Significance
No current therapies prevent recurrence in patients with glioblastoma multiforme (GBM). Extracellular matrix (ECM) remodeling and epithelial mesenchymal transition (EMT) are important processes regulating GBM progression and acquisition of radiation and temozolomide (radio/TMZ) resistance. Fli-1 choreographs ECM remodeling and EMT in GBM via transcriptional regulation of HSPB1. Lumefantrine, an antimalarial drug, inhibits the transcriptional activities of Fli-1 and its downstream targets HSPB1, ECM remodeling, and EMT. Lumefantrine is selectively cytotoxic and promotes apoptosis in GBM and reverses radio/TMZ resistance in GBM. This drug can easily be repurposed for management of GBM. Identification of drugs like lumefantrine from FDA-approved therapeutic agents and uncommon sources provides opportunities to broaden the breadth and versatility of current therapeutic regimens for GBM.
Keywords: glioblastoma, HSPB1, Fli-1, radioresistance, TMZ resistance
Abstract
Glioblastoma multiforme (GBM) is an aggressive cancer without currently effective therapies. Radiation and temozolomide (radio/TMZ) resistance are major contributors to cancer recurrence and failed GBM therapy. Heat shock proteins (HSPs), through regulation of extracellular matrix (ECM) remodeling and epithelial mesenchymal transition (EMT), provide mechanistic pathways contributing to the development of GBM and radio/TMZ-resistant GBM. The Friend leukemia integration 1 (Fli-1) signaling network has been implicated in oncogenesis in GBM, making it an appealing target for advancing novel therapeutics. Fli-1 is linked to oncogenic transformation with up-regulation in radio/TMZ-resistant GBM, transcriptionally regulating HSPB1. This link led us to search for targeted molecules that inhibit Fli-1. Expression screening for Fli-1 inhibitors identified lumefantrine, an antimalarial drug, as a probable Fli-1 inhibitor. Docking and isothermal calorimetry titration confirmed interaction between lumefantrine and Fli-1. Lumefantrine promoted growth suppression and apoptosis in vitro in parental and radio/TMZ-resistant GBM and inhibited tumor growth without toxicity in vivo in U87MG GBM and radio/TMZ-resistant GBM orthotopic tumor models. These data reveal that lumefantrine, an FDA-approved drug, represents a potential GBM therapeutic that functions through inhibition of the Fli-1/HSPB1/EMT/ECM remodeling protein networks.
Major pathological features that impose impediments to the management of glioblastoma (GBM) include infiltrative growth behavior, intratumoral heterogeneity, and propensity for tumor recurrence (1–5). Conventional approaches using radiation and temozolomide (TMZ; chemotherapy) have proven unsuccessful in treating GBM because of acquired therapeutic resistance and eventual disease recurrence. Unraveling the molecular mechanisms involved in GBM development and progression provide a potential path forward for developing effective GBM therapies (1, 3, 5). Omics studies in GBM have reported elevated expression of heat shock proteins (HSPs) (1, 6) that promote tumor growth by activating GBM cell proliferation and inhibiting death pathways (1). A positive correlation exists between HSP expression and ECM remodeling that contributes to the infiltrative potential of GBM via binding to MMPs (7). The HSPs also facilitate epithelial-mesenchymal transition (EMT) in cancer (6). Up-regulation of HSPs is evident in GBM and radiation (radio)/TMZ-resistant GBM (6, 8). In these contexts, HSPs provide prospective targets that may be amenable for developing improved clinical strategies for anti-GBM drug development.
HSPB1 is the most up-regulated HSP in GBM and radio/TMZ-resistant GBM (8). Progression and acquisition of radio/TMZ resistance in GBM correlates with elevated expression of HSPB1 mediated through the Ets family of transcriptional regulator(s) (8). In GBM, previous studies confirm a link between hypoxia and up-regulation of HSP expression (9, 10) and regulation of hypoxia and HSPB1 by Ets family proteins (11, 12). Additional evidence in multiple cancer indications highlight important connections between HSPB1 and the Ets family of transcription factors (12–16). The 5-kb upstream region of the HSPB1 gene contains genomic elements that bind to the Friend leukemia integration factor 1 (Fli-1) (8). A member of the ETS family, Fli-1 is a target of insertional activation by Friend murine leukemia virus (F-MuLV) (17). It is expressed in vascular endothelial cells and hematopoietic tissues (18), affecting cellular proliferation and tumorigenesis in Ewing sarcoma and primitive neuroectodermal tumors (13–16, 19, 20). Several oncology studies have reported Fli-1 overexpression as a cancer biomarker (21–25). The present study now defines a relationship between Fli-1 protein and HSPB1 expression that correlates with radio/TMZ resistance in GBM.
Based on the connection between Fli-1 expression and radio/TMZ resistance in GBM cells (8), we screened for Fli-1 inhibitors and identified an antimalarial FDA-approved drug, lumefantrine, as a putative therapeutic agent targeting radio/TMZ resistance in GBM. We have scrutinized the therapeutic actions of lumefantrine in vitro using parental, radioresistant, and TMZ-resistant GBM and in vivo using U87MG orthotopic parental, radioresistant, and TMZ-resistant GBM models. We also endeavored to assess the molecular interactions of lumefantrine with Fli-1 signaling axes, EMT (β-catenin, vimentin, and Snail) and ECM (MMP-2 and MMP-9) remodeling, and apoptosis. In addition, molecular docking studies were performed to understand the interaction pattern of lumefantrine at the binding site of Fli-1, and this interaction was further confirmed by isothermal titration calorimetry (ITC). Toxicity profiling of lumefantrine on major organs, including liver, spleen, lungs, heart, and kidney, was also performed. Taken together, these studies provide compelling evidence that targeting Fli-1 using lumefantrine has potential as an effective therapy for treating both primary GBM and radio/TMZ-resistant GBM.
Results and Discussion
GBM accounts for the majority of brain tumors with a poor patient prognosis, despite current interventional therapeutic strategies (surgery, radiation, and/or chemotherapy) (1–5). In principle, defining promising GBM-specific therapeutic targets could serve as a basis for developing strategies to advance effective therapies for GBM by elucidating the molecular pathways involved in gliomagenesis (2–5). Heat shock proteins (HSPs) provide a primary protein signature for GBM progression serving as a nexus for ECM remodeling and EMT (6). We previously demonstrated that specific HSPs, particularly HSPB1, are up-regulated in GBM and radio/TMZ-resistant GBM (8). Accordingly, inhibiting the transcriptional regulation of HSPB1 is an appealing strategy for creating a therapeutic approach to selectively target therapy-resistant GBM cells. We reported a potentially relevant transcription factor, Fli-1, located in the 5-kb upstream region of the HSPB1 gene as an important mediator of HSPB1 expression (8). Fli-1 was shown to functionally regulate HSPB1 expression, and using an innovative drug screening strategy, a selective inhibitor of Fli-1 was identified.
Isolation of Fli-1 Inhibitors from Small-Molecule Libraries.
Fli-1 and HSPB1 expression were evaluated by Western blot analysis in five GBM cell lines: U87MG, LN229, LN18, T98G, and A172 (Fig. 1 A and B). Based on these initial studies, we focused on U87MG and T98G cells. To identify small molecules capable of inhibiting Fli-1 transactivation ability, luciferase assays were performed using an engineered Fli-1 vector (Fli-1 Ets DNA-binding site cloned in front of a minimal promoter, upstream of a luciferase reporter gene), designated Fli-1-Lu (Fig. 1C). Exogenous expression of Fli-1 produces more robust luciferase activity compared with endogenous Fli-1 expression. Accordingly, an Fli-1 expression vector, murine stem cell virus (MSCV) neo-Fli-1, was used in screening for Fli-1 inhibitors. T98G cells were cotransfected with an Fli-1-Lu construct and an MSCV neo-Fli-1 vector (Fig. 1C). The negative controls were T98G cells cotransfected with Fli-1-Lu and MSCV empty vector to exclude drugs that could inhibit luciferase activity through an Fli-1–independent pathway.
Fig. 1.
Fli-1 drug screening. (A and B) Relative expression of Fli-1 and HSPB1 in five different GBM cell lines determined by Western blot analysis (A) and normalization of expression (based on β-actin expression) (B). (C) Schematic representation of the Fli-1 drug-screening strategy. Fli-1-Lu: multiple copies of the Fli-1 consensus-binding site (FB site) cloned upstream of a minimal promoter immediately upstream of the luciferase gene. MSCV neo-Fli-1: Fli-1 gene cloned into the MSCV vector. Luciferase gene driven by the Luc promoter: Fli-1-Lu was cotransfected with either MSCV neo-Fli-1 or MSCV empty vector into T98G cells. Cells were treated with various drugs at 36 h posttransfection and screened for efficient down-regulation of luciferase activity. (D) Relative expression by Western blot analysis of Fli-1 and HSPB1 in T98G and U87MG cells treated with positive inhibitory candidates identified using the Fli-1 drug screening protocol. (E) Corresponding densitometry plot for D showing normalization of expression (based on β-actin expression). Each bar represents average of three independent experiments. The level of significance is indicated as *P < 0.05, **P < 0.01, or ***P < 0.001.
Screening experiments were performed using biologically and pharmacologically active compounds, natural products and marketed drugs. In Ewing’s sarcoma (EWS), a chromosomal translocation generating a fusion of the transactivation domain of EWS with the Ets domain of Fli-1 has been well documented (14). Thus, the drugs used for screening were selected on the basis of the structure-activity relationship from the EWS class of inhibitors, including naphthoquinones, monochlorobenzenes, nitrofuran, and benzamidines. Library screening identified 17 compounds that efficiently reduced Fli-1–mediated luciferase activity by 50% (SI Appendix, Table S1). The lead compounds were further screened by docking them with Fli-1 DNA-binding domains resulting in the selection of four compounds for further investigation.
Four lead compounds (furamidine, nifurtimox, lumefantrine, and atovaquone) were assayed for cytotoxicity in SVGp12 cells (normal astroglial cells). These four compounds showed minimal cytotoxicity with IC50 values of 289.7 μM for furamidine, 319.1 μM for nifurtimox, 732.2 μM for lumefantrine, and 450.9 μM for atovaquone (SI Appendix, Fig. S1A). These molecules displayed antiproliferative effects in U87MG and T98G cells (GBM cells). The IC50 values for furamidine, nifurtimox, lumefantrine, and atovaquone were 94.6 μM, 151.2 μM, 77.1 μM, and 50.7 μM, respectively, in U87MG cells and 128.1 μM, 101.2 μM, 37.4 μM, and 92.6 μM, respectively, in T98G cells (SI Appendix, Fig. S1 B and C). Among these four lead compounds, the Fli-1 inhibitor chosen for further studies was determined by evaluating the activity on Fli-1 and HSPB1 protein at a translational level. The T98G and U87MG cells were exposed to IC50 doses for 12 h and were lysed at 24 h after drug treatment, followed by Western blot analysis (Fig. 1 D and E). This approach identified lumefantrine as a potential therapeutic agent inhibiting Fli-1 transactivating activity. Lumefantrine is a Food and Drug Administration (FDA)-approved aryl-amino alcohol antimalarial drug used in combination with artemether to treat malaria (26).
Lumefantrine-Binding Affinity to the Fli-1 Protein.
Phase-contrast microscopy images of lumefantrine-treated U87MG and T98G cells showed morphological changes suggestive of apoptotic cell death (SI Appendix, Fig. S2A, arrows). Lumefantrine inhibited wound healing, migration, infiltration, and anchorage-independent growth in U87MG and T98G cells (SI Appendix, Fig. S2 B–D). In addition, lumefantrine significantly reduced the enzymatic activity of MMP-2 and MMP-9 in U87MG and T98G cells (SI Appendix, Fig. S2E). A dose-dependent antiproliferative effect was also observed in lumefantrine-treated radio/TMZ-resistant cells (SI Appendix, Fig. S1 D and E). Half-maximal inhibitory concentration (IC50) values of lumefantrine were 137.2 μM in U87MG RR cells, 73.1 μM in T98G RR cells, 185.3 μM in U87MG TMZR cells, and 120.2 μM in T98G TMZR cells.
A direct interaction between lumefantrine and Fli-1 was confirmed by docking of lumefantrine with the DNA-binding domain of the Fli-1 protein (Fig. 2 A–C) and ITC. The chemical structure of lumefantrine is shown in Fig. 2B. The ITC thermogram and fitted binding isotherm (Fig. 2D) displayed the following thermodynamic parameters of lumefantrine’s binding to Fli-1 protein: ΔH (enthalpy), −1.349E4 ± 1,349 cal/mol; ΔS (entropy), −27.8 cal/mol/degree; Kd (dissociation constant), 6.5 μM; and N (stoichiometry), 0.687 ± 0.0496. These studies confirm that lumefantrine affects the in vitro properties of GBM and radio/TMZ-resistant GBM, and that it directly binds to the Fli-1 protein.
Fig. 2.
Interaction between lumefantrine and Fli-1 protein. (A) The three-dimensional structure of Fli-1 protein using I-TASSER. (B) Chemical structure of lumefantrine. (C) Docked conformation of lumefantrine with the DNA-binding domain of Fli-1 protein. (D) ITC thermogram and fitted binding isotherm. Direct interaction between lumefantrine and Fli-1 was measured by ITC. The thermodynamic parameters of binding of lumefantrine to Fli-1 are as follows: ΔH (enthalpy), −1.349E4 ± 1,349 cal/mol; ΔS (entropy), −27.8 cal/mol/deg; Kd (dissociation constant), 6.5 μM; and N (stoichiometry), 0.687 ± 0.0496.
Lumefantrine Mediates Apoptosis and Inhibits Fli-1/HSPB1, MMP-2/-9, and EMT in Radio/TMZ-Resistant GBM Cells.
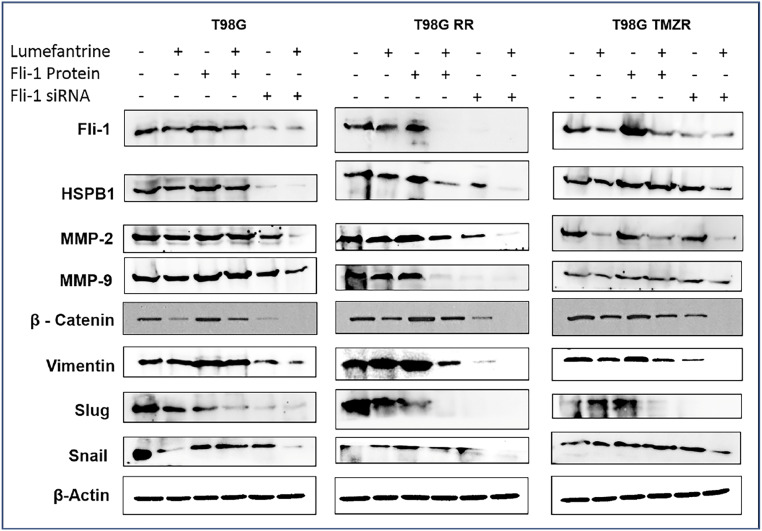
To determine the effect of lumefantrine on GBM and GBM-resistant cells at a molecular level, T98G, T98G RR, and T98G TMZR cells were treated with Fli-1 protein for 3 h, transfected with Fli-1 siRNA and then treated with lumefantrine individually and in combination. Fli-1 siRNA, as well as lumefantrine treatment, robustly reduced expression of Fli-1, HSPB1, MMP-2, MMP-9, β-catenin, Vimentin, Slug, and Snail protein in all three cell lines (Fig. 3 and SI Appendix, Fig. S3). These results demonstrate comparable efficiencies of Fli-1 inhibitor and Fli-1 siRNA in terms of reduction of Fli-1 and HSPB1 protein expression. In addition, the inhibitor also significantly reduced Fli-1 and HSPB1 expression in the presence of Fli-1 protein.
Fig. 3.
Lumefantrine-mediated inhibition of Fli-1 for the treatment of radio/TMZ-resistant GBM. Lumefantrine inhibits Fli-1, HSPB1, MMP-2/-9, and EMT markers in Fli-1 protein and Fli-1 siRNA-treated T98G, T98G RR, and T98G TMZR cells. Densitometry plots are shown in SI Appendix, Fig. S3.
Lumefantrine induced apoptosis in a time-dependent manner, as demonstrated by DNA content-based cell cycle analysis of U87MG, U87MG RR, and U87MG TMZR cells (SI Appendix, Fig. S4A). Cells were grown in 60-mm tissue culture plates and treated with an IC50 dose of lumefantrine for 24, 36, and 48 h. The sub-G1 population (apoptotic cells) was increased due to lumefantrine treatment (SI Appendix, Fig. S4A). Similar increases in the sub-G1 population (apoptotic cells) were observed based on DNA content-based cell cycle analysis in T98G, T98G RR, and T98G TMZR cells exposed to lumefantrine (SI Appendix, Fig. S4B). To provide further confirmation that lumefantrine mediates apoptosis in parental and radio/TMZ-resistant GBM cells, annexin V/propidium iodide assays were performed in T98G, T98G RR, and T98G TMZR cells. As shown in SI Appendix, Fig. S5, lumefantrine caused a substantial increase in cell death by inducing apoptosis in all three cell lines, which was significantly reduced in the presence of a pan-caspase inhibitor Z-VAD (SI Appendix, Fig. S5).
Radio/TMZ-resistant GBM cells were grown and treated with lumefantrine (IC50 dose) for 12 h and after 24, 36, and 48 h, the cells were lysed and examined for molecular changes (Fig. 4 and SI Appendix, Fig. S6). In U87MG cells, Fli-1 and HSPB1 were down-regulated at 36 h, and in T98G cells, these genes were down-regulated at 24 h. In radiation-resistant cells, Fli-1 was down-regulated at 36 h and HSPB1 was vigorously down-regulated at 24 h in U87MG RR cells, while Fli-1 expression was extinguished at 48 h and HSPB1 expression was dramatically ablated at 36 h in T98G RR cells. In TMZ-resistant cells, Fli-1 and HSPB1 expression were significantly reduced at 48 h in U87MG TMZR cells, and Fli-1 expression was reduced at 24 h and HSPB1 expression was reduced at 48 h in T98G TMZR cells. These data provide solid evidence that lumefantrine is effective in suppressing Fli-1 and HSPB1 expression in radio/TMZ-resistant GBM after 24 h or 36 h of exposure, respectively. Expression of the antiapoptotic proteins Bcl-2 and Bcl-xL and EMT proteins β-catenin and vimentin were down-regulated in a time-dependent manner. Caspase activation and release of cytochrome c were evident in lumefantrine-treated GBM and radio/TMZ-resistant GBM cells.
Fig. 4.
Lumefantrine triggered apoptosis in GBM cells. Western blot analysis of Fli-1, HSPB1, EMT markers, and various proapoptotic and antiapoptotic proteins in U87MG, U87MG RR, U87MG TMZR, T98G, T98G RR, and T98G TMZR cells. GBM cells were grown and treated with lumefantrine (IC50 dose) for 24, 36, and 48 h. Expression of Bcl-2 and Bcl-xL were down-regulated, caspase activation was increased, and cytochrome c release was observed in lumefantrine-treated parental and resistant GBM cells. Densitometry plots are shown in SI Appendix, Fig. S6.
Lumefantrine Inhibits Tumor Growth in U87MG, Radioresistant U87MG, and TMZ-Resistant U87MG Orthotopic Animal Models.
Antitumor effects of lumefantrine were tested in U87MG radio/TMZ-resistant orthotopically injected animal models. A representative group of animals containing the various U87 GBM cell lines injected intracranially and either untreated or treated with lumefantrine were sacrificed at 2 and 4 wk. Tumor size and weight were lower at both time points in the lumefantrine-treated animals (SI Appendix, Fig. S7). After 28 d of treatment with lumefantrine (20 mg/kg for U87MG, 40 mg/kg for U87MG RR, and 50 mg/kg for U87MG TMZR), immunohistochemical analysis confirmed decreased expression levels of Fli-1 and HSPB1 in lumefantrine-treated U87MG and U87MG radio/TMZ-resistant animal models (Fig. 5A). Histopathological analyses of major organs, including brain, heart, lung, liver, spleen, and kidney, were performed after 28 d of treatment of U87MG, U87MG RR, and U87MG TMZR cells in mouse models. Hematoxylin and eosin (H&E) staining of organs from the different groups indicated no significant morphological changes following lumefantrine treatment in U87MG, U87MG RR, and U87MG TMZR animals. Only the organ toxicity H&E images for U87MG TMZR cells are provided, since these cells had received the highest dose of lumefantrine (50 mg/kg) among the three GBM models (Fig. 5B). The Fli-1 inhibitor displayed no toxic effects in these major organs. However, some toxic effects at this higher dose were found in liver and spleen based on morphological observations.
Fig. 5.
Lumefantrine mediates inhibition of Fli-1 in vivo in U87MG, U87MG RR, and U87MG TMZR orthotopic GBM models. (A) Immunohistochemistry of tissue sections derived from different groups of orthotopic GBM models. Expression of Fli-1 and HSPB1 were significantly reduced in the treated groups. (B) H&E staining of major organs, including brain, heart, lungs, liver, spleen, and kidney, in the U87MG TMZR GBM model to detect any organ-specific toxicity induced by lumefantrine.
Protein expression in tissue lysates derived from tumors demonstrated that Fli-1, HSPB1, antiapoptotic protein Bcl-2, EMT markers, and ECM remodeling proteins were down-regulated, while the apoptotic protein Bax was up-regulated in the lumefantrine-treated groups (Fig. 6). These data support the in vitro observations and demonstrate antitumor effects and similar biochemical/molecular effects of lumefantrine on Fli-1/HSPB1/EMT markers/ECM remodeling protein networks in vivo in U87MG, U87MG RR, and U87MG TMZR mouse models.
Fig. 6.
Inhibition of Fli-1/HSPB1 axes by lumefantrine in an in vivo orthotopic GBM model. (A and B) Western blot analysis of tissue lysates derived from tumors confirming down-regulation of Fli-1, HSPB1, Bcl-2, β-catenin, Vimentin, Snail, MMP-9, and MMP-2 as well as up-regulation of Bax in lumefantrine-treated groups compared with controls. Each bar represents the average of three independent experiments. The level of significance is indicated by *P < 0.05, **P < 0.01, or ***P < 0.001.
In summary, the present study documents the importance and relevance of HSPB1 and Fli-1 in the regulation of radio/TMZ-resistance of GBM cells. They confirm that targeted inhibition of Fli-1/HSPB1–mediated EMT and ECM remodeling signaling axes by lumefantrine provides a novel therapeutic reagent for radio/TMZ resistance in glioblastoma (Fig. 7). These preclinical studies provide a solid rationale for Fli-1/HSPB1 inhibition with lumefantrine as a potentially selective approach for glioblastoma management. Although the safety profile of lumefantrine (an FDA-approved drug) is appropriate for clinical utility, further studies are needed to determine the efficiency of this new antiglioblastoma drug to pass the blood-brain barrier (using an authentic primary intracranial GBM model) and also to evaluate additional combinatorial approaches for enhancing further therapeutic outcomes. Preliminary analysis using a computational blood-brain barrier algorithm (27) indicates that lumefantrine should pass through the blood-brain barrier (SI Appendix, Fig. S8). In addition, the screening strategy described in the paper using additional transcriptional targets and a diverse array of natural products, marketed drugs, and existing and new therapeutic agents offers potential for identifying additional drugs or drug combinations that may be of value in treating GBM, an aggressive and fatal cancer without any effective therapies (with recurrence at 7 mo after surgery; ref. 2). Further studies in other cancers in which Fli-1 expression is relevant would also be of value in defining additional applications of lumefantrine for potential cancer therapy.
Fig. 7.
Schematic outline of lumefantrine-mediated inhibition of Fli-1, a transcription factor in the upstream region of HSPB1, targeting radioresistance and TMZ resistance (RR/TMZR) in glioblastoma.
Materials and Methods
Cell Cultures.
The human GBM cell lines U87MG, A172, and LN18 were generously provided by Ellora Sen, National Brain Research Centre. LN229, T98G (human GBM), and SVGp12 cells (normal astroglial cells) were provided by Annapoorni Rangarajan, Indian Institute of Science. Cells were grown as described previously (7). Radio/TMZ-resistant GBM cell lines were selected as described previously (8). The GBM cell lines were authenticated by cell repository (National Centre for Cell Science).
Immunohistochemistry.
Immunohistochemistry (IHC) analysis and hematoxylin and eosin (H&E) staining of orthotopic GBM tumors grown in nude mice were performed with different antibodies, including Fli-1 and HSPB1 (28), using a super-sensitive polymer-HRP (horseradish peroxidase) IHC detection system (QD420-YIKE; BioGenex). The secondary antibody poly-HRP reagent and peroxide-DAB (3,3′-diaminobenzidine) staining (for protein expression) provided in the kit was performed, followed by hematoxylin counterstaining for nuclear staining.
Wound Healing Assay.
The migration potential of GBM cells was analyzed by wound-healing assays or scratch assays as described previously (29). The cells were seeded in a six-well plate, and a wound was drawn using a pipette tip on attaining ∼80% confluency. Washing in PBS (phosphate-buffered saline) was done to remove debris and suspended cells. The microscopic images were obtained at 0, 12 and 24 h under an inverted phase-contrast microscope (Leica Microsystems).
Boyden Chamber Assay.
Boyden chamber assays were performed to check the invasive property of GBM cells as described previously (7). A total of 1 × 105 cells were suspended in serum-free medium and then added to the upper chamber. Chemoattractant (vascular endothelial growth factor and serum medium) was added in the lower compartment of the Boyden chamber, followed by incubation for 16 h at 37 °C. The nonmigrated cells were then removed by cotton swab, and migrated cells were fixed with 100% methanol. Bright-field images of migrated cells stained with H&E were obtained by microscopy.
Western Blot Analysis.
Western blotting was performed to assess protein expression in human GBM and radio/TMZ-resistant cells as described previously (28). The cells were grown in six-well plates. After attaining 70% confluence, cells were treated with specific experimental reagents, harvested, and lysed with Nonidet P-40 lysis buffer (Invitrogen) according to the manufacturer’s protocol after a stipulated treatment schedule. An equal amount of protein was separated by SDS/PAGE (sodium dodecyl sulfate/polyacrylamide gel electrophoresis), and separated proteins were transferred to a nitrocellulose membrane, followed by 3% BSA (bovine serum albumin) membrane blocking. Blocked membranes were washed and incubated with specific primary antibodies overnight at 4 °C. The membranes were then washed and exposed to specific secondary HRP-conjugated antibodies, followed by washing and developing in an ImageQuant LAS 4000 biomolecular imager (GE Healthcare).
Transfection Experiments.
Transfection studies were performed to determine the effect of Fli-1 shRNA/siRNA on T98G/T98G RR/T98G TMZR cells. Cells were grown in Petri dishes and serum-starved overnight. The respective cells were transfected with Fli-1 shRNA/siRNA using a previously reported method (30). After 24 h of transfection, cells were lysed with Nonidet P-40 lysis buffer, and Western blot analysis was performed.
Cell Proliferation Assay.
The antiproliferative effects of TMZ and lumefantrine on SVGp12, U87MG, T98G, U87MG RR, U87MG TMZR, T98G RR, and T98G TMZR cells were evaluated by MTT assays as described previously (31).
Binding Assay.
To determine lumefantrine binding to the Fli-1 protein, we performed ITC using a MicroCal iTC200 system (GE Healthcare) as described previously (32). Initially, ITC was performed using a wide range of proteins (1 to 50 µM) in the instrument cell and 10× drug in the syringe. Recombinant human Fli-1 protein was obtained from Abcam and diluted to various concentrations with DNase/RNase-free water. The optimum binding curve was obtained at 20 µM Fli-1 protein and 180 µM lumefantrine. The cell and syringe in the calorimeter contained 2% DMSO (dimethyl sulfoxide). Titration was performed using 20 μM Fli-1 protein in sample cell and 180 μM lumefantrine in the injection. Experiments were performed at 25 °C as described previously (28).
Flow Cytometry Analysis.
DNA content-based cell cycle analyses of lumefantrine-treated cells were performed by flow cytometry as described previously (32). In brief, U87MG, T98G, U87MG RR, U87MG TMZR, T98G RR, and T98G TMZR cells were seeded, grown and treated with an IC50 dose of lumefantrine. Treated cells were then processed and subjected to flow cytometry analysis.
In Vivo Orthotopic GBM Model.
The antitumor effect of lumefantrine was assessed using U87MG cells in a nude mouse orthotopic model. The mice were housed at the Department of Biotechnology, Indian Institute of Technology in accordance with institutional guidelines. The experiments were approved by the Institutional Ethical Committee (project no. IE-6/MM-SMST/3.15) and were conducted observing all animal ethics regulations streamlined by the Institutional Animal Ethics Committee under guidance of the Indian government’s Committee for the Purpose of Control and Supervision of Experiments on Animals.
A 5-μL Hamilton syringe fitted with a 26-gauge needle was used to inject 2.5 μL of complete culture medium containing 3 × 104 U87MG, U87MG RR, or U87MG TMZR cells as reported previously (28). After 12 d, tumors developed, and animals were divided at random into three main groups each with two subgroups (control and lumefantrine-treated): U87MG (control and treated with 20 mg/kg), U87MG RR (control and treated with 40 mg/kg), and U87MG TMZR (control and treated with 50 mg/kg). The treatment (via tail vein injection) regimens were followed with each group of six animals for 4 wk on alternate days. Western blot and immunohistochemistry analyses of tumor tissue and histopathology of major organs (brain, heart, liver, lungs, and spleen) were performed.
Statistical Analysis.
Data are presented as mean ± SEM unless stated otherwise. Statistical significance was determined by two-way ANOVA followed by Student’s t test.
Data Availability Statement.
All data obtained for this study are presented in the main text and SI Appendix.
Supplementary Material
Acknowledgments
Support for this work was provided by the Indian Department of Science and Technology’s Innovation in Science Pursuit for Inspired Research (INSPIRE) Fellowship (IF130658), the Indian Institute of Technology Kharagpur and Ministry of Human Resource Development, the Indian Science and Engineering Research Board Fellowship (JCB/2019/000008), the Indian Council of Medical Research Grant (5/13/7/2019- NCD-III), the Indian Council of Scientific and Industrial Research Grant (27(0347)/19/EMR-II), and the Genetics Enhancement Fund of the Department of Human and Molecular Genetics and the VCU Institute of Molecular Medicine of VCU School of Medicine (S.K.D., L.E., and P.B.F.). P.B.F. is holder of the Thelma Newmeyer Corman Chair in Oncology at the VCU Massey Cancer Center.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921531117/-/DCSupplemental.
References
- 1.Rajesh Y., et al. , Insights into molecular therapy of glioma: Current challenges and next-generation blueprint. Acta Pharmacol. Sin. 38, 591–613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das S. K., Sarkar D., Cavenee W. K., Emdad L., Fisher P. B., Rethinking glioblastoma therapy: MDA-9/Syntenin-targeted small molecule. ACS Chem. Neurosci. 10, 1121–1123 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kegelman T. P., et al. , MDA-9/syntenin is a key regulator of glioma pathogenesis. Neuro-oncol. 16, 50–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kegelman T. P., et al. , Inhibition of radiation-induced glioblastoma invasion by genetic and pharmacological targeting of MDA-9/Syntenin. Proc. Natl. Acad. Sci. U.S.A. 114, 370–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S. K., Sarkar D., Emdad L., Fisher P. B., MDA-9/Syntenin: An emerging global molecular target regulating cancer invasion and metastasis. Adv. Cancer Res. 144, 137–191 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Rajesh Y., Biswas A., Mandal M., Glioma progression through the prism of heat shock protein-mediated extracellular matrix remodeling and epithelial to mesenchymal transition. Exp. Cell Res. 359, 299–311 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Rajesh Y., et al. , Delineation of crosstalk between HSP27 and MMP-2/MMP-9: A synergistic therapeutic avenue for glioblastoma management. Biochim. Biophys. Acta Gen. Subj. 1863, 1196–1209 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Rajesh Y., et al. , Transcriptional regulation of HSPB1 by Friend Leukemia Integration-1 factor modulates radiation and temozolomide resistance in glioblastoma. Oncotarget 11, 1097–1108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C., Hong C. S., Zhuang Z., Hypoxia and glioblastoma therapy. Aging (Albany NY) 7, 523–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerer-Lercher A., et al. , Hypoxia induces heat shock protein expression in human coronary artery bypass grafts. Cardiovasc. Res. 50, 115–124 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Qiao N., et al. , Ets-1 as an early response gene against hypoxia-induced apoptosis in pancreatic β-cells. Cell Death Dis. 6, e1650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster C. S., et al. , Hsp-27 expression at diagnosis predicts poor clinical outcome in prostate cancer independent of ETS-gene rearrangement. Br. J. Cancer 101, 1137–1144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroulakou I. G., Bowe D. B., Expression and function of ets transcription factors in mammalian development: A regulatory network. Oncogene 19, 6432–6442 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Oikawa T., Yamada T., Molecular biology of the Ets family of transcription factors. Gene 303, 11–34 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Davidson B., et al. , Ets-1 messenger RNA expression is a novel marker of poor survival in ovarian carcinoma. Clin. Cancer Res. 7, 551–557 (2001). [PubMed] [Google Scholar]
- 16.Oikawa T., ETS transcription factors: Possible targets for cancer therapy. Cancer Sci. 95, 626–633 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-David Y., Giddens E. B., Letwin K., Bernstein A., Erythroleukemia induction by Friend murine leukemia virus: Insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 5, 908–918 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Ben-David Y., Bernstein A., Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell 66, 831–834 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Liu F., Walmsley M., Rodaway A., Patient R., Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr. Biol. 18, 1234–1240 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Spyropoulos D. D., et al. , Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell Biol. 20, 5643–5652 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torlakovic E. E., et al. , Fli-1 expression in malignant melanoma. Histol. Histopathol. 23, 1309–1314 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Song W., et al. , Oncogenic Fli-1 is a potential prognostic marker for the progression of epithelial ovarian cancer. BMC Cancer 14, 424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song W., et al. , Overexpression of Fli-1 is associated with adverse prognosis of endometrial cancer. Cancer Invest. 33, 469–475 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Scheiber M. N., et al. , FLI1 expression is correlated with breast cancer cellular growth, migration, and invasion and altered gene expression. Neoplasia 16, 801–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang X., et al. , Friend leukemia virus integration 1 expression has prognostic significance in nasopharyngeal carcinoma. Transl. Oncol. 7, 493–502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byakika-Kibwika P., et al. , Artemether-lumefantrine combination therapy for treatment of uncomplicated malaria: The potential for complex interactions with antiretroviral drugs in HIV-infected individuals. Malar. Res. Treat. 2011, 703730 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., et al. , AlzPlatform: An Alzheimer’s disease domain-specific chemogenomics knowledgebase for polypharmacology and target identification research. J. Chem. Inf. Model. 54, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajesh Y., et al. , Targeting NFE2L2, a transcription factor upstream of MMP-2: A potential therapeutic strategy for temozolomide-resistant glioblastoma. Biochem. Pharmacol. 164, 1–16 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Dey G., et al. , Therapeutic implication of “Iturin A” for targeting MD-2/TLR4 complex to overcome angiogenesis and invasion. Cell. Signal. 35, 24–36 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Dey G., et al. , Marine lipopeptide Iturin A inhibits Akt-mediated GSK3β and FoxO3a signaling and triggers apoptosis in breast cancer. Sci. Rep. 5, 10316–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parida S., et al. , Gold nanorod embedded reduction responsive block copolymer micelle-triggered drug delivery combined with photothermal ablation for targeted cancer therapy. Biochim. Biophys. Acta Gen. Subj. 1861, 3039–3052 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Bharti R., et al. , Diacerein-mediated inhibition of IL-6/IL-6R signaling induces apoptotic effects on breast cancer. Oncogene 35, 3965–3975 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data obtained for this study are presented in the main text and SI Appendix.