Abstract
Studying multiple post-translational modifications (PTMs) of proteins is a crucial step to understand PTM crosstalk and gain more holistic insights into protein function. Despite the importance of multi-PTM enrichment studies, few studies investigate more than one PTM at a time, due partially to the expenses, time, and large protein quantities required to perform multiple global proteomic analysis of PTMs. The “one-pot” affinity enrichment detailed in this protocol overcomes these barriers by permitting the simultaneous identification and quantification of peptides with lysine residues containing acetylation and succinylation PTMs with low amounts of sample input. The protocol involves preparation of protein lysate from mouse livers of SIRT5 knockout mice, performance of trypsin digestion, enrichment for PTMs, and performance of mass spectrometric analysis using a data-independent acquisition (DIA) workflow. Because this workflow allows for the enrichment of two PTMs from the same sample simultaneously, it provides a practical tool to study PTM crosstalk without requiring large amounts of samples, and it greatly reduces the time required for sample preparation, data acquisition, and analysis. The DIA component of the workflow provides comprehensive PTM-specific information. This is particularly important when studying PTM site localization, as DIA provides comprehensive sets of fragment ions that can be computationally deciphered to differentiate between different PTM localization isoforms.
Keywords: Biology, Issue 156, post-translational modifications, immunoaffinity enrichment, crosstalk, site localization, mass spectrometry, data-independent acquisition, acylation, acetylation, succinylation
Introduction
A myriad of post-translational modifications dynamically regulate proteins and pathways through effects on the activity1, signaling2, and turnover3,4. For example, protein kinases are activated or deactivated by the addition of phosphate groups5, and histone acetylation and other modifications provide a mechanism to change the chromatin structure and serve as transcriptional regulatory mechanisms6,7. In recent years, evidence has mounted that multiple PTMs work in concert or compete to regulate protein function or activity8,9,10,11. Therefore, understanding PTM crosstalk is an emerging need in PTM research. However, most available proteomic workflows to identify and quantify PTM sites focus on single modifications, rather than the interplay of multiple modifications. The described workflow correlates specific protein modification “hotspots” and lysine residues that are modified by multiple different PTMs.
There is a growing need in the scientific community for feasible methods to study multiple PTMs simultaneously12. Most methods to globally identify and quantify the sites of multiple types of PTMs are challenging due to the high costs and amount of tissue required12,13. Not only are multi-PTM enrichment experiments time-consuming in terms of sample preparation, data acquisition, and data analysis, but these studies typically require large and often prohibitive amounts of protein11. Described here is a protocol for simultaneous enrichment and analysis of multiple PTMs, which also addresses several of these barriers and enables large-scale PTM profiling and assessing crosstalk between various PTMs14. This one-pot workflow outlines a practical way for biomedical researchers to globally profile multiple PTMs, identify co-modified peptides, and study PTM crosstalk in an efficient and cost-effective way14,15.
Here, this method is showcased by examining mitochondrial protein acylation, which was first studied over 50 years ago16. It specifically concentrates on lysine acetylation17 and succinylation18, including the co-occurrence of these modifications on proteins and even co-modification at the peptide level. Since the study uses a sirtuin 5 (SIRT5) knockout mouse model, it was chosen to focus on enrichment of acetylation and succinylation sites. This decision was made because succinylation sites are targets of SIRT5 desuccinylase and are thus expected to show significant upregulation in KO mice, making them the most relevant PTMs in this case. Both PTMs are biologically relevant as recently summarized by Carrico et al.19. In general, acetylation shows important effects on gene expression and metabolism, and succinylation has been reported to regulate heart metabolism and function20.
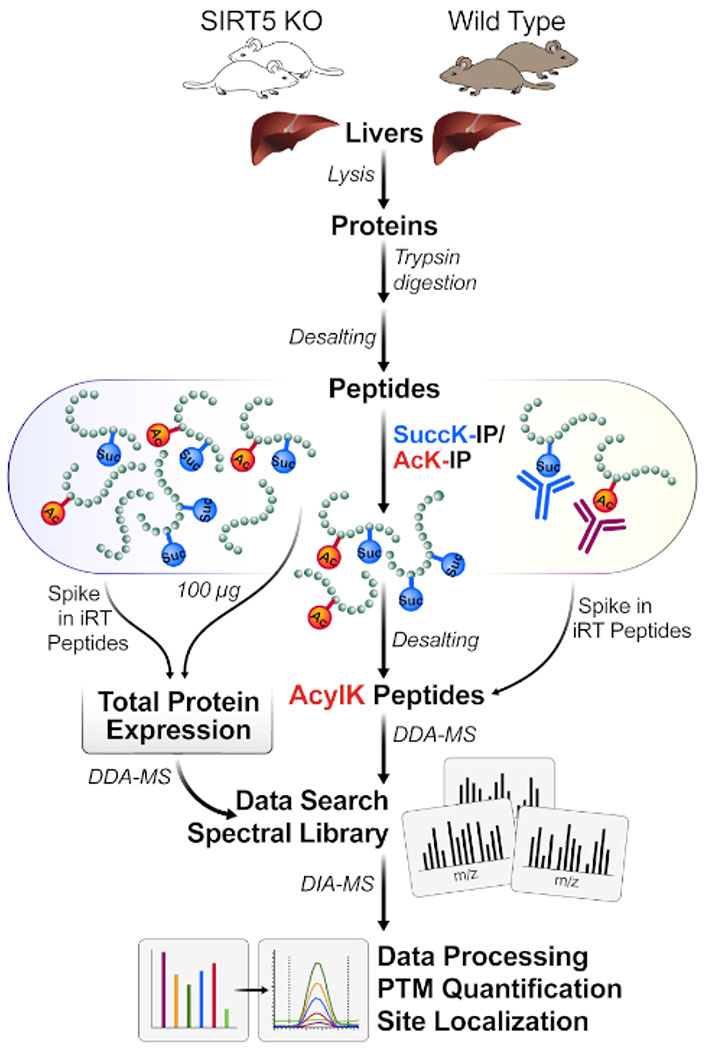
The described protocol can be performed with a low amount of protein input material (e.g., 1 mg of protein lysate) and reduces the total duration of the experiment by reducing the time spent for sample processing, MS acquisition, and data analysis. A workflow schematic is provided in Figure 1. We have also used even lower amounts of starting material (down to 100 μg of protein lysate, scaling down the amounts of beads used accordingly), which as expected, reduces the overall yield of identified acylated peptides; however, it still provides highly valuable results and quantifiable acylated peptides.
Figure 1: Typical workflow for one-pot enrichment of PTMs.
Tissue (here, livers) are harvested from SIRT5 KO and wild-type (WT) mice, and proteins are lysed, trypsin-digested into peptides, and desalted. Peptides are then enriched by immunoaffinity with combinations of succinyl- and acetyl-antibody beads. Parallel MS workflows measure both 1) small aliquots of whole lysate protein expression changes (for protein normalization) and 2) enriched acyl-containing peptides for acylation site identification (DDA-MS) and site localization, followed by quantification (DIA-MS).
While so-called top-down or middle-down workflows typically do not use proteolytic digestion approaches (and thus maintain the connectivity of multiple PTMs within one protein), this protocol focuses on a peptide-based affinity enrichment approach to gain extra depth and sensitivity for PTM identification and quantification (Figure 1). In addition, this peptide-centric workflow utilizes modern mass spectrometry methods, including 1) a combination of data-dependent acquisition (DDA) to generate spectral libraries, and 2) data-independent acquisition (DIA) for accurate PTM quantification in a label-free workflow.
DIA workflows overcome sampling stochasticity of typical DDA scan schemes by comprehensively fragmenting all peptide signals within the sampled m/z range21. This feature is also extremely beneficial in terms of site localization, because it is easier to gain information regarding which specific fragment ions are modified within the peptide. In addition, DIA workflows allow identification and quantification of minor PTM-peptide isoforms with identical precursor ions. DIA methods can also determine a specific PTM site localization within a peptide based on specific corresponding fragment ions that are comprehensively measured at all times. However, DDA approaches often utilize “dynamic exclusion” features that exclude multiple sampling of MS/MS for the same precursor ion, thus missing minor PTM isoforms.
The simultaneous enrichment strategy described here is ideally suited for studies that will benefit from the global profiling and quantification of multiple PTMs, examining PTM crosstalk, and understanding the dynamic interactions of post-translational modifications. Identification of multiple enriched PTMs in one combined workflow have been described by: global, serial or parallel enrichment of PTM containing proteolytic peptides, or alternatively by the analysis of intact proteins. In direct comparison with serial enrichment of acetylation and succinylation, the efficiency of the one-pot methodology was established as being very similar14. These alternative protocols require significant amounts of starting material and time and can be prohibitively expensive. In contrast, the one-pot protocol provides an inexpensive and efficient method for enrichment of more than one PTM with subsequent analysis and identification.
Mouse liver tissues are obtained from SIRT5 knockout mice and are used here as starting material. This protocol can also be performed for protein lysates from different tissues or cell culture experiments. This protocol can be applied to protein lysates obtained from tissues or cell culture pellets.
Protocol
All experiments described in the protocol follow the guidelines of the Buck Institute Institutional Animal Research Committee.
1. Extraction of protein from homogenized tissue and digestion with protease
-
1
Freshly prepare the lysis buffer to a final concentration of 8 M urea, 50 mM triethylammonium bicarbonate (TEAB), 1 μM trichostatin A (TSA), 20 mM nicotinamide, 75 mM sodium chloride (NaCI), 1x protease/phosphatase inhibitor cocktail (PIC) with HPLC-grade water.
-
2
Add one sterile steel bead to a 2 mL tube compatible with a bead mill homogenizer, then place the tissue piece (pea-sized) into the tube. Add lysis buffer (500 μL) and vortex briefly to ensure the buffer covers the tissue piece (add more lysis buffer, if necessary). Place tubes on prechilled adaptor sets, ensuring that the tubes are balanced between the homogenizer’s two adaptors.
-
3
Homogenize samples 2x at 25 Hz for 3 min each at 4 °C. If the tissue is not fully homogenized, spin the tubes briefly and transfer the homogenized lysate into a freshly labeled 1.5 mL tube. Then, add additional lysis buffer to the remaining tissue piece, and repeat the homogenization 2x at 25 Hz for 3 min each. Two rounds of homogenization are typically sufficient to break up the tissue.
-
4
Remove the metal bead with a tweezer. In between each sample, rinse the tweezer with HPLC-grade water, HPLC-grade methanol, and HPLC-grade water again.
-
5
Combine the homogenized lysates if two rounds of homogenization were applied and sonicate samples with an ultrasonicator for 10 cycles at 30 s on/30 s off and 4 °C on high power.
-
6
Centrifuge the homogenized lysates at 14,000 x g for 10 min at 4 °C to clear the lysate. Transfer the supernatant (cleared lysate) to a new 1.5 mL microcentrifuge tube, avoiding any fat layer that may sit on top of the supernatant and any debris at the bottom of the tube.
-
7
Perform a bicinchoninic acid (BCA) assay to measure the protein concentration of the cleared lysate with a proper dilution (for example, 1:20 and/or 1:200). According to BCA results, aliquot 1 mg of protein from each sample.
-
8
Reduce proteins in 4.5 mM dithiothreitol (DTT) at 37 °C for 30 min with agitation at 1,400 rpm. Subsequently, alkylate proteins in 10 mM iodoacetamide (IAA) with incubation in the dark (place in drawer or cabinet) at room temperature (RT) for 30 min.
NOTE: Alternative reagents may be used, such as N-ethylmaleimide (NEM) instead of IAA.
-
9
Add 50 mM TEAB to the samples to dilute the urea concentration to below 2 M. Spot 1 μL of the sample onto a pH paper to ensure the pH of the diluted sample is between 7.0-8.5. Add trypsin to each sample at a ratio of 1:50 (trypsin-to-protein, wt/wt) and digest the protein with agitation at 37 °C overnight (about 14-16 h).
-
10
Quench the digests with 10% formic acid to reach 1% formic acid the next day. Vortex and spin briefly. Spot 1 μL of the sample onto pH strips to ensure the digest is pH = 2-3. Centrifuge samples at 1,800 x g for 15 min at RT to pellet any insoluble material.
2. Desalting of non-enriched proteolytic peptides via large-scale solid-phase extraction
Obtain cartridges containing C18 resin that can bind up to 10 mg of protein. Fit these cartridges into a vacuum apparatus to use vacuum suction that can pull the liquid through the cartridge during each of the following steps. Designate one cartridge for each peptide sample.
Add 800 μL of 80% acetonitrile (ACN) in 0.2% formic acid to cartridges with 19.8% water and use vacuum suction to pull the liquid through. Repeat this step 1x, avoiding drying of the cartridges completely.
Equilibrate cartridges by adding 800 μL of 0.2% formic acid in water with vacuum suction to pull the entire volume through the filter. Repeat this 2x. Load peptides onto the cartridges with vacuum suction. Wash peptides 2x with 800 μL of 0.2% formic acid in water under vacuum suction.
Arrange 1.5 mL microcentrifuge tubes beneath each cartridge to collect peptide eluting in the final step. Under vacuum suction elute peptides from cartridges, first with 800 μL of 80% ACN in 0.2% formic acid and 19.8% water, then with 400 μL of the same solution. Dry the desalted peptide samples completely in a vacuum concentrator (2-3 h).
3. Simultaneous enrichment of K-acetylated and K-succinylated peptides with immunoaffinity beads
-
1
Resuspend the dried peptides in 1.4 mL of cold 1x immuno-affinity purification (IAP) buffer. Vortex to mix and ensure a pH of ~7. Centrifuge samples at 10,000 x g for 10 min at 4 °C. A small pellet may appear.
-
2
Set peptides aside on ice while preparing antibody-beads. To a tube of K-acetyl and tube of K-succinyl antibody bead slurry (volume: 100 μL of slurry per tube), add 1 mL of cold 1x phosphate-buffered saline (PBS) and mix by pipetting. Transfer the entire solution to a new 1.5 mL tube and spin in a miniature centrifuge for 30 s at RT.
-
3
Aspirate the PBS, taking care to avoid aspirating off any beads. Repeat the wash 3x by washing again with 1 mL of cold 1x PBS, centrifuging for 30 s at RT and aspirating off the PBS.
-
4
Re-suspend the beads in about 440 μL of PBS. One-quarter of the number of beads in each PTM tube (100 μL, provided by the manufacturer) is used for the one-pot enrichment (around 62.5 μg of each immobilized antibody for both K-acetyl and K-succinyl antibody beads). Pipette the beads several times to mix well. Remove 100 μL of acetyl-lysine antibody beads and transfer into one tube with 200 μL precut tips, ensuring that the master mix stays consistently well-mixed.
-
5
Do the same with the succinyl-lysine antibody beads by removing 100 μL of succinyl-lysine antibody beads to the tube to reach equal parts of the acetyl-lysine antibody and succinyl-lysine antibody in each tube. Spin down for 30 s in a miniature centrifuge and aspirate off media with gel loader tip (beads will turn white).
NOTE: Each tube should have a similar bead pellet at the bottom.
-
6
Pipette the resuspended peptide directly onto the washed beads. Be careful to avoid any pellets (add quickly to avoid drying out the beads). Incubate the peptides with beads at 4 °C overnight with agitation.
4. Elution of the peptides bound to antibody beads
Spin the peptide samples at 2,000 x g for 30 s at 4 °C. Remove the supernatant containing unbound peptides and save for future experiments if desired. Add 1 mL of cold 1x IAP buffer to the beads to wash and mix by inverting 5x. Spin at 2,000 x g for 30 s at 4 °C. Aspirate off the IAP solution and repeat the IAP wash step 1x for a total of two washes.
Add 1 mL of cold HPLC water to the beads and mix by inverting 5x. Spin at 2,000 x g for 30 s at 4 °C. Aspirate off the water. Repeat the water wash step twice for a total of three washes. Spin an additional time at 2,000 x g for 30 s at 4 °C to collect any remaining water in the bottom ofthe tube. Aspirate off any extra water that may have collected with flat-tipped gel loading tips. Be careful to avoid aspirating the beads.
Add 55 μL of 0.15% trifluoroacetic acid in water to the beads. Incubate the beads for 10 min at RT while occasionally tapping the bottom of the tube to mix. Spin the beads at RT for 30 s in a miniature centrifuge. Remove the eluted peptides and set aside using a flat-tipped gel-loading tip.
Add 45 μL of 0.15% trifluoroacetic acid in water to the beads. Incubate for 10 min at RT while tapping the bottom of the tube occasionally to mix. Spin the beads at RT for 30 s in a miniature centrifuge. Remove the second elution using a flat-tipped gel-loading tip and combine with the first elution.
Spin the combined eluted peptides at 12,000 x g for 5 min at RT to pellet any beads that carry over. Transfer the peptides sample solution into a new 500 μL tube.
NOTE: The protocol can be paused here.
5. Desalting of enriched peptides
NOTE: The pH of samples should be less than 4 for optimal binding to the tip containing C18 resin. Use a 10 μL pipette tip containing 0.6 μL of C18 resin and a 10 μL pipette to control the tip.
Pre-wet the C18 tip by pipetting 10 μL of acetonitrile into the tip. Dispense acetonitrile into waste. Repeat this step two more times.
Equilibrate the tip by washing 3x with 10 μL of 0.2% formic acid in water. Dispense the solution into waste after each equilibration.
Load peptides from the sample onto the tip by continuously pipetting and dispensing enriched peptide sample with the tip. Pipette up and down repeatedly at least 20x and dispense the remaining solution from the tip.
Wash the tip containing bound peptide with 10 μL of 0.2% formic acid in water. Dispense solution into waste. Repeat this step 9x.
Elute peptides into a new tube using 10 μL of an elution buffer containing 0.2% formic acid, 50% acetonitrile, and 49.8% water. Repeatedly pipette and dispense the 10 μL of solution within the new tube 15x. Repeat this step with an additional 10 μL of elution buffer, repeatedly pipetting into the same tube as the previous elution.
Dry peptides completely in vacuum concentrator (approximately 20 min). Resuspend the dried peptides in appropriate volume of 0.2% formic acid in water.
NOTE: The protocol can be paused here.
6. Data acquisition using DDA and DIA
NOTE: Analyze the samples with DDA and DIA LC-MS/MS methods, which can be adjusted depending on the available mass spectrometric instrument. Here, the samples were analyzed using a nano-LC 2D HPLC system coupled to a high-resolution mass spectrometer.
-
1
Use a HPLC system combined with a chip-based platform HPLC system directly connected to a mass spectrometer (many LC-MS configurations and systems can also be used).
-
2Analysis of samples using reverse-phase HPLC-ESI-MS/MS
- After injection, transfer the peptide mixtures onto a C18 pre-column chip and desalt the peptides by washing with mobile phase A at 2 μL/min for 10 min. Then, transfer the peptides to an analytical column and elute at a flow rate of 300 nL/min with a 2-3 h gradient using mobile phases A and B. Specifically, use a linear gradient from 5% mobile phase B to 35% mobile phase B over 80 min.
- Subsequently, ramp the mobile phase B to 80% over 5 min, then hold at 80% B for 8 min before returning to 5% B for a 25 min re-equilibration.
-
3Build an MS instrument method for DDA and define the following instrument scan experiments
- Experiment 1: MS1 precursor ion scan from m/z 400-1,500 (accumulation time of 250 ms). Set the intensity threshold to trigger MS/MS scans for ions of charge states 2-5 to 200 counts. Set the dynamic exclusion of precursor ions to 60 s.
- Experiment 2: MS/MS product ion scan with a MS2 scan range from m/z 100-1,500 (accumulation time of 100 ms per each 30-product ion scan per cycle). Set the collision energy spread to CES = 5, then select the “high sensitivity product ion scan mode”.
NOTE: The DDA method will acquire MS/MS spectra for the 30 most abundant precursor ions after each survey MS1 scan per cycle, and the total cycle time will be ~3.3 s. DDA acquisitions will be used to build spectral libraries as described in section 7.
-
4Building a MS instrument method for DIA and defining the following instrument scan experiments.
- Experiment 1: perform MS1 precursor ion scan from m/z 400-1,250 (accumulation time of 250 ms).
- Experiment 2: perform MS/MS product ion scans for 64 variable SWATH segments with a MS2 scan range from m/z 100-1,500 (accumulation time of 45 ms per each 64-product ion scan per cycle). Set the collision energy spread to CES = 10, then select the “high sensitivity product ion scan mode”.
- Use the 64 variable window DIA/SWATH acquisition strategy as described by Schilling et al.22 to obtain label-free quantification with a total cycle time of ~3.2 s. Briefly, in this acquisition, instead of the Q1 quadrupole transmitting a narrow mass range through to the collision cell, a wider window of variable window width (5 -90 m/z) is passed in incremental steps over the full mass range (m/z 400-1,250 with 64 SWATH segments, each with a 45 ms accumulation time, yielding a cycle time of 3.2 s, which includes one MS1 scan with a 250 ms accumulation time).
NOTE: The variable window width is adjusted according to the complexity of the typical MS1 ion current observed within a certain m/z range using a variable window calculator algorithm22 (more narrow windows are chosen in “busy” m/z ranges, wide windows in m/z ranges with few eluting precursor ions). On other MS instrument platforms, other DIA window strategies may be implemented.
7. Data analysis
NOTE: Some data analysis settings should be changed and tailored to the specific experiment. For example, the protein database (FASTA file) selected depends on the species from which the sample was prepared (here, Mus musculus). Below, the data analysis for mouse samples enriched for acetylated and succinylated peptides is described.
-
1Use an MS database search engine to analyze DDA acquisitions. Create a database search engine method as follows:
- For Sample Description parameters: select “Identification” under Sample Type, select “Iodoacetic Acid” under “Cysteine Alkylation”, select “Trypsin” under “Digestion” (assuming C-terminal cleavage at lysine and arginine), select “TripleTOF 6600” under Instrument, under Special Factors, check Acetylation emphasis and Succinylation enrichment, and select Mus musculus under Species.
- For Specific Processing parameters: select “Biological modifications” under ID Focus, select “SwissProt” under Database, check “Thorough ID” under Search Effort, select “0.05 (10%)” under “Detected Protein Threshold”, and check “Run False Discovery Rate Analysis” under “Results Quality”. Save the search engine method and submit the mass spectrometric raw files for processing by the database search engine using the generated method.
NOTE: In an iterative process, all MS and MS/MS scans are automatically recalibrated by the search engine based on initial annotations and results.
-
2
Click on “Export Peptide Summary” upon completion of the search and filter all peptide identification results by a “confidence threshold” of 99 in a spreadsheet software (e.g., Excel; false discovery rate [FDR] of 1%).
-
3
In the “Peptide Summary” spreadsheet file, filter for all peptides that contain the PTM annotation “acetylation” and “succinylation” in the modification column to generate a results report to present exclusively the acylated peptides and their corresponding proteins.
-
4
To build MS/MS spectral libraries for further processing the DIA raw file and further relative quantification, open the DIA Quantitative Analysis Software. Select the “Library” tab, then (at the bottom of the page) click “Generate Spectral Library” from “Database Search Engine” and open a Database Search Engine FDR report (the *FDR.xlsx file), which was automatically generated as part of the DDA raw file database search process. Then, click “next” and select the “LibrarySettings Schema” and “next”. Select “Uniprot_mouse_proteome” as the database, then click “next” and “goa_mouse” as the gene annotation (ontology) file. Finally, click “finish”, and the spectral library will be generated.
NOTE: Information about the acetylated and succinylated peptides from theDDA raw data files, precursor ion scans from MS1, and fragment ion scans from MS2 will be included in the spectral libraries (this includes retention time, MS/MS fragmentation pattern, etc.).
-
5Use DIA quantitative proteomics analysis software to perform relative quantification of acetylation and succinylation levels and create spreadsheets of candidate PTM-containing peptides that can be used for further data analysis.
-
1To analyze and quantify PTM-containing peptides open the DIA Quantitative Analysis Software, use the template analysis schema. The template schema is available in the software by selecting the option “Settings Perspective” | “DIA Analysis” | “BGS PTMs” (either significant or sparse, depending on the experiment).
-
1
NOTE: Instead of using the BGS PTM analysis template in the DIA Quantitative Analysis Software, all PTM-specific settings can also be manually set up following these instructions: 1) under “identification”, select “PTM localization” (probability cutoff = 0.75); 2) in “quantification”, select “minor peptide grouping” | “modified sequence”; and 3) under “post-analysis”, select “differential abundance grouping” | “minor group” (quantification settings). These settings will activate the PTM localization feature so that modified peptides are listed as individual entries and differential analysis are performed at the peptide level.
-
2
To start the Quantification Analysis, select the “Pipeline” tab, then click “Set up a DIA Analysis from File”, open the MS DIA raw files of interest for relative quantification. Select “Assign Spectral Library” and select the library that was built above, click “load” | “next”. Select the “BGS PTMs” analysis schema and click “next”. Select the appropriate database FASTA file “Uniprot_mouse_proteome” | “next”. Define the condition set-up, which assigns the different conditions to the samples, and click “next”. Select “goa_mouse” as the gene annotation (ontology) file and click “next”. Review the analysis overview (summary of the experiment set-up) and select “output directory” | “finish”. Finally, click “Run Pipeline” to perform the label-free quantitative analysis.
NOTE: Statistical modules in the DIA Quantitative Analysis Software automatically perform FDR analysis, generate heat maps and volcano plots comparing the different conditions, generate lists of identified and quantified peptides and proteins, and provide Q-values along with relative fold changes comparing different conditions.
-
6
As an alternative, use software for interference removal of DIA datasets for processing data and performing statistical analysis after exporting the extracted peak areas for acetylation and succinylation sites.
8. Data visualization of modified peptides and assessment of PTM site localization
To open a generated DIA Quantitative Analysis select the “Analysis” tab followed by “load the quantitative Analysis Software Experiment” (from a saved experiment opening a *.SNE file). Navigate to the *.SNE results file and select “open”.
In the left panel, click on the arrow to the left of the third raw data *.wiff file from the top to expand and visualize all 64 SWATH segments. Click on the arrow to the left of the segment [428.7-437.3] to expand the specific segment and display the modified peptides identified for this mass range. Click on the triply charged peptide KQYGEAFEK[Acetyl]R.
NOTE: By default, the right upper panel typically displays the MS2 XIC, and the lower panel displays MS1 Isotope Envelope XIC
In the upper panel, select “PTM Localization Plot”. In the lower panel, select “PTM Localization Plot”.
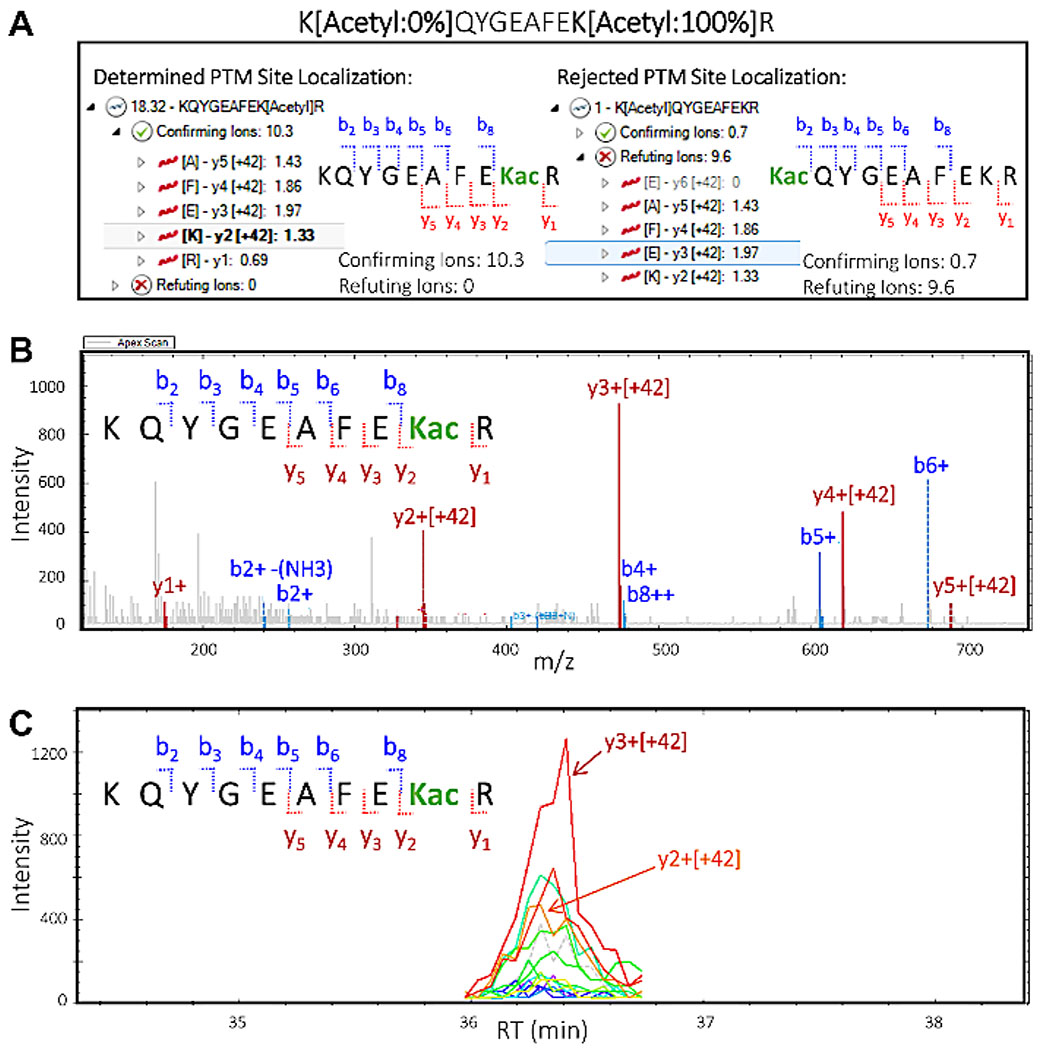
In the PTM Localization Plot, select the top peptide sequence “KQYGEAFEK[Acetyl]R”, with a total PTM localization score of 18.32. Click on the arrow to the left to expand the view and visualize the confirming and refuting ions. Click on the arrow next to the “Confirming Ions”, then select the ion “y3 [+42]” to highlight that particular ion, which confirms that the acetylation site is located on K2 of the peptide sequence.
In the PTM Localization Plot select the second peptide sequence “K[Acetyl]QYGEAFEKR” with a total (very low) PTM localization score of 1. Click on the arrow to the left to expand the view and visualize the confirming and refuting ions. Click on the arrow next to the “Confirming Ions”, then on the arrow next to the “Refuting Ions”, then select some fragment ions to visualize and assess the extracted ion chromatogram and MS/MS spectra (Figure 5).
Figure 5: Quantitative proteomics software deciphers the peptide site localization of PTMs.
Based on MS/MS fragmentation of the peptide, it is possible to provide information about the specific lysine residue the acetyl group is modifying. This showcases the ability of the software to offer valuable information on site localization of PTMs. (A) Two possibilities of lysine residue modification and PTM site localization: KQYGEAFEKacR (left) and KacQYGEAFEKR (right). “Confirming” and “refuting” fragment ions are shown for each of the potential site localization isoforms of the peptide. Based on this information, confirming scores and refuting scores are assigned, confirming the presence of isoform KQYGEAFEKacR in the sample. (B) MS/MS spectrum corresponding to the confirmed isoform KQYGEAFEKacR indicating that all y ions including the acetylated lysine residue (y2 and higher) carry an increment mass of +42 m/z, which corresponds to the acetyl modification. Observed b ions do not contain the modification. (C) Extracted ion chromatogram (XIC) with abundant peak areas resulting from y2 and y3 ions, both of which make up the acetylation site in the confirmed isoform KQYGEAFEKacR.
NOTE: The assigned PTM Localization Score and visual inspection indicate that the correct PTM isomer is KQYGEAFEK[Acetyl]R, while no or minimal evidence exists for the other possible PTM isomer K[Acetyl]QYGEAFEKR.
Representative Results
Figure 1 shows a general diagram of the workflow, including harvesting the tissue from mouse livers, using 1 mg of protein for digesting the protein lysate with trypsin, incubating peptides with antibody-conjugated beads, acquiring the samples on the MS, and finally performing DIA/SWATH analysis of the data using various quantitative proteomics software packages (academic and commercial).
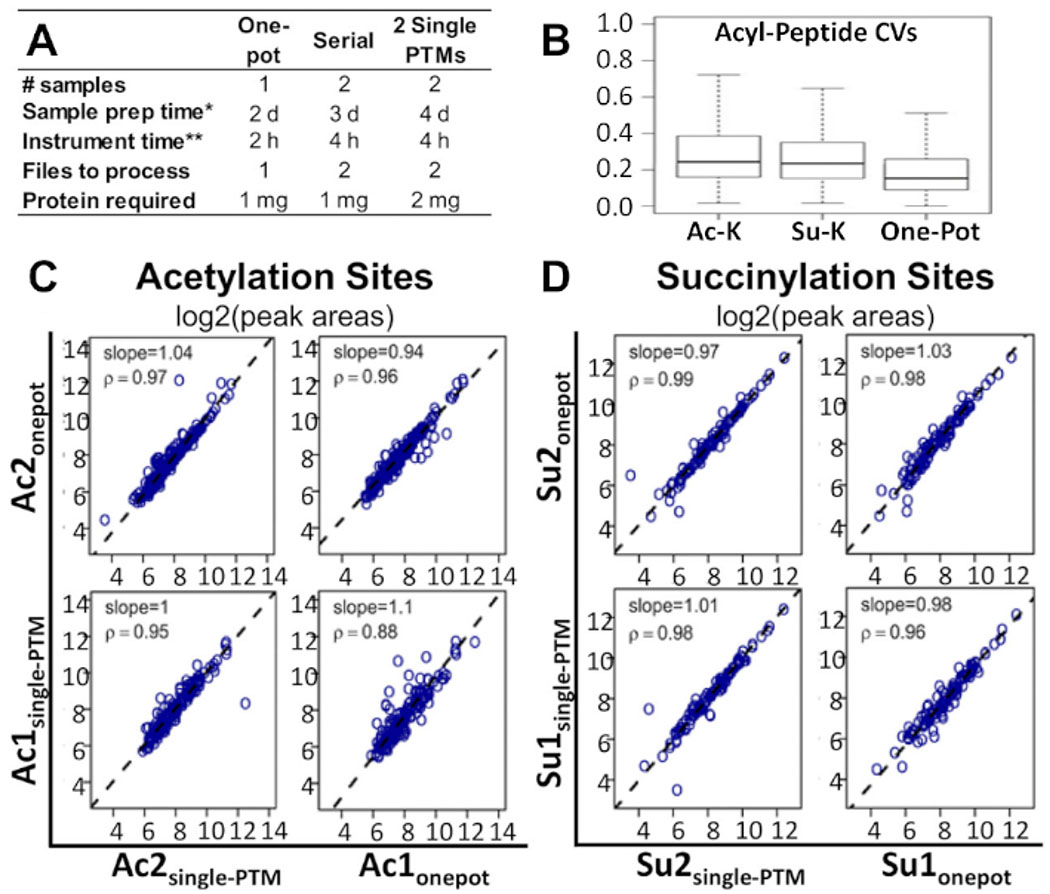
Figure 2A shows how the timeline of the workflow and the amounts of sample and protein required, compared to alternative methods currently being used for multi-PTM enrichment studies. The one-pot method can be performed in half as much time and with half the number of samples as these alternative methods. Compared to the two single-PTM enrichment method, the one-pot protocol also requires half the amount of protein.
Figure 2: Comparison of one-pot workflow with alternative methods.
(A) Comparison of time, costs, and materials required for the one-pot workflow, serial-PTM enrichment, and two single-PTM enrichments. (B) Comparison of CVs between the one-pot workflow, single acetyl-lysine PTM enrichment, and single succinyl-lysine PTM enrichment. Spearman correlation analysis comparing the acyl peptide peak areas obtained from the one-pot workflow, and the single-PTM enrichments: corresponding plots of the log2 peak area results for (C) acetylation sites and (D) succinylation sites. Regression slopes and correlation factors are indicated in the individual panels14. Two independent biological replicates were processed for each of the conditions. This figure has been modified from Basisty et al.14. .
This protocol has been shown to be a feasible and cost-effective alternative. Figure 2B shows that the median coefficient of variation (CV) for modified peptide areas was lower in the one-pot method than in the single-PTM and serial-PTM enrichments. Figure 2C,D shows that, while comparing the one-pot PTM and single-PTM enrichment methods, no noteworthy differences were apparent between the correlations of site-level quantifications for the two modifications. This was also true for the peptide-level and fragment-level correlations. The same observation held for all three correlations when comparing the one-pot and serial-PTM enrichments. All underlying MS raw data and processed Excel results sheets associated with a recent report by Basisty et al.14 are available and can be downloaded from MassIVE (MSV00081906) and ProteomeXchange (PXD008640).
In general, while antibody enrichment strategies may show certain limitations, such as potential epitope occlusion or limited specificity, the antibodies used in this study are mixtures of independently generated clones and thus provide wider ranges of specificities.
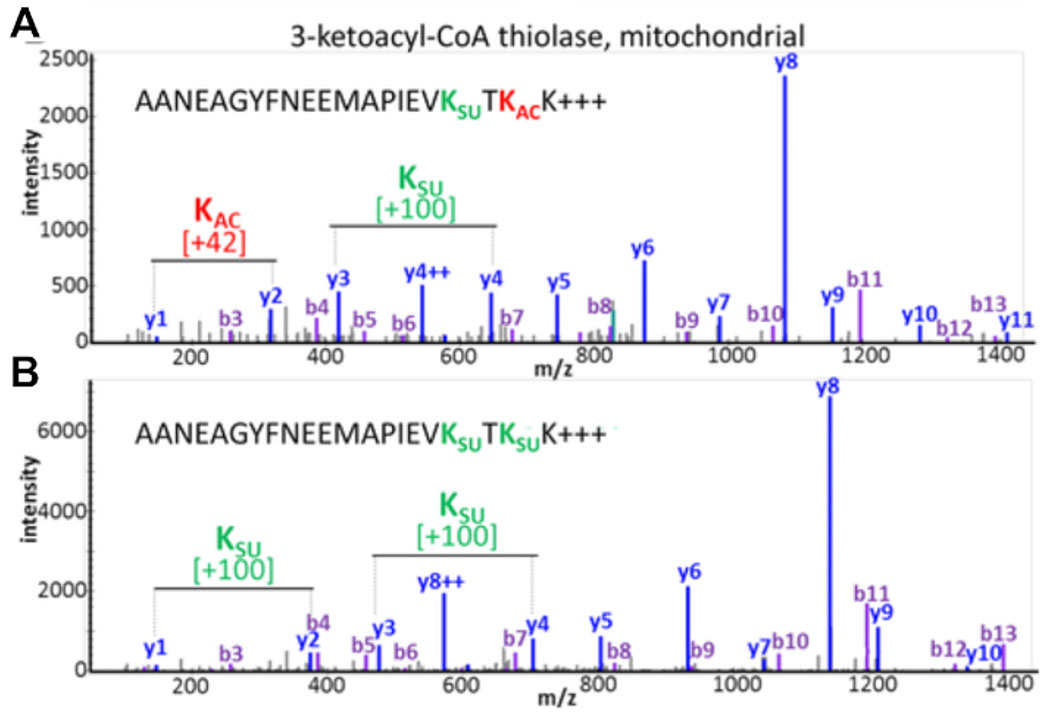
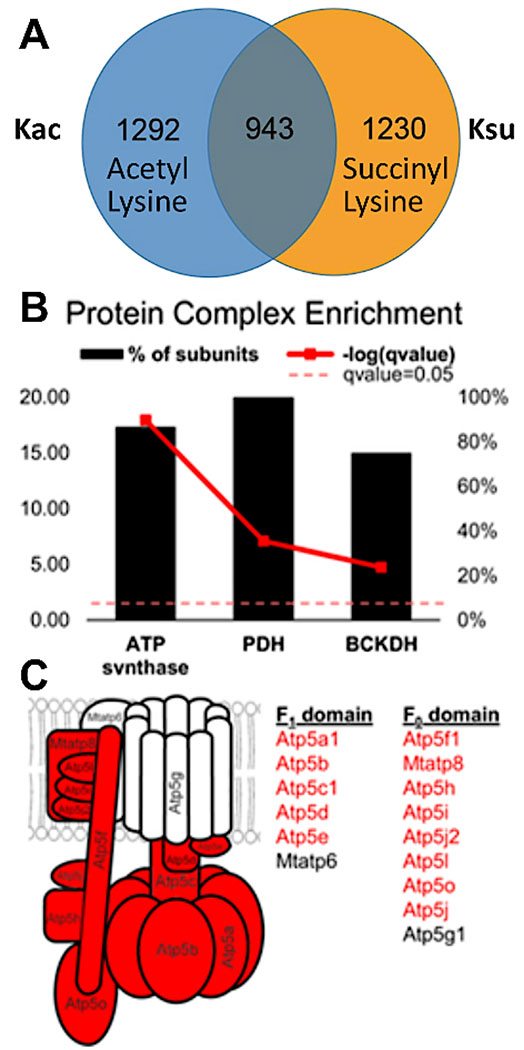
Experimental results document the possibility of detecting and assessing PTM crosstalk. Figure 3 displays data from a successful enrichment and illustrates an example for a peptide containing multiple and different acyl modifications visualizing PTM crosstalk. Figure 3A shows a peptide that is acetylated on one lysine residue and succinylated on the other, and Figure 3B shows the same peptide succinylated at both lysines. This demonstrates that the same lysine residue can be modified with both acylation groups, and there is a possibility of crosstalk occurring at that site. Figure 4 displays the number of lysine residues that were identified as described in sections 7.1-7.3 to carry both modifications, also pointing towards possible PTM crosstalk.
Figure 3: Crosstalk between acetylation and succinylation modifications of lysine residues.
MS/MS spectra of tryptic peptides from mitochondrial 3-ketoacyl-CoA thiolase that show the same amino acid sequence but have been modified at two lysine residues with different PTMs. (A) MS/MS of peptide AANEAGYFNEEMAPIEVKsuccTKacK and (B) MS/MS of peptide AANEAGYFNEEMAPIEVKsuccTKsuccK. This figure has been modified from Basisty et al.14.
Figure 4: Overlap and crosstalk between the acetylated and succinylated lysine residues–specific examples in protein complexes.
(A) Venn diagram displaying overlap between 2,235 acetylation and 2,173 succinylation sites. Of these, 943 sites were both acetylated and succinylated. Liver from a SIRT5 (de-succinylase) knockout mouse was analyzed, and many succinylation sites were identified. In fact, they were more abundant than normally observed in mouse liver (modified peptides were filtered at a Q value of <0.05). (B) Protein complexes showing the percentage of their subunits containing both acetylated and succinylated sites (bold red line represents the significance as determined by Fisher’s exact test). (C) Diagram of ATP synthase complex: protein subunits in red depict the subunits that contain both acetylated and succinylated sites. This figure has been modified from Basisty et al.14.
As Figure 5 demonstrates, processing DIA PTM datasets with quantitative proteomics software allows us to pinpoint which specific lysine residues are modified. This is a concept known as site localization, which is an essential step for any analysis for determination of possible PTM crosstalk. Figure 5 displays two potential isoforms along with the confirming and refuting ions for each that could be visualized and assessed as described in sections 8.1-8.3 (specifically steps 8.3.2 and 8.3.3). Based on this information, we were able to confidently identify which of the two isoforms was present in the original sample. The MS/MS spectrum of the confirmed isoform KQYGEAFEKacR demonstrates clearly that the y ions (y2-y5) containing the acetylated lysine residue, which shifted by 42 m/z (the increment mass of an acetyl group), confirmed the specific lysine residue in the peptide that was modified.
Discussion
This protocol describes a novel technique for simultaneous multiple PTM enrichment to more effectively understand PTM crosstalk. Alternative methods of reaching this objective tend to be prohibitively time-consuming and expensive, and they require large amounts of protein to be successful11,13. This protocol presents a multi-PTM enrichment workflow that involves incubation in antibody-conjugated beads for two PTMs at once to improve the overall efficiency of the experiment. This method also involves the use of DDA for spectral library generation and DIA MS acquisitions to detect and quantify the peptides present with reduced interference from fragment ions22,23. Software programs, such as MS database search engines are used to analyze and quantify data from DDA acquisitions, whereas Quantitative Proteomics Software24 and specific DIA Quantitative Analysis Software25 are necessary to interpret the complex spectra produced by DIA acquisitions.
There are several critical steps within this protocol that should be followed carefully. As the main goal of the protocol is to enrich for multiple PTMs simultaneously, the antibody-affinity enrichment step (section 2) is critical to success of the experiment. When performing washes on the beads, it is necessary to ensure none of the beads are aspirated accidentally. Ensuring the urea concentration has been diluted to 1 M prior to digestion with trypsin (step 1.8) is also necessary. Although 8 M urea is required earlier in the protocol for protein solubilization, urea concentrations above 1 M will inhibit trypsin’s enzymatic activity. In addition, it is important to consistently check the pH of the sample throughout the protocol. This is especially important prior to digestion. If the pH of the sample and the trypsin solution are not neutralized appropriately prior to the incubation, it can result in an inefficient digestion wherein many cleavage sites may be missed, resulting in fewer peptide identifications.
A few modifications to the protocol may be helpful when preparing samples. For a protein lysate procured from 1 mg of starting material, a quarter of the antibody beads provided in each PTM scan tube can be used as a cost-effective alternative. A greater amount of starting material can be used for better results, as long as the amount of antibody beads used are increased proportionately. Another modification that can enhance the workflow is to digest samples with another protease in addition to trypsin. This modification would result in more variability in the peptides cleaved, providing increased coverage of protein residues. Although not necessary, it is recommended that trypsin be one of the enzymes used for PTM analysis due to its high cleavage specificity26.
A limitation of this protocol is that the PTMs being studied need to have similar chemistries in order to be enriched simultaneously14. The procedures for the different antibody-conjugated beads must be similar, utilizing similar solvents and solutions, including similar elution conditions, and preferably from the same vendor. For this reason, the method outlined here consistently uses acetylation and succinylation as an example, both of which use antibody-conjugated beads (Cell Signaling Technology, Inc). Although this method can theoretically be applied to any number of PTMs, additional studies would be needed to assess the exact limitation of the protocol in this respect. Furthermore, as this is an antibody-based enrichment method, the method can only provide a relative quantification of PTM sites.
In comparison with existing multi-PTM enrichment methods, this workflow is a more feasible and cost-effective alternative. From this experiment, it was observed that the efficacy of this method compares extremely well with alternative methods, such as individual or serial enrichments. Figure 2B shows that the median CV for modified peptide peak areas was actually decreased in the one-pot method in comparison to single-PTM enrichments and serial-PTM enrichments13. We further analyzed the experimental results assessing site-level quantifications for acetylation or succinylation. Additionally, Spearman correlation analysis (Figure 2C,D) demonstrated that the one-pot PTM enrichment performed similarly to the single-PTM enrichment workflows. This was also true for the peptide- and fragment-level correlations. The same observation held for all three correlations when comparing one-pot with serial-PTM enrichment.
This protocol allows researchers to make fascinating biological insights into PTM crosstalk in a quick and cost-effective manner. The DIA component of the workflow allows researchers to understand more about PTMs, as it provides information about site localization and overcomes challenges such as low site occupancy of PTMs. Precursor ions tend to be excluded with DDA, which is particularly important when studying PTMs, as site occupancy is often low to the point where these peptides do not get selected for MS/MS but still contain crucial information. Follow-up experiments could be performed to assess the upper limit of how many PTMs can be enriched simultaneously using this method. A future improvement of this workflow may include development of more advanced software platforms to further automate the analysis of site localization and PTM site occupancy.
Acknowledgments
We acknowledge the support from the NIH shared instrumentation grant for the TripleTOF system at the Buck Institute (1S10 OD016281). This work was also supported by the National Institute of Allergy and Infectious Disease (R01 AI108255 to B.S.) and the National Institute of Diabetes and Digestive and Kidney Diseases (R24 DK085610 to Eric Verdin; R01 DK090242 to Eric Goetzman). X.X. was supported by a grant from the National Institutes of Health (NIH grant T32GM8806, to Judith Campisi and Lisa Ellerby), N.B. was supported by a postdoctoral fellowship from the Glenn Foundation for Medical Research.
Footnotes
The video component of this article can be found at https://www.jove.com/video/60780/
Disclosures
The authors have nothing to disclose.
References
- 1.Christensen DG et al. Post-translational Protein Acetylation: An Elegant Mechanism for Bacteria to Dynamically Regulate Metabolic Functions. Frontiers in Microbiology. 10, 1604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deribe YL, Pawson T, Dikic I Post-translational modifications in signal integration. Nature Structural Molecular Biology. 17 (6), 666–672 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Sadoul K, Boyault C, Pabion M, Khochbin S Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 90 (2), 306–312 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Swaney DL et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nature Methods. 10 (7), 676–682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P The regulation of protein function by multisite phosphorylation--a 25 year update. Trends in Biochemical Sciences. 25 (12), 596–601 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Grunstein M Flistone acetylation in chromatin structure and transcription. Nature. 389 (6649), 349–352 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Struhl K Histone acetylation and transcriptional regulatory mechanisms. Genes and Development. 12 (5), 599–606 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Mocciaro A, Rape M Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. Journal of Cell Sciences. 125 (Pt 2), 255–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Otin C, Hunter T The regulatory crosstalk between kinases and proteases in cancer. Nature Reviews in Cancer. 10 (4), 278–292 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Du Z et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science Signalling. 3 (146), ra80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McManus FP, Lamoliatte F, Thibault P Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach. Nature Protocols. 12 (11), 2342–2358 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Venne AS, Kollipara L, Zahedi RP The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 14 (4-5), 513–524 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Mertins P et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nature Methods. 10 (7), 634–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basisty N, Meyer JG, Wei L, Gibson BW, Schilling B Simultaneous Quantification of the Acetylome and Succinylome by ‘One-Pot’ Affinity Enrichment. Proteomics. 18 (17), e1800123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G et al. Regulation of UCP1 and Mitochondrial Metabolism in Brown Adipose Tissue by Reversible Succinylation. Molecular Cell. 74 (4), 844–857 e847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdin E, Ott M 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nature Reviews Molecular Cell Biology. 16 (4), 258–264 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Rardin MJ et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Science U. S. A 110 (16), 6601–6606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rardin MJ et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metabolism. 18 (6), 920–933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrico C, Meyer JG, He W, Gibson BW, Verdin E The Mitochondrial Acylome Emerges: Proteomics, Regulation by Sirtuins, and Metabolic and Disease Implications. Cell Metabolism. 27 (3), 497–512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadhukhan S et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proceedings of the National Academy of Science U. S. A 113 (16), 4320–4325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins BC et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature Communications. 8 (1), 291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schilling B, Gibson BW, Hunter CL Generation of High-Quality SWATH((R)) Acquisition Data for Label-free Quantitative Proteomics Studies Using TripleTOF((R)) Mass Spectrometers. Methods in Molecular Biology. 1550, 223–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer JG, Schilling B Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Reviews in Proteomics. 14 (5), 419–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26 (7), 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sticker A, Martens L, Clement L Mass spectrometrists should search for all peptides, but assess only the ones they care about. Nature Methods. 14 (7), 643–644 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Olsen JV, Ong SE, Mann M Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Molecular Cell Proteomics. 3 (6), 608–614(2004). [DOI] [PubMed] [Google Scholar]