–18. doi:13093977 [pii] [DOI] [PubMed] [Google Scholar]
- Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, …Tanaka T (2017a). Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet, 49(6), 953–958. doi:ng.3842 [pii] 10.1038/ng.3842 [DOI] [PubMed] [Google Scholar]
- Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, …Tanaka T (2017b). Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet. doi:ng.3842 [pii] 10.1038/ng.3842 [DOI] [PubMed] [Google Scholar]
- Lubitz SA, Brody JA, Bihlmeyer NA, Roselli C, Weng LC, Christophersen IE, …Lin H (2016). Whole Exome Sequencing in Atrial Fibrillation. PLoS Genet, 12(9), e1006284. doi: 10.1371/journal.pgen.1006284 PGENETICS-D-16-01184 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc IM, & Freed AE (1955). The Wolff-Parkinson-White syndrome; report of a case occurring in a mother and infant. AMA Am J Dis Child, 89(6), 743–7. [PubMed] [Google Scholar]
- Miyake CY, Del Nido PJ, Alexander ME, Cecchin F, Berul CI, Triedman JK, …Walsh EP (2011). Cardiac tumors and associated arrhythmias in pediatric patients, with observations on surgical therapy for ventricular tachycardia. J Am Coll Cardiol, 58(18), 1903–9. doi:S0735-1097(11)02869-5 [pii] 10.1016/j.jacc.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, …Bennett V (2003). Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature, 421(6923), 634–9. doi: 10.1038/nature01335 nature01335 [pii] [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, …Bennett V (2004). A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A, 101(24), 9137–42. doi: 10.1073/pnas.04025461010402546101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella J, DeBiasi RM, Coplan NL, Suri R, & Keller S (2014). Noninvasive risk stratification for sudden death in asymptomatic patients with Wolff-Parkinson-White syndrome. Rev Cardiovasc Med, 15(4), 283–9. [DOI] [PubMed] [Google Scholar]
- Obeyesekere MN, Leong-Sit P, Massel D, Manlucu J, Modi S, Krahn AD, …Klein GJ (2012). Risk of arrhythmia and sudden death in patients with asymptomatic preexcitation: a meta-analysis. Circulation, 125(19), 2308–15. doi: 10.1161/CIRCULATIONAHA.111.055350 [DOI] [PubMed] [Google Scholar]
- Packard JM, Graettinger JS, & Graybiel A (1954). Analysis of the electrocardiograms obtained from 1000 young healthy aviators; ten year follow-up. Circulation, 10(3), 384–400. [DOI] [PubMed] [Google Scholar]
- Park H, Staehling K, Tsang M, Appleby MW, Brunkow ME, Margineantu D, …Iritani BM (2012). Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity, 36(5), 769–81. doi:S1074-7613(12)00181-1 [pii] 10.1016/j.immuni.2012.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VV, Arad M, Moskowitz IP, Maguire CT, Branco D, Seidman JG, …Berul CI (2003). Electrophysiologic characterization and postnatal development of ventricular pre-excitation in a mouse model of cardiac hypertrophy and Wolff-Parkinson-White syndrome. J Am Coll Cardiol, 42(5), 942–51. doi:S0735109703008507 [pii] [DOI] [PubMed] [Google Scholar]
- Perez-Serra A, Campuzano O, & Brugada R (2017). Update about atrial fibrillation genetics. Curr Opin Cardiol. doi: 10.1097/HCO.0000000000000387 [DOI] [PubMed] [Google Scholar]
- Pietersen AH, Andersen ED, & Sandoe E (1992). Atrial fibrillation in the Wolff-Parkinson-White syndrome. Am J Cardiol, 70(5), 38A–43A. [DOI] [PubMed] [Google Scholar]
- Purevjav E, Varela J, Morgado M, Kearney DL, Li H, Taylor MD, …Towbin JA (2010). Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. J Am Coll Cardiol, 56(18), 1493–502. doi:S0735-1097(10)03498-4 [pii] 10.1016/j.jacc.2010.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, …Boerwinkle E (2014). Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics, 15, 30. doi: 10.1186/1471-2105-15-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, …Rehm HL (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–24. doi:gim201530 [pii] 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santinelli V, Radinovic A, Manguso F, Vicedomini G, Ciconte G, Gulletta S, …Pappone C (2009a). Asymptomatic ventricular preexcitation: a long-term prospective follow-up study of 293 adult patients. Circ Arrhythm Electrophysiol, 2(2), 102–7. doi: 10.1161/CIRCEP.108.827550 [DOI] [PubMed] [Google Scholar]
- Santinelli V, Radinovic A, Manguso F, Vicedomini G, Gulletta S, Paglino G, …Pappone C (2009b). The natural history of asymptomatic ventricular pre-excitation a long-term prospective follow-up study of 184 asymptomatic children. J Am Coll Cardiol, 53(3), 275–80. doi: 10.1016/j.jacc.2008.09.037 [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, & Seelow D (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods, 11(4), 361–2. doi:nmeth.2890 [pii] 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- Sharma AD, Klein GJ, Guiraudon GM, & Milstein S (1985). Atrial fibrillation in patients with Wolff-Parkinson-White syndrome: incidence after surgical ablation of the accessory pathway. Circulation, 72(1), 161–9. [DOI] [PubMed] [Google Scholar]
- Siggs OM, Stockenhuber A, Deobagkar-Lele M, Bull KR, Crockford TL, Kingston BL, …Cornall RJ (2016). Mutation of Fnip1 is associated with B-cell deficiency, cardiomyopathy, and elevated AMPK activity. Proc Natl Acad Sci U S A, 113(26), E3706–15. doi:1607592113 [pii] 10.1073/pnas.1607592113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne LA, Murphy NP, Asuri S, Chen L, Xu X, McIntosh S, …Arbour LT (2017). Novel Variant in the ANK2 Membrane-Binding Domain Is Associated With Ankyrin-B Syndrome and Structural Heart Disease in a First Nations Population With a High Rate of Long QT Syndrome. Circ Cardiovasc Genet, 10(1). doi:CIRCGENETICS.116.001537 [pii] 10.1161/CIRCGENETICS.116.001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylva M, van den Hoff MJ, & Moorman AF (2014). Development of the human heart. Am J Med Genet A, 164A(6), 1347–71. doi: 10.1002/ajmg.a.35896 [DOI] [PubMed] [Google Scholar]
- Van Driest SL, Jaeger MA, Ommen SR, Will ML, Gersh BJ, Tajik AJ, & Ackerman MJ (2004). Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol, 44(3), 602–10. doi: 10.1016/j.jacc.2004.04.039 S0735109704009568 [pii] [DOI] [PubMed] [Google Scholar]
- Vidaillet HJ Jr., Pressley JC, Henke E, Harrell FE Jr., & German LD (1987). Familial occurrence of accessory atrioventricular pathways (preexcitation syndrome). N Engl J Med, 317(2), 65–9. doi: 10.1056/NEJM198707093170201 [DOI] [PubMed] [Google Scholar]
- Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, & Martin JF (2010). Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A, 107(21), 9753–8. doi:0912585107 [pii] 10.1073/pnas.0912585107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf CM, Arad M, Ahmad F, Sanbe A, Bernstein SA, Toka O, Konno T, …Berul CI (2008). Reversibility of PRKAG2 glycogen-storage cardiomyopathy and electrophysiological manifestations. Circulation, 117(2), 144–54. doi:CIRCULATIONAHA.107.726752 [pii] 10.1161/CIRCULATIONAHA.107.726752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YQ, Xu YJ, Li RG, Qu XK, Fang WY, & Liu X (2013). Prevalence and spectrum of PITX2c mutations associated with familial atrial fibrillation. Int J Cardiol, 168(3), 2873–6. doi:S0167-5273(13)00579-2 [pii] 10.1016/j.ijcard.2013.03.141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.