Significance
Transposable elements (TEs) are genomic parasites that are found in all genomes. Here, we show that viral infections impact TE transcript amounts in Drosophila somatic tissues. This is of major importance in the understanding of the tempo and mode of genetic diversification, suggesting that viral infections act as a significant factor determining TE activity. In addition, our results also suggest that the amounts of the maternally transmitted small RNAs that control TEs in the progeny may also be altered, which could have long-term, evolutionary impacts.
Keywords: transposon, piRNA, siRNA, SINV, insect
Abstract
Transposable elements (TEs) are genomic parasites that are found in all genomes, some of which display sequence similarity to certain viruses. In insects, TEs are controlled by the Piwi-interacting small interfering RNA (piRNA) pathway in gonads, while the small interfering RNA (siRNA) pathway is dedicated to TE somatic control and defense against viruses. So far, these two small interfering RNA pathways are considered to involve distinct molecular effectors and are described as independent. Using Sindbis virus (SINV) in Drosophila, here we show that viral infections affect TE transcript amounts via modulations of the piRNA and siRNA repertoires, with the clearest effects in somatic tissues. These results suggest that viral acute or chronic infections may impact TE activity and, thus, the tempo of genetic diversification. In addition, these results deserve further evolutionary considerations regarding potential benefits to the host, the virus, or the TEs.
The genomes of virtually all organisms contain genomic parasites called transposable elements (TEs) (1–4). They are mainly vertically transmitted and, along the branches of the tree of life, they display variable amounts and activities. TEs constitute 15% of Drosophila melanogaster’s genome (5), at least half of Homo sapiens’s (6), and up to 85% of maize’s (7). Among TEs, endogenous retroviruses (ERVs) share features with some viruses, like sequences and life cycles (8). Most of the times, TE insertions are neutral or slightly deleterious (9–12). Certain are known to be associated with diseases, because they disrupt coding or regulatory sequences (13, 14). Nevertheless, TE sequences are occasionally recruited in adaptive processes (15). Different factors have been identified that trigger TE mobilization. The most spectacular and best understood is hybrid dsygenesis (16, 17). Environmental changes and abiotic stress are also frequently evoked (18–21), while biotic factors are more scarcely reported (22). Nevertheless, the list of TE mobilization stimuli is not exhaustive, and the mechanisms are still poorly understood.
The potentially harmful activity of TEs is silenced at the transcriptional and posttranscriptional levels by the Piwi-interacting small interfering RNAs (piRNAs). In Drosophila, piRNAs are 23- to 30-nt-long single-stranded RNA molecules that recognize TE transcripts by sequence complementarity and trigger their slicing due to the RNase activity of the PIWI proteins they are associated with. Some of these loaded piRNAs also translocate into the nucleus and induce the heterochromatic silencing of the corresponding TE sequences (23–25). The so-called secondary piRNA pathway involves the Ago3 and Aub proteins and displays the characteristic “ping-pong signature,” which corresponds to an enrichment in 10-nt overlaps between sense and antisense piRNAs (26). In Drosophila, piRNAs are restricted to gonads (27); however, somatic piRNAs are often described (28) (and even with evidence of efficient ping-pong production; ref. 26), but are potentially restricted to certain cell types or tissues (for instance, somatic piRNAs are found in D. melanogaster heads, ref. 29; but not in thorax; ref. 27). In somatic tissues, TEs are also controlled by small interfering RNAs (siRNAs), a distinct class of small interfering RNAs (30–34). siRNAs are 21-nt-long single-stranded RNA molecules, which are processed by Dicer-2 from double-stranded RNAs (dsRNAs). These siRNAs are then loaded onto the Ago2 protein, which cleaves RNA fragments sharing sequence complementarity to the siRNAs. Additionally, the siRNA pathway is the first line of immune defense against viruses in insects (35–37). Considering that they rely on distinct molecular effectors, the piRNA and siRNA pathways are described as independent. However, another connection between TE control and antiviral immunity has recently been discovered: Upon viral infection, the reverse transcriptase of certain TEs make DNA copies of RNA viruses, which then boosts the production of antiviral siRNAs and allows a stronger immune response (38, 39).
Here, we investigated the impacts of viral infections on TE activity. We hypothesize that there is 1) either a trade-off between TE control and antiviral immunity, 2) or a synergistic interaction between both. According to the trade-off scenario, a given organism can either control the activity of its TEs or fight against viruses but cannot do both well at the same time. This first hypothesis is suggested by the previous observation that the siRNA productor Dicer-2 may end up saturated (40). Accordingly, there should exist cases in which the siRNA pathway cannot handle antiviral immunity and TE control at the same time, resulting in a trade-off between both processes. On the contrary, according to the synergistic scenario, an organism displaying strong TE control would also display strong antiviral immunity, and inversely, an organism displaying weak TE control would also display weak antiviral immunity. This second hypothesis is hinted at by results suggesting that the siRNA pathway may cooperate with the piRNA pathway for the control of TEs (41).
We used an experimental system made of Makindu, a Drosophila simulans WT strain that we previously deeply characterized for its TEs (42–44), which we infected with Sindbis virus (SINV). SINV is an arbovirus of the Togaviridae family, which naturally infects mosquitoes (45). It has a single-stranded RNA genome of positive polarity. While it is not a natural pathogen of Drosophila, SINV is frequently used for experimental infections in flies and brings the advantage that the fly genome is totally naive to it. Indeed, we do not expect SINV sequences to be integrated within the fly genome and, therefore, to be involved in the production of endogenous small RNAs. While many viruses are known to encode suppressors of RNA interference (RNAi) (35, 36, 46), no such proteins were reported for SINV so far (47, 48). Here, we provide observation that TE transcript amounts are modulated upon viral infection. This modulation is associated with changes in TE-derived small RNA repertoires and was further investigated using D. melanogaster mutants. This work identifies viral infections as a biotic factor, which TE activity is sensitive to. It suggests that viral infections may affect TE mobilization rates and, thus, the speed of somatic genetic diversification as well as that of genome evolution. In addition, these results deserve further evolutionary considerations regarding potential benefits to the host, the virus, or the TEs.
Results
SINV Replicates and Triggers an siRNA Response in the D. simulans Makindu Strain.
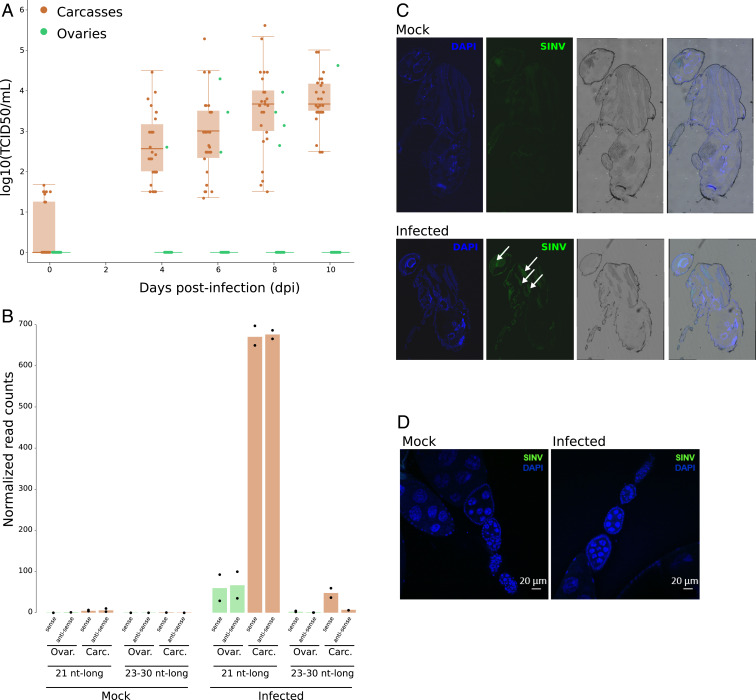
We infected female adults with SINV by intrathoracic injections, and we assessed SINV replication in Makindu through TCID50 assays. In carcasses (whole bodies without ovaries), SINV titers increased from 2.6 to 3.7 log10 (TCID50/mL) between day 4 to day 10 post infection (dpi) (Fig. 1A). We did not observe fly mortality induced by SINV infection (96% vs. 98% mortality at 10 dpi). Importantly, we detected the production of SINV-derived 21-nt RNAs, equally targeting the sense and antisense viral strands, as expected (35) (Fig. 1B). Using an anti-SINV capsid antibody, we could clearly detect SINV in thorax (Fig. 1C). However, in ovaries, the tissue where the piRNA pathway is described to control TEs in Drosophila, SINV barely replicated: We found SINV infectious particles in the ovaries of only a few infected flies (3 of 30 at 6 dpi; Fig. 1A), and we could not detect SINV capsid protein in our immunofluorescent assays (Fig. 1D). Nevertheless, we detected SINV-derived 21-nt RNAs in ovaries (Fig. 1B), consistent with the systemic spread of the antiviral RNAi response (49). However, we cannot completely rule out the presence of somatic tissue from carcasses in the samples used for sequencing.
Fig. 1.
SINV replicates in D. simulans Makindu flies and triggers siRNA production. (A) Boxplots for viral titration assessed by TCID50 assays in ovaries (green) and carcasses (brown). (B) SINV-derived small RNA production as assessed from small RNA-seq data (biological duplicates). As expected, upon SINV infection, SINV-derived 21-nt-long small RNAs are detected in carcasses, and in ovaries to a lesser extent. (C and D) SINV tropism as revealed by an anti-capsid antibody (green labeling, white arrows). (C) SINV is mainly detected in thorax. (D) SINV is not detected in ovaries.
SINV Infection Modulates TE Transcript Amounts and TE-Derived Small RNAs in the D. simulans Makindu Strain.
In order to assess TE transcriptional activity and control, we decided to perform RNA-sequencing (seq) and small RNA-seq from samples extracted at 6 dpi, which corresponds to the exponential phase of viral replication in carcasses (Fig. 1A). Samples were produced from ovaries and carcasses. As previously reported (50), we used a small RNA preparation protocol that ensures the isolation of small RNAs involved in nucleoproteic complexes and, therefore, excludes mRNA degradation products. The strategy is based on the adsorption and reversible binding of negatively charged biomolecules to a positively charged insoluble matrix. RISC (RNA induced silencing complexes) can thus be recovered using mild salt concentration elution, whereas contaminant RNAs (such as ribosomal RNAs or degradation products) remain fixed to the column (50). We confirmed that this protocol only provided us with 20- to 31-nt-long RNA molecules (SI Appendix, Fig. S1), which is the expected size for piRNAs, siRNAs, and micro RNAs (miRNAs). The nucleotide composition of the reads displayed the expected patterns (SI Appendix, Fig. S2). The very low proportions of sequences mapping to protein coding genes among our small RNA pools also discard the presence of mRNA degradation products (SI Appendix, Table S3). We then separated reads corresponding to 23- to 30-nt-long RNAs and 21-nt-long RNAs and considered them to be piRNAs and siRNAs, respectively. Small RNA read counts were normalized relative to miRNA amounts (Methods).
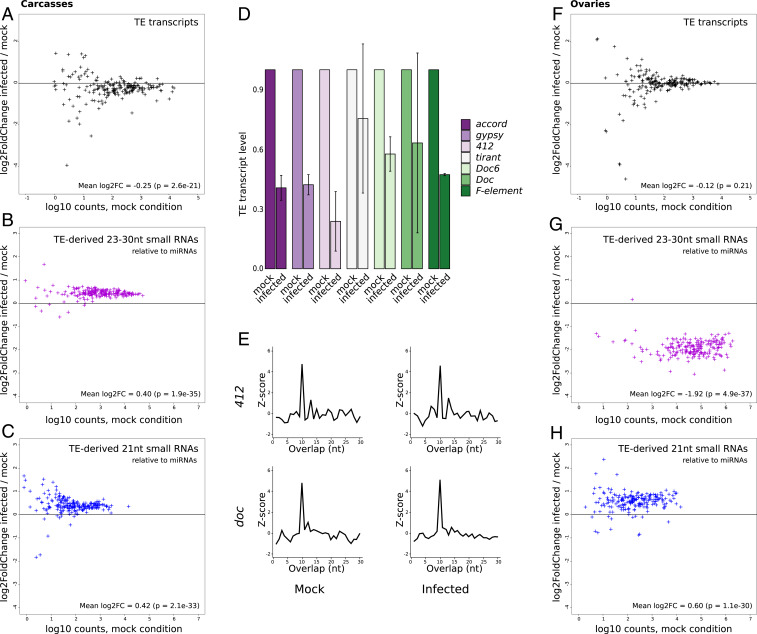
Here, the analysis of 237 annotated TE families and subfamilies revealed a significant global reduction of TE transcript read counts in carcasses upon infection compared to the control samples (mock) (15% mean decrease, Fig. 2A). These results were confirmed using RT-qPCR (Fig. 2D). All TE classes behaved in the same way (Kruskal–Wallis test; P = 0.57; SI Appendix, Fig. S4). This reduction may appear modest; however, it is larger than the modulation observed for immune genes (SI Appendix, Fig. S5). The global decrease in TE transcript amounts was associated with a significant increase in TE-derived 23- to 30-nt (32% mean increase, Fig. 2B) and 21-nt-long RNAs (34% mean increase, Fig. 2C). We could observe the 1U enrichment expected for piRNAs when we analyzed the 23- to 30-nt small RNAs that aligned against TEs (SI Appendix, Fig. S6), and, interestingly, ping-pong signatures were found for TE-derived 23- to 30-nt RNAs in carcasses (Fig. 2E and SI Appendix, Figs. S7 and S8). We statistically confirmed that the TE families that display transcript amount decreases upon infection are significantly enriched in TE families that also display small RNA amount increases (Fisher exact tests; P = 0.001 and 0.030 for 21-nt and 23- to 30-nt small RNAs, respectively, SI Appendix, Fig. S9).
Fig. 2.
SINV infection induces TE modulation in D. simulans Makindu flies. (A and F) TE transcript modulation upon SINV infection, expressed as log2 of the ratio (counts in infected condition/counts in mock condition) for each annotated TE family, in carcasses and ovaries, respectively. TE-derived 23- to 30-nt small RNA modulation upon SINV infection, in carcasses and ovaries, respectively (B and G), and TE-derived 21-nt small RNA modulation upon SINV infection, in carcasses and ovaries, respectively (C and H). Small RNA amounts were normalized relative to miRNAs. (D) TE transcript levels, as assessed by RT-qPCR, are expressed as fold changes relative to rp49 transcript levels. The majority of TE families show a reduction of transcript amounts upon infection. Error bars are SDs. (E) Ping-pong signatures for 23- to 30-nt RNAs in Makindu carcasses. Significant enrichment in 10-nt overlaps (i.e., ping-pong signatures) could be observed, as illustrated here by the 412 and doc families.
On the contrary, in ovaries, no significant shift in TE transcript amounts upon infection could be detected (8% mean decrease, Fig. 2F). However, we found that TE-derived small RNAs were largely modulated: TE-derived 23- to 30-nt small RNAs were less abundant (74% mean decrease, Fig. 2G) while TE-derived 21-nt small RNA amounts increased (53% mean increase; Fig. 2H). This mirror-like response between the two siRNA classes may explain the resulting absence of modulation of TE transcript amounts.
Altogether, our results show that TE transcripts and TE-derived small RNAs are modulated upon viral infection in Drosophila. However, our RNA-seq analysis also revealed that the Makindu strain was chronically infected by Nora virus (SI Appendix, Fig. S10), which may interfere with SINV immune response. Therefore, in order to control for chronic infections, and go deeper into the mechanisms allowing such a modulation in TE transcript amounts upon infection, we decided to switch to the D. melanogaster model, which has available a wealth of mutants. We decided to bleach the embryos from all D. melanogaster strains used thereafter to remove viral particles resulting from chronic infections (51).
The dsRNA Uptake Pathway Is Involved in TE-Derived Small RNA Modulation in Carcasses.
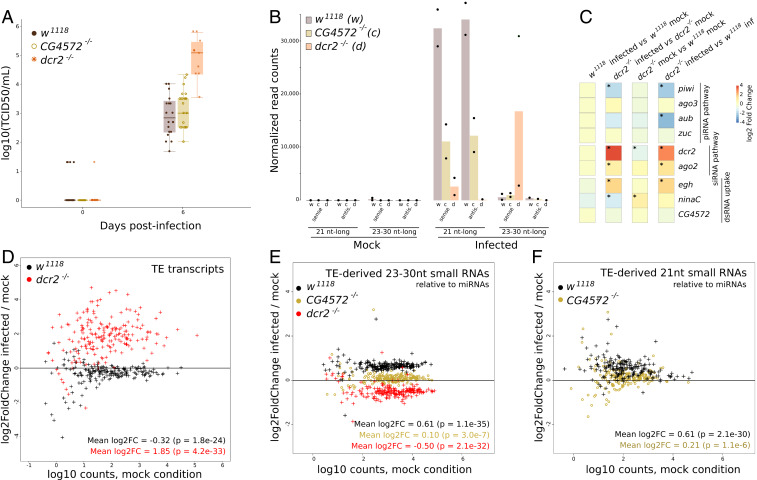
We infected w1118 adult females with SINV by intrathoracic injections and found results comparable to those we observed using Makindu carcasses, although using a smaller infectious viral dose (2,300 plaque-forming units [pfu]). Similar to what we observed in Makindu, SINV replicated in carcasses (2.83 log10 [TCID50/mL]) at 6 dpi, Fig. 3A) and triggered the production of virus-derived 21-nt RNAs (Fig. 3B). Genes of the siRNA pathway were not differentially transcribed in w1118 upon infection (Fig. 3C), suggesting that this machinery is constitutively produced and immediately efficient against the virus. Similar to what was reported by Petit et al. (52), SINV-derived 23- to 30-nt small RNAs were virtually absent in w1118 upon infection (Fig. 3B). TE transcript amounts decreased upon infection (20% mean decrease, Fig. 3D, black dots), and this TE modulation was associated with an increase in TE-derived 23- to 30-nt small RNAs (53% mean increase, Fig. 3E, black dots) and 21-nt small RNAs (52% mean increase, Fig. 3F, black dots).
Fig. 3.
SINV infection in D. melanogaster carcasses. (A) Boxplots of viral titration assessed by TCID50 assays in the three strains. (B) SINV-derived small RNAs, as assessed from small RNA-seq data (biological duplicates). (C) Modulation of transcript levels for a subset of genes involved in the piRNA, siRNA, and dsRNA uptake pathways, as assessed by DESeq2. Significant shifts (adjusted P < 0.05) are indicated by asterisks. (D) TE transcript modulation upon infection expressed as log2 of the ratio (counts in infected condition/counts in mock condition) for each annotated TE family. (E) TE-derived 23- to 30-nt small RNA modulation upon infection. (F) TE-derived 21-nt small RNA modulation upon infection. Small RNA amounts were normalized relative to miRNAs.
It has been previously demonstrated that the viral response through siRNAs depends on the systemic spread of dsRNA viral intermediates, which triggers the production of siRNAs (49). The systemic spread relies on the dsRNA uptake pathway, of which CG4572 is an effector (49, 53). We used the CG4572c05963 mutation that was obtained from a w1118 genetic background (49); therefore, our CG4572−/− strain harbors the same pool of TE insertions as w1118. In this mutant strain, the SINV titers obtained at 6 dpi are similar to what we observed in w1118 flies (3 log10 [TCID50/mL], Fig. 3A). The analysis revealed that the modulation in TE-derived 23- to 30-nt and 21-nt small RNAs upon infection virtually disappeared in the CG4572−/− mutant carcasses, compared to w1118 (7% mean increase, and 16% mean increase, respectively, Fig. 3 E and F, brown dots). This indicates that the TE-derived small RNA modulation in carcasses relies on the dsRNA uptake pathway.
The Production of siRNAs Is Involved in the TE Modulation Observed in Carcasses.
We next investigated the consequences of the impairment of siRNA production, using a mutant of dcr2, which is responsible for siRNA formation. As expected, SINV replicated more abundantly in this mutant, reaching 5.27 log10 (TCID50/mL) at 6 dpi (Fig. 3A). Notably, this abundant SINV replication was associated with a high production of SINV-derived 23- to 30-nt small RNAs (SI Appendix, Fig. S11), however only in the sense orientation (Fig. 3B). These 23- to 30-nt small RNAs thus cannot be involved in a ping-pong loop (SI Appendix, Fig. S12). In the dcr2−/− genotype, SINV infection led to an increase in TE transcript amounts (261% mean increase, Fig. 3D, red dots), and a decrease in TE-derived 23- to 30-nt RNAs (29% mean decrease, Fig. 3E, red dots). This therefore indicates that the siRNA pathway is involved in the observed pattern. We propose that the reduction in TE-derived 23- to 30-nt RNAs is due to the piRNA machinery being saturated by the particularly abundant viral RNAs. In addition, in the infection context, we observed that the dcr2−/− mutant also displayed a significant repression of the piwi gene (Fig. 3C; log2 fold change = –2.12; P = 0.001), associated with a strong induction of dcr2 transcription (which, however, does not lead to more Dcr-2 function since the coding sequence is mutated) compared to w1118 (Fig. 3C; log2 fold change = 2.80; P = 6.6e−34).
In addition, we note that while the general trend for 23- to 30-nt small RNA is a strong decrease upon infection in the dcr2−/− mutant (Fig. 3E), 11 TE families (over 237) display a positive log2 fold change. Consistently with observations by others (41, 54–56), this suggests that the control of the different TE families depends in a variable way on the different small RNA pathways.
In order to test further the involvement of siRNA production, we retrieved an already published RNA-seq dataset, corresponding to D. melanogaster flies infected with the Drosophila C virus (DCV) (57). DCV belongs to the Dicistroviridae family; it is a natural pathogen of D. melanogaster, and its genome is made of single-stranded RNA of positive polarity. DCV encodes an RNAi suppressor that blocks Dcr-2 (36). Merkling et al. produced RNA-seq data in mock and infected conditions, from whole flies or only fat bodies, and in a w1118 genetic background either or not mutated for the gene encoding the G9a H3K9 methyltransferase (57). We will not discuss further data from the mutant strain in the present study. Similar to what we did above, focusing the analysis on TE sequences only, we found that DCV infection leads to an increase in TE transcript amounts in the w1118 background (SI Appendix, Fig. S13). The largest effect is observed in fat bodies (65% mean increase) compared to whole flies (21% mean increase). Although there are significant differences in the experimental setups (particularly since DCV may not fully suppress RNAi), we found that these results using DCV in WT flies are similar to those we obtained using dcr2−/− flies infected with SINV, i.e., a strong increase in TE transcript amounts.
D. melanogaster Ovaries Are Protected Against TE Transcript Modulation.
In accordance with our observations in Makindu, SINV barely replicated in w1118 and CG4572−/− ovaries (SI Appendix, Fig. S14). However, viral particles were detected in dcr2−/− (SI Appendix, Fig. S14A). Although viral particles did not enter ovaries (SI Appendix, Fig. S15), it has to be noted that we could observe the production of SINV-derived 21-nt RNAs in this tissue (SI Appendix, Fig. S14B). Like carcasses, ovaries did not display differential transcription of genes of the siRNA pathway nor the piRNA pathway (SI Appendix, Fig. S14C). Like Makindu, w1118 ovaries barely displayed TE transcript modulation (6% mean decrease, SI Appendix, Fig. S14D, black dots). However, w1118 was clearly different from Makindu because TE-derived small RNA amounts were virtually not modulated upon infection (15% mean decrease for 23- to 30-nt small RNAs, 8% mean decrease for 21-nt small RNAs, SI Appendix, Fig. S14 E and F, black dots), which contrasted with Makindu patterns (Fig. 2 G and H). CG4572−/− and w1118 log2 fold change patterns largely overlapped (SI Appendix, Fig. S14 E and F), suggesting that the dsRNA uptake pathway is not involved in TE-derived small RNA patterns in ovaries. On the contrary, TE-derived 23- to 30-nt RNA amounts increased in the dcr2−/− mutant (19% mean increase, SI Appendix, Fig. S13E, red dots).
Discussion
Viral Infection Impacts TE-Derived Small RNA Amounts in D. simulans Ovaries.
In both D. melanogaster and D. simulans ovaries, we could not detect variation in TE transcript amounts upon infection. This suggests that there are strong selective pressures to maintain TE transcriptome homeostasis in the ovaries. Indeed, this is where the genetic material for the next generation lies.
Nevertheless, what is very interesting from an evolutionary perspective and deserves future investigation is that while TE transcript amounts were virtually stable in ovaries upon infection in both Makindu and w1118, TE-derived small RNAs showed completely different patterns: strong mirror-like pattern in Makindu versus virtually no change in w1118. This suggests that the observed maintenance of TE transcriptome stability in ovaries is achieved by distinct mechanisms in D. simulans and D. melanogaster. However, we cannot exclude the possibility that there is considerable within-species variation between different strains. In addition, future investigations are needed to explore the impacts of the strong decrease in ovarian piRNA amounts, which are transmitted to the next generation in D. simulans (e.g., Akkouche et al.; ref. 43). Indeed, the large reduction in TE-derived piRNAs in the ovaries of infected Makindu flies may lead to a reactivation of TEs in the zygote and, therefore, be responsible for an increase in transposition rates. If this were confirmed, it would suggest that viral infections have an impact on genome evolution through the enhancement of zygotic transposition. However, as long as resolution is not cell-scaled, we cannot exclude the possibility that the observed effects do not occur in the oocytes but in the adjacent cells.
Viral Infection Impacts TE Transcript Amounts in Carcasses.
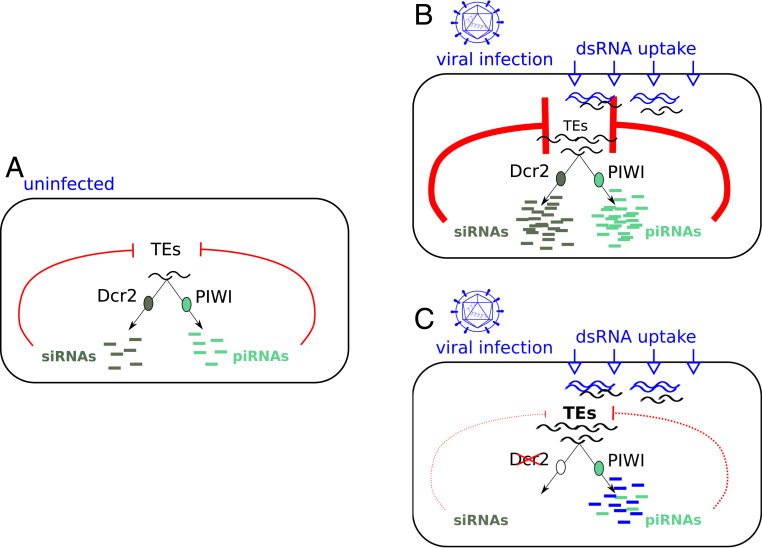
On the contrary, in D. simulans and D. melanogaster carcasses, we found that TE transcript amounts decrease upon SINV infection. This decrease is associated with an increase in TE-derived small RNA production, which is abolished in the dsRNA uptake mutant (CG4572−/−). Therefore, we propose that the dsRNA uptake pathway allows the opportunistic systemic spread of TE dsRNA intermediates as a by-product of viral dsRNA intermediates systemic spread. The spread of viral dsRNAs allows a systemic production of virus-derived siRNAs. Similarly, the spread of TE RNAs leads to a boosted production of TE-derived small RNAs, both by the piRNA and the siRNA pathways (Fig. 4). This idea is strengthened by the strong positive correlations that we observe between TE-derived siRNA and piRNA amounts (SI Appendix, Fig. S16). Indeed, it was previously shown that TEs produce sense and antisense transcripts (allowing the formation of dsRNAs) and that mixed ping-pong pairs of piRNAs and siRNAs exist (58, 59).
Fig. 4.
Model for viral infection effects on TEs in carcasses. (A) In uninfected conditions, TEs are controlled by piRNAs and siRNAs. (B) Upon infection, the dsRNA uptake pathway allows the systemic spread of viral RNAs, and this machinery also opportunistically carries TE dsRNA intermediates and allows for their spread. This triggers the systemic production of virus-derived siRNAs as well as TE-derived siRNAs, which subsequently leads to the reduction of TE transcript amounts in carcasses. The uptaken TE RNAs are also substrates for piRNA synthesis, which strengthen TE posttranscriptional silencing. (C) In case the siRNA pathway is impaired (e.g., dcr2−/− mutant, or viral RNAi suppressor targeting Dcr-2), viral replication increases, and we propose that viral RNAs saturate the piRNA pathway, which is no longer available for TEs. This results in an increase in TE transcript amounts.
Upon SINV infection, in the dcr2−/− mutant, we observe a strong signal for viral sense piRNAs, a decrease in TE-derived 23- to 30-nt small RNAs, and an increase in TE transcript amounts. Therefore, when the siRNA pathway is impaired, we propose that the piRNA pathway is saturated with viral RNAs (and viral RNAs are even more abundant when the siRNA pathway is impaired). Saturation and competition between RNAi-related gene silencing pathways are well documented in mammals (60, 61), and competition for Dcr-2 loading was demonstrated in Drosophila (40). However, further experimental validation will be needed to rule out alternative scenarios, such as the role of the cellular stress induced by viral infection (20, 62). Considering that SINV-derived 23- to 30-nt RNAs are only found in the sense orientation, they are not expected to play any antiviral role. The presumably saturated piRNA machinery is less available for the production of TE-derived piRNAs, resulting in TE transcript amount increase. In the case of DCV, which encodes a Dcr-2 inhibitor, we also observed a clear increase in TE transcript amounts (unfortunately, no small RNA-seq data are available to confirm this scenario in this study). These results mean that either a mutation in the coding sequence of dcr2, or an RNAi suppressor targeting Dcr-2, both lead to an increase in TE transcript amounts, strengthening our model (Fig. 4).
Therefore, our results show that viral infections do impact TE transcript amounts in somatic tissues. This is of fundamental importance and highlights that viral infections have to be considered as a biotic factor involved in the modulation of TE activity. The results that we gather here using different viruses and different genetic backgrounds suggest that the outcome of the viral infection from the TE side—either increase or decrease of TE transcript amounts—depends on the immune pathways that are triggered upon infection and whether or not the siRNA pathway is a major player of the immune response. Indeed, it is known that triggered immune response pathways vary a lot according to the viruses (63). In addition, it is known that genes of the siRNA pathway and dcr2, in particular, display a high rate of sequence evolution between and within species (64, 65). Therefore, we may predict that Dcr-2 efficiency displays natural variability, so that the effects of viral infections on TEs may also vary depending on the genetic background of the host.
Somatic TE activity has recently been the focus of many studies (29, 66, 67). Its functional relevance is still a matter of debate; however, it is suggested to be associated with neuronal functions and aging (29, 66, 67). Therefore, if the modulation of TE transcript amounts leads to a modulation of TE transposition rates, we suggest that these TE-associated physiological processes may be affected as well.
Evolutionary Perspective: The Tripartite Interaction Host/Virus/TEs.
We cannot exclude that the phenomenon we observe here is simply a by-product of the immune response through small RNA pathways. However, we may wonder whether it could be beneficial to any one of the three partners and, thus, be subject to natural selection. For instance, we may envision that some chronic infections may be beneficial to the host because they participate in reinforcing the inhibition of TEs. Otherwise, we may also propose that it is at the virus’s benefits that TE expression is reduced because it may be associated with a reduction of reverse transcriptase activity, thus a reduction in the production of the DNA copies of RNA viruses, and therefore a weaker antiviral response. Similarly, considering the involvement of TE reverse transcriptases in the production of viral DNA copies (38, 39), we may propose that there is a balance to reach for the host: Allowing TE transcription leads to the production of these beneficial viral DNA copies, but at the same time, it is a threat for genome stability because it may be associated with transposition. Testing these scenarios will require the investigation of a larger set of viruses and host genetic backgrounds.
Conclusion
Here, we demonstrate that viral infection affects TE transcript amounts in the soma via a modulation of TE-derived small RNA amounts. Regarding our starting expectations, our results in carcasses seem to support the synergistic scenario: Indeed, the antiviral response is accompanied by a strengthened TE control. On the contrary, when Dcr-2 function is impaired, both the control of viral replication and the control of TEs are less efficient. We propose that this synergistic scenario relies on the fact that the siRNA and piRNA pathways may share the same RNA substrates, at least in part.
The next step is clearly to investigate whether transposition rates are affected by these variations in TE transcript amounts. In particular, it would be interesting also to test the effects of viruses that are able to replicate in ovaries, such as the D. melanogaster Sigma virus (68). These results are of fundamental importance because they indicate that viral infections may affect mutation frequencies through the modulation of TE activity and, therefore, the rate of genetic diversification.
Methods
Fly Strains and Husbandry.
Flies were reared on a standard corn medium at 25 °C. We first treated flies with tetracycline to get rid of the Wolbachia endosymbiont, which is known to interfere with viral replication (69, 70). Wolbachia infections were eliminated after two generations on standard agarose medium, 0.25 mg/mL tetracycline hydrochloride. The absence of Wolbachia was validated using Wolbachia 16S primers (5′ TTG TAG CCT GCT ATG GTA TAA CT 3′ and 5′ GAA TAG GTA TGA TTT TCA TGT 3′), and Wolbachia wsp primers (5′ TGG TCC AAT AAG TGA TGA AGA AAC 3′ and 5′ AAA AAT TAA ACG CTA CTC CA 3′), as described previously (71). D. melanogaster chronic infections were eliminated following egg dechorionation, as described previously (51): 3- to 6-h-old eggs were collected, treated with 50% (vol/vol) household bleach solution (2.6% active chlorine) for 10 min, washed with water, and transferred to fresh medium for adults to hatch. One generation was treated. The analyses of viral contents in small RNA reads are provided SI Appendix, Fig. S17 and Table S18. The absence of viruses was confirmed using RT-PCR (see below) and primers already described (51) (DAV: 5′ AGG AGT TGG TGA GGA CAG CCC A 3′ and 5′ AGA CCT CAG TTG GCA GTT CGC C 3′; DCV: 5′ AAA ATT TCG TTT TAG CCC AGA A 3′ and 5′ TTG GTT GTA CGT CAA AAT CTG AG 3′; NV: 5′ ATG GCG CCA GTT AGT GCA GAC CT 3′ and 5′ CCT GTT GTT CCA GTT GGG TTC GA 3′; act42A: 5′ ATG GCG CCA GTT AGT GCA GAC CT 3′ and 5′ CTT CTC CAT GTC GTC CCA GT 3′; rp49: 5′ CGG ATC GAT ATG CTA AGC TGT 3′ and 5′ GCG CTT GTT CGA TCC GTA 3′) (SI Appendix, Fig. S19).
All experiments were performed using 3- to 6-d-old females. The CG4572−/− and dcr2−/−mutants have a w1118 genetic background.
Virus Production and Titration.
SINV stocks were produced by reverse genetics. Viral RNA was in vitro transcribed using mMessage mMachine SP6 kit (Thermo Fisher Scientific), and 10 µg of RNA were electroporated in BHK-21 cells. Viral titrations were determined in pfu/mL by a plaque assay on Vero E6 cell line.
Fly Infections and Viral Titration.
Flies were individually intrathoracically injected (Nanoject II, Drummond Scientific) with either 32.2 nL containing 2,300 SINV infectious particles (pfu) (D. melanogaster), or 207 nL containing 14,900 SINV pfu (Makindu), in the BHK-21 cell culture medium (Dulbecco's Modified Eagle Medium [DMEM]).
For viral titration, tissues were crushed in DMEM, 4% fetal calf serum, 2.5 µg/mL B amphotericin, 100 U/mL nystatin, 50 g/mL gentamicin, 50 g/mL penicillin/streptomycin. Viral titers in flies were determined by end-point dilution analysis on individual flies (n = 10) on Vero E6 cell line using the Reed and Muench’s method (72). SINV injection experiments were performed independently, in triplicates, and using three different stocks of SINV.
RT-qPCR.
Total RNAs of 30 pairs of ovaries or carcasses (biological duplicates) were extracted at 6 dpi by TRIzol (Thermo Fisher Scientific) coupled with the RNeasy Mini Kit method (Qiagen). Purified RNAs were treated using Turbo DNase (Ambion DNAfree kit). Reverse transcription was performed from 200 ng (carcasses) or 500 ng (ovaries) of total RNAs using SuperScript IV VILO Master Mix (Thermo Fisher Scientific). qPCR was performed using the LightCycler 480 SYBR Green I Master. rp49 and adh were used for normalization purposes (rp49: 5′ CGG ATC GAT ATG CTA AGC TGT 3′ and 5′ GCG CTT GTT CGA TCC GTA 3′; adh: 5′ CCG TGG TCA ACT TCA CCA GCT C 3′ and 5′ TCC AAC CAG GAG TTG AAC TTG TGC 3′). We quantified transcript levels for a set of TE families (accord, gypsy, 412, tirant, Doc6, Doc, F element), using the following primers: accord: 5′ TTC ACC CGT CGA AAG ACT TC 3′ and 5′ GCC GTG AAA GAG TTC GAA AG 3′; gypsy: 5′ GGC TCA TTG CCG TTA AAC AT 3′ and 5′ TCT TCC TTC TTT CGC TGA GG 3′; 412: 5′ TTG ATG GGC AAA AGA TCC AT 3′ and 5′ TTG CTG GAA TTG TCG TTT CA 3′; tirant: 5′ ACA CGT TCC CTG AAC AGA CG 3′ and 5′ GAA CGT TAC CAA TCC GAG CA 3′; Doc6: 5′ ACA TCA TGT GCC CAT TGT GC 3′ and 5′ AAA TTC GGC ATG GGC ATG TC 3′; Doc: 5′ GCA CCT GGT TTC GAA GTG AT 3′ and 5′ GTA GCC AGG CGA TTT TCT TG 3′; F-element: 5′ ACG TCG CTT ACG CAG AGA AT 3′ and 5′ GTT TTG TGC CTG TTG GTG TG 3′.
Immunostaining Experiments.
Immunostaining experiments were performed at 6 dpi. Ovary pairs were fixed for 4 h in 4% paraformaldehyde and permeabilized using phosphate-buffered saline (PBS), 0.5% Triton X-100 for 1 h. Whole flies were fixed for 4 h in 2% paraformaldehyde, then incubated for 16 h in 12% sucrose at 4 °C and arranged in OCT blocs. Fourteen-micrometer-thick slices were made using a cryostat. A second PFA fixation (0.5%, 20 min) and permeabilization (PBS, 0.3% Triton X-100, 30 min) for slices were performed using a SINV anti-capsid (73) and a secondary A488 goat anti-mouse (A-11029, Invitrogen).
RNA-seq.
Female flies were carefully, manually dissected: Pairs of ovaries were separated from the rest of the bodies, which will further be called “carcasses.” Ovaries had to remain intact for the samples to be kept. Principal component analysis of RNA-seq data further confirmed that ovary and carcass samples were clearly different (SI Appendix, Fig. S20), which allows us to feel confident about the absence of tissue contamination associated with our dissection protocol. Total RNAs from 30 pairs of ovaries or 30 carcasses were extracted at 6 dpi using TRIzol (Thermo Fisher Scientific) and RNeasy Mini Kit (Qiagen). Purified RNAs (10 µg) were treated using DNase TurboDNase (Ambion DNAfree kit). Library preparation was done using the TruSeq RNA Sample Prep Kit v2, after polyadenylated RNA selection, and paired-end sequencing was performed (Hiseq 4000; Illumina). Sequencing adapters were removed using UrQt (74) and trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were aligned on reference genes retrieved from FlyBase (ftp://ftp.flybase.net/releases/FB2019_01/dsim_r2.02/fasta/dsim-all-gene-r2.02.fasta.gz and ftp://ftp.flybase.net/releases/FB2019_01/dmel_r6.26/fasta/dmel-all-gene-r6.26.fasta.gz) using TopHat2 (75). Gene count tables were generated using eXpress (76). TE count tables were generated using the TEcount module of TEtools (77), and the list of TE sequences available at ftp://pbil.univ-lyon1.fr/pub/datasets/Roy2019/. Gene and TE count tables were concatenated to make the complete count table, which was further analyzed using the DESeq2 R package (version 1.18) (78) (SI Appendix, Table S21).
Data retrieved from Merkling et al. (57) corresponded to accession no. GSE56013 (NCBI Gene Expression Omnibus) and were treated the same way.
Small RNA-seq.
Female flies were carefully, manually dissected: pairs of ovaries were separated from the rest of the bodies, which will further be called “carcasses.” Ovaries had to remain intact for the samples to be kept. Small RNAs were isolated at 6 dpi from 50 pairs of ovaries or 50 carcasses as previously described (50). Briefly, this strategy is based on the adsorption and reversible binding of negatively charged biomolecules to a positively charged insoluble matrix. RISC can thus be recovered using mild salt concentration elution, whereas contaminant RNAs (such as ribosomal RNAs or degradation products) remain fixed to the column (50). Size selection of small RNAs (18–50 bp) was performed on gel at the GenomEast sequencing platform. Purified small RNAs were used for library preparation (SQ00/SIL-04-SR) and sequenced on a Hiseq 4000 apparatus (Illumina). Adapter sequences were removed using cutadatp (79) -a TGGAATTCTCGGGTGCCAAGGAACTCCAGTCACTTA. Per base nucleotide composition plots were then built using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Read count numbers were normalized to miRNA read count numbers (miRNA sequences retrieved from FlyBase: dsim-all-miRNA-r2.02.fasta.gz for Makindu, and dmel-all-miRNA-r6.25.fasta.gz for D. melanogaster samples). Alternatively, in the w1118 and CG4572−/− samples, read count numbers were normalized to endogenous, Ago2-bound siRNA read count numbers (54), which provided similar results (SI Appendix, Fig. S22). We could not use these endogenous siRNAs as normalizators for D. simulans samples because these loci were not annotated in this species. Similarly, we could not use siRNA normalizators for dcr2−/− samples because of the impairment of the siRNA pathway.
Using PRINSEQ lite version 0.20.4 (80), we filtered reads of size 23–30 nt and considered these as piRNAs, and reads of size 21 nt were considered as siRNAs. Using a modified version of the TEcount module of TEtools (44), we mapped the reads against the above list either in the sense or antisense directions. We then computed the sums of sense and antisense counts. To calculate the proportion of reads mapping to protein-coding genes, we used the same reference sequences as above (RNA-seq), which we previously masked using RepeatMasker (81). Right after the adapter trimming step, we mapped small RNA reads using bowtie --best (82), and the numbers of alignments were calculated using SAMtools (83), samtools view -F 4 -c. To estimate the proportion of reads corresponding to viral sequences, we kept 20- to 35-nt reads. We aligned reads against SINV sequence (accession no. GM893992) using bowtie --best (82). For Drosophila natural viruses, we used the reference sequences from Obbard laboratory (http://obbard.bio.ed.ac.uk/data/Updated_Drosophila_Viruses.fas.gz, downloaded in 2017) and ran the TEcount module of TEtools (77).
We looked for ping-pong signatures using signature.py with the options min_size = 23 and max_size = 30 (84). Alignments were performed using bowtie --best (82) on doc (accession no. X17551), 412 (chrU:15,457,641–15,461,149 from WUGSC mosaic 1.0/droSim1), and SINV (accession no. GM893992). We analyzed base composition at each position using SAMStat (85) on the output alignments obtained from TEcount.
Statistical Analyses.
We tested whether the modulations of TE counts upon infection were different from zero using Wilcoxon paired test comparing TE normalized counts in the infected versus mock conditions, for the considered 237 TE families. The null hypothesis corresponds to an equal number of dots above and below the 0 horizontal line.
Differential expression of genes was tested using the DESeq2 R package (version 1.18) (78) and adjusted P values were considered.
Data Availability.
RNA-seq and small RNA-seq data were deposited under the accession no. PRJNA540249.
Supplementary Material
Acknowledgments
We acknowledge the contribution of Structure Fédérative de Recherche Biosciences (UMS3444/CNRS, US8/Inserm, École Normale Supérieure de Lyon, Université Claude Bernard Lyon 1) facility: Arthro-tools platform. This work was performed using the computing facilities of Laboratoire de Biométrie Biologie Évolutive / Pôle Rhône-Alpes de Bioinformatique (CC LBBE/PRABI), and the Qubit Invitrogen of Institut de Génomique Fonctionnelle de Lyon (managed by Benjamin Gillet, who we thank for his availability). Sequencing was performed by the GenomEast platform, a member of the “France Génomique” consortium (ANR-10-INBS-0009). We thank Maria Carla Saleh, who provided us with w1118 and Dcr-2[L811fsX] D. melanogaster strains. We thank Carine Maisse-Paradisi, who provided us with SINV plasmid and antibody. We thank Cristina Vieira, Rita Rebollo, Séverine Chambeyron, Matthieu Boulesteix, Hervé Seitz, and Hélène Henri for helpful discussions. We thank Judit Salces-Ortiz, Justine Picarle, Vincent Mérel, Angélique Champavère, Nelly Burlet, Margot Enguehard, Sarah Bouzidi, and Biologie Fonctionnelle, Insectes et Interactions (BF2I) for technical help. We also thank Clément Gilbert, Maria del Pilar García Guerriero, Émilie Brasset, Fabrice Vavre, Thierry Dupressoir, François Leulier, and Philippe Marianneau for helpful discussions. We thank Christophe Terzian who initiated this work but left us too early. This work was supported by the Agence Nationale de la Recherche Laboratory of Excellence LABEX ECOFECT (ANR-11-LABX-0048 of the Université de Lyon, within the program Investissements d’Avenir [ANR-11-IDEX-0007] operated by the French National Research Agency Grant ERMIT) and Agence Nationale de la Recherche Grants ExHyb and TEMIT.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The original datasets are available via the National Center for Biotechnology Information’s Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA540249).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006106117/-/DCSupplemental.
References
- 1.Wichman H. A., Van den Bussche R. A., Hamilton M. J., Baker R. J., Transposable elements and the evolution of genome organization in mammals. Genetica 86, 287–293 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Robertson H. M., Lampe D. J., Distribution of transposable elements in arthropods. Annu. Rev. Entomol. 40, 333–357 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Sultana T., Zamborlini A., Cristofari G., Lesage P., Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Biémont C., Vieira C., What transposable elements tell us about genome organization and evolution: The case of Drosophila. Cytogenet. Genome Res. 110, 25–34 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Bergman C. M., Quesneville H., Anxolabéhère D., Ashburner M., Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 7, R112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Nature411, 720 (2001) and Nature412, 565 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Schnable P. S., et al. , The B73 maize genome: Complexity, diversity, and dynamics. Science 326, 1112–1115 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Wicker T., et al. , A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Golding G. B., Aquadro C. F., Langley C. H., Sequence evolution within populations under multiple types of mutation. Proc. Natl. Acad. Sci. U.S.A. 83, 427–431 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlesworth B., Lapid A., Canada D., The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. II. Inferences on the nature of selection against elements. Genet. Res. 60, 115–130 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Biémont C., Population genetics of transposable DNA elements. A Drosophila point of view. Genetica 86, 67–84 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Barrón M. G., Fiston-Lavier A.-S., Petrov D. A., González J., Population genomics of transposable elements in Drosophila. Annu. Rev. Genet. 48, 561–581 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kahyo T., et al. , Identification and association study with lung cancer for novel insertion polymorphisms of human endogenous retrovirus. Carcinogenesis 34, 2531–2538 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Moyes D., Griffiths D. J., Venables P. J., Insertional polymorphisms: A new lease of life for endogenous retroviruses in human disease. Trends Genet. 23, 326–333 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Casacuberta E., González J., The impact of transposable elements in environmental adaptation. Mol. Ecol. 22, 1503–1517 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Kidwell M. G., Reciprocal differences in female recombination associated with hybrid dysgenesis in Drosophila melanogaster. Genet. Res. 30, 77–88 (1977). [DOI] [PubMed] [Google Scholar]
- 17.Picard G., Non-mendelian female sterility in Drosophila melanogaster: Hereditary transmission of I factor. Genetics 83, 107–123 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todeschini A.-L., Morillon A., Springer M., Lesage P., Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 7459–7472 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogasawara H., Obata H., Hata Y., Takahashi S., Gomi K., Crawler, a novel Tc1/mariner-type transposable element in Aspergillus oryzae transposes under stress conditions. Fungal Genet. Biol. 46, 441–449 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Horváth V., Merenciano M., González J., Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 33, 832–841 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Fablet M., Vieira C., Evolvability, epigenetics and transposable elements. Biomol. Concepts 2, 333–341 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Melayah D., Bonnivard E., Chalhoub B., Audeon C., Grandbastien M. A., The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 28, 159–168 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Tóth K. F., Pezic D., Stuwe E., Webster A., The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol. 886, 51–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siomi M. C., Sato K., Pezic D., Aravin A. A., PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Czech B., Hannon G. J., One loop to rule them all: The ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 41, 324–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke J., et al. , Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Lewis S. H. et al., Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2, 174–181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirkovic-Hösle M., Förstemann K., Transposon defense by endo-siRNAs, piRNAs and somatic pilRNAs in Drosophila: Contributions of Loqs-PD and R2D2. PLoS One 9, e84994 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrat P. N., et al. , Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340, 91–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czech B., et al. , An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamura Y., et al. , Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Chung W.-J., Okamura K., Martin R., Lai E. C., Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau N. C., et al. , Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 19, 1776–1785 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghildiyal M., et al. , Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J. A., Imler J.-L., Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 7, 590–597 (2006). [DOI] [PubMed] [Google Scholar]
- 36.van Rij R. P., et al. , The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X.-H., et al. , RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goic B., et al. , RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 14, 396–403 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Tassetto M., Kunitomi M., Andino R., Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169, 314–325.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durdevic Z., Mobin M. B., Hanna K., Lyko F., Schaefer M., The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 4, 931–937 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Barckmann B., et al. , The somatic piRNA pathway controls germline transposition over generations. Nucleic Acids Res. 46, 9524–9536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akkouche A., et al. , tirant, a newly discovered active endogenous retrovirus in Drosophila simulans. J. Virol. 86, 3675–3681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akkouche A., et al. , Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO Rep. 14, 458–464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fablet M., et al. , Dynamic interactions between the genome and an endogenous retrovirus: Tirant in Drosophila simulans wild-type strains. G3 (Bethesda) 9, 855–865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor R. M., Hurlbut H. S., Work T. H., Kingston J. R., Frothingham T. E., Sindbis virus: A newly recognized arthropodtransmitted virus. Am. J. Trop. Med. Hyg. 4, 844–862 (1955). [DOI] [PubMed] [Google Scholar]
- 46.van Mierlo J. T., et al. , Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 10, e1004256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell C. L., et al. , Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 8, 47 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J., Cherry S., Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 42, 67–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saleh M.-C., et al. , Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458, 346–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grentzinger T., et al. , A user-friendly chromatographic method to purify small regulatory RNAs. Methods 67, 91–101 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Merkling S. H., van Rij R. P., Analysis of resistance and tolerance to virus infection in Drosophila. Nat. Protoc. 10, 1084–1097 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Petit M., et al. , piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 113, E4218–E4227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saleh M.-C., et al. , The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malone C. D., et al. , Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senti K.-A., Jurczak D., Sachidanandam R., Brennecke J., piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 29, 1747–1762 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Beek M., et al. , Dual-layer transposon repression in heads of Drosophila melanogaster. RNA 24, 1749–1760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merkling S. H., et al. , The epigenetic regulator G9a mediates tolerance to RNA virus infection in Drosophila. PLoS Pathog. 11, e1004692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shpiz S., Ryazansky S., Olovnikov I., Abramov Y., Kalmykova A., Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline. PLoS Genet. 10, e1004138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olovnikov I., et al. , De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res. 41, 5757–5768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haussecker D., et al. , Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gestel M. A., et al. , shRNA-induced saturation of the microRNA pathway in the rat brain. Gene Ther. 21, 205–211 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Arnold P. A., Johnson K. N., White C. R., Physiological and metabolic consequences of viral infection in Drosophila melanogaster. J. Exp. Biol. 216, 3350–3357 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Kemp C., et al. , Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obbard D. J., Jiggins F. M., Halligan D. L., Little T. J., Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16, 580–585 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Kolaczkowski B., Hupalo D. N., Kern A. D., Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol. Biol. Evol. 28, 1033–1042 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Faulkner G. J., Garcia-Perez J. L., L1 mosaicism in mammals: Extent, effects, and evolution. Trends Genet. 33, 802–816 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Chang Y.-H., Keegan R. M., Prazak L., Dubnau J., Cellular labeling of endogenous retrovirus replication (CLEVR) reveals de novo insertions of the gypsy retrotransposable element in cell culture and in both neurons and glial cells of aging fruit flies. PLoS Biol. 17, e3000278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.L’Heritier P., The hereditary virus of Drosophila. Adv. Virus Res. 5, 195–245 (1958). [PubMed] [Google Scholar]
- 69.Teixeira L., Ferreira A., Ashburner M., The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6, e2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharya T., Newton I. L. G., Hardy R. W., Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathog. 13, e1006427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riegler M., Sidhu M., Miller W. J., O’Neill S. L., Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15, 1428–1433 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 73.Greiser-Wilke I., Moenning V., Kaaden O. R., Figueiredo L. T., Most alphaviruses share a conserved epitopic region on their nucleocapsid protein. J. Gen. Virol. 70, 743–748 (1989). [DOI] [PubMed] [Google Scholar]
- 74.Modolo L., Lerat E., UrQt: An efficient software for the unsupervised quality trimming of NGS data. BMC Bioinformatics 16, 137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roberts A., Trapnell C., Donaghey J., Rinn J. L., Pachter L., Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lerat E., Fablet M., Modolo L., Lopez-Maestre H., Vieira C., TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 45, e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 80.Schmieder R., Edwards R., Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smit A. F., Hubley R., Green P., RepeatMasker Open-4.0. (2013) http://www.repeatmasker.org. Accessed 2018.
- 82.Langmead B., Aligning short sequencing reads with Bowtie. Curr. Protoc. BioinformaticsChapter 11, Unit 11.7 (2010). [DOI] [PMC free article] [PubMed]
- 83.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Antoniewski C., Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 1173, 135–146 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Lassmann T., Hayashizaki Y., Daub C. O., SAMStat: Monitoring biases in next generation sequencing data. Bioinformatics 27, 130–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and small RNA-seq data were deposited under the accession no. PRJNA540249.