Significance
Regulatory T cells (Tregs) expressing the Treg-specific transcription factor Foxp3 are indispensable for suppressing hazardous immune responses such as autoimmune disease and allergy. Their stable function requires DNA hypomethylation at specific regions of Treg function-associated genes such as Foxp3. This report shows that, in the course of in vitro Treg generation from conventional T cells by antigenic stimulation in the presence of TGF-β and IL-2, deprivation of CD28 costimulatory signal can induce Treg-specific DNA hypomethylation in developing Tregs. Additional in vitro culture with IL-2 alone further stabilizes their Treg-type hypomethylation status, enabling their in vivo transfer to effectively suppress immune responses. These findings would help in producing functionally stable Tregs from disease-mediating T cells for treatment of various immunological diseases.
Keywords: Treg, Foxp3, Treg-DR, DNA hypomethylation, iTreg
Abstract
Foxp3-expressing regulatory T cells (Tregs) can be generated in vitro by antigenic stimulation of conventional T cells (Tconvs) in the presence of TGF-β and IL-2. However, unlike Foxp3+ naturally occurring Tregs, such in vitro induced Tregs (iTregs) are functionally unstable mainly because of incomplete Treg-type epigenetic changes at Treg signature genes such as Foxp3. Here we show that deprivation of CD28 costimulatory signal at an early stage of iTreg generation is able to establish Treg-specific DNA hypomethylation at Treg signature genes. It was achieved, for example, by TCR/TGF-β/IL-2 stimulation of CD28-deficient Tconvs or CD28-intact Tconvs without anti-CD28 agonistic mAb or with CD80/CD86-blocked or -deficient antigen-presenting cells. The signal abrogation could induce Treg-type hypomethylation in memory/effector as well as naive Tconvs, while hindering Tconv differentiation into effector T cells. Among various cytokines and signal activators/inhibitors, TNF-α and PKC agonists inhibited the hypomethylation. Furthermore, CD28 signal deprivation significantly reduced c-Rel expression in iTregs; and the specific genomic perturbation of a NF-κB binding motif at the Foxp3 CNS2 locus enhanced the locus-specific DNA hypomethylation even in CD28 signaling-intact iTregs. In addition, in vitro maintenance of such epigenome-installed iTregs with IL-2 alone, without additional TGF-β or antigenic stimulation, enabled their expansion and stabilization of Treg-specific DNA hypomethylation. These iTregs indeed stably expressed Foxp3 after in vivo transfer and effectively suppressed antigen-specific immune responses. Taken together, inhibition of the CD28-PKC-NF-κB signaling pathway in iTreg generation enables de novo acquisition of Treg-specific DNA hypomethylation at Treg signature genes and abundant production of functionally stable antigen-specific iTregs for therapeutic purposes.
Naturally occurring regulatory T cells (nTregs), which specifically express the transcription factor Foxp3, play indispensable roles in the maintenance of immunological self-tolerance and homeostasis (1). While a majority of nTregs present in the immune system are thymus-derived (thymus-derived Tregs [tTregs]), a proportion of Foxp3+ Tregs appear to be generated in the periphery (peripherally derived Tregs [pTregs]) from conventional T cells (Tconvs) under certain conditions (2). Foxp3+ Tregs phenotypically similar to tTregs or pTregs can also be generated in vitro (induced Tregs [iTregs]) from Tconvs by antigen stimulation in the presence of TGF-β and IL-2 (3). Both tTregs and pTregs possess epigenetic signatures distinct from those of Tconvs (reviewed in ref. 9). The Treg-specific epigenetic changes reinforce the Treg lineage determination/stability and acquisition of stable immunosuppressive function. In particular, Treg-specific DNA demethylation at specific genomic regions (hereafter referred to as Treg-specific demethylated regions or Treg-DRs) of Treg signature genes such as Foxp3, Cd25, and Ctla4 contributes to continued high expression of these genes and, consequently, leads to robust and stable Treg phenotype and function (4–10). In contrast with tTregs and pTregs, iTregs are unstable in the expression of Foxp3 and other Treg signature genes mainly because of incomplete epigenetic changes at Treg-DRs, and can be driven, under certain in vivo conditions, to differentiate into effector T cells (7). Key questions are, therefore, how nTregs acquire Treg-specific DNA hypomethylation in the course of their physiological development in the thymus and the periphery (10) and how Treg-specific DNA hypomethylation can be generated de novo in iTregs for their clinical use to suppress immune responses stably.
Previous studies have demonstrated that TCR signaling is an essential requirement for Treg-type DNA hypomethylation as well as the expression of Foxp3 and other Treg function-associated genes in developing tTregs (7, 11–13). However, specific contribution of costimulatory signaling in pTreg or iTreg development has been controversial. For example, while costimulatory signals generated by the engagement of CD80/86 and CD28 is indispensable for tTreg development (14–17), pTreg generation in the intestine was reportedly normal in CD28-deficient mice (18). In iTreg generation, CD28 stimulation has been shown to induce IL-2 production from antigen-stimulated Tconvs, thereby indirectly enhancing iTreg generation (19). Excessive CD28 stimulation, however, reportedly limits iTreg generation (20, 21). These apparently contradictory findings have prompted us to revisit the role of costimulatory signaling for iTreg development, in particular, for the establishment of Treg-specific DNA hypomethylation required for stable expression of Treg signature genes in iTregs (9).
Here we show that in iTreg generation, CD28 costimulation inhibits Treg-type DNA hypomethylation in TGF-β/IL-2-stimulated Tconvs and that the abrogation of CD28 signaling suffices to induce the hypomethylation by attenuating the intensity of intracellular CD28 signaling via protein kinase C (PKC) to NF-κB. CD28 signal deprivation is thus able to generate functionally stable iTregs in vitro from effector/memory as well as naive Tconvs. The results help our understanding of how Treg-specific epigenetic changes are established in developing tTregs and pTregs, and are instrumental in preparing a large number of functionally stable antigen-specific iTregs for therapeutic use in diverse immunological diseases.
Results
Removal of CD28 Costimulation Induces Treg-Specific Hypomethylation in iTregs.
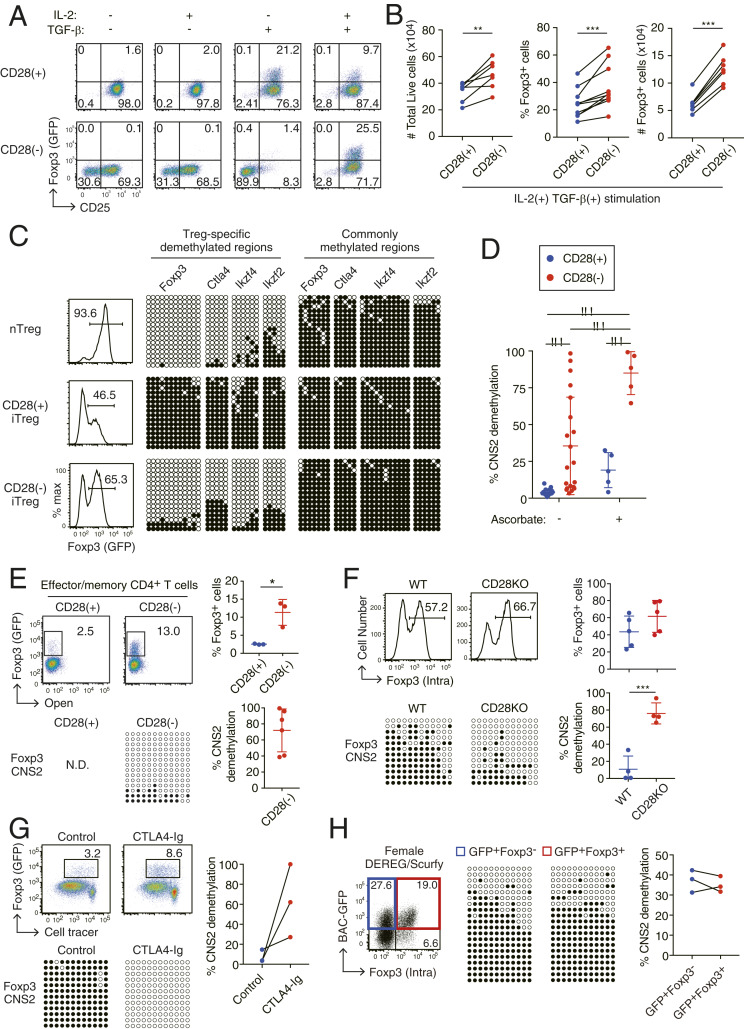
In order to examine the effects of CD28 signal deprivation on iTreg generation, we prepared Foxp3−CD44loCD62Lhigh naive CD4+ Tconvs from eFox reporter mice, which express a Foxp3-eGFP fusion protein (7), and stimulated them in vitro under an iTreg polarizing condition for 3 d using plate-bound anti-CD3 mAb with or without soluble agonistic CD28 mAb (Fig. 1A). In the presence of anti-CD28 costimulation, TGF-β alone (at 1 ng/mL) or together with IL-2 (at 50 U/mL) was sufficient to induce Foxp3+ T cells [designated as CD28(+) iTregs] (3). In contrast, in the absence of CD28 costimulation, TGF-β alone failed to induce Foxp3, whereas the presence of both TGF-β and IL-2 generated Foxp3+ T cells [designated as CD28(−) iTregs] more efficiently than the generation of CD28(+) iTregs. We found that activation and proliferation of CD4+ Tconv cells was independent of CD28 costimulation in the presence of sufficient IL-2 and TGF-β (Fig. 1B and SI Appendix, Fig. S1). However, under TGF-β-sufficient but IL-2-deficient conditions, CD28 stimulation was necessary for T cell activation and proliferation. Notably, the addition of TGF-β inhibited early and late phases of cell apoptosis in CD28(−) iTreg but not CD28(+) iTreg induction. CD28(−) iTregs were indeed higher than CD28(+) iTregs in the total numbers of live cells and Foxp3+ cells, and in the ratio of Foxp3+ cells among live CD4+ T cells (Fig. 1B and SI Appendix, Fig. S1).
Fig. 1.
Generation of Treg-DR hypomethylation in developing iTregs by deprivation of CD28 costimulation. (A) CD25 and Foxp3 expression of CD4+ T cells stimulated in the presence or absence of CD28 stimulation in the presence of IL-2 and/or TGF-β. CD4+ T cells from eFox reporter mice were cultured for 3 d under indicated conditions. A representative result of 3 independent experiments. See also SI Appendix, Fig. S1. (B) Total numbers of live and Foxp3+ cells (n = 7), percentages of Foxp3+ (i.e., GFP+) cells among CD4+ T cells (n = 10) under the CD28(+) or CD28(−) iTreg-inducing condition in the presence of IL-2 and TGF-β. **P < 0.01 and ***P < 0.001 (paired Student’s t test). (C) CpG methylation status of CD28(+) or CD28(−) iTregs or nTregs at Treg-DRs in Foxp3 CNS2 intron 1, Ctla4 exon 2, Eos intron 1, and Helios intron 3a, and at commonly methylated regions adjacent to these Treg-DRs. White and black circles indicate hypomethylated or methylated CpGs, respectively. A representative result of 16 independent experiments. (D) Foxp3 CNS2 hypomethylation in CD28(+) and CD28(−) iTregs in the absence (n = 16) or presence (n = 5) of 10 µg/mL ascorbate. Bars: mean ± SD ***P < 0.001 (Sidak’s multiple comparison test). (E) iTreg induction from effector/memory T cells. CD4+CD44+CD62Llow T cells collected from lymph nodes of Foxp3-eGFP mice were used for iTreg generation. A representative result (Left) and total results of 6 (for methylation analysis) and 3 (for %Foxp3 assessment) independent experiments (Right). *P < 0.05 (unpaired Student’s t test). (F) Foxp3 expression and Foxp3 CNS2 hypomethylation in CD28KO Tconvs subjected to CD28(+) iTreg generation. Representative Foxp3 expression (assessed by intracellular staining) and Foxp3 CNS2 demethylation (Left); and percentages of Foxp3+ cells among CD4+ T cells (n = 5) and %demethylation of CpGs in Foxp3 CNS2 (n = 4) (Right). ***P < 0.001 (unpaired Student’s t test). (G) iTreg induction from DO11.10 TCR Tg naive CD4+ T cells cocultured with OVA-loaded bone marrow-derived CD11c+ antigen-presenting cells in the presence or absence of CTLA4-Ig (Abatacept). A representative result (Left) and %demethylation of CpGs in Foxp3 CNS2 (n = 3) (Right). (H) Foxp3 CNS2 methylation status of iTregs generated from CD4+ naive T cells in female mice heterozygous for Scurfy Foxp3 mutation and crossed with DEREG transgenic mice (expressing BAC-transgenic GFP-fused diphtheria toxin receptor under the control of Foxp3 promoter). A representative result of BAC-GFP+ Foxp3− cells (Foxp3-mutated iTregs) and GFP+Foxp3+ cells (Foxp3 WT iTregs) in such a female mouse (Left) and %demethylation of CpGs in Foxp3 CNS2 (n = 3) (Right). In C–H, iTregs were induced in the presence of IL-2 and TGF-β. DNA methylation analysis was performed on Foxp3+ cells purified by cell sorting. N.D., not determined.
We next assessed Treg-specific DNA hypomethylation in CD28(+) and CD28(−) iTregs. Notably, in contrast with CD28(+) iTregs, which were devoid of Treg-specific DNA hypomethylation (7), CD28(−) iTregs possessed specific hypomethylation in Treg-DRs of Foxp3, Ctla4, Ikzf2, and Ikzf4 genes, but not in commonly methylated regions adjacent to the respective genes, to a similar extent as observed in nTregs (7) (Fig. 1C). The degree of Treg-DR hypomethylation in iTregs was inversely dependent on the dose of anti-CD28 mAb (SI Appendix, Fig. S2A), and, in the absence of CD28 costimulation, dependent on the strength of anti-CD3 stimulation (SI Appendix, Fig. S2B). At the Foxp3 mammalian conserved noncoding sequence 2 (CNS2) enhancer region, a typical Treg-DR, DNA hypomethylation was significantly higher in CD28(−) iTreg induction than in CD28(+) iTregs (Fig. 1D). Ascorbic acid (vitamin C), a coenzyme for Tet enzymes that nonspecifically facilitates de novo demethylation of Foxp3 CNS2 in iTreg cells (22–24), enhanced the hypomethylation, enabling preparation of a homogenous population of Foxp3 CNS2 hypomethylated CD28(−) iTregs (Fig. 1D). Removal of CD28 stimulation was also able to induce both Foxp3 expression and Treg-type DNA demethylation in CD62LlowCD44high effector/memory CD4+ T cells, which are generally refractory to iTreg generation (19, 25) (Fig. 1E).
To confirm the above effects of CD28 signal deprivation on iTreg epigenetic changes, we used CD28 knockout (KO) CD4+ T cells for iTreg induction and found that iTregs generated from CD28-deficient T cells similarly exhibited Treg-DR hypomethylation (Fig. 1F). Consistent with the results, blockade of CD80/CD86 costimulatory molecules on antigen-presenting cells (APCs) with CTLA4-Ig similarly facilitated Treg-DR hypomethylation in iTreg generation (Fig. 1G). Since activated CD4+ Tconv cells also expressed CD80 and CD86 (SI Appendix, Fig. S3A), which could provide CD28 costimulation to other T cells, we used CD80/CD86-deficient CD4+ Tconvs for CD28(−) iTreg induction and observed an equivalent or more efficient induction of Foxp3 CNS2 demethylation compared with the use of CD80/CD86-intact CD4+ T cells (SI Appendix, Fig. S3B).
Throughout these CD28(−) iTreg induction experiments, we consistently observed that while the Foxp3+ cell population possessed the hypomethylation, the Foxp3− cell population did not (SI Appendix, Fig. S4), showing a close association of Foxp3 protein expression with Foxp3 gene hypomethylation in CD28(−) iTregs. To determine whether the transcription factor Foxp3 should play a role in regulating Treg-type hypomethylation in CD28(−) iTregs, we generated CD28(−) iTregs from naive CD4+ T cells in female Foxp3 reporter mice (DEREG mice) on the heterozygous Foxp3-mutant Scurfy background, in which a half of GFP+ cells express Foxp3 protein (7). The methylation ratio and pattern were comparable between Foxp3-deficient and Foxp3-intact iTregs in these DEREG/Scurfy mice, suggesting that Foxp3 protein was dispensable for the demethylation process in CD28(−) iTregs (Fig. 1H).
Taken together, our results indicate that CD28 signaling inhibits Treg-DR hypomethylation in the course of iTreg generation from CD4+ Tconvs, that abrogation of this costimulatory signaling by blocking/deleting CD28 on CD4+ T cells or CD80/CD86 on APCs is sufficient to establish the hypomethylation in developing iTregs by a Foxp3-independent mechanism, and that the abrogation is able to induce iTregs with Treg-type DNA hypomethylation not only from naive Tconvs but also from effector/memory Tconvs.
The Effects of Cytokines on Treg-Type DNA Hypomethylation in iTreg Generation.
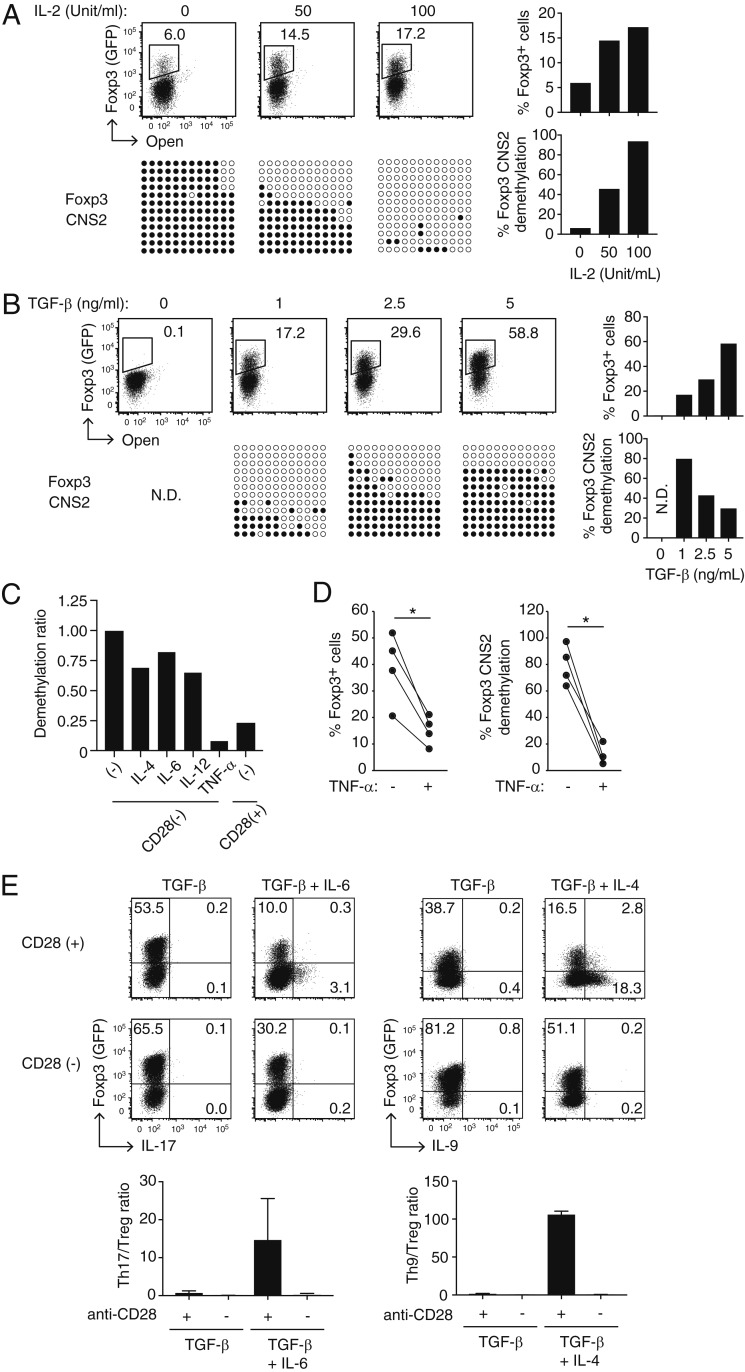
To further study the roles of IL-2 and TGF-β for Foxp3 expression and Treg-type DNA hypomethylation establishment in Tconvs, we examined the dose effects of each signal. In CD28(−) iTreg generation, IL-2 was required for Foxp3 induction, cell proliferation, and cell survival as well as for Foxp3 CNS2 hypomethylation in a dose-dependent manner (Fig. 2A). Conversely, IL-2 neutralization during CD28(+) iTreg induction did not affect TGF-β-dependent Foxp3 expression (SI Appendix, Fig. S1). This indicated that IL-2 was not required for iTreg generation if CD28 costimulation was provided and that the role of CD28 signal in iTreg induction could not be attributed solely to its ability to trigger IL-2 production in Tconv cells (19).
Fig. 2.
Effects of inflammatory cytokines on Treg-specific demethylation in CD28(−) iTregs. (A and B) Dose–response of IL-2 (A) and TGF-β (B) in CD28(−) iTreg generation. Representative patterns of Foxp3 induction and Foxp3 CNS2 hypomethylation are shown at graded concentrations of IL-2 or TGF-β in the presence of TGF-β at 1 ng/mL or IL-2 at 50 U/mL, respectively (Left figures), with the percentages of Foxp3+ cells and the degree of their Foxp3 CNS2 hypomethylation (Right figures) in at least two independent experiments. (C) Effects of indicated inflammatory cytokines (10 ng/mL) on Foxp3 CNS2 demethylation in CD28(−) iTreg induction in the presence of IL-2 and TGF-β. A representative result of two independent experiments is shown. The degree of Foxp3 CNS2 hypomethylation was normalized to cytokine nontreated cells. (D) Foxp3 CNS2 hypomethylation and percentage of Foxp3 induction with TNF-α under the same conditions as in C (n = 4, *P < 0.05, paired Student’s t test). (E) Foxp3+ iTreg and helper T cell differentiation in the indicated Th17/Th9 skewing conditions. Th17 or Th9 cells were induced with 10 ng/mL IL-6 or IL-4, respectively, in the presence of IL-2 and TGF-β. Representative staining (Upper figures) and ratios of the percentage of Foxp3+IL-17− cells (Treg) vs. Foxp3−IL-17+ cells (Th17) or Foxp3+IL-9− cells (Treg) vs. Foxp3−IL-9+ cells (Th9) are shown (Lower figures). Bars: mean ± SD, n = 3. N.D., not determined.
Although both Foxp3 protein expression and Foxp3 gene hypomethylation appeared to require TGF-β in CD28(−) iTreg generation (Fig. 1), TGF-β exerted opposing effects on the respective events: the higher the dose of TGF-β, the stronger the inhibition of hypomethylation and the more effective enhancement of Foxp3 expression (Fig. 2B). Assuming that T cell activation via TCR signaling is necessary for Treg-type DNA hypomethylation in iTreg generation (7), TGF-β at a high dose might suppress T cell activation, thereby hindering hypomethylation of Treg-DRs. We therefore used TGF-β at 1 ng/mL and IL-2 at 50 U/mL, if not indicated otherwise, for CD28(−) or CD28(+) iTreg induction in the present experiments.
We next examined possible effects of inflammatory cytokines (e.g., IL-4, IL-6, IL-12, and TNF-α) on the development of CD28(−) iTregs and on their Treg-type DNA hypomethylation at a dose sufficient to alter Tconv function or drive them to differentiate into Th subsets (Fig. 2C). Among these cytokines, TNF-α potently suppressed the generation of Foxp3+ cells and their hypomethylation of Foxp3 CNS2 (Fig. 2D). In combination with TGF-β, IL-4 and IL-6 are known to drive Tconv differentiation into Th9 or Th17 cells, respectively, but not toward iTregs (26–29). This was indeed the case with CD28(+) iTreg generation. In contrast, addition of these cytokines to the CD28(−) iTreg generation culture augmented iTreg differentiation while hampering Th9 or Th17 cell development, yielding high ratios of Foxp3+ cells to IL-17- or IL-9-forming cells when compared with the respective CD28(+) culture (Fig. 2E).
Thus, IL-2 and TGF-β are required not only for Foxp3 induction but also for the generation of Treg-type hypomethylation in CD28(−) iTregs. TNF-α profoundly suppresses the generation of CD28(−) iTregs and their Treg-type hypomethylation. Furthermore, in the presence of inflammatory cytokines such as IL-4 and IL-6, CD28 signal deprivation enhanced TGF-β-dependent iTreg differentiation, while inhibiting Tconv differentiation into relevant Th populations.
The CD28-PKC-NF-κB Signaling Axis Suppresses Foxp3 CNS2 Demethylation.
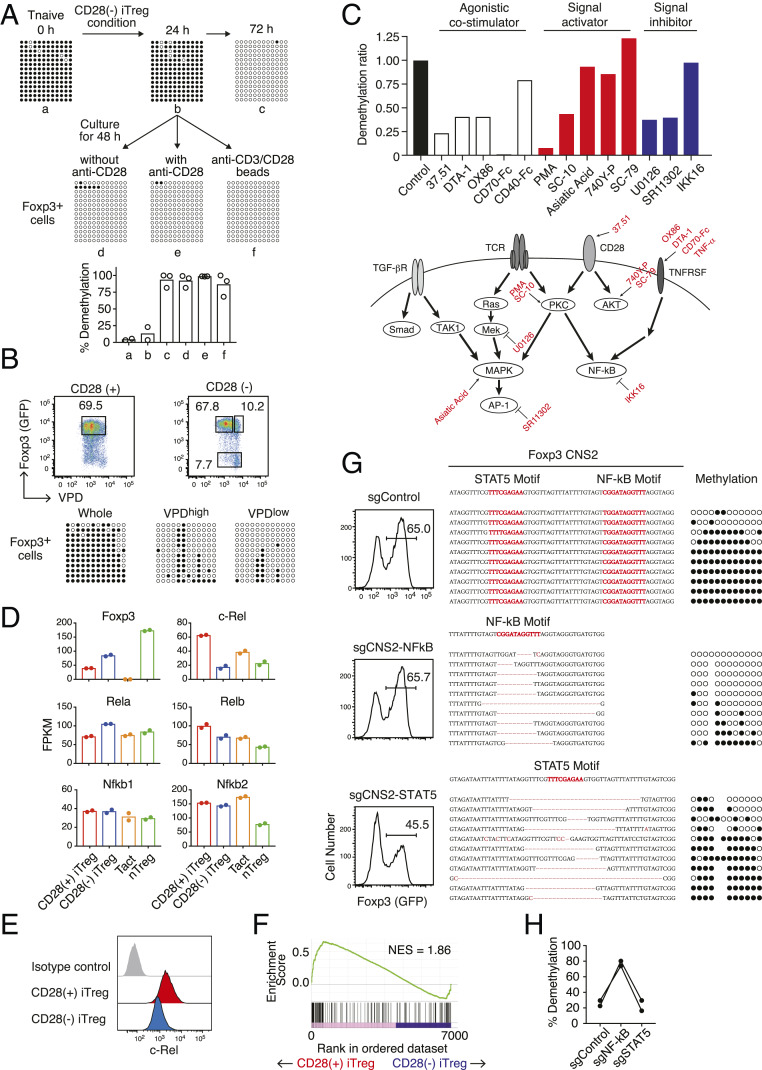
A time point analysis of Treg-DR hypomethylation in generated CD28(−) iTregs showed that Foxp3 CNS2 hypomethylation occurred between 24 and 72 h of their generation (Fig. 3A). Notably, addition of anti-CD28 agonistic mAb after 24 h of CD28 signal deprivation did not hamper the progression of Treg-DR hypomethylation, irrespective of the presence or absence of anti-CD3 stimulation. Furthermore, both dividing and nondividing Foxp3+ cells similarly acquired Treg-type hypomethylation under these conditions (Fig. 3B). The results indicate that CD28 signal deprivation at an early stage for a limited period (less than 24 h) is sufficient for generating and maintaining Treg-DR hypomethylation in iTregs regardless of cell proliferation, and that continuous deprivation of CD28 signal is not required for maintaining the hypomethylation.
Fig. 3.
Inhibition of Treg-DR hypomethylation by NF-κB signaling. (A) Kinetics of Foxp3 CNS2 methylation in CD28(−) iTregs. After CD28(−) iTreg generation (i.e., plate-bound anti-CD3 stimulation in the presence of IL-2 and TGF-β, without anti-CD28) for 24 h from naive CD4+ T cells in eFox reporter mice, some cells continued to be cultured for an additional 48 h (Upper Right); other cells were transferred to anti-CD3 noncoated wells and cultured without soluble anti-CD28 mAb (Lower Left), or to anti-CD3 coated wells and cultured with 1 µg/mL soluble anti-CD28 mAb (Lower Center), or to noncoated wells and cultured with anti-CD3/CD28 Dynabeads (Lower Right) for an additional 48 h. GFP+ (i.e., Foxp3+) cells purified from each culture were assessed for the degree of Foxp3 CNS2 methylation. A representative result (Upper figure) and total results of two (for group a and b) or three (for groups c–f) independent experiments (Lower figure) are shown. (B) Foxp3 CNS2 hypomethylation in highly proliferated (CTVlow) or nonproliferated (CTVhigh) Foxp3+ cells in the indicated iTreg conditions. Representative of three independent experiments. (C) Screening of inhibitors of Foxp3 CNS2 hypomethylation in CD28(−) iTregs. CD28(−) iTregs were induced in the presence of the indicated antibodies or chemical compounds for 3 d. “Control” indicates without the use of any additional reagents. “37.51” indicates the clone number of anti-CD28 mAb used as positive control. Values are shown as a ratio normalized to nontreated control. A representative result from two independent experiments. (D) FPKM of Foxp3 and NF-κB signaling-related genes from RNA-Seq data. Tact, CD3/28-stimulated naive T convs; nTreg, CD3/28-stimulated peripheral Foxp3+ Tregs. Results from two independent experiments are shown. (E) Histogram representation of intracellular c-Rel staining in CD28(+) or CD28(−) iTregs. A representative result of three independent experiments. (F) Ranked enrichment plot of “HALLMARK_TNFA_SIGNALING_VIA_NFKB” (Molecular Signature Database) in gene set enrichment analysis of CD28(+) vs. CD28(−) iTregs. (G and H) CRISPR/Cas9 targeting of STAT5 or NF-κB binding motifs in Foxp3 CNS2. gRNAs were retrovirally induced in Cas9-expressing CD4+ T naive cells followed by iTreg induction (see Materials and Methods for details). A representative result from two independent experiments.
To determine then what intracellular signals mediate Treg-DR hypomethylation upon CD28 signal deprivation, we examined whether known costimulatory reagents, cytokines, and signaling activators or inhibitors could affect Foxp3 CNS2 hypomethylation during CD28(−) iTreg induction at the doses known to exhibit biological effects on Tconvs without affecting cell viability (Materials and Methods) (Fig. 3C). These experiments revealed that in addition to CD28 agonist (37.51 agonistic mAb), other costimulatory signals (DTA-1 mAb as a GITR agonist, OX86 mAb as an OX-40 agonist, CD70-Fc as a CD27 agonist) also substantially inhibited CNS2 hypomethylation in CD28(−) iTregs while CD40-Fc failed to do so. Consistent with this observation, CD28(−) iTregs after 24 h in culture indeed expressed GITR, OX-40, and CD27, but not CD154 (CD40L) (SI Appendix, Fig. S5). Among signaling activators and inhibitors, PKC activators such as phorbol 12-myristate 13-acetate and 5-Chloro-N-heptylnaphthalene-1-sulfonamide (SC-10) abolished or substantially reduced Foxp3 CNS2 hypomethylation, while activation of PKC-downstream MAPK-AP1 signaling by asiatic acid did not. NF-κB inhibition by IKK16 did not hinder hypomethylation; and inhibition of other signaling pathways including cytokine-induced JAK-STAT, PI3-Akt, and p38 did not exhibit a substantial effect. Notably, the specific inhibition of MAPK or AP-1 signaling by U0126 and SR11302, respectively, suppressed hypomethylation. Taken together, TNF-α and other TNFSF members as well as PKC-activating signals, which commonly activate NF-κB signaling, inhibited Treg-type DNA hypomethylation in CD28(−) iTregs, while signaling through MAPK/AP-1 was required for inducing the hypomethylation.
To assess then the transcriptomic differences of CD28(+) and CD28(−) iTregs, we performed RNA-sequencing (RNA-Seq) analysis. We found that while Foxp3 expression was increased in CD28(−) iTregs compared to CD28(+) iTregs, the expression of other Treg-DR-possessing genes such as Ctla4, Ikzf2, and Ikzf4 was not. This suggested that Treg-DR hypomethylation was not simply correlated with mRNA expression levels in these iTregs (SI Appendix, Fig. S6A). Notably, CD28 signal deprivation down-regulated Rel (encoding c-Rel) expression, but not other components of the NF-κB complex such as Rela/p65, Relb, Nfkb1, and Nfkb2, in CD28(−) iTregs compared with CD28(+) iTregs (Fig. 3D). Expression of c-Rel at the protein level was also lower in CD28(−) iTregs compared with CD28(+) iTregs (Fig. 3E). Consistently, gene set enrichment analysis (30) of CD28(+) versus CD28(−) iTregs revealed significant enrichment of “TNF alpha signaling via NF-κB”-related genes in CD28(+) iTregs (normalized enrichment score [NES] = 1.86, false discovery rate [FDR] < 0.01), suggesting that the NF-κB signaling pathway was less activated in CD28(−) iTregs (Fig. 3F). In addition, we observed that nTregs expressed c-Rel at lower levels compared with Tconvs or CD28(+) iTregs upon in vitro TCR stimulation (Fig. 3D). A hierarchal clustering analysis based on Fragments per kilo base per million mapped reads (FPKM) values indeed showed a proximity of the expression patterns between CD28(−) iTregs and in vitro stimulated nTregs in the expression of NF-κB signaling-related genes (SI Appendix, Fig. S6B). Thus, suppression of NF-κB signaling appeared to be a commonality between CD28(−) iTregs and nTregs, suggesting that NF-κB signaling might play an inhibitory role for the generation of DNA hypomethylation in Treg-DRs. To address this possibility, we disrupted a NF-κB binding motif in the Foxp3 CNS2 region of primary Tconvs (13) by retroviral delivery of specific sgRNA using the CRISPR/Cas9 gene-editing system in Cas9 knockin mice and examined the effect of the deletion on Foxp3 CNS2 hypomethylation in CD28(+) iTregs. Deletion of the NF-κB motif indeed enhanced Treg-DR hypomethylation, while it did not affect Foxp3 expression (Fig. 3 G and H). In contrast, deletion of a nearby STAT5 binding motif, an essential transcription factor to induce Foxp3 expression (31, 32), impaired Foxp3 expression but did not affect Foxp3 CNS2 hypomethylation.
Collectively, NF-κB, a common downstream effector of TNF-α-stimulated signaling and CD28-PKC signaling, binds to Foxp3 CNS2 and is a specific inhibitory factor for Foxp3 CNS2 hypomethylation in iTreg induction. Thus, reduction of the NF-κB signaling, while retaining MAPK-AP-1 signaling, is required for installing Treg-specific DNA hypomethylation in developing iTregs.
Functional Stabilization and Expansion of CD28(−) iTregs by IL-2.
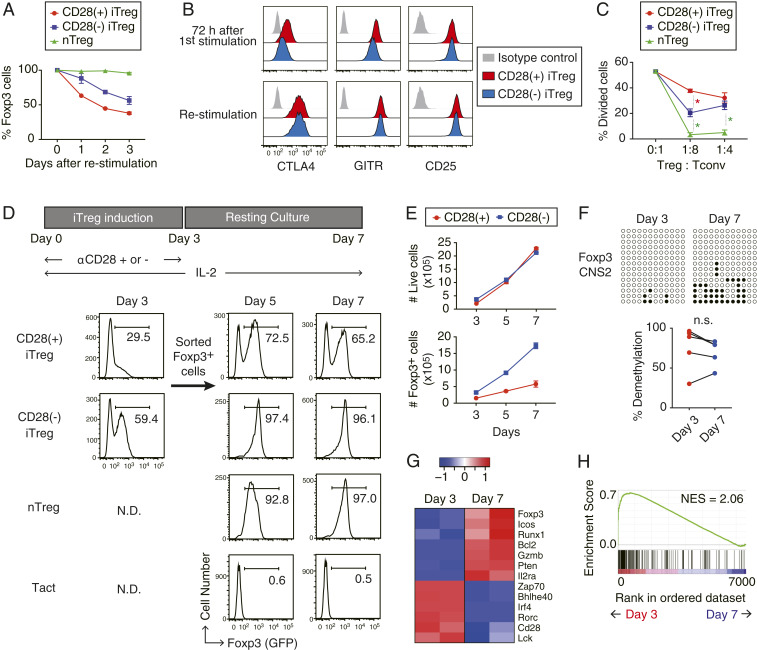
Next, we examined how Foxp3 expression and suppressive activity of CD28(−) iTregs could be maintained in vitro. Upon restimulation with anti-CD3/CD28 mAb and IL-2 in vitro, CD28(−) iTregs exhibited more stable Foxp3 expression compared with CD28(+) iTregs (Fig. 4A), with comparable levels of Treg function-associated molecules such as CD25, CTLA-4, and GITR (Fig. 4B). Consistently, CD28(−) iTregs displayed more potent suppressive activity than CD28(+) iTregs in in vitro suppression assays (Fig. 4C).
Fig. 4.
In vitro expansion and functional stabilization of iTregs by IL-2. (A) Kinetics of Foxp3 retention after restimulation of CD28(+) or CD28(−) iTregs or nTregs using anti-CD3 and anti-CD28 mAbs. Data shown with mean ± SD of three independent experiments. (B) CTLA-4, GITR, and CD25 expression in CD28(+) iTregs (red) and CD28(−) iTregs (blue) were analyzed by FACS, before (Upper) and after (Lower) restimulation with anti-CD3/CD28 mAbs and IL-2 for 3 d. (C) Suppressive activity of nTreg and iTregs. Respective Treg populations were cocultures with cell trace violet (CTV)-labeled CD4+ Tconvs at indicated ratios in the presence of CD3− splenocytes. Percentages of CTV-diluting cells were monitored as divided cells. Data shown as mean ± SD of three independent experiments. Red asterisk means significance between CD28(−) iTregs and CD28(+) iTregs, green asterisk between CD28(−) iTregs and nTregs. *P < 0.0001 (ANOVA). (D) Foxp3 expression by iTregs after resting culture. After 3 d of anti-CD3 stimulation with or without CD28 costimulation, Foxp3+ cells were sorted and subjected to resting culture in the presence of 100 U/mL IL-2. Foxp3-eGFP expression was then analyzed by FACS at the indicated time points. Representative data of three independent experiments. (E) Number of live or Foxp3+ cells during resting culture at the indicated time points. Data shown as mean with SD (n = 3). (F) Hypomethylation status of Foxp3 CNS2 in CD28(−) iTreg cells before and after resting culture. n.s.; not significant (paired Student’s t test). (G and H) RNA-Seq analysis of CD28(−) iTregs before (day 3) and after (day 7) resting culture (n = 2). Representative Treg signature or TCR signal-related genes are shown in a z-scored heat map (G) and as ranked enrichment plot of “HALLMARK_TNFA_SIGNALING_VIA_NFKB” gene set in the Molecular Signature Database (H). N.D., not determined.
When Foxp3+ cells were purified from 3-d in vitro culture of CD28(+) or CD28(−) iTregs and further cultured for 4 more days with only IL-2 (referred as “resting culture”), CD28(+) iTregs readily lost Foxp3 expression, whereas CD28(−) iTregs stably maintained the expression in a similar manner as nTregs (Fig. 4D). CD28(−) iTregs expanded similarly as CD28(+) iTregs during the 4-d resting culture, but yielded 6- to 10-fold higher numbers of total Foxp3+ cells than the latter (Fig. 4E), while maintaining Treg-DR hypomethylation (Fig. 4F). Moreover, RNA-Seq analysis revealed that the additional resting culture step up-regulated in CD28(−) iTregs several Treg signature genes, such as Foxp3 and Il2ra, as well as Bcl2, Gzmb, and Pten (Fig. 4G). Concomitantly, TCR-signaling genes, such as Zap70 and Lck, and effector T cell program-related genes, such as Rorc and Bhlhe40, were down-regulated. Gene set enrichment analysis (30) of 3-d versus 7-d culture of CD28(−) iTregs revealed significant reduction of TNF alpha signaling via NF-κB-related genes in the latter (NES = 2.06, FDR < 0.01), suggesting decreased NF-κB signaling during the resting culture (Fig. 4H). Protein expression levels of CD25 and CTLA-4 in CD28(−) iTregs were initially similar to that of activated Foxp3− Tconvs in the same culture on day 3, but were further increased by day 7, while Foxp3− Tconvs reduced the expression presumably because of the lack of continued TCR stimulation (SI Appendix, Fig. S7 A and B). The resting culture method therefore enabled better separation of Foxp3+ iTregs from Foxp3−-activated Tconvs by cell surface molecules such as CD25.
Taken together, resting culture with IL-2 alone is able to efficiently expand CD28(−) iTregs while stably maintaining their Treg-type DNA hypomethylation and enhance their acquisition of a gene expression profile more akin to that of nTregs.
In Vivo Foxp3 Stability and Suppressive Function of CD28(−) iTregs.
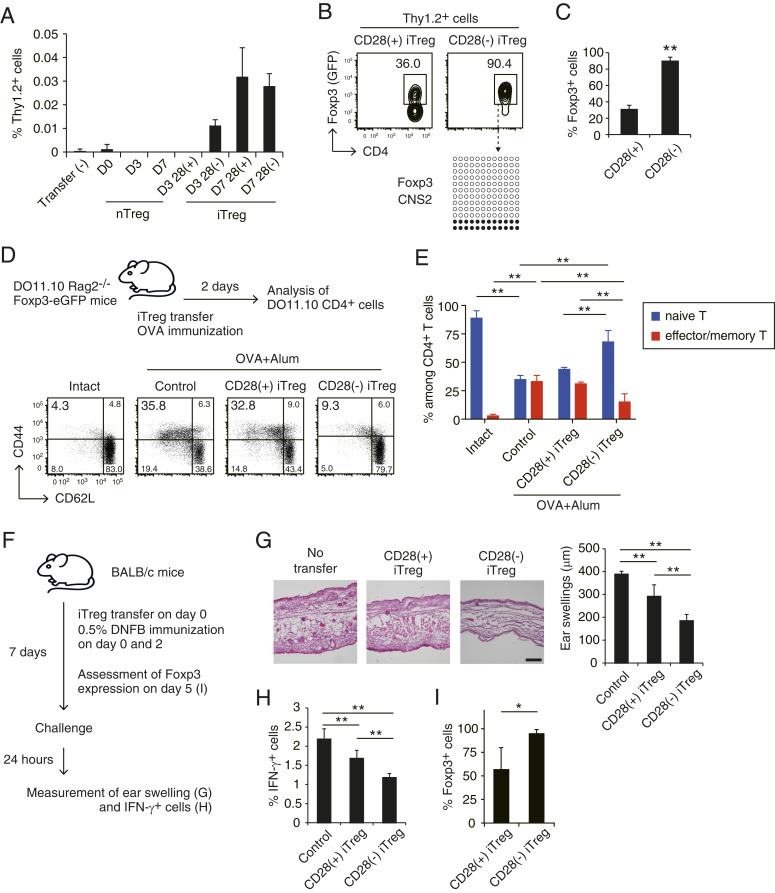
To evaluate the in vivo survivability of CD28(−) iTregs and the stability of their Foxp3 expression, we purified Foxp3+ cells from rested or nonrested CD28(−) or CD28(+) iTregs as shown in Fig. 4D and transferred an equal number of each Foxp3+ population into syngeneic nonlymphopenic mice. We then assessed the number of Foxp3+ and Foxp3− cells among the surviving transferred cells as well as their Foxp3 CNS2 methylation status 2 wk later. Notably, compared with the nonrested counterparts, rested CD28(+) and CD28(−) iTregs showed better survival in the recipients while transferred nTregs, regardless of being rested or nonrested, were poor in survival (Fig. 5A). In addition, CD28(−) iTregs displayed superior stability of Foxp3 expression with over 90% of transferred CD28(−) iTregs maintaining Foxp3 expression and Foxp3 CNS2 hypomethylation, compared with a substantial loss (by ∼60%) of Foxp3 expression in CD28(+) iTregs, after cell transfer (Fig. 5 B and C). Furthermore, CD28(−) iTregs after resting culture exhibited stable Foxp3 expression in vitro even in the presence of an inflammatory cytokine IL-6 (SI Appendix, Fig. S8).
Fig. 5.
Therapeutic effects of CD28(−) iTregs in an animal model of contact hypersensitivity. (A) Survival of in vivo transferred iTregs. The same number (2 × 105) of BALB/c-derived CD28(+) or CD28(−) iTregs, or nTregs, stimulated in vitro for 3 or 7 d (as shown in Fig. 4D) were transferred i.v. into Thy1.1-BALB/c mice; and percentages of Thy1.2+ cells among lymphocytes in inguinal lymph nodes were assessed 14 d after cell transfer. Data shown are a representative result of two independent experiments with three mice for each cell transfer. Data shown as mean ± SD. (B) Foxp3 expression and Foxp3 CNS2 methylation status of transferred 7-d cultured CD28(+) or CD28(−) iTregs 14 d after cell transfer as shown in A. (C) Percentages of Foxp3+ cells among Thy1.2+CD4+ T cells derived from transferred CD28(+) or CD28(−) iTregs as shown in B (n = 3). **P < 0.01 (Student’s t test). (D) In vivo suppressive activity of CD28(+) or CD28(−) iTregs after IL-2 resting culture. CD4+ T cells in draining lymph node of DO11.10 Tg Rag2−/− mice were analyzed for CD44 and CD62L expression 2 d after iTreg transfer and simultaneous OVA immunization. Representative data of three independent experiments. (E) Percentages of effector/memory (CD44+CD62L−) or naive (CD44−CD62L+) CD4+ T cells in the mice analyzed in D (n = 3). Data shown as mean ± SD **P < 0.01. (F–I) Suppressive activity of iTreg cells in DNFB-sensitized contact hypersensitivity responses. CD28(+) or CD28(−) iTregs were generated from CD4+ Tconvs in DNFB-immunized BALB/c mice. Foxp3+ cells (2 × 105) from CD28(+) or CD28(−) iTregs prepared by resting culture as shown in Fig. 4D were transferred i.v. into BALB/c mice and then sensitized with 0.5% DNFB painting of the skin (F). Histology of the ear (H&E staining) (Scale bar, 100 µm.) and the degree of ear swelling (G), IFN-γ production by CD4+ T cells in the regional lymph nodes (H) were assessed 24 h after DNFB challenge performed 1 wk after sensitization (n = 4 to 6). When cells were similarly transferred to Thy1.1 BALB/c mice, the percentages of Foxp3+ cells among transferred Thy1.2+ cells in the regional lymph node cells were assessed 5 d after immunization (n = 3) (I). **P < 0.01 (ANOVA), *P < 0.05 (Student’s t test).
Based on these results, we then examined in vivo suppressive ability and therapeutic potential of rested CD28(+) or CD28(−) iTregs prepared from DO11.10 transgenic (Tg) mice on the RAG2-deficient background, which are devoid of Foxp3+ nTregs in the periphery (33). Transfer of DO11.10-derived rested CD28(+) or CD28(−) iTregs into RAG2−/−DO11.10 mice immediately after Ovalbumin (OVA) immunization revealed that transferred CD28(−) iTregs inhibited the expansion and differentiation of host DO11.10 Tconvs into CD44+CD62Llow effector/memory T cells with preservation of CD44−CD62Lhigh naive T cells, whereas CD28(+) iTregs were hardly suppressive (Fig. 5 D and E). This potent suppressive activity of CD28(−) iTregs was further confirmed using a skin contact hypersensitivity (CHS) model (Fig. 5F), in which CD28(−) iTregs exerted better therapeutic potential than CD28(+) iTregs in suppressing ear swelling and IFN-γ production by effector T cells in the regional lymph nodes (Fig. 5 G and H). The transferred CD28(−) iTregs indeed maintained Foxp3 expression more stably than CD28(+) iTregs (Fig. 5I).
Thus, CD28(−) iTregs are more resilient in vivo than CD28(+) iTregs upon antigenic stimulation and possess both phenotypic and functional stability with sustained Foxp3 CNS2 hypomethylation. These inherent properties of CD28(−) iTregs, along with in vitro resting and expansion, ensure stronger and more stable immune suppression in vivo, when compared with CD28(+) iTregs.
Discussion
The main finding in this report is that CD28 costimulation inhibits Treg-type DNA hypomethylation in iTregs generated by TCR/TGF-β/IL-2 stimulation of Tconvs, and that abrogation of CD28 signaling is able to induce Treg-specific hypomethylation in developing iTregs.
Using pharmacological screening, we uncovered that the CD28-PKC-NF-κB axis represses Foxp3 CNS2 demethylation in activated Tconvs and that the attenuation or abrogation of this signaling, for example, by deletion of a single NF-κB binding site in Foxp3 CNS2, is sufficient to enable CNS2 demethylation in developing iTregs. We also found the computationally predicted NF-κB binding motif not only in Foxp3 CNS2 but also in other Treg-DRs, for example, in the Ctla4, Ikzf2, and Ikzf4 gene loci (Fig. 1C). These results collectively suggest possible binding of NF-κB family transcription factors, such as c-Rel, to Treg-DRs in Treg signature genes and the contribution of NF-κB to the negative regulation of Treg-type DNA demethylation in developing iTregs. Other costimulatory signals, such as those from GITR, OX-40, and CD27 stimulation or signals from TNF-α binding appeared to similarly hamper the establishment of Treg-type DNA hypomethylation in iTregs via NF-κB.
In contrast with such an inhibitory role for epigenetic changes in iTregs, it has been demonstrated that strong signaling from these CD28 and other signaling pathways are required for the development of tTregs possessing Treg-type DNA hypomethylation (14–16, 34–37). This difference between tTregs and iTregs in their developmental requirement of CD28-PKC-NF-κB signaling can be attributed, at least in part, to distinct roles of costimulatory signaling in tTreg and iTreg development. For example, thymic immature T cells are much more dependent on the CD28-PKC-NF-κB signal than mature peripheral T cells for their survival in the thymus (38). CD28 deficiency has been reported to result in a substantial decrease of tTreg cells in the thymus and constitutively active Stat5 expression reportedly rescues the loss of tTregs in the thymus of CD28−/− mice (31). c-Rel has also been considered as an indispensable factor for Treg generation because c-Rel-deficient mice showed a drastic reduction of Foxp3+ Tregs in the thymus (34–36). Generation of iTregs from c-Rel-deficient CD4+ Tconvs was also defective but rescued by the addition of IL-2 to in vitro iTreg generation (39). The IL-2 promoter is reportedly a direct target of c-Rel (40). These previous findings collectively indicate that developing tTreg cells that fail to receive CD28-PKC-NF-κB signal may undergo apoptosis because of the paucity of available IL-2 in the thymus where the major source of IL-2 is T cells themselves (41). In contrast, in the periphery where IL-2 and TGF-β are sufficiently supplied by the various sources such as dendritic cells (DCs) and innate lymphoid cells (42–44), CD28 signaling is dispensable for the generation of pTregs; further, CD28 signal reduction might be able to induce Treg-type DNA hypomethylation in developing pTregs (18).
It is known that various costimulatory signals facilitate Tconv differentiation into particular Th subsets. For example, CD28 costimulation facilitates Th1 differentiation while GITR costimulation enhances Th9 differentiation (45–47). With our observation of a lower level of NF-κB signaling-related genes expressed in both stimulated CD28(−) iTregs and nTregs, it remains to be investigated whether excessive NF-κB signaling may preferentially polarize Tconvs toward effector T cell differentiation and away from pTregs (48, 49). In line with this possibility, we provided compelling evidence that CD28 signal deprivation is able to induce iTregs with Treg-type hypomethylation from both effector/memory and naive Tconvs, whereas CD28 signal-intact TGF-β/IL-2 stimulation could only generate iTregs from the latter but not from the former (50). Whether this difference can be attributed to possible differences in NF-κB signaling between naive and effector/memory Tconv populations needs to be determined.
Developing tTregs and pTregs physiologically possess Treg-type DNA hypomethylation (7), indicating that a reduction of CD28 signaling could bear a physiological significance in tTreg and pTreg development and function in vivo. In the thymus, developing tTreg cells might interact with immature DCs or thymic medullary epithelial cells, which are low in CD80/CD86 expression, to acquire Treg-type DNA hypomethylation (51). In the periphery, the degree of CD28 signal reduction, together with local provision of TGF-β, IL-2, and other costimulations, may determine the cell fate of antigen-stimulated Tconvs: whether they die by apoptosis, become anergic, or differentiate into functionally stable or unstable Tregs. For example, immature tolerogenic DCs, which are low in CD80/CD86 expression and able to secrete TGF-β (50), may not only induce pTregs but also contribute to stabilizing their function by mediating the establishment of Treg-type DNA hypomethylation. In addition, assuming that nTregs expressing CTLA-4 are able to down-modulate CD80/CD86 expression by APCs (52), they may help render self-reactive T cells anergic (53) and also contribute to the generation of functionally stable pTregs with Treg-type DNA hypomethylation in a manner of infectious tolerance (54). These possible effects of CD28 signal reduction in vivo are under investigation.
In order to prepare functionally potent and stable iTregs for therapeutic use, it is necessary to generate iTregs with high Foxp3 expression and stable Treg-type DNA hypomethylation, which is independent of Foxp3 expression (7, 55) (Fig. 1H). Foxp3 induction can be achieved in antigen-specific naive Tconvs by TGF-β, and even in antigen-specific effector Tconvs by inhibition of CDK8/19 (56). Here we have shown that Treg-type DNA hypomethylation can be installed by CD28 signal blockade. Vitamin C, neutralization of inflammatory cytokines such as TNF-α, blockade of other costimulatory signals, and resting culture with IL-2 alone, can intensify and further stabilize the specific hypomethylation. Thus, a combination of such procedures for Foxp3 induction and installation of Treg-type DNA hypomethylation will enable preparation of functionally stable antigen-specific iTregs in a large quantity from disease-mediating Tconvs in order to treat autoimmune and other immunological diseases and to prevent graft rejection in organ transplantation.
Materials and Methods
Mice.
C57BL/6, BALB/c mice were purchased from SLC or CLEA. DO.11.10 TCR transgenic mice, Rag2 KO mice, Scurfy mice, BAC-transgenic Foxp3 promoter-DTR-GFP (DEREG) mice, Foxp3-eGFP (eFox) reporter mice, CD28KO mice, and CD80/CD86KO mice were previously described (7, 57–60). H11-LSL-Cas9, CD4-Cre, and Foxp3-IRES-DTR-GFP mice were purchased from The Jackson Laboratory. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (61) and approved by the Committee on Animal Research of Osaka University.
Antibodies and Reagents.
Antibodies are listed in SI Appendix, Table S1. Purchases include 740 Y-P, SC-10, SC-79, SB202190, U0126, and SR11302 from Tocris Bioscience; asiatic acid from Cayman Chemical; Ikk 16 from Calbiochem; CTLA4-Ig (Abatacept [Orencia]) from Ono Pharm Co.; and OVA (323 to 339) peptides from MBL. Recombinant mouse IL-4, IL-6, IL-12, and TNF-α were purchased from PeproTech. Mouse CD4 T Lymphocyte Enrichment Set (BD) was used for the enrichment of CD4+ T cells, and Cell Stimulation Mixture (plus protein transport inhibitors) (eBioscience) for intracellular cytokine staining.
Cell Sorting and Flow Cytometry Analysis.
Cell staining by fluorescence-conjugated antibodies and flow cytometry analysis were performed as previously described (10). To prepare cells for culture experiments, FACSAriaII (BD) was used for collecting particular population. The definition of cell populations used are as follows: naive T cells, CD4+GFP−CD44lowCD62Lhigh; nTreg cells, CD4+GFP+; and effector/memory T cells, CD4+GFP−CD44highCD62Llow. When intracellular staining was needed, cell fixation and permeabilization were performed using the Foxp3/Transcription Factor Staining Buffer Set (eBioscence) or BD Pharmingen Transcription Factor Buffer Set (BD) in accordance with the manufacturer’s instructions.
Cell Culture, iTreg Induction, and Resting Culture.
For cell culture, we used RPMI1640 culture medium supplemented with 10% FCS (vol/vol), 60 µg/mL penicillin G, 100 µg/mL streptomycin, and 0.1 mM 2-mercaptoethanol. For the induction of CD28(−) iTregs, sorted 2 × 105 naive CD4+ T cells were stimulated with plate-bound anti-CD3 mAb (clone 2C11, BD) (coating at 10 μg/mL overnight) in the presence of 50 U/mL of human IL-2 (Imunace 35, Shionogi Pharm Co.) and 1 to 5 ng/mL of human TGF-β1 (R&D), in 96-well flat-bottom plates (Thermo Scientific, 167008). Soluble anti-CD28 mAb (clone 37.51, BD) at 1 µg/mL was additionally used to generate CD28(+) iTregs. For resting culture, CD28(+) or CD28(−) iTregs prepared by 3-d culture were collected, washed once, and further cultured at 1 × 106/mL cell concentration with fresh culture medium containing 100 U/mL of IL-2. Cells were split every 2 d and fresh IL-2-containing medium was added. In coculture assays using bone marrow-derived APCs and T cells, 2 × 105 DO11.10 CD4+ T cells were cocultured with bone marrow-derived CD11c+ APCs (bone marrow cells stimulated with 20 ng/mL GM-CSF for 10 d) in the presence of 5 μM of OVA peptide.
Suppression Assay.
CD4+ naive T cells were labeled with carboxyfluorescein diacetate succinimidyl ester or violet proliferation dye (VPD; Cell Tracer Violet, Thermo Scientific) by the following protocol: cells were incubated at 1 × 106 cells/mL with 5 μM reagents at room temperature (RT) for 5 min. Labeling reaction was quenched by adding 5 volumes of cold medium and incubated further for 20 min on ice. After washing cells once, 2 × 105 labeled cells were cocultured with 4 × 104 T cell-depleted splenocytes and graded numbers of Tregs in the presence of 5 μM OVA peptide. Percentages of cells dividing more than once were assessed by fluorescence-activated cell sorting (FACS) after 72 h.
Cell Proliferation Analysis.
Cells (1 × 106) were washed by fetal calf serum (FCS)-free phosphate-buffered saline (PBS) at least twice, and then incubated with 1 mL of 1 µM CellTrace Violet Cell Proliferation Kit (Thermo Fisher) at RT or 37 °C for 15 min. Cells were then washed with 10 times volume of 10% FCS RPMI 1640 for neutralization of dye, followed by flow cytometry analysis.
Analysis of Apoptosis.
Cells were stained by surface-staining antibodies and with LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher) according to the manufacture’s protocol. After washing cells with ethylenediaminetetraacetic acid-free medium once, cells were incubated with 0.125 µg/mL of PE-conjugated AnnexinV (BioLegend) in AnnexinV binding buffer (10 µM Hepes, 140 mM NaCl, 2.5 mM CaCl2) at RT for 15 min, followed by flow cytometry analysis.
Retroviral CRISPR/Cas9-Mediated Targeting on Foxp3 CNS2.
Naive CD4+ T cells were isolated from H11-LSL-Cas9/CD4-Cre/FDG mice. Cells were then activated using 5 μg/mL anti-CD3 mAb and 2.5 µg/mL anti-CD28 mAb at a density of 1.25 × 106 cells/mL overnight and supplemented with 50 U/mL IL-2. Cells were infected with sgRNA-containing retroviruses by spin inoculation (1,600 × g, 32 °C for 90 min) at 24 h postactivation. Cells were then put into resting 72 h postactivation by addition of exogenous 100 U/mL IL-2 and expanded for a week before Foxp3 induction. For iTreg induction, infected cells were sorted and activated with 10 µg/mL anti-CD3 mAb at a density of 5 × 105 cells/mL supplemented with 100 U/mL IL-2, 2.5 ng/mL TGF-β, 1 µg/mL ascorbate, 5 µg/mL anti-IL4 mAb, 5 µg/mL anti-IFN-γ, 1 µg/mL anti-IL6 mAb, and 10 µg/mL anti-FasL mAb. Foxp3+ cells were sorted and analyzed 72 h postinduction.
CpG Methylation Analysis by Bisulfite Sequencing.
Bisulfite sequencing analysis was performed as previously described (7). Cells were collected by FACSAriaII and DNA was extracted by phenol extraction followed by ethanol precipitation. When cells were fixed for intracellular staining, reverse-crosslinking reaction was performed overnight prior to gDNA extraction. The bisulfite conversion was carried out using the MethylEasy Xceed Rapid DNA Bisulphite Modification Kit (Human Genetic Signatures) by following the manufacturer’s protocol. PCR primer sequences of commonly methylated regions or Treg-specific demethylated regions are available in ref. 7.
RNA-Sequencing and Analysis.
RNA-sequencing was performed as previously described (10). Briefly, cells were lysed in RLT RNA lysis buffer (Qiagen) containing 1% 2-mercaptoethanol, followed by RNA reverse transcription by SMART-seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech). After fragmentation of cDNA using the Covaris sonication system, sequencing libraries were prepared using the Kapa Library preparation kit for IonTorrent (KAPA) by following the manufacturer’s protocol. Sequencing of cDNA libraries was performed on a IonS5 (Thermo Scientific). Acquired sequencing results were mapped to the reference mouse genome (mm9) using Tophat2, and unmapped sequences were analyzed again by bowtie2. Normalized FPKM values were acquired using Cuffnorm of the Cufflinks package (version 2.2.1, Trapnell Lab) under default settings. Gene set enrichment analysis (30) was performed with the following settings: collapse = true, permutation type = gene_set, scoring = weighted, metric = log2_ratio_of_classes. Hierarchical clustering was performed using the heatmap.2 function in R package gplots.
In Vivo Cell Transfer into Nonlymphopenic Mice.
Cells (2 × 105) prepared from BALB/c mice, which are Thy1.2, were i.v. transferred into congenic Thy1.1-BALB/c mice. Fourteen days after transfer, the percentage of transferred Thy1.2 cells in inguinal lymph nodes was assessed.
OVA Immunization.
DO11.10/Rag KO mice were immunized s.c. with 100 μL of an emulsion containing 200 µg of OVA (323–339) in 50 µL of PBS, and 50 μL of complete Freund’s adjuvant (CFA). OVA-specific iTreg cells (2 × 105) were transferred just before immunization. Draining lymph nodes were collected for flow cytometry analysis 2 d later.
2,4-Dinitrofluorobenzene-Induced CHS.
Mice were sensitized epicutaneously on days 0 and 2 by applying 100 µL of 0.5% 2,4-dinitrofluorobenzene (DNFB) diluted in acetone on the abdominal skin and challenged on day 7 by applying 20 μL 0.5% DNFB on the ear. For generating DNFB-specific iTregs, effector T cells were collected from DNFB-sensitized mice, converted into iTregs, and transferred to other mice before DNFB immunization.
Statistics.
Values were expressed as mean ± SD. Statistical significance was assessed by paired or unpaired Student’s t test (two groups), nonrepeated-measures analysis of variance (ANOVA) followed by the Bonferroni test (versus control), Dunnett’s test, or Student-Newman-Keuls test (multiple comparisons). A probability of less than 5% (P < 0.05) was considered statistically significant.
Data Availability.
All data discussed in this paper are included in this article and SI Appendix. The next generation sequencing data are deposited in DDBJ Sequence Read Archive under accession number DRA008294. Detailed information on antibodies and reagents used in this study is provided in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Y. Kitagawa for advice on DNA methylation and RNA-sequencing analysis; A. Kawasaki, Y. Nakamura, M. Hata, S. Tomita, H. Lin, N. Mizoguchi, and R. Ishii for technical assistance. This work was supported by Grants-in-Aid by Japan Society for Promotion of Science (JSPS) for Research Activity Start-Up (25893115) to N.M. and Young Scientists (B) (15K21125) to N.M., JSPS Fellowship (16J03628) to R.K., Specially Promoted Research (16H06295) to S.S., Core Research for Evolutional Science and Technology of the Japan Agency for Medical Research and Development (JP 19gm0010005) to S.S., and Leading Advanced Projects for Medical Innovation to S.S.
Footnotes
Competing interest statement: This study was funded by RegCell Co., Ltd (Kyoto, Japan), which had no control over the interpretation, writing, or publication of this work.
Data deposition: All data discussed in this paper are included in this article and SI Appendix. The next generation sequencing (NGS) data are deposited in DNA Data Bank of Japan (DDBJ) Sequence Read Archive under accession number DRA008294. Detailed information on antibodies and reagents used in this study is provided in SI Appendix, Table S1.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922600117/-/DCSupplemental.
References
- 1.Sakaguchi S., Yamaguchi T., Nomura T., Ono M., Regulatory T., Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Curotto de Lafaille M. A., Lafaille J. J., Natural and adaptive foxp3+ regulatory T cells: More of the same or a division of labor? Immunity 30, 626–635 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Chen W., et al. , Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floess S., et al. , Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5, e38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H.-P., Leonard W. J., CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: A role for DNA methylation. J. Exp. Med. 204, 1543–1551 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polansky J. K., et al. , DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38, 1654–1663 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Ohkura N., et al. , T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Toker A., et al. , Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 190, 3180–3188 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Ohkura N., Kitagawa Y., Sakaguchi S., Development and maintenance of regulatory T cells. Immunity 38, 414–423 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Kitagawa Y., et al. , Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y., et al. , Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y., et al. , Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Liang Y., LeBlanc M., Benner C., Zheng Y., Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158, 734–748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salomon B., et al. , B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity 12, 431–440 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Tai X., Cowan M., Feigenbaum L., Singer A., CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 6, 152–162 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Guo F., Iclozan C., Suh W.-K., Anasetti C., Yu X.-Z., CD28 controls differentiation of regulatory T cells from naive CD4 T cells. J. Immunol. 181, 2285–2291 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coquet J. M., et al. , Epithelial and dendritic cells in the thymic medulla promote CD4+Foxp3+ regulatory T cell development via the CD27-CD70 pathway. J. Exp. Med. 210, 715–728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakamatsu E., et al. , CD28 co-stimulation is dispensable for the steady state homeostasis of intestinal regulatory T cells. Int. Immunol. 30, 171–180 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Davidson T. S., DiPaolo R. J., Andersson J., Shevach E. M., Cutting edge: IL-2 is essential for TGF-β-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 178, 4022–4026 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Benson M. J., Pino-Lagos K., Rosemblatt M., Noelle R. J., All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204, 1765–1774 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semple K., et al. , Strong CD28 costimulation suppresses induction of regulatory T cells from naive precursors through Lck signaling. Blood 117, 3096–3103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue X., et al. , Control of Foxp3 stability through modulation of TET activity. J. Exp. Med. 213, 377–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasidharan Nair V., Song M. H., Oh K. I., Vitamin C., Vitamin C facilitates demethylation of the Foxp3 enhancer in a tet-dependent manner. J. Immunol. 196, 2119–2131 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Kasahara H., et al. , Generation of allo-antigen-specific induced Treg stabilized by vitamin C treatment and its application for prevention of acute graft versus host disease model. Int. Immunol. 29, 457–469 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kim B.-S., et al. , Conversion of Th2 memory cells into Foxp3+ regulatory T cells suppressing Th2-mediated allergic asthma. Proc. Natl. Acad. Sci. U.S.A. 107, 8742–8747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veldhoen M., Hocking R. J., Atkins C. J., Locksley R. M., Stockinger B., TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Bettelli E., et al. , Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Dardalhon V., et al. , IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 9, 1347–1355 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veldhoen M., et al. , Transforming growth factor-β “reprograms” the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9, 1341–1346 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burchill M. A., et al. , Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chinen T., et al. , An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 17, 1322–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh M., et al. , Thymus and autoimmunity: Production of CD25+CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunologic self-tolerance. J. Immunol. 162, 5317–5326 (1999). [PubMed] [Google Scholar]
- 34.Isomura I., et al. , c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J. Exp. Med. 206, 3001–3014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long M., Park S.-G., Strickland I., Hayden M. S., Ghosh S., Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity 31, 921–931 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Ruan Q., et al. , Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity 31, 932–940 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmud S. A., et al. , Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat. Immunol. 15, 473–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M. O., Rudensky A. Y., T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visekruna A., et al. , c-Rel is crucial for the induction of Foxp3(+) regulatory CD4(+) T cells but not T(H)17 cells. Eur. J. Immunol. 40, 671–676 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Bunting K., et al. , Genome-wide analysis of gene expression in T cells to identify targets of the NF-κ B transcription factor c-Rel. J. Immunol. 178, 7097–7109 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Hemmers S., et al. , IL-2 production by self-reactive CD4 thymocytes scales regulatory T cell generation in the thymus. J. Exp. Med. 216, 2466–2478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coombes J. L., et al. , A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen D. L., et al. , Identification of cellular sources of IL-2 needed for regulatory T cell development and homeostasis. J. Immunol. 200, 3926–3933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L., et al. , Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao X., Constant S., Jorritsma P., Bottomly K., Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4+ T cell differentiation. J. Immunol. 159, 5956–5963 (1997). [PubMed] [Google Scholar]
- 46.Chen L., Flies D. B., Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao X., et al. , GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat. Commun. 6, 8266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molinero L. L., Miller M. L., Evaristo C., Alegre M.-L., High TCR stimuli prevent induced regulatory T cell differentiation in a NF-κB-dependent manner. J. Immunol. 186, 4609–4617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh H., Ghosh S., NF-κB: Roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 252, 41–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki S., et al. , Dendritic cells are specialized accessory cells along with TGF- for the differentiation of Foxp3+ CD4+ regulatory T cells from peripheral Foxp3 precursors. Blood 110, 4293–4302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garg G., et al. , Unique properties of thymic antigen-presenting cells promote epigenetic imprinting of alloantigen-specific regulatory T cells. Oncotarget 8, 35542–35557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wing K., et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Maeda Y., et al. , Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science 346, 1536–1540 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Andersson J., et al. , CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-β-dependent manner. J. Exp. Med. 205, 1975–1981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morikawa H. et al.; FANTOM Consortium , Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc. Natl. Acad. Sci. U.S.A. 111, 5289–5294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akamatsu M., et al. , Conversion of antigen-specific effector/memory T cells into Foxp3-expressing Treg cells by inhibition of CDK8/19. Sci. Immunol. 4, eaaw2707 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Shahinian A., et al. , Differential T cell costimulatory requirements in CD28-deficient mice. Science 261, 609–612 (1993). [DOI] [PubMed] [Google Scholar]
- 58.Lahl K., et al. , Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wing J. B., Ise W., Kurosaki T., Sakaguchi S., Regulatory T., Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41, 1013–1025 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ito Y., et al. , Detection of T cell responses to a ubiquitous cellular protein in autoimmune disease. Science 346, 363–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this paper are included in this article and SI Appendix. The next generation sequencing data are deposited in DDBJ Sequence Read Archive under accession number DRA008294. Detailed information on antibodies and reagents used in this study is provided in SI Appendix, Table S1.