Significance
Carotenoids are essential molecules in oxygenic photoautotrophs, and they fulfill essential requirements for human and animal nutrition. How carotenoid accumulation is regulated in the chloroplast, a cyanobacterium-derived organelle, remains poorly understood, despite significant advancements in identifying enzymes of the carotenoid biosynthetic pathway. This study identifies a role of chloroplast Sec14-like 1 (CPSFL1), a CRAL-TRIO protein of eukaryotic origin, in modulation of carotenoid biosynthesis and accumulation in the chloroplast. The CPSFL1 protein represents an isoprenoid- and carotenoid-binding protein that associates with membranes through interactions with the phospholipid phosphatidic acid. These findings have implications for understanding carotenoid biosynthesis and optimizing algal carotenoid nutritional quality.
Keywords: carotenoids, CRAL-TRIO domain, phosphatidic acid, photosynthesis, phytoene
Abstract
Plastid isoprenoid-derived carotenoids serve essential roles in chloroplast development and photosynthesis. Although nearly all enzymes that participate in the biosynthesis of carotenoids in plants have been identified, the complement of auxiliary proteins that regulate synthesis, transport, sequestration, and degradation of these molecules and their isoprenoid precursors have not been fully described. To identify such proteins that are necessary for the optimal functioning of oxygenic photosynthesis, we screened a large collection of nonphotosynthetic (acetate-requiring) DNA insertional mutants of Chlamydomonas reinhardtii and isolated cpsfl1. The cpsfl1 mutant is extremely light-sensitive and susceptible to photoinhibition and photobleaching. The CPSFL1 gene encodes a CRAL-TRIO hydrophobic ligand-binding (Sec14) domain protein. Proteins containing this domain are limited to eukaryotes, but some may have been retargeted to function in organelles of endosymbiotic origin. The cpsfl1 mutant showed decreased accumulation of plastidial isoprenoid-derived pigments, especially carotenoids, and whole-cell focused ion-beam scanning-electron microscopy revealed a deficiency of carotenoid-rich chloroplast structures (e.g., eyespot and plastoglobules). The low carotenoid content resulted from impaired biosynthesis at a step prior to phytoene, the committed precursor to carotenoids. The CPSFL1 protein bound phytoene and β-carotene when expressed in Escherichia coli and phosphatidic acid in vitro. We suggest that CPSFL1 is involved in the regulation of phytoene synthesis and carotenoid transport and thereby modulates carotenoid accumulation in the chloroplast.
The chloroplast is a cyanobacterium-derived organelle that plays a central role in the metabolism of photosynthetic eukaryotes. In addition to harboring the photosynthetic apparatus for the conversion of light energy into chemical energy, the chloroplast is also the site of synthesis for a variety of essential metabolites, such as amino acids, nucleotides, fatty acids, and isoprenoids (1). The main groups of plastid isoprenoids are derived from the methylerythritol phosphate (MEP) biosynthetic pathway and include photosynthesis‐related metabolites, such as carotenoids and the side chain of chlorophylls, tocopherols, phylloquinone, and plastoquinone (2, 3). Carotenoids are key structural components of the photosynthetic apparatus in thylakoid membranes (TM), where they are subject to continuous turnover in the light (4). Molecules of β-carotene are found in both photosystems and in the cytochrome (cyt) b6f complex, whereas oxygenated carotenoids, known as xanthophylls, are found primarily in the light-harvesting complexes (LHCs) (5, 6). Carotenoids function as accessory light-harvesting pigments and are essential for photoprotection, playing a crucial role in nonphotochemical quenching of excess light energy, quenching of excited triplet chlorophyll and singlet oxygen, and preventing lipid peroxidation (7, 8).
Because of their multiple functions, carotenoids are widely distributed in the chloroplast. They are present in the chloroplast envelope, TM, and plastoglobules, as well as the eyespot of green algae (9–12). The chloroplast envelope is generally regarded as the major site of carotenoid biosynthesis, and many carotenoid biosynthetic enzymes are exclusively located in the envelope (13, 14), raising a question of how carotenoid synthesis, transport, sequestration, and degradation are coordinated in the chloroplast in response to dynamic developmental and environmental cues.
The unicellular green alga Chlamydomonas reinhardtii is an excellent model organism for studying the genetics of chloroplast development and photosynthesis (15, 16). In addition to the well-established approaches for mutant generation and characterization, C. reinhardtii is particularly useful for studying photosynthetic mutants because cells are able to grow heterotrophically in the presence of acetate, therefore bypassing the necessity of photosynthesis. Furthermore, the study of light-sensitive mutants is possible in C. reinhardtii, because cells grown in the dark are able to synthesize chlorophyll in a light-independent manner, enabling the full assembly of TM protein complexes in the dark (17, 18).
Here we describe the genetic, biochemical, and chloroplast 3D ultrastructural characterization of cpsfl1, a nonphotosynthetic and extremely light-sensitive C. reinhardtii mutant with impaired carotenoid accumulation. We show that lack of the chloroplast Sec14-like 1 (CPSFL1) protein, a CRAL-TRIO hydrophobic ligand-binding (Sec14) domain protein predicted to be present in the chloroplast, causes the cpsfl1 phenotype. We find that CPSFL1 binds carotenoids and phosphatidic acid (PA) and affects the synthesis of phytoene, the committed precursor in carotenoid biosynthesis.
Results
The cpsfl1 Mutant Exhibits a Nonphotosynthetic and Extreme Light-Sensitivity Phenotype.
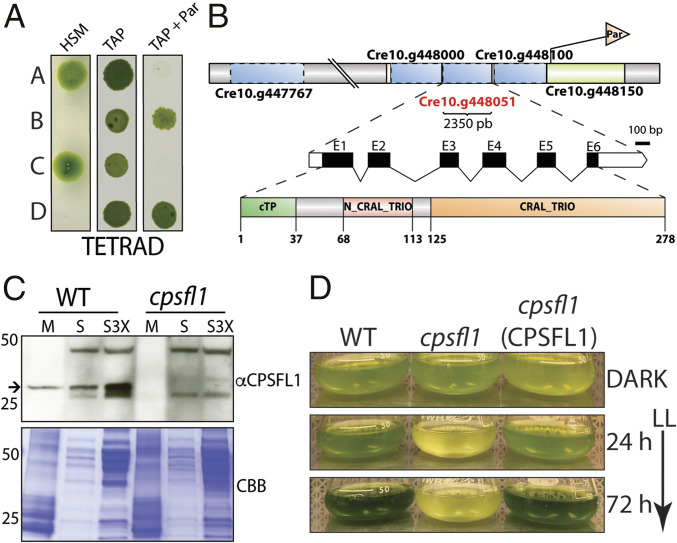
To discover genes that are necessary for the optimal functioning of oxygenic photosynthesis in C. reinhardtii, we generated a collection of DNA insertional mutants and screened for acetate-requiring mutants with impaired photosynthesis (17). One such mutant, CAL028_01_06, was renamed cpsfl1, because the mutant was found to lack a gene encoding a Sec14-like protein (see below). This mutant was not only unable to grow photoautotrophically (Fig. 1A and SI Appendix, Fig. S1D), but it also exhibited an extreme light sensitivity under mixotrophic growth conditions in the presence of acetate. Indeed, shifting the dark-grown mutant cells to constant low light (65-μmol photons m−2 s−1) caused the culture to bleach completely after 72 h (Fig. 1D).
Fig. 1.
Genetic analysis and light sensitivity of the cpsfl1 mutant. (A) Analysis of a representative tetrad from a back-cross of the mutant (mating type +) to the WT (mating type −). Cells were spotted onto minimal (HSM) and acetate-containing (TAP) solid media, in the presence or absence of paromomycin, and grown under constant light (80-μmol photons m−2 s−1) and in the dark, respectively. (B) Schematic representations showing the paromomycin-resistance gene insertion site within chromosome 10 in the mutant, the complete annotation of the CPSFL1 gene, Cre10.g448051, as well as the C. reinhardtii CPSFL1 protein showing the N-terminal chloroplast transit peptide (cTP), the N-terminal N-CRAL-TRIO, and the C-terminal CRAL-TRIO protein domains. (C) Immunoblot analysis from WT and cpsfl1 fractionated cells showing the absence of the CPSFL1 protein in the mutant. The membrane fraction (M) corresponds to 4 μg of chlorophyll and supernatant fractions (S and S3X) correspond to loading of 10 and 30 μg of protein, respectively. Coomassie brilliant blue-stained gel (CBB). The arrow indicates immunoblot signals specific to CPSFL1. (D) Growth phenotypes of C. reinhardtii dark-grown WT, cpsfl1 mutant and cpsfl1 (CPSFL1) complemented line in TAP medium shifted to constant light (65-μmol photons m−2 s−1) for 72 h.
To assess the degree of light sensitivity experienced by the cpsfl1 mutant under mixotrophic growth conditions, we spotted dark-grown WT, cpsfl1, and fud7 cells onto solid medium containing acetate (SI Appendix, Fig. S1D). The fud7 mutant is a nonphotosynthetic mutant that lacks the D1 subunit of photosystem II (PSII) protein complexes, thus exhibiting no PSII activity as assessed by measuring PSII quantum efficiency (Fv/Fm = 0) (19). Dark-grown cells were shifted to 2-, 10-, 40-, or 80-μmol photons m−2 s−1 of light for 6 d. Remarkably, the cpsfl1 mutant exhibited an extremely light-sensitive phenotype as it was unable to grow under light irradiances higher than 2-μmol photons m−2 s−1, while WT and fud7 strains showed normal growth on acetate under all tested conditions (SI Appendix, Fig. S1D). Notably, dark-grown cpsfl1 mutant cells displayed similar PSII activity as compared to WT cells (SI Appendix, Fig. S1D).
Genetic Analysis and Complementation of cpsfl1.
To determine the genetic basis of the cpsfl1 phenotype, we performed mating and tetrad dissection of progeny from a back-cross between the cpsfl1 mutant and a WT strain of the opposite mating type (Fig. 1A and SI Appendix, Fig. S1A). Analysis of five complete tetrads showed Mendelian inheritance of the light-sensitive and the nonphotosynthetic phenotype of cpsfl1 with a 2:2 segregation pattern. The selectable marker introduced by the DNA insertion conferred resistance to the antibiotic paromomycin, and the paromomycin resistance assessed on acetate-containing plates, cosegregated with the lack of photoautotrophic growth on minimal medium (Fig. 1A and SI Appendix, Fig. S1A). These results suggest that insertion of the paromomycin-resistance marker into the nuclear genome causes the acetate-requiring phenotype of cpsfl1.
PCR-based mapping of the genomic DNA flanking the inserted paromomycin-resistance marker in cpsfl1 indicated an insertion site on chromosome 10 (18) (Fig. 1B and SI Appendix, Fig. S1C and Dataset S1) and a genomic deletion of 26.3 kb. The deletion disrupted the expression of multiple genes (Cre10.g447767; Cre10.g447800; Cre10.g447850; Cre10.g447900; Cre10.g447950; Cre10.g448000; Cre10.g448051, and Cre10.g448100) (Fig. 1B). Of these eight genes, only Cre10.g448051 was found to be homologous to a chloroplast-localized protein in Arabidopsis thaliana. The complete cDNA for Cre10.g448051 was predicted by the Augustus gene prediction software (20) and cloned. It differed from the Phytozome v.12 annotated gene model and included two extra exons at its 5′ end (Fig. 1B and SI Appendix, Dataset S2). Complementation of the cpsfl1 mutation was successfully performed with either CPSFL1 cDNA or genomic DNA under the control of the PSAD promoter (SI Appendix, Fig. S1B). The cpsfl1(CPSFL1) strain complemented with genomic DNA exhibited restored photoautotrophic growth and rescue of the light-sensitivity phenotype (SI Appendix, Fig. S1D). Successful transformation was confirmed by PCR (SI Appendix, Fig. S1C).
To test the abundance and location of the CPSFL1 protein, immunoblot analyses were performed with an anti-CPSFL1 polyclonal antibody on membrane and soluble fractions prepared from WT and mutant whole cells. As expected, the CPSFL1 protein was completely absent in the cpsfl1 mutant (Fig. 1C and SI Appendix, Fig. S1E). In the WT, the CPSFL1 protein was detected in both soluble and membrane fractions (Fig. 1C). Furthermore, we quantified the content of CPSFL1 protein by comparing recombinant purified CPSFL1 protein dilutions with WT dark-grown whole-cell extracts by protein immunoblot analysis and found ∼1 ng (∼0.032 pmol) of CPSFL1 per 106 cells (SI Appendix, Fig. S1E).
The CPSFL1 Gene Encodes a CRAL-TRIO Domain-Containing Protein.
The complete coding sequence of CPSFL1 translates into a 31.5-kDa protein (278 amino acids). The protein possesses a predicted N-terminal chloroplast transit peptide of 37 amino acids (Fig. 1B). Searching for protein motifs within the CPSFL1 protein using ScanProsite (21) revealed a single CRAL-TRIO domain (PS50191), located between amino acids 125 to 278. An additional CRAL-TRIO-N motif (pfam03765), located between amino acids 68 and 113, was detected by searching the National Center for Biotechnology Information Conserved Domain Database (22) (Fig. 1B). The CRAL-TRIO is a structurally conserved domain found exclusively in eukaryotic organisms. It is comprised of alternating α-helices and β-strands, which together constitute a hydrophobic ligand-binding pocket. The CRAL-TRIO domain in various proteins has been shown to bind a diverse set of small hydrophobic molecules as ligands, including α-tocopherol, retinaldehyde, squalene, and phosphatidylinositol (PI) (23, 24). The predicted protein structures of A. thaliana and C. reinhardtii CPSFL1, modeled by I-TASSER (25), yielded a protein with 5 β-sheets and 14 α-helices with a C-score of 1.03 and 0.81, respectively (SI Appendix, Fig. S2A), and showed high similarity to the structure of other CRAL-TRIO proteins, such as cellular retinaldehyde-binding protein (CRALBP), human α-tocopherol transfer protein (α-TTP), and yeast Sec14 homolog 1 (Sfh1) (protein structures in the Protein Data Bank: 3HY5, 1R5L, and 3B7Z, respectively) (SI Appendix, Fig. S2A).
To better understand the relationships between the CRAL-TRIO proteins of photosynthetic organisms, we constructed a phylogenetic tree of all CRAL-TRIO proteins present in the model plant A. thaliana and C. reinhardtii, along with yeast Sfh1 and human α-TTP (Fig. 2A and SI Appendix, Dataset S3). In C. reinhardtii, 12 proteins were found to contain a CRAL-TRIO domain in comparison to 32 in A. thaliana. The phylogenetic analysis clearly resolved that the CPSFL1 A. thaliana ortholog is the protein encoded by the gene At5g63060, which exhibits 37.2% amino acid sequence identity to CPSFL1. In comparison, the amino acid sequence identity between CPSFL1 and yeast Sfh1, CRALBP, and human α-TTP was determined to be 20.8%, 21.2%, and 18.3%, respectively (SI Appendix, Fig. S2B). The CRAL-TRIO domain may form the majority of the mature polypeptide as a standalone domain or as part of a multidomain protein (26, 27). In plants, CRAL-TRIO domain-containing proteins may also possess a Golgi dynamics domain, which is involved in Golgi function and secretion, or a nodulin domain that is involved in phosphoinositide molecule binding (26), but the C. reinhardtii and A. thaliana CPSFL1 proteins lack any of these additional motifs.
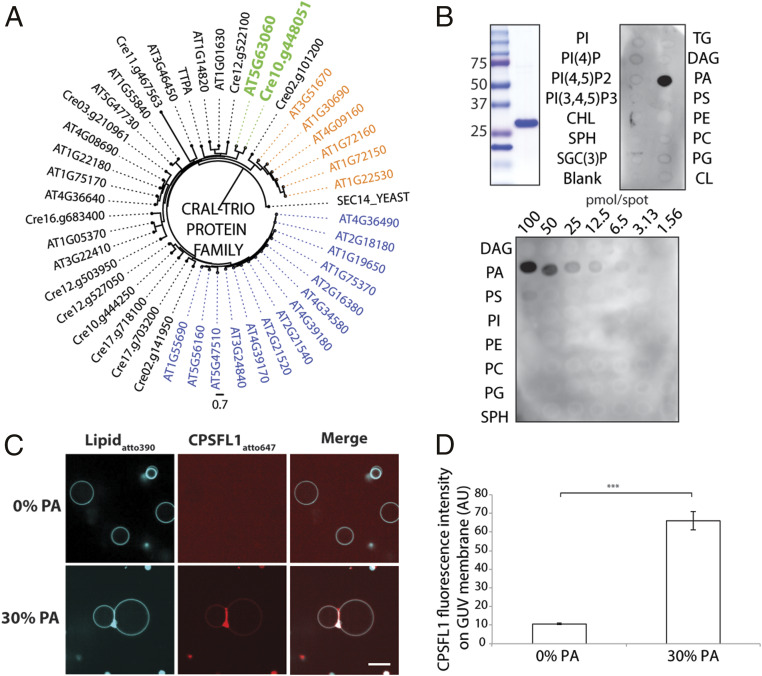
Fig. 2.
CPSFL1 encodes a chloroplast-targeted CRAL-TRIO domain protein that binds PA. (A) Phylogenetic analysis of A. thaliana and C. reinhardtii CRAL-TRIO proteins. The gene At5g63060 encodes the A. thaliana chloroplast-localized ortholog of C. reinhardtii CPSFL1 (colored in green). Genes with an additional Golgi dynamics domain or nodulin domain are color-coded in orange and blue, respectively. (B) CPSFL1 protein lipid overlay assay. Stained Coomassie gel of purified CPSFL1 protein is shown on the left. Lipid abbreviations are detailed in Methods. (C) Fluorescently labeled CPSFL1 proteins bind to GUVs containing 30% DOPA. (Scale bar, 10 μm.) (D) CPSFL1 fluorescence intensity quantification on GUV membranes (Right). Data represent means ± SE. Asterisk (***) marks significant difference relative to the 0% PA GUVs at P < 0.001, derived using a two-sample Student’s t test.
The CPSFL1 Protein Binds to PA and Promotes Membrane Adhesion.
Several CRAL-TRIO domain proteins have been shown to bind phospholipids in addition to their hydrophobic ligand (27, 28), prompting us to test whether the CPSFL1 protein is capable of binding phospholipids. Purified recombinant CPSFL1 proteins were assayed in vitro for overlay binding to lipids that had been prespotted on membranes (Fig. 2B). CPSFL1 exhibited specific binding to PA molecules and, notably, these interactions were detected even under very low quantities of spotted PA lipids as low as 3.1 pmol per spot (Fig. 2B). To characterize the binding of CPSFL1 to PA within a lipid bilayer, we visualized the binding of fluorescently labeled CPSFL1 protein to giant unilamellar vesicles (GUVs) by confocal microscopy. The GUV bilayers were prepared with phosphatidylcholine (PC) alone or PC containing 30% PA (Fig. 2C). As expected, CPSFL1 protein bound more strongly to PA-containing GUVs than PC-only GUVs (Fig. 2C). We also observed the formation of GUV dimers and multimers, indicating that the bound protein exhibits adhesive behavior (Fig. 2C). As is characteristic for adhesion proteins (29), the CPSFL1 protein showed significant enrichment at the interface of the GUV’s in the presence of PA versus its absence with an enrichment index of 2.63 ± 0.74 (Fig. 2D).
TM Architecture and Lipid Composition of Dark-Grown Cells Are Not Altered in cpsfl1.
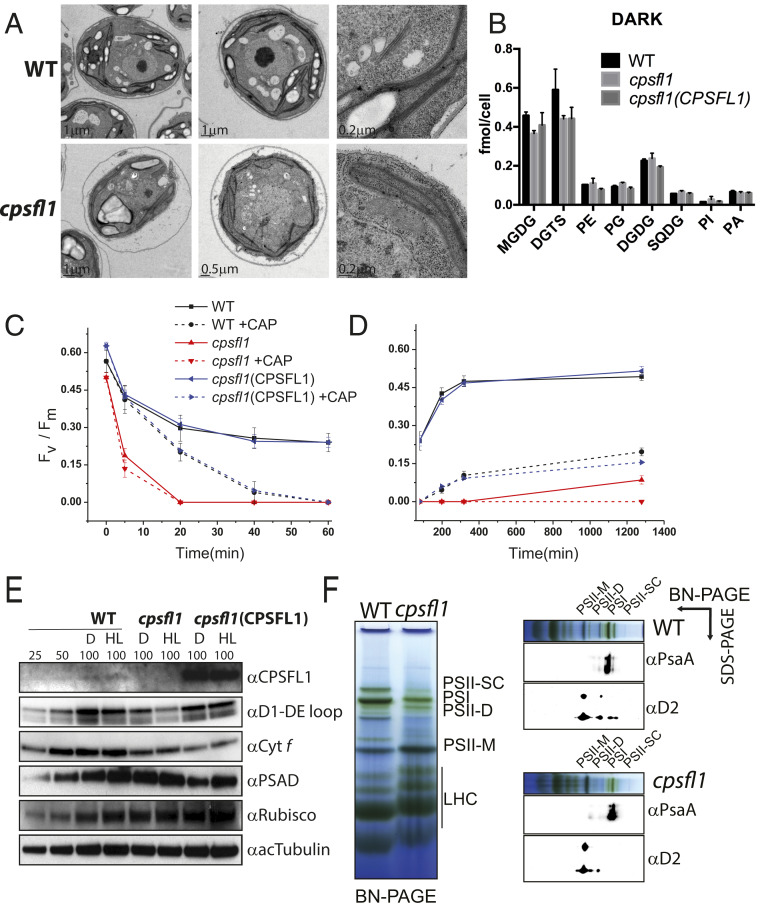
TM constitute the scaffolding matrix in which photosynthetic protein complexes are assembled in the chloroplast. The formation of TM requires complex and concerted actions of multiple biosynthetic processes, including the synthesis and trafficking of lipids, pigments, and proteins (30). In C. reinhardtii cells, the formation of TM can take place under heterotrophic growth conditions, making it possible to study chloroplast and thylakoid formation in the dark (16). Because yeast Sfh1 has been shown to be involved in lipid transfer (31), we tested whether a lack of CPSFL1 protein alters either chloroplast TM formation or cellular lipid content in the dark. The TM architecture of dark-grown cells was visualized by transmission electron microscopy (TEM). Images of WT (Fig. 3 A, Upper) and cpsfl1 mutant cells (Fig. 3 A, Lower) displayed no apparent differences in TM network formation or starch granules accumulation. Furthermore, whole-cell lipid analyses of WT, cpsfl1, and cpsfl1(CPSFL1) complemented strain showed no significant changes in lipid content and their specific acyl composition for the chloroplast galactolipids digalactosyldiacylglycerol and sulfoquinovosyldiacylglycerol, and also for phospholipids, such as phosphatidylethanolamine (PE), PI, phosphatidylglycerol (PG), and PA (Fig. 3B and SI Appendix, Figs. S3C, S5, and S6). In contrast, minor reduction of monogalactosyldiacylglycerol and about 25% reduction in diacylglyceryltrimethylhomoserine (DGTS) lipid content were measured in the mutant in comparison to WT. We found a similar reduction in DGTS in the complemented line in comparison to the mutant (Fig. 3B), suggesting that these differences are unrelated to the cpsfl1 mutation. Consistent with this suggestion, we also found the lipid content of dark-grown cells shifted to low light for 12 h to be comparable among the different lines examined while still exhibiting a reduced content of DGTS in both the mutant and the complemented line (SI Appendix, Fig. S3C). Thus, the CPSFL1 protein appears to exert no direct role in chloroplast glycerolipid metabolism.
Fig. 3.
Analyses of TM structure, lipid composition, and photosystem organization in cpsfl1. (A) TEM images of different cell sections of dark-grown WT (Upper) and cpsfl1 mutant cells (Lower). (B) Lipid profiles of WT, cpsfl1, and cpsfl1(CPSFL1) complemented cells grown in the dark. Error bars represent means ± SD (n = 3). DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol. (C–E) Photoinhibition analysis of cpsfl1. Dark-grown WT, cpsfl1, and complemented cpsfl1(CPSFL1) cells were shifted to HL (800-µmol photons m−2 s−1) for 1 h (C) and recovered in the dark for 20 h (D) in the absence or presence of chloramphenicol (CAP). PSII activity as assessed by measuring PSII maximum quantum efficiency (Fv/Fm) after 5, 20, 40, and 60 min during the HL treatment and after 2, 4, and 20 h during recovery in the dark. Error bars represent means ± SD (n = 3). (E) Immunoblot analyses of dark-grown (D) and 1 h HL-treated cells (HL). Loading of 100% corresponds to ∼1 × 106 cells. (F) First-dimension BN-PAGE analysis of solubilized TM protein complexes isolated from dark-grown cells and immunoblot analyses against PSII-D2 and PSI-PsaA subunits in the second denaturing dimension. PSII-D, PSII dimer; PSII-M, PSII monomer; PSII-SC, PSII-LHCII supercomplexes.
The cpsfl1 Mutant Exhibits Severe Photoinhibition and Impaired Recovery of PSII Activity.
Because of the light sensitivity of cpsfl1, we next examined whether the mutant was also susceptible to enhanced photoinhibition: That is, the inhibition of the activity of PSII under strong light. During a 1-h exposure of dark-grown mutant cells to high light (HL) (800-μmol photons m−2 s−1), the PSII activity declined very rapidly during the first 5 min in the mutant (from an initial Fv/Fm value of ∼0.5 to a value of 0.18), and it was completely abolished after 20 min (Fig. 3C). On the other hand, the WT and cpsfl1(CPSFL1) complemented line experienced only a moderate decline in Fv/Fm from 0.56 and 0.6, respectively, to ∼0.42 in the first 5 min and exhibited a final Fv/Fm value of ∼0.25, which was maintained until the end of the HL treatment. Immunoblot analyses were performed at the beginning and at the end of the 1-h HL treatment (Fig. 3E). Concomitant with the decrease in PSII activity, the abundance of the PSII-D1 subunit in the cpsfl1 mutant was reduced by half at the end of the HL treatment as compared to the initial D1 levels of dark-grown cells. In contrast, relatively little or no changes were observed in the mutant at the end of the HL treatment for tubulin or other chloroplast proteins, such as cytochrome f (Cyt f), Rubisco large subunit (RbcL), or the PSAD subunit of PSI, although Cyt f was lower in the dark-grown mutant. Reduced amounts of Cyt f and PSAD were measured in the complemented line in the dark in comparison to WT (Fig. 3E).
To test whether the enhanced PSII degradation observed in cpsfl1 mutant cells is caused by an altered PSII repair process, we measured PSII activity in the presence of the antibiotic chloramphenicol (CAP) during the HL treatment (Fig. 3C). CAP treatment inhibits chloroplast protein translation, thereby blocking the repair of PSII. We found that the CAP-treated cpsfl1 mutant cells showed rapid reduction of PSII activity as observed in untreated mutant cells, whereas WT and complemented cells exhibited a more gradual reduction of PSII activity that led to complete abolishment only after 60 min of CAP treatment (Fig. 3C). We further measured the ability of HL-treated cells to recover PSII activity in the dark (Fig. 3D). Notably, we found that WT and cpsfl1(CPSFL1) complemented cells recovered PSII activity after 4 h, whereas the mutant cells failed to completely recover PSII activity even after 20 h (Fig. 3D). WT and complemented strains treated with CAP exhibited minor recovery of PSII activity. Taken together, these results indicate that the cpsfl1 mutant is susceptible to enhanced photoinhibition and exhibits impaired repair of PSII protein complexes.
PSII Macroorganization Is Altered in Dark-Grown cpsfl1 Cells.
Given the nonphotosynthetic growth and extreme light sensitivity exhibited by the cpsfl1 mutant, we examined whether a lack of CPSFL1 protein affects the assembly and stability of TM protein complexes. We therefore isolated TM from the dark-grown WT and cpsfl1 cells and studied the native organization of TM protein complexes by first-dimension blue-native polyacrylamide gel electrophoresis (BN-PAGE) followed by second-dimension denaturing SDS/PAGE and immunoblot analyses (Fig. 3F). We found the assembly of PSI protein complexes to be comparable in both WT and mutant cells; however, PSII macroorganization was severely altered in the mutant. In particular, the mutant exhibited a drastic reduction in PSII dimers and a complete absence of PSII-LHCII supercomplexes, while the accumulation of PSII monomers was enriched in comparison to the WT (Fig. 3F). These results indicate that although the assembly of PSII monomers takes place in cpsfl1 mutant membranes in the dark, these fail to stably assemble into larger complexes, such as PSII dimers and PSII-LHCII supercomplexes.
Dark-Grown cpsfl1 Mutant Cells Exhibit Reduced Carotenoid Content.
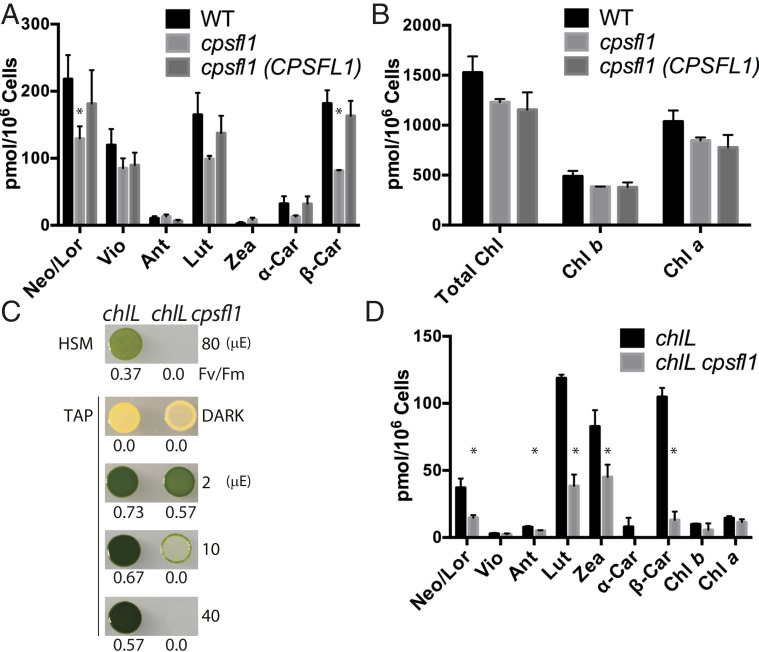
Because the cpsfl1 mutant is unable to grow mixotrophically and exhibits bleaching under continuous low light (Fig. 1D), the pigment content and composition of WT, cpsfl1, and the cpsfl1(CPSFL1) complemented strain were measured from dark-grown cells and cells shifted to HL for 1 h (Fig. 4 A and B and SI Appendix, Fig. S3 A and B). We found that the total carotenoid content in the mutant was reduced by ∼42% and 49% compared to the dark-grown and HL-treated WT (P < 0.05, two-tailed Student’s t test), respectively, whereas the complemented line did not significantly differ from the WT. In particular, β-carotene was significantly reduced by ∼55% and 63% in the mutant under dark and after HL treatment, respectively, as compared to WT (Fig. 4A and SI Appendix, Fig. S3A). Despite these dramatic changes in carotenoid content in the mutant, total chlorophyll content in both the mutant and its complemented line were only slightly reduced in comparison to that of WT levels (Fig. 4B and SI Appendix, Fig. S3B), suggesting that the difference in chlorophyll content is unrelated to the cpsfl1 mutation.
Fig. 4.
The cpsfl1 mutant shows reduced carotenoid content. HPLC pigment analysis of carotenoids (A) and chlorophylls (B) of dark-grown WT, cpsfl1, and cpsfl1(CPSFL1) complemented cells. (C) Growth phenotypes and PSII activity (Fv/Fm) of chlL and chlL cpsfl1 double mutant. Cells were spotted onto minimal (HSM) and acetate-containing (TAP) solid media and grown under indicated light intensities. The term “μE” represents micromole photons m−2 s−1. (D) HPLC pigment analysis of chlL and chlL cpsfl1 cells grown in the dark. Error bars represent means ± SD (n = 3). Significantly changed pigments relative to the WT (two-tailed Student’s t test; P < 0.05) are marked with asterisks. Abbreviations: α-Car, α-Carotene; β-Car, β-Carotene; Ant, Antheraxanthin; Chl, Chlorophyll; Lor, Loroxanthin; Lut, Lutein; Neo, Neoxanthin; Vio, Violaxanthin; Zea, Zeaxanthin.
Carotenoid Content in the chlL cpsfl1 Double Mutant Is Reduced.
Carotenoids and chlorophylls follow coordinated synthesis as both molecules are required for the assembly of the photosynthetic apparatus during chloroplast biogenesis (32). To separate chlorophyll from carotenoid biosynthesis and to further understand the role of CPSFL1 in promoting carotenoid accumulation, we generated a yellow-in-the-dark chlL and cpsfl1 double mutant, chlL cpsfl1, by mating and tetrad dissection. The chlL mutant is deficient in the chloroplast-encoded CHLL protein subunit, which is necessary for activity of dark-operative protochlorophyllide oxidoreductase, a key enzyme in the light-independent chlorophyll biosynthesis pathway in C. reinhardtii (33, 34). Thus, C. reinhardtii cells with the chlL mutation are unable to synthesize chlorophyll when grown in the dark, and they exhibit a characteristic yellow phenotype due to the presence of carotenoids in these cells. The plastids of dark-grown chlL mutant cells contain a large number of starch granules but lack TM (35). Upon shifting dark-grown chlL cells to light, light-dependent protochlorophyllide oxidoreductase is activated, and chlorophyll synthesis resumes. The latter process leads to the formation of TM, and concomitantly, the assembly of the pigmented photosynthetic apparatus. This is known as “greening” and provides an algal analog to the light-controlled etioplast-to-chloroplast development seen in plants. This greening process was severely altered in the chlL cpsfl1 double mutant, as dark-grown chlL cpsfl1 cells spotted on Tris acetate-phosphate (TAP) agar plates were unable to sustain mixotrophic growth under the constant light irradiances of 10- and 40-μmol photons m−2 s−1 (Fig. 4C). Similar to the cpsfl1 single mutant, the double mutant exhibited a light-sensitive phenotype as cell growth and greening were only observed under a very low-light irradiance of less than 2-µmol photons m−2 s−1 (Fig. 4C). Notably, the carotenoid content of dark-grown chlL cpsfl1 was reduced by 68% as compared to the single chlL mutant, while only residual amounts of chlorophylls were detected (Fig. 4D). Like cpsfl1, the chlL cpsfl1 double mutant also exhibited a remarkable ∼87% reduction of β-carotene in comparison to chlL (Fig. 4D).
Impaired Formation of Carotenoid-Rich Structures in the Chloroplast of cpsfl1 Mutant Cells.
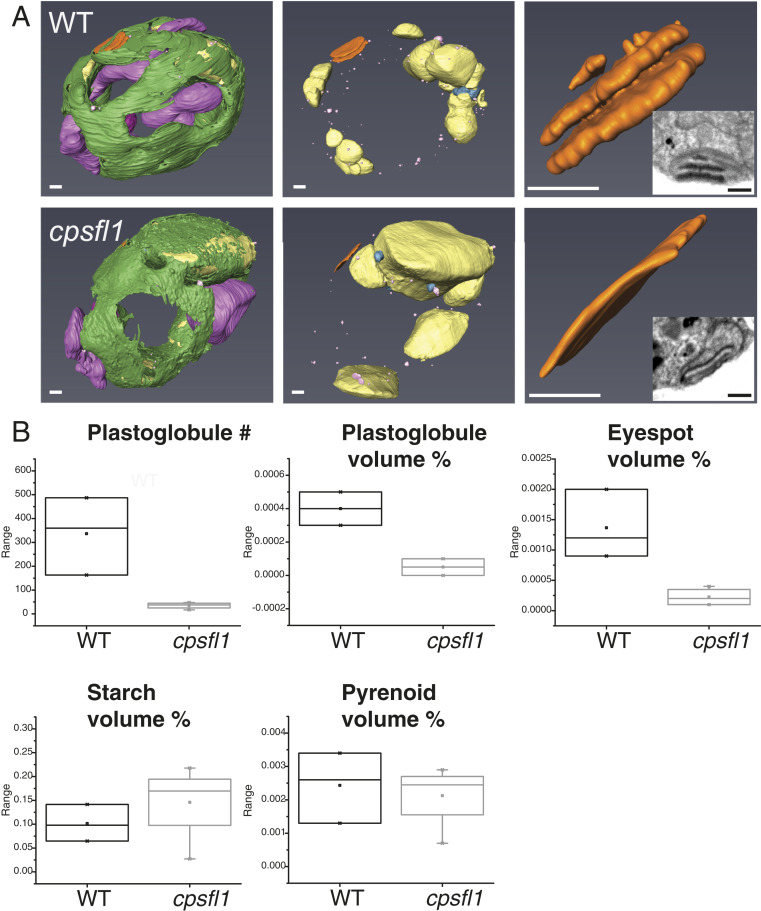
Given the reduced carotenoid content observed in the mutant, we investigated whether it had an effect on the organization of carotenoid-rich structures in the chloroplast. Since carotenoid-containing structures are distributed throughout the chloroplast, we used a 3D visualization technique providing high resolution to describe these structures. We used focused ion beam scanning-electron microscopy (FIB-SEM) (36) (see details in Methods) on groups of whole, fixed, and heavy metal-stained C. reinhardtii cells to obtain consecutive series of 8-nm × 8-nm resolution 2D images every 2 nm and thus generated 3D maps encompassing a large number of cells.
We visually inspected the internal structure of a total of 111 WT and 81 mutant cells and segmented the 3D densities for three representative WT and four cpsfl1 mutant cells (Fig. 5A and Movies S1 and S2). From the resulting 3D models, we calculated the percentage of total cell volume of carotenoid-rich structures, such as the eyespot and plastoglobules, but also of other noncarotenoid-rich structures, such as the pyrenoid and starch granules (Fig. 5B). As already observed by TEM of thin cell sections (Fig. 3A), the formation of the TM network was not significantly altered, but the mutant exhibited a reduced number and relative volume of plastoglobules (as percent of total cell volume) as well as aberrant eyespot formation (Fig. 5 A, Lower Right Inset, and Fig. 5B). The WT eyespot was observed as a multilayered structure of carotenoid-filled granules organized in slightly curved parallel planes, strongly stained by osmium in our fixed specimens (Fig. 5 A, Upper Right Inset). In contrast, all mutant cells had only small, single-layered or undetectable eyespot structure, displaying poor osmium staining when present (Fig. 5 A, Lower Right Inset). Additionally, mutant cells exhibited a relative increase in starch content (percent cell volume), while no major changes were measured for the pyrenoid formation in comparison to WT cells (Fig. 5B). The latter is also supported by the WT levels measured for the RbcL in the mutant (Fig. 3E).
Fig. 5.
FIB-SEM 3D ultrastructural analysis of WT and cpsfl1 cells. (A) Three-dimensional models based on the FIB-SEM densities of dark-grown representative WT (Upper) and cpsfl1 mutant (Lower) cells. In green, TM; purple, mitochondria; yellow, starch granules; pink, plastoglobules; and orange, eyespot. (See also Movies S1 and S2). (B) Box plots of, from left to right and top to bottom, number, and relative volume (in percent of total cell volume) of plastoglobules, eyespot, starch granules, and pyrenoid, measured in the FIB-SEM densities. (Scale bars, 500 nm.)
The CPSFL1 Protein Binds Carotenoids.
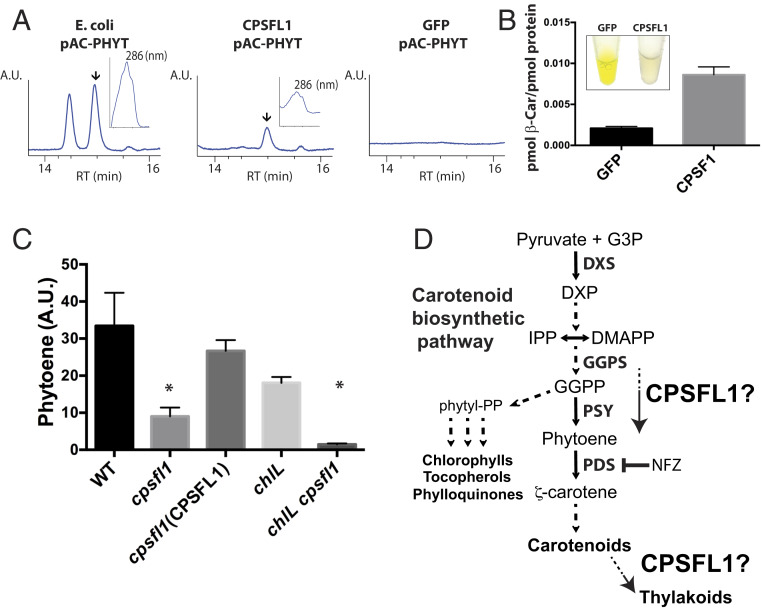
Because of the low abundance of CPSFL1 protein in the chloroplast and difficulty in studying membrane-associated carotenoid biosynthetic enzymes, we used carotenoid-producing Escherichia coli strains as heterologous hosts (37) to test whether CPSFL1 binds carotenoids. We purified recombinant CPSFL1 proteins lacking the chloroplast transit peptide from E. coli cells that constitutively produced phytoene (pAC-PHYT) or β-carotene (pAC-BETA). Remarkably, the purified CPSFL1 proteins were found to bind both phytoene and β-carotene (Fig. 6 A and B), and the binding of these carotenoids to the protein was found to be resistant to the E. coli cell fractionation, centrifugation, washes, and concentration steps during the protein purification. The CPSFL1 proteins bound to phytoene were colorless, whereas the CPSFL1 proteins bound to β-carotene were light orange (Fig. 6B). Since GFP protein has molecular mass and native solubility comparable to those of CPSFL1, it was used as a negative control. Analyses of natively purified GFP proteins isolated from E. coli cells producing phytoene (pAC-PHYT) showed that GFP has no binding activity toward phytoene and exhibited approximately fourfold less binding to β-carotene than CPSFL1 (Fig. 6 A and B). Taken together, these results suggest that the CRAL-TRIO domain of CPSFL1 can bind carotenoids as a hydrophobic ligand, but is not a strong binder.
Fig. 6.
Impaired accumulation of phytoene in the cpsfl1 mutant and carotenoid binding by recombinant CPSFL1 protein. HPLC analysis of affinity-purified CPSFL1 and GFP proteins expressed from phytoene- (pAC-PHYTO) and β-carotene– (pAC-BETA) producing E. coli cells. (A) HPLC chromatogram of E. coli whole cells expressing phytoene as well as recombinant purified CPSFL1 and GFP proteins from these cells. Specific pigment peaks are indicated by arrows in the HPLC chromatograms. Inset plots show the absorption spectra for phytoene peak with its absorbance maxima at 286 nm. (B) HPLC β-carotene quantification of recombinant purified CPSFL1 and GFP proteins from E. coli cells overexpressing β-carotene. Inset are pictures of purified proteins at ∼50 mg/mL Error bars represent means ± SD (n = 3). (C) HPLC analysis of phytoene accumulation of NFZ-treated WT, cpsfl1, cpsfl1(CPSFL1) complemented line, chlL, and chlL cpsfl1 double-mutant cells. Error bars represent means ± SD (n = 3). Significantly changed pigments (two-tailed student’s t test; P < 0.05) are marked with asterisks. (D) Schematic diagram showing the carotenoid biosynthetic pathway and the putative roles of CPSFL1 protein in promoting synthesis and/or flux, or transport, of carotenoids (or carotenoid biosynthesis substrates) within the chloroplast. Abbreviations: DMAPP, dimethylallyl pyrophosphate; DXP, 1-deoxy-d-xylulose-5-phosphate; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; G3P, glyceraldehyde 3-P; GGPS, geranylgeranyl pyrophosphate synthase; IPP, isopentenyl pyrophosphate; Pyr, pyruvate.
CPSFL1 Protein Modulates Phytoene Accumulation.
Phytoene synthase (PSY) catalyzes the first committed and flux-regulating step of carotenoid biosynthesis: the condensation of two molecules of the C20 isoprenoid, geranylgeranyl pyrophosphate (GGPP), leading to the formation of C40 phytoene (38) (Fig. 6D). GGPP is synthesized by the plastidial MEP pathway. To assess how the carotenoid biosynthetic pathway functions in the mutant, we measured the rate of phytoene accumulation in dark-grown cells treated with the herbicide norflurazon (NFZ) (39). The herbicide inhibits carotenoid biosynthesis at the step of phytoene desaturation, consequently allowing the phytoene precursor to accumulate in these cells (Fig. 6D). We incubated dark-grown WT and cpsfl1 mutant cells with NFZ (3.5 μM) for up to 48 h in the dark (Fig. 6C) and analyzed the levels of different pigments and the accumulation of phytoene after 0, 6, 24, and 48 h of NFZ treatment (SI Appendix, Fig. S4). As NFZ blocks carotenoid synthesis, the levels of carotenes, xanthophylls, and chlorophylls are gradually decreased with the prolonged NFZ treatment, by degradation or by dilution through cell division (SI Appendix, Fig. S4 A–C), whereas the accumulation of phytoene increases during the treatment (SI Appendix, Fig. S4D). Strikingly, the NFZ-treated mutant contained ∼73% less phytoene per cell than WT after 48 h (Fig. 6C and SI Appendix, Fig. S4D), while the reduction rates of chlorophylls, carotenes, and xanthophylls was similar in both the WT and the cpsfl1 mutant (SI Appendix, Fig. S4 A–C). In contrast, the cpsfl1(CPSFL1) complemented line exhibited a minor reduction of ∼21% of WT phytoene content (Fig. 6B). Furthermore, we also assessed the phytoene accumulation of NFZ-treated chlL and chlL cpsfl1 dark grown cells for 48 h and measured a reduction of ∼91% for the latter as compared to the chlL single mutant (Fig. 6C). The low accumulation of phytoene in the NFZ-treated cpsfl1 mutant backgrounds suggests a critical role for CPSFL1 in modulating the flux toward phytoene accumulation in the chloroplast.
Discussion
This study characterizes the function of CPSFL1, an evolutionarily conserved protein of photosynthetic eukaryotes. Along with a recently identified protein that functions in α-tocopherol metabolism in tomato (40), CPSFL1 is a eukaryotic CRAL-TRIO domain-containing protein that is proposed to function in the chloroplast, an organelle of endosymbiotic origin. The C. reinhardtii cpsfl1 mutant is a nonphotosynthetic and extremely light-sensitive mutant that is susceptible to rapid photoinhibition and photobleaching, even in the presence of acetate (Figs. 1 A and D and 3 C–E and SI Appendix, Fig. S1D). Here we have exploited the ability of C. reinhardtii to grow and assemble a photosynthetic apparatus in the presence of acetate in the dark, which enables the analysis of the cpsfl1 phenotype in the absence of photo-oxidative stress. Loss of CPSFL1 resulted in pleiotropic phenotypes that are attributable to a severe reduction in the overall carotenoid content (Fig. 4 A and D and SI Appendix, Fig. S3A). The mutant exhibited impaired PSII macroorganization with reduced accumulation of PSII dimers and undetectable levels of PSII-LHCII supercomplexes in the dark with enhanced photoinhibition under HL (Fig. 3 C–F). The mutant also showed abnormal chloroplast carotenoid-rich structures, such as plastoglobules and the eyespot, as revealed by 3D chloroplast ultrastructure visualization using FIB-SEM (Fig. 5 and Movie S1 and S2). These phenotypes are qualitatively similar to those of a knockout mutant affecting the CPSFL1 ortholog in A. thaliana, At5g63060 (Fig. 2A). Homozygous cpsfl1 plants are unable to grow in soil and display a seedling-lethal phenotype with albino cotyledons (41). Thus, CPSFL1 has an essential role in chloroplast development and growth of both C. reinhardtii and A. thaliana.
The Loss of CPSFL1 Affects Carotenoid Biosynthesis and Accumulation in the Chloroplast.
Carotenoids are not only important molecules for photoprotection, but they also participate in the assembly, stabilization, and repair of the photosynthetic apparatus (42, 43). This explains the light sensitivity and defects in dark PSII assembly and enhanced photoinhibition that we observed in the cpsfl1 mutant (Fig. 3 C–F). Unlike other carotenoid mutants that exhibit complete blockage for a specific carotenoid precursor, leading to arrested chloroplast development phenotypes (44, 45), synthesis of all major carotenoid types was detected in all studied cpsfl1 mutant backgrounds (Fig. 4 A and D and SI Appendix, Fig. S3A).
The defective carotenoid accumulation and extreme light sensitivity observed in cpsfl1 was also seen in the yellow-in-the-dark chlL cpsfl1 double mutant (Fig. 4 C and D). Yellow-in-the-dark mutants, such as chlL, can accumulate carotenoids but contain only residual amounts of chlorophylls. These cells exhibit a complete absence of TM and photosynthetic apparatus when grown heterotrophically (33, 35). The accumulation of carotenoids in chlL supports the idea that carotenoid biosynthesis must take place within chloroplast envelope membranes, where the majority of the carotenoid biosynthetic machinery has been detected by proteomic analysis (11, 14, 46). Furthermore, the severe impairment of carotenoid and phytoene accumulation in the chlL cpsfl1 double mutant suggests that CPSFL1 might function in the stroma and in association with the envelope membrane (Fig. 4 C and D).
Taken together, these results indicate that the reduced carotenoid content observed in these mutants did not result from a complete blockage in the biosynthetic carotenoid pathway, but rather a decreased synthesis and flux toward carotenoid biosynthesis. In agreement with the latter, treatment of cpsfl1 cells with NFZ in the dark showed that the reduction in carotenoid content is due to a limitation in the synthesis rate of phytoene rather than enhanced degradation of carotenoids (Figs. 6C and SI Appendix, Fig. 4 A, B, and D). Phytoene serves as the first committed molecule of the carotenoid biosynthetic pathway and is produced by the PSY enzyme (Fig. 6D) (47, 48). Indeed, treatment of WT C. reinhardtii cells with NFZ to block carotenoid biosynthesis in HL has been shown to cause photobleaching of β-carotene molecules, which triggers the rapid destabilization and degradation of PSII complexes (49). Similarly, C. reinhardtii lts1 mutants lacking PSY (42, 50) and pds1 mutants lacking phytoene desaturase (PDS) (51) are unable to grow photoautotrophically and exhibit extreme light sensitivity. The latter mutant accumulates phytoene, as the blockage occurs at the second step in carotenoid biosynthesis, the conversion of phytoene to ζ-carotene catalyzed by the PDS enzyme (51). Dark-grown lts1 cells still accumulate about 20% of WT levels of chlorophyll a, despite a complete lack of phytoene and, consequently, carotenoids. These cells are still able to functionally assemble and accumulate significant levels of PSI protein complexes, as well as minor amounts of Cyt b6f; however, they accumulate neither PSII nor LHC complexes (42). Unlike pds1 but like lts1, the cpsfl1 mutant does not accumulate phytoene in the dark, and only accumulates phytoene when mutant cells are treated with NFZ, although at a much-reduced rate compared to the WT (Fig. 6C and SI Appendix, Fig. S4D). These results suggest that the cpsfl1 mutant phenotypically resembles a partial loss-of-function mutant affecting PSY.
How Does CPSFL1 Modulate Carotenoid Accumulation?
Based on our genetic, biochemical, and 3D chloroplast ultrastructural analyses, we hypothesize that CPSFL1 is necessary for efficient synthesis and flux of carotenoid biosynthesis at the chloroplast envelope. CPSFL1 contains a CRAL-TRIO domain, which constitutes a hydrophobic ligand-binding pocket. Indeed, we show that recombinant CPSFL1 binds PA in vitro (Fig. 2 B–D), and carotenoids, such as phytoene and β-carotene, when expressed in carotenoid-producing E. coli strains (Fig. 6 A and B). CPSFL1 shows protein structural similarity to several standalone CRAL-TRIO proteins that exhibit diverse hydrophobic ligand binding and, in some cases, the ability to transfer ligands between membranes. These include human α-TTP, the yeast Sfh1, and human CRALBP (SI Appendix, Fig. S2 A and B). For example, α-TTP protein binds α-tocopherol and regulates its secretion from liver cells (23, 28, 52). Interestingly, α-TTP exhibits a secondary phospholipid interaction domain, which provides specific binding to the PI phosphates (PIPs) from target membranes. The interaction of α-TTP with membrane PIPs promotes the intermembrane release and transfer of its ligand, α-tocopherol (28). The yeast Sfh1 has been shown to transfer phospholipids PI and PC between membranes in vitro, although neither PI nor PC transfer is absolutely required for the essential function of this protein in yeast, leaving open the identity of its true ligand in vivo (31, 53). It is worth noting that C. reinhardtii cells lack PC (54). We also show that the phospholipid, acyl lipid composition, and TM network formation of dark-grown cpsfl1 mutant are not significantly altered as compared to WT cells (Figs. 3 A and B and 5A and SI Appendix, Figs. S5 and S6). CRALBP is a cytosolic protein localized in the vertebrate retinal pigment epithelium, where it appears to play a role in the vertebrate visual process as a substrate-routing protein, influencing the enzymatic partitioning of 11-cis-retinol at a key branch point in the visual cycle (55). Similar to α-TTP, release of bound-intermediates from CRALBP is triggered by binding to phospholipids like PA (56). Thus, many CRAL-TRIO domain-containing proteins are involved in metabolism of isoprenoid-derived molecules by a mechanism that requires interaction and facilitated transport to targeted membranes.
Based on the functions of these related CRAL-TRIO proteins and current results presented here, we hypothesize two possible mechanisms of action of CPSFL1 in the chloroplast (Fig. 6D). First, CPSFL1 could be involved in the substrate routing of isoprenoid intermediates at the chloroplast envelope, the site of carotenoid biosynthesis. This substrate chaperone function would be similar to that of CRALBP, which binds intermediates and enhances flux through the visual cycle. Like CRALBP, CPSFL1 also binds PA, suggesting that the release of intermediates from CPSFL1 might be controlled by PA binding. Because CPSFL1 occurs in both soluble and membrane-bound forms (Fig. 1C), it is tempting to speculate that it might be involved in delivery of a relatively hydrophilic substrate to a membrane-bound enzyme. For example, the immediate isoprenoid precursor to phytoene synthesis, GGPP, is a versatile compound that must be partitioned between several chloroplast biosynthetic pathways, including those that lead to the synthesis of carotenoids, chlorophylls, plastoquinone, phylloquinone, tocopherols, and gibberellins (3). A putative function of CPSFL1 could be in specifically routing GGPP molecules from the stroma to the membrane sites where carotenoid biosynthesis takes place (Fig. 6D). Alternatively, we also show that CPSFL1 is able to bind carotenoids such as phytoene and β-carotene, so it is conceivable that CPSFL1 facilitates transfer of hydrophobic carotenoids between biosynthetic enzymes, for example phytoene routing from PSY to PDS. These two mechanisms are not mutually exclusive and could explain the reduced phytoene synthesis observed in the cpsfl1 mutant background strains. Accordingly, a lack of CPSFL1 would result in a decreased flux into carotenoid biosynthesis by limiting delivery of GGPP to PSY or phytoene to PDS. Additionally, mutation of CPSFL1 might be expected to result in accumulation of GGPP (and possibly other precursors), leading to the feedback inhibition of earlier steps in isoprenoid biosynthesis as described above, which could further limit flux to phytoene.
A second proposed hypothetical mode of action for CPSFL1 would involve, directly or indirectly, transfer of carotenoids from their sites of synthesis in the chloroplast envelope to other destinations in the chloroplast, such as thylakoids, plastoglobules, and the eyespot (Fig. 6D). Direct transfer would be analogous to the function of α-TTP, which transfers α-tocopherol (28). Similar to CRALBP (56), PA binding could be a mechanism to trigger release of bound carotenoids from CPSFL1 to a target membrane. Alternatively, CPSFL1 might be involved in a vesicle transport pathway of carotenoids from the chloroplast envelope (57). Such a function would be consistent with our results on the cpsfl1 mutant of A. thaliana, which lacks chloroplast vesicles that are detectable in the WT (58). A defect in carotenoid transport could result in an accumulation of carotenoids or biosynthetic intermediates in the envelope, which could cause feedback inhibition of carotenoid biosynthesis as described above. This would manifest as a decreased rate of phytoene synthesis, as observed in cpsfl1. Thus, it is possible that the C. reinhardtii and A. thaliana CPSFL1 proteins have a conserved function in carotenoid transport. On the other hand, there are differences such as the binding of phosphatidylinositides by A. thaliana CPSFL1 (58) but not C. reinhardtii CPSFL1 (Fig. 2B) that might point to different functions. Testing these hypotheses will require the future development of suitable biochemical assays for delivery and/or transfer of carotenoids, GGPP, and other isoprenoid precursors.
Methods
C. reinhardtii Strains and Culture Conditions.
Cell cultures were grown in TAP liquid or solid (agar) media (59), heterotrophically (dark plus acetate), or mixotrophically (light plus acetate) at 25 °C. For photoautotrophic growth conditions, high-salt minimal (HSM) medium was used (60). The cpsfl1 mutant (CAL028.01.06) was generated by insertional mutagenesis of the 4A+ WT strain with linearized pBC1 plasmid encoding paromomycin resistance, as described previously (17). Other C. reinhardtii mutant strains used in this study are D1-less (fud7) (19) and the yellow-in-the-dark chlL mutant (61, 62). SiteFinding PCR (63) was performed to map the DNA insertion. Gamete autolysin preparation and mating and tetrad analyses were done as described in Harris (60). Mating and tetrad analyses of cpsfl1 mutant were performed between cpsfl1 (mating type +) and 4Ax5.2 (mating type −). The double-mutant chlL cpsfl1 was isolated from a mating and tetrad analysis between cpsfl1 (mating type −) and chlL (mating type +).
Complementation.
The C. reinhardtii complete gene model was predicted by using Augustus software (20). RNA isolation for RT-PCR and cDNA synthesis were performed as described (51). The complete CPSFL1 genomic DNA and CDS sequences annotations are found in SI Appendix, Dataset S2. Primers used in this study are listed in SI Appendix, Table S1. PCR products were cloned into pJET2.1 (CloneJET PCR Cloning Kit, ThermoFisher Scientific) and sequenced. Complementation of the cpsfl1 mutant was performed with either CPSFL1 genomic DNA (Cre10.g448051) or the complete annotated cDNA. Their constitutive gene expression was driven by the PSAD promoter and terminator of the pSL18 plasmid (64). Both constructs included a C-terminal FLAG-tag. Transformation of C. reinhardtii was performed by electroporation from logarithmic-phase grown cells resuspended with MAX Efficiency transformation medium and following manufacturer’s instructions (ThermoFisher Scientific) (65). Complemented lines were obtained by rescuing photoautotrophic growth in the presence of paromomycin (25 μg/mL) on solid (agar) HSM medium under constant light of 80-μmol photons m−2 s−1 for 2 wk.
Light Sensitivity and Photoinhibition Experiments.
Cell numbers were determined with Multisizer3 Coulter Counter (Beckman Coulter). If cells aggregated, cells were harvested by centrifugation with 1,000 × g, washed with water once, and pretreated with autolysin for 10 min prior to assessing their cells numbers. Cells were diluted in TAP liquid media and adjusted to the indicated cell concentrations. For light-sensitivity experiments in liquid, dark-grown cells were diluted to about 0.5 × 106 cells mL−1 in TAP media and then shifted to constant light for 48 h. For cell growth analyses on solid agar plates, cells were diluted with water to about 1 × 106 cells mL−1. Subsequent cell dilutions were spotted (5 μL) onto solid plates and growth was studied, either under photoautotrophic or mixotrophic conditions under the indicated light irradiance, or under heterotrophic conditions in the dark for 6 d. For photoinhibition studies, dark-grown cells were concentrated to about 1 × 107 cells mL−1 in liquid TAP and incubated either in complete darkness or shifted to HL (800-µmol photons m−2 s−1) for 1 h in a temperature-controlled (25 °C) HL chamber (Percival). TAP medium was supplemented with 100 μg/mL of CAP or equivalent volume of 100% ethanol. Whole-cell samples were collected at different time intervals for protein, pigment, and chlorophyll fluorescence analyses. For characterization of cell recovery from photoinhibition treatment, 5 μL of HL treated cells were spotted onto fresh TAP-agar plates and recovered in complete dark up to 48 h. The minimal fluorescence (Fo) and the PSII activity (66) were monitored by measuring the maximum quantum yield of PSII (Fv/Fm), determined after 15 min of dark acclimation, with a pulse-amplitude–modulated chlorophyll flurescence imaging system (MAXI-IMAGING-PAM, Heinz Walz).
Membrane Preparations and Protein Analyses.
C. reinhardtii cells were harvested by centrifugation at 2,500 × g for 10 min at 4 °C and resuspended in lysis buffer (20 mM Hepes-KOH pH 7.5, 5 mM MgCl2) supplemented with protease inhibitor mixture complete (Roche). Cells were disrupted by passage through a French Press (SLM Aminco) at 10K psi at 4 °C. Membranes were isolated from soluble samples by centrifugation at 20,000 × g for 10 min at 4 °C. Pelleted membranes were resuspended with TM buffer (20 mM Hepes-KOH pH 7.5, 300 mM Sorbitol, 10 mM KCl and 5 mM MgCl2). BN-PAGE analyses were performed as described (67). Briefly, one volume of membranes at a chlorophyll concentration of 1 mg mL−1 was resuspended in 25 mM BisTris pH 7.0 and 20% glycerol buffer and solubilized with equal volume of 2% n-dodecyl-β-d-maltoside (β-DM) detergent (Anatrace) for 10 min at 4 °C. Solubilized membrane protein complexes were centrifuge at 20,000 × g for 1 min at 4 °C, and the supernatant was resolved on a NativePAGE 4 to 16% Bis-Tris Protein Gel (1.0 mm, 10-well; ThermoFisher Scientific) on a first dimension. For a second SDS-denaturing dimension, a solution of 2% SDS, 1 M urea, 5% β-mercaptoethanol in in 100 mM Tris pH 6.8 was applied to the first-dimension native-PAGE slabs to denature protein complexes, which were subsequently resolved on Novex 10% Tris-Glycine Protein Gels (1.0 mm, 2D-well; ThermoFisher Scientific). Protein immunoblots analyses were performed as in García-Cerdán et al. (67). Denatured proteins were resolved on precast 10 to 20% gradient SDS/PAGE (ThermoFisher Scientific) and transferred to a polyvinylidene difluoride membrane (Immobilon-FL 0.45 μm; Millipore) via a tank electro-transfer system. Polyclonal antibodies against RbcL, D2, D1-DE loop, Cyt f, PsaA, and PSAD were obtained from Agrisera. The antibodies against acetylated tubulin and anti-FLAG were obtained from Sigma. Protein immunoblot signals were visualized by a Supersignal West Pico Chemiluminescent substrate detection system (Thermo Scientific) in a ChemiDoc MP imager and quantitated with the Image Lab v3.0 software (Bio-Rad). Several milligrams of recombinant purified CPSFL1 protein lacking the chloroplast transit peptide were used to generate specific rabbit polyclonal antibodies (ProSci). CPSFL1 cDNA coding for the protein lacking the N-terminal chloroplast transit peptide (amino acids 1 to 21) and containing an additional C-terminal FLAG epitope was PCR-amplified with primers oeCPSFL1 forward and oeCPSFL1 reverse. These primers included a forward NdeI and reverse BamHI restriction sites. PCR products were restriction enzyme digested and subcloned into the pET28 (a+) vector (Novagen). Protein expression and affinity purification under native conditions with Ni-NTA agarose (Qiagen) were performed as described (68). Purified proteins were concentrated with ultra4 centrifugal filters with 10-kDa cutoff (Amicon, MilliporeSigma), and protein concentrations were determined by using a nanodrop instrument with the calculated extinction coefficient (Protparam, Expasy) and by reading the absorbance at 280 nm. Fluorescent-labeled CPSFL1 proteins were generated with atto647N dye (Sigma-Aldrich) following the manufacturer’s directions for labeling. Unlabeled dye was removed by gel-filtration chromatography via a Superdex 200 column. Proteins were concentrated, aliquoted, and snap frozen. Protein purity was verified by SDS/PAGE and Coomassie staining analyses. For carotenoid binding analyses, recombinant CPSFL1 or GFP proteins were expressed and purified from carotenoid-producing E. coli BL21 (DE3) cells harboring pAC-PHYTO or pAC-BETA plasmids (37). Native purified His-tagged proteins were extracted with 100% acetone, and their carotenoid content was analyzed by high-performance liquid chromatography (HPLC).
Pigment Analyses.
Pigments were extracted and analyzed by HPLC as described previously (51). Briefly, pigments were extracted from whole cells by pipetting with 200 µL of pure acetone for 15 s. After centrifugation at 20,000 × g for 1 min, the supernatant was filtered through a 0.45-µM nylon filter and analyzed by reverse-phase HPLC with a C18 Spherisorb S5 ODS1 4.6 × 250-mm cartridge column (Waters) at 30 °C. The carotenoids and chlorophylls were identified by their absorbance at 445 and 296 nm using a diode array detector. For NFZ experiments, dark-grown cells (1 × 106 cells mL−1) were either supplemented with 3.5 μM of NFZ dissolved in ethanol or an equal volume of ethanol in control cultures. Treated cultures were grown as indicated for up to 48 h in the dark. Culture cell numbers were determined, and their pigments were extracted in 100% acetone and resolved by HPLC. A standard curve of known concentrations of each purified compound was used for calculating chlorophyll and carotenoid concentrations (51). Since no commercially purified phytoene was available to create a standard curve, phytoene levels were compared using peak areas derived from HPLC analysis divided by the 106 cell number. Pigment composition abbreviations are as follows: Chl, Chlorophyll; Neo, Neoxanthin; Lor, Loroxanthin; Vio, Violaxanthin; Ant, Antheraxanthin; Lut, Lutein; Zea, Zeaxanthin; α-Car, α-Carotene;β-Car, β-Carotene; α-Toc, α-Tocopherol.
Protein–Lipid Overlay Assay.
Membrane lipid arrays (Echelon Biosciences) were tested to determine specific lipid binding of recombinant purified CPSFL1 proteins lacking the chloroplast transit peptide by following the manufacturer’s instructions. Briefly, the strips were blocked with a buffer that contained 3% BSA fatty acid free (Sigma), 10 mM 2-(N-morpholino) ethanesulfonic acid (MES) pH 6.5, 150 mM NaCl, and 0.05% Tween-20 buffer. Incubation was performed for 1 h at room temperature. A total of 0.5 µg/mL of FLAG-tagged CPSFL1 proteins were incubated. A rabbit anti-FLAG epitope antibody (Sigma) at a dilution 1:5,000 was incubated for 1 h, washed, and immunostained with a horseradish peroxidase conjugated-secondary donkey anti-rabbit antibody at 1:20.000 and detected by using chemiluminiscence substrate solution (ThermoFisher Scientific). Membrane-spotted lipid abbreviations are as follows: CHL, cholesterol; CL, cardiolipin; DAG, 1,2-diacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PI, phoshatidylinositol; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI- (4)P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; PS, phosphatidylserine; SGC(3)P, 3-sulfogalactosylceramide; SPH, sphingomyelin; TG, triglyceride.
Preparation and Microscopy Analysis of GUVs.
GUVs were prepared by electroformation (69). To ensure mixing of all lipid components, we performed electroformation at ∼55 °C. Vesicles were electroformed in solution containing ∼350 mM sucrose (∼350 mOsm). Lipid composition was either 99.7% DOPC, 0.3% Atto 390-DOPE or 69.7% DOPC, 30% POPA, 0.3% Atto 390-DOPE. After electroformation, 1 μL GUV solution was gently transferred to a home-made polydimethylsiloxane (Sylgard) chamber containing 100 μL of 100 nM protein solution (in 25 mM Hepes buffer with 150 mM NaCl, and 1 mM DTT, osmotically balanced to 320 mOsm). The mixture was incubated on room temperature for 10 min and imaged immediately thereafter. GUVs were imaged on a spinning-disk confocal microscope (AxioObserverZI, Zeiss, with motorized nosepiece and spinning disk confocal, Yokogawa CSU-10) for confocal microscopy with a cooled EMCCD camera (Cascade II, Photometrics). Images were acquired using a 63× objective (Zeiss, Plan Apochromat 1.4 NA, oil), and analyzed using ImageJ (National Institutes of Health) and Matlab (Mathworks). To quantify the relative proportion of proteins at membrane interfaces containing both binding and nonbinding proteins, we measured fluorescence intensity along a line bisecting the GUV–GUV pair and calculated an enrichment index, the intensity ratio between the interface and the sum of the individual vesicle intensities. Enrichment index values greater than 1 indicate an enrichment of molecules (increased surface density, #/µm2) at the membrane interface.
TEM and FIB-SEM.
For TEM cell fixation, 7 mL of cell culture was first supplemented with 1 mL 8% glutaraldehyde and then put on a rocker for 1 min. After the addition of another 1 mL of aqueous 2% osmium tetroxide and 1.5% potassium ferricyanide, cells were rocked for 7 min. Cells were then spun down at 1,500 × g for 1 min, supernatant containing fixatives was discarded, and the pellet containing the cells was washed three times with distilled water. The final resuspended pellet was warmed to 37 C for 5 min, complemented with 1 volume of 2% low melting agarose at 37 C, centrifuged at 20,000 × g for 10 s, and transferred to 4°C until processing. After three washes with distilled water, the cell pellet was cut into pieces (1 mm3), dehydrated using increasing concentrations of acetone for 5 min (25%, 35%, 50%, 70%, 75%, and 95%), and left in pure acetone for 15 min. For infiltration, cell pellet was incubated for 15 min with increasing concentrations of 25%, 50%, and 75% acetone/epon resin mixture. Cells were sedimented at 2,500 × g between changes followed by a final three changes for 10 min in pure Epon resin. Finally, samples were placed in BEEM capsules for polymerization at 60 °C for 2 d. Ultrathin sections (60 to 70 nm) were cut with a diamond knife (Diatome, type ultra 358) on an EM UC6 ultramicrotome (Leica) and mounted on single-slot Pioloform coated copper grids (Plano). Sections were stained with uranyl acetate and lead citrate (70) and viewed with an EM 902A (Carl Zeiss) TEM (both operated at 80 kV). Micrographs were taken using a 4k × 4k or 1,350 × 1,040-pixel camera (UltraScan 4000 or Erlangshen ES500W, respectively; Gatan) and Gatan Digital Micrograph software (v1.70.16). Image brightness and contrast were adjusted, and figures assembled using Adobe Photoshop 8.0.1. For FIB-SEM visualization, cell cultures were pelleted and then fixed with glutaraldehyde (1%). After fixation, the cells were poststained using potassium ferrocyanide, osmium tetraoxide, and uranyl acetate and then postfixed again with glutaraldehyde. Before resin embedding in hard Durcupan resin, the samples were dehydrated with rinses of ethanol and acetone. Resin-embedded samples were initially screened by TEM to select the samples for FIB-SEM data collection. Each embedded sample was sectioned using a Leica EM UC 7 Ultramicrotome.
Next, 70-nm-thick sections were deposited onto nitrocellulose-coated copper grids for visualization with a Tecnai 12 TEM equipped with a TVIPS TemCam F416 camera. Once well-preserved samples with the best contrast were selected, the targeted regions were trimmed into block faces of 200 × 200 µm2 or less and mounted onto a metal stud for FIB-SEM imaging. A thin layer of 10-nm gold followed by 100 nm of carbon was used to coat the mounted sample. FIB-SEM imaging of both WT and mutant samples was performed as described (36). A customized Zeiss NVision40 FIB-SEM system with a 3-nA electron beam of 1.5-keV landing energy at 1.25 MHz was used to acquire raw images at 8 × 8 × 2-nm3 voxel resolution. The samples were biased at +400V to filter out secondary electrons. The initial images were then aligned and binned by a factor of 4 along the z-axis to form final 3D stacks of 8 × 8 × 8-nm3 isotropic voxel resolution. Total volumes of 60 × 30 × 73 µm3 for WT and 60 × 30 × 44 µm3 for the cpsfl1 mutant cells were imaged, containing 111 and 81 whole cells, respectively. The resulting 3D density data were further scaled down to 32 × 32 × 32-nm3 resolution for visual inspection of the cell internal features in 3dmod (IMOD) and to select the coordinates of a few representative cells to be extracted at 8-nm resolution. Those cells were then segmented and further analyzed using the Amira software (FEI). The appropriate density range was determined to visualize the organelle of interest in Amira. The organelle was then semiautomatically (“magic wand” tool) or manually (“brush” tool) selected plane by plane until the entirety of the organelle in the data were segmented and added to the model. For organelle densities that were denser and more continuous, only one of every five slices was highlighted, and the interpolation tool was used in between. Once the segmentation was finished, each organelle volume was smoothed in Amira (four passes) before generating the final model surfaces used for visualization and movie recording.
Data Availability.
All data presented in the paper are available in the main text and SI Appendix. Biological materials are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Erika Erickson for critical reading of the manuscript. This work was supported by the US Department of Energy, Office of Science, through the Photosynthetic Systems program in the Office of Basic Energy Sciences. T.T.’s contribution was supported by a grant from the National Science Foundation (MCB-1515169) to Christoph Benning and by the Plant Biotechnology for Health and Sustainability Training Program at Michigan State University (NIH T32 GM110523). E.N. and K.K.N. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916948117/-/DCSupplemental.
References
- 1.Wise R. R., Hoober J. K., The Structure and Function of Plastids (Springer, Dordrecht, 2006), vol. xl. [Google Scholar]
- 2.Ruiz-Sola M. A., Rodríguez-Concepción M., Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book 10, e0158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Sola M. A., et al. , Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 209, 252–264 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Beisel K. G., et al. , Continuous turnover of carotenes and chlorophyll a in mature leaves of Arabidopsis revealed by 14CO2 pulse-chase labeling. Plant Physiol. 152, 2188–2199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H., Huang D., Cramer W. A., Stoichiometrically bound beta-carotene in the cytochrome b6f complex of oxygenic photosynthesis protects against oxygen damage. J. Biol. Chem. 274, 1581–1587 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Bassi R., Pineau B., Dainese P., Marquardt J., Carotenoid-binding proteins of photosystem II. Eur. J. Biochem. 212, 297–303 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Baroli I., Niyogi K. K., Molecular genetics of xanthophyll-dependent photoprotection in green algae and plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1385–1394 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramel F., et al. , Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158, 1267–1278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank H. A., Cogdell R. J., Carotenoids in photosynthesis. Photochem. Photobiol. 63, 257–264 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Niyogi K. K., Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Austin J. R. 2nd, Frost E., Vidi P. A., Kessler F., Staehelin L. A., Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18, 1693–1703 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreimer G., The green algal eyespot apparatus: A primordial visual system and more? Curr. Genet. 55, 19–43 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Ferro M., et al. , AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyard J., et al. , Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2, 1154–1180 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Dent R. M., Han M., Niyogi K. K., Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii. Trends Plant Sci. 6, 364–371 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Rochaix J. D., Assembly, function, and dynamics of the photosynthetic machinery in Chlamydomonas reinhardtii. Plant Physiol. 127, 1394–1398 (2001). [PMC free article] [PubMed] [Google Scholar]
- 17.Dent R. M., Haglund C. M., Chin B. L., Kobayashi M. C., Niyogi K. K., Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137, 545–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dent R. M., et al. , Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82, 337–351 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Bennoun P., et al. , Characterization of photosystem II mutants of Chlamydomonas reinhardii lacking the psbA gene. Plant Mol. Biol. 6, 151–160 (1986). [DOI] [PubMed] [Google Scholar]
- 20.Stanke M., Diekhans M., Baertsch R., Haussler D., Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008). [DOI] [PubMed] [Google Scholar]
- 21.de Castro E., et al. , ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchler-Bauer A., et al. , CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panagabko C., et al. , Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42, 6467–6474 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Sha B., Phillips S. E., Bankaitis V. A., Luo M., Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh R., Bankaitis V. A., Phosphatidylinositol transfer proteins: Negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors 37, 290–308 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Saito K., Tautz L., Mustelin T., The lipid-binding SEC14 domain. Biochim. Biophys. Acta 1771, 719–726 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Kono N., et al. , Impaired α-TTP-PIPs interaction underlies familial vitamin E deficiency. Science 340, 1106–1110 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Schmid E. M., et al. , Size-dependent protein segregation at membrane interfaces. Nat. Phys. 12, 704–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rast A., Heinz S., Nickelsen J., Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847, 821–830 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Phillips S. E., et al. , Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell 4, 187–197 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Herrin D. L., Battey J. F., Greer K., Schmidt G. W., Regulation of chlorophyll apoprotein expression and accumulation. Requirements for carotenoids and chlorophyll. J. Biol. Chem. 267, 8260–8269 (1992). [PubMed] [Google Scholar]
- 33.Cahoon A. B., Timko M. P., Yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12, 559–568 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki J. Y., Bauer C. E., Light-independent chlorophyll biosynthesis: Involvement of the chloroplast gene chlL (frxC). Plant Cell 4, 929–940 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohad I., Siekevitz P., Palade G. E., Biogenesis of chloroplast membranes. II. Plastid differentiation during greening of a dark-grown algal mutant (Chlamydomonas reinhardi). J. Cell Biol. 35, 553–584 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu C. S., et al. , Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6, e25916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham F. X. Jr, Gantt E., A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth. Res. 92, 245–259 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Cunningham F. X., Gantt E., Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 557–583 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Chamovitz D., Sandmann G., Hirschberg J., Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 268, 17348–17353 (1993). [PubMed] [Google Scholar]
- 40.Bermúdez L., et al. , A tomato tocopherol-binding protein sheds light on intracellular α-tocopherol metabolism in plants. Plant Cell Physiol. 59, 2188–2203 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Savage L. J., Imre K. M., Hall D. A., Last R. L., Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS One 8, e73291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santabarbara S., et al. , The requirement for carotenoids in the assembly and function of the photosynthetic complexes in Chlamydomonas reinhardtii. Plant Physiol. 161, 535–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tóth T. N., et al. , Carotenoids are essential for the assembly of cyanobacterial photosynthetic complexes. Biochim. Biophys. Acta 1847, 1153–1165 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Dong H., et al. , The Arabidopsis spontaneous cell death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 17, 458–470 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Qin G., et al. , Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Schmidt M., et al. , Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18, 1908–1930 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cazzonelli C. I., Pogson B. J., Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15, 266–274 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Rodríguez-Villalón A., Gas E., Rodríguez-Concepción M., Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 60, 424–435 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Trebst A., Depka B., Role of carotene in the rapid turnover and assembly of photosystem II in Chlamydomonas reinhardtii. FEBS Lett. 400, 359–362 (1997). [DOI] [PubMed] [Google Scholar]
- 50.McCarthy S. S., Kobayashi M. C., Niyogi K. K., White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 168, 1249–1257 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran P. T., Sharifi M. N., Poddar S., Dent R. M., Niyogi K. K., Intragenic enhancers and suppressors of phytoene desaturase mutations in Chlamydomonas reinhardtii. PLoS One 7, e42196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Min K. C., Kovall R. A., Hendrickson W. A., Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency. Proc. Natl. Acad. Sci. U.S.A. 100, 14713–14718 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tahotna D., Holic R., Poloncova K., Simockova M., Griac P., Phosphatidylcholine transfer activity of yeast Sec14p is not essential for its function in vivo. Biochim. Biophys. Acta 1771, 83–92 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Vieler A., Wilhelm C., Goss R., Süss R., Schiller J., The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids 150, 143–155 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Intres R., Goldflam S., Cook J. R., Crabb J. W., Molecular cloning and structural analysis of the human gene encoding cellular retinaldehyde-binding protein. J. Biol. Chem. 269, 25411–25418 (1994). [PubMed] [Google Scholar]
- 56.Saari J. C., Nawrot M., Stenkamp R. E., Teller D. C., Garwin G. G., Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Mol. Vis. 15, 844–854 (2009). [PMC free article] [PubMed] [Google Scholar]
- 57.Khan N. Z., Lindquist E., Aronsson H., New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: An Arabidopsis model. PLoS One 8, e59898 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hertle A. P., et al. , A Sec14 domain protein is required for photoautotrophic growth and chloroplast vesicle formation in Arabidopsis thaliana. 10.1073/pnas.1916946117 (3 April 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorman D. S., Levine R. P., Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris E. H., The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use (Academic Press, San Diego, CA, 1989). [DOI] [PubMed] [Google Scholar]
- 61.García-Cerdán J. G., et al. , A thylakoid membrane-bound and redox-active rubredoxin (RBD1) functions in de novo assembly and repair of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 116, 16631–16640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gutman B. L., “DNA repair in the chloroplast”, PhD thesis, University of California, Berkeley, California: (2007). [Google Scholar]
- 63.Tan G., et al. , SiteFinding-PCR: A simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 33, e122 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park S., Khamai P., Garcia-Cerdan J. G., Melis A., REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143, 1547–1560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kindle K. L., High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maxwell K., Johnson G. N., Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51, 659–668 (2000). [DOI] [PubMed] [Google Scholar]
- 67.García-Cerdán J. G., et al. , The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65, 368–381 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Kirst H., Garcia-Cerdan J. G., Zurbriggen A., Ruehle T., Melis A., Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol. 160, 2251–2260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmid E. M., Richmond D. L., Fletcher D. A., Reconstitution of proteins on electroformed giant unilamellar vesicles. Methods Cell Biol. 128, 319–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reynolds E. S., The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in the paper are available in the main text and SI Appendix. Biological materials are available from the corresponding authors upon request.