Significance
The biosynthetic genes of some specialized plant metabolites appear to be clustered in the genomes of higher plants. Momilactones are defense compounds produced in rice and barnyard grass by family-conserved biosynthetic gene clusters (BGCs). We sequenced the genome of Calohypnum plumiforme, a momilactone-producing nonvascular bryophyte, and showed that it also contains a functionally similar momilactone BGC distinguished by its lack of synteny with the clusters found in vascular plants. The expression of the Calohypnum biosynthetic genes in tobacco demonstrated their role in momilactone A production. This is the first report of a BGC for a specialized metabolite in bryophytes. Our findings indicate that the momilactone clusters present in three different plant species may have evolved independently via convergent evolution.
Keywords: specialized metabolites, gene cluster, Bryophytes, convergent evolution, diterpene momilactones
Abstract
Momilactones are bioactive diterpenoids that contribute to plant defense against pathogens and allelopathic interactions between plants. Both cultivated and wild grass species of Oryza and Echinochloa crus-galli (barnyard grass) produce momilactones using a biosynthetic gene cluster (BGC) in their genomes. The bryophyte Calohypnum plumiforme (formerly Hypnum plumaeforme) also produces momilactones, and the bifunctional diterpene cyclase gene CpDTC1/HpDTC1, which is responsible for the production of the diterpene framework, has been characterized. To understand the molecular architecture of the momilactone biosynthetic genes in the moss genome and their evolutionary relationships with other momilactone-producing plants, we sequenced and annotated the C. plumiforme genome. The data revealed a 150-kb genomic region that contains two cytochrome P450 genes, the CpDTC1/HpDTC1 gene and the “dehydrogenase momilactone A synthase” gene tandemly arranged and inductively transcribed following stress exposure. The predicted enzymatic functions in yeast and recombinant assay and the successful pathway reconstitution in Nicotiana benthamiana suggest that it is a functional BGC responsible for momilactone production. Furthermore, in a survey of genomic sequences of a broad range of plant species, we found that momilactone BGC is limited to the two grasses (Oryza and Echinochloa) and C. plumiforme, with no synteny among these genomes. These results indicate that while the gene cluster in C. plumiforme is functionally similar to that in rice and barnyard grass, it is likely a product of convergent evolution. To the best of our knowledge, this report of a BGC for a specialized plant defense metabolite in bryophytes is unique.
Momilactones, including the two major structurally diverged forms momilactone A and B, originally isolated from the hulls of cultivated rice Oryza sativa (Os), are specialized diterpenoid metabolites, with the potential to competitively inhibit the growth of other plants and microorganisms, and thought to function as a chemical defense against pathogens (1–6). In addition to their original physiological roles in plants, it is also reported that these molecules have cytotoxic and antitumor activity in cancer cell lines (7). Therefore, momilactones exhibit a broad range of growth-inhibitory effects in various organisms, such as animals, plants, fungi, and bacteria.
In addition to O. sativa, some wild-rice species in the genus Oryza are also known to produce momilactones (8). Moreover, we previously reported that the moss Calohypnum plumiforme (Cp) (Hypnum plumaeforme [Hp] prior to 2019 in the National Center for Biotechnology Information) produces momilactone as a chemical defense compound and the CpDTC1/HpDTC1 gene, by encoding a stress-inducible bifunctional diterpene cyclase (DTC) (syn-pimaradiene synthase), is key in the first committed step of the momilactone biosynthetic pathway (9). Feeding experiments using uniformly labeled 13C-syn-pimaradiene have demonstrated the existence of an in planta biosynthetic route of momilactone production from syn-pimaradiene in C. plumiforme (10). Thus, momilactones are specialized diterpenoid metabolites produced in evolutionally highly diverged plant species.
The biosynthetic genes of a number of specialized metabolites produced in plants have been shown to be clustered in the genome of the plants in which they are found. For example, the biosynthetic gene cluster (BGC) for DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one, a naturally occurring hydroxamic acid) located on maize chromosome 4, was the first reported example of operonic gene clustering in higher plants associated with allelochemical production (11). Oat avenacin, a specialized triterpenoid saponin, is also the product of clustered genes in the oat genome (12). More recent examples of plant BGCs are those forming diterpenoid phytoalexins, including momilactones in rice. Initially it was noted that the terpene cyclase genes for diterpenoid phytoalexins are located within a narrow genomic region of the rice chromosome (13). Subsequently it was found that genes encoding enzymes functioning at later steps of the momilactones biosynthetic pathway are within this region and their enzymatic functions were confirmed. In fact the majority of the momilactone A biosynthetic genes, including OsCPS4 (syn-CDP synthase), OsKSL4 (syn-pimaradiene synthase), CYP99A2, CYP99A3 (Cytochrome P450 monooxygenase [CYP] for syn-pimaradiene oxidations), and OsMAS (dehydrogenase momilactone synthase) are clustered on chromosome 4 in rice and coordinately regulated under stress conditions (14, 15). While the biosynthetic genes involved momilactone A formation are known, the genes that encode the enzymes mediating the conversion steps of momilactone A to B are still unknown among all momilactone-producing plants.
Recently, Echinochloa crus-galli (Ec, barnyard grass) has been shown to possess one copy of the momilactone gene cluster in its genome. Thus, all of the momilactone-producing higher plants reported thus far appear to have a family-conserved BGC in their genomes. In addition, the expression of the orthologous genes EcKSL and EcMAS in the E. crus-galli momilactone A gene cluster are coordinately induced upon pathogen infection (16). As mentioned above, the nonvascular moss C. plumiforme also produces momilactones. However, the clustering (or lack thereof) of momilactone biosynthesis genes in the moss genome was not previously examined since the only available moss genome sequence was that of Physcomitrella patens, reported by Rensing et al. (17) over a decade ago, with no evidence that P. patens produces any momilactones. Moreover, Physcomitrella species are phylogenetically very distant from the Calohypnum mosses (17–19).
The evolution of gene clusters has been hypothesized to proceed by recruitment of genes from elsewhere in plant genomes via gene duplication and neofunctionalization, rather than by horizontal gene transfer from other organisms, such as fungi and microbes. It has also been suggested that partial clustering of plant genes occurs in different metabolic pathways by the duplication of functionally related gene pairs and modules (20). At present, there is little evidentially supported information to aid in understanding the evolution of gene clustering among plant species and how this leads to the formation of specialized metabolites. Moreover, there are no studies that describe the existence and nature of biosynthetic gene clustering in bryophytes and other nonseed-forming plants.
In this study, to examine the evolution of BGCs for momilactone formation, we sequenced the C. plumiforme genome and provide evidence of a momilactone BGC in this nonvascular plant. We further show that the genes in this cluster are inducible and their encoded products are responsible for momilactone formation. Phylogenetic analysis of the clustered genes and syntenic comparisons of the gene cluster organization in the genomes of species producing momilactones indicate that these clusters might have evolved independently via convergent evolution in each plant.
Results
Genome Sequencing, Assembly, and Annotation.
The genome survey of C. plumiforme suggested that it is a typical simple diploid genome with low heterozygosity and repeat content, with an estimated genome size of ∼434 Mb (Table 1 and SI Appendix, Fig. S1). The C. plumiforme genome sequencing was performed through the combination of Illumina and PacBio Single Molecule Real Time (SMRT) technology (Table 1 and SI Appendix, Table S1A). For Illumina sequencing, 350-bp to 20-kb insertion libraries were constructed with one PacBio sequencing cell data. In order to identify the candidate momilactone gene cluster, we tested several strategies to assemble the C. plumiforme genome and determined that the best genome assembly v5.0 (V5.0) was achieved based on the data from the hybrid approach with both Illumina and PacBio sequencing reads (Table 1 and SI Appendix, Table S2). The final V5.0 assembly has an accumulated length of 335 Mb, accounting for 77% of the estimated genome size (SI Appendix, Table S3). Benchmarking universal single-copy orthologs (BUSCO) were applied to evaluate the completeness of the C. plumiforme assembly. Based on the Viridiplantae BUSCO datasets (V3) (21), we found that 93.9% of the gene space was covered. The gene space coverage for the C. plumiforme assembly is similar to that reported for another moss genome, P. patens (94.2%).
Table 1.
Summary of the C. plumiforme genome assembly
| Sequence and annotation | ||||
| Sequencing and assembly | ||||
| Sequencing | Insert libraries: 350bp ∼ 20 kb | Illumina∼203× | Pacbio RS II: ∼3.5× | Total reads ∼90 Gb |
| Assembly (scaffold) | N50 size: 790.02 kb | N90 size: 241.57 kb | Longest: 3.38 Mb | Total size: 335 Mb |
| Genome annotation | ||||
| Protein-coding gene | Gene models: 32,195 | Swiss-Prot hits: 12,342 | BUSCO 93.9% | Gene size: 2,948 bp |
| Repetitive elements (%) | LTR: 16.1 | LINEs and SINEs: 0.96 | DNA transposon: 3.98 | Total: 49.17 |
LINE, long-interspersed retrotransposable element; LTR, long-terminal repeat; SINE, short-interspersed retrotransposable element.
To annotate repeat elements in the C. plumiforme genome, we first built the de novo repeat library from the assembled genome. We identified 164.8 Mb of repetitive elements (16.41% long terminal-repeat elements) covering 49.17% of the assembled genome (Table 1 and SI Appendix, Table S4). De novo gene-structure predictions were carried out based on combined evidence from ab initio prediction using AUGUSTUS/GeneMark.hmm/FGENESH and similarity searches (for details, see Materials and Methods and SI Appendix, Fig. S2). The annotated coding sequences of moss P. patens (v3.3), liverwort Marchantia polymorpha (v3.1), and RNA-sequencing (RNA-seq) reads obtained from C. plumiforme in our previous study (9) were used to perform the homology predictions. All candidate gene models from the above evidence were combined using EVidenceModeler (22), with a higher weight for the expression evidence (RNA-seq). After removing gene structures with low quality or those containing repeat elements, 32,195 protein-coding genes were finally annotated in the genome assembly (Table 1).
Evolution of the Calohypnum Genome.
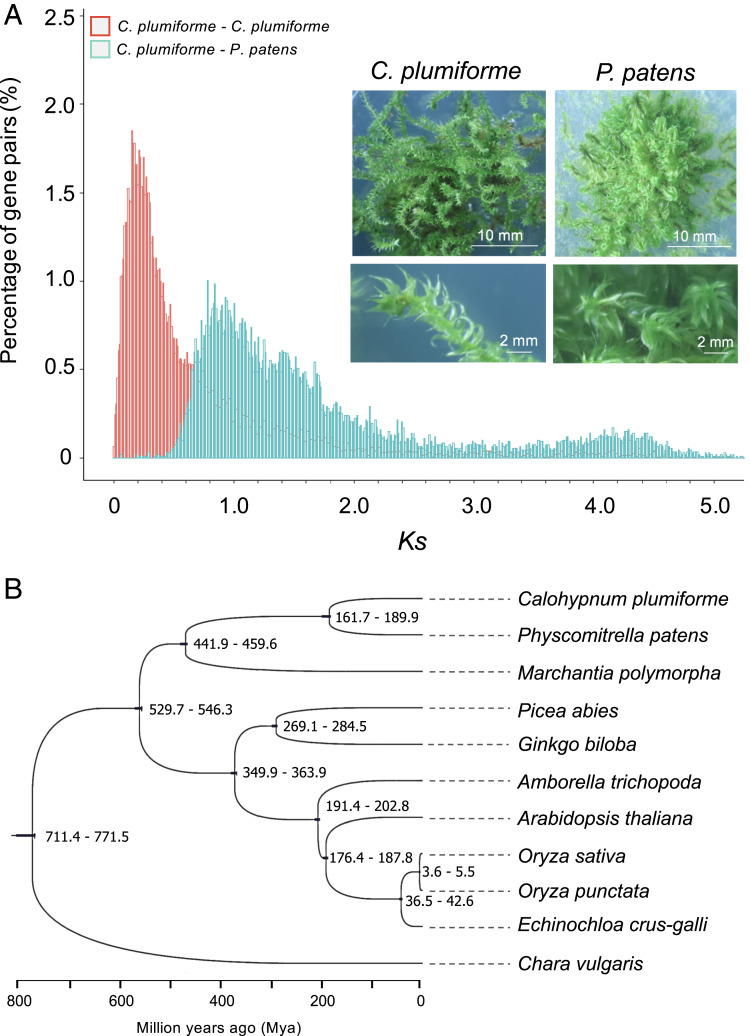
Mosses are important basal members of the plant kingdom that have diversified to inhabit a wide range of environments worldwide. The genome sequence of only one moss, P. patens, had been reported over a decade ago (17). Based on annotated genes of the Calohypnum genome assembly and the Ks (rate of synonymous substitution) distribution of paralogous gene pairs, at least one whole-genome duplication event could be identified (Fig. 1A). The event dates to approximately 8 Mya (Ks peak located at 0.125 to 0.175), modeled after the predicted time of divergence from another moss, P. patens.
Fig. 1.
Evolution of the Calohypnum genome. (A) Graph showing the Ks (rate of synonymous substitution) distribution of paralogous gene pairs of C. plumiforme and orthologous gene pairs between C. plumiforme and P. patens. The Inset in the panel is a photograph of the two moss species under analysis in this study. (B) Phylogenetic trees of showing the relation between of bryophytes and seed plants based on single-copy genes of their chloroplast genomes. The alga Chara (a nonland plant) was used as outgroup. The divergence time periods (Mya) at branches and time scales are shown.
We also assembled the Calohypnum chloroplast genome using our sequencing data, and with these data, we were able to carry out evolutionary analysis with more species at key evolutionary nodes. Using Chara as outgroup, the phylogenetic tree based on single-copy genes in their chloroplast genomes indicated that the two mosses with available genome sequences separated ∼162 to 190 Mya, similar to the time at which divergence occurred between dicot and monocot plants (∼176 to 189 Mya) (Fig. 1B). Unsurprisingly, no apparent genomic synteny was found between the two mosses P. patens and C. plumiforme examined in this study. This is reasonable, given their ancient divergence time. Therefore, the genome sequence of C. plumiforme reported in this study provides an important reference point for species in Bryidae (13 orders) and a valuable comparison on a new lineage. Recently, the genome sequence of liverwort M. polymorpha has been reported (23). We also found that the split between the mosses and other early land plants, such as liverworts, is estimated to have occurred over 450 Mya (Fig. 1B).
Identification of a Momilactone Gene Cluster in the Calohypnum Genome.
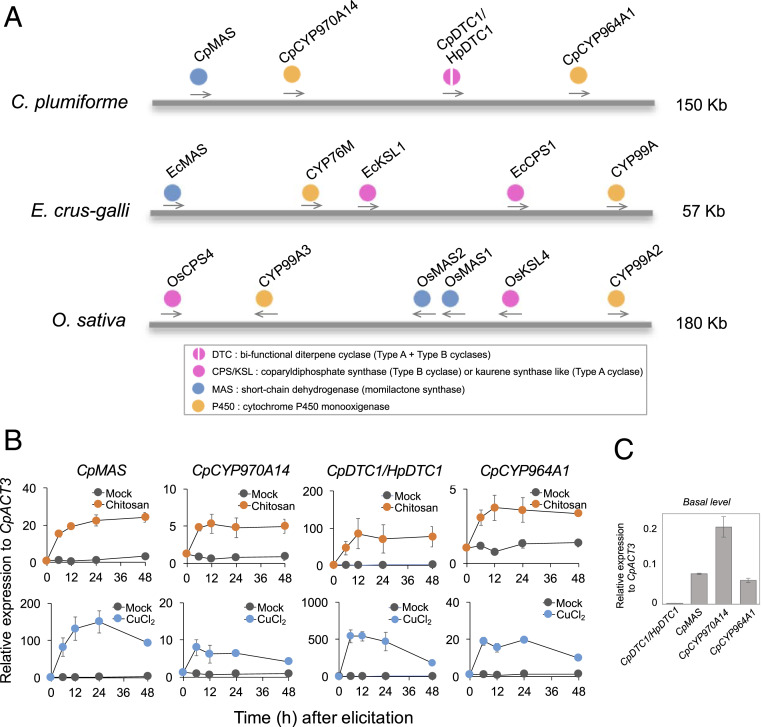
In order to identify any putative momilactone gene clusters in the Calohypnum genome, we first searched for orthologs of the O. sativa terpene cyclase genes (OsCPS4 and OsKSL4), CYP genes (OsCYP99A2/3), and dehydrogenase momilactone A synthase (MAS) (OsMAS) in the Calohypnum genome assembly. A putative momilactone gene cluster, which includes one terpene synthase gene (CpDTC1/HpDTC1), two CYP homologs, and one MAS homolog within a 150-kb region, was eventually identified in one scaffold (Scaffold38) of the V5.0 assembly built from several contigs (Fig. 2A and SI Appendix, Fig. S3). The putative cluster was located at a contig (i.e., no gaps in the 150-kb region).
Fig. 2.
The putative gene cluster for momilactone biosynthesis in C. plumiforme. (A) The genomic synteny of the BGCs for momilactone formation in plants. (B) Time course gene-expression profiles of the four genes (CpDTC1/HpDTC1, CpMAS, CpCYP970A14, and CpCYP964A1) found in the momilactone BGC after the treatment of CuCl2 or chitosan. CpMAS: Unigene_5051, LC494432; CpCYP970A14: Unigene_16484, LC494433; CpDTC1/HpDTC1: LC128408; CpCYP964A1: Unigene_12783, LC494434. The data shown were normalized by comparison to the expression of the housekeeping gene CpACT3. (C) Comparison of basal expression levels of the four genes in the cluster. The expression data represent the mean ± SD of three biological replicates with independent elicitor treatments to C. plumiforme gametophytes.
We previously reported that the biosynthesis of momilactones and relevant gene expression in C. plumiforme is induced by biotic and abiotic stresses, such as fungal infection and heavy metal treatment (9). Therefore, we examined the expression profiles of the clustered genes in response to environmental stresses. To determine whether there was a coordinated transcription of the clustered momilactone biosynthesis genes in the C. plumiforme genome or not, RNA-seq data obtained from C. plumiforme CuCl2-treated foliage bodies were reanalyzed.
Significant transcriptional induction was found in the four candidate genes clustered in the 150-kb region of Scaffold38 following CuCl2-induced stress in our RNA-seq analysis (SI Appendix, Fig. S4). We further verified the time course of gene expression for the four clustered genes (designated as CpDTC1/HpDTC1, CpCYP970A14, CpCYP964A1, and CpMAS based on their amino acid identities, described later) following both CuCl2 and chitosan treatment using qRT-PCR. Treatment with both stress inducers resulted in significantly increased transcript levels of the four tandemly arranged genes by 6 to 12 h posttreatment (Fig. 2B). These results suggest high induction of the expression of all clustered genes in response to both abiotic and biotic stress, thereby inductively producing the bioactive momilactones for chemical defense. In addition, compared to their basal level of expression, the levels of CpCYP970A14, CpCYP964A1, and CpMAS genes, which would be responsible for the latter step of the pathway, were 30- to 100-fold higher than that of CpDTC1/HpDTC1 under no treatment condition (Fig. 2C), suggesting that different types of transcriptional regulation are involved in the expression of each momilactone biosynthetic gene in the moss cluster.
Enzymatic Functions of the Clustered Genes for Momilactones.
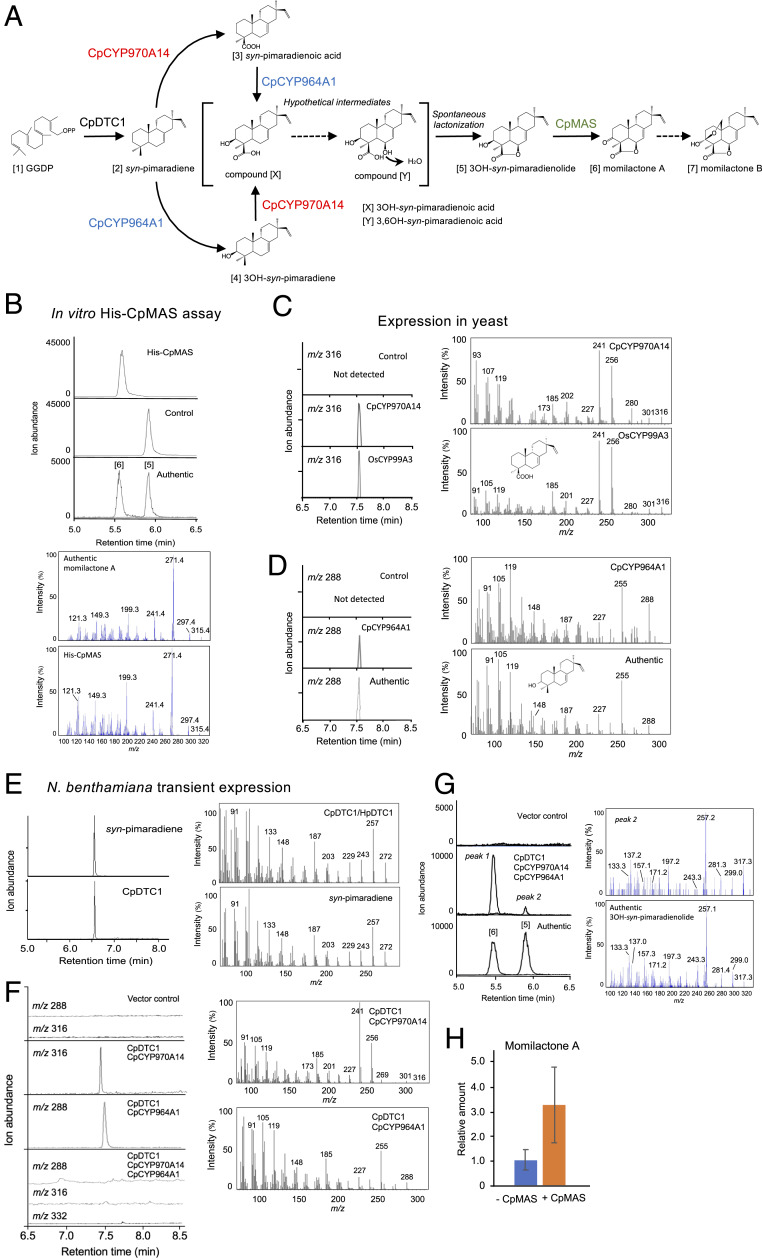
To provide direct evidence that the clustered genes identified in the Calohypnum genome encode active enzymes involved in momilactone synthesis, we tested their enzymatic function. The predicted full-length 834 bp CpMAS cDNA was generated using RT-PCR. The cDNA encodes a 277-amino acid protein (LC494432) with 52.3% and 55.7% identity to rice OsMAS and barnyard grass EcMAS, respectively. Recombinant His-tagged CpMAS protein (His-CpMAS) was expressed in Escherichia coli, affinity-purified, and analyzed for enzymatic function in vitro by incubation with 3β-hydroxy-9β-pimara-7,15-dien-19,6β-olide (3OH-syn-pimaradienolide) and NAD+ as substrates. As shown in Fig. 3A, the recombinant His-CpMAS protein catalyzed the formation of momilactone A [6] synthesis that could be easily identified following LC-MS/MS analysis (Fig. 3B). The identity of the momilactone A reaction product was further validated by GC-MS analysis (SI Appendix, Fig. S5). The result suggests that the CpMAS catalyzes the same enzymatic function as that done by the previously reported orthologous rice enzyme, OsMAS (8).
Fig. 3.
Functional analyses of clustered genes in Scaffold38. (A) The products and corresponding genes involved are indicated in the putative biosynthetic pathway of momilactones in C. plumiforme. Hypothetical intermediates are also shown in parentheses. (B, Upper) Momilactone A [6] converted from the substrate 3β-hydroxy-9βH-pimara-7,15-dien-19,6β-olide [5] by His-CpMAS protein is analyzed by LC-MS/MS. (Top) His-CpMAS products; (Middle) negative control without enzyme; (Bottom) authentic standards. (B, Lower) Product ion spectra of authentic momilactone A [6] and His-CpMAS product. (C) Mass chromatogram (scanned on m/z 316) of the reaction product (methyl ester derivative) obtained using GC-MS after incubation of syn-pimaradiene [2] with CpCYP970A14 (unigene16484), and OsCYP99A3. The mass spectra of the peaks on GC at 7.65 min are shown (Right). (D) Mass chromatogram (scanned on m/z 288) of the reaction product obtained using GC-MS after incubation of syn-pimaradiene [2] with CpCYP964A1 (Unigene12783). The mass spectra of the peaks on GC are shown (Right). (E) GC-MS mass chromatogram (scanned on m/z 272) of 9β-pimara-7,15-diene [2] produced by N. benthamiana in planta reaction. The mass spectra of the peaks on GC at 7.65 min are shown (Right). (F) Mass chromatograms and spectra of the reaction products from N. benthamiana leaves coexpressed CpDTC1/HpDTC1, CpCYP970A14, and CpCYP964A1. (G) LC-MS/MS analysis of the reaction products from N. benthamiana leaves coexpressed CpDTC1/HpDTC1, CpCYP970A14, and CpCYP964A1. The product ion spectra of the peak2 and authentic 3OH-pimaradienolide [5] are shown (Right). (H) Enhanced production of momilactone A in the N. benthamiana leaves coexpressed CpDTC1/HpDTC1, CpCYP970A14, CpCYP964A1 with or without CpMAS. The relative amount of momilactone A to 3OH-pimaradienolide are shown. The data represent the mean ± SE of three biological replicates of independently agroinfiltrated N. benthamiana leaves.
To analyze the two putative CYPs in the cluster, full-length cDNAs (Unigene_16484 and Unigene_12783) corresponding to CpCYP970A14 and CpCYP964A1, respectively, were amplified by RT-PCR and the amplification products were cloned into a Pichia pastoris yeast expression vector. The names of two CYPs were formally named by David Nelson according to the cytochrome P450 nomenclature system (24). The full-length cDNA of CpCYP970A14 and CpCYP964A1 were determined to be 1,527 bp encoding 508 amino acids (LC494433), and 1,404 bp encoding 467 amino acids (LC494434), respectively. CpCYP970A14 has 39.1% identity to rice CYP71A1 and 41.8% identity to Arabidopsis thaliana CYP75B1, while CpCYP964A1 has 39.5% identity to rice CYP707A7 and 43.8% identity to Selaginella moellendorffii CYP707C1. Liquid cultures of the transformed yeast cells independently expressing the CpCYP970A14 and CpCYP964A1 cDNAs were fed syn-pimara-7,15-diene [2] and the enzymatic conversion products were analyzed by GC-MS. Pichia cells expressing the CpCYP970A14 cDNA exhibited the ability of the rice to produce syn-pimara-7,15-dien-19-oic acid [3] based on the direct comparison to the spectrum of the syn-pimara-7,15-dien-19-oic acid (as methyl ester derivative) produced by rice CYP99A3, which has previously been shown as syn-pimara-7,15-diene oxidase (25) (Fig. 3C). Negative control yeast harboring only vector plasmid showed no corresponding signal. This result indicated that CpCYP970A14 unequivocally catalyzes oxidation of the C19 methyl group of syn-pimara-7,15-diene [2] to form the momilactone biosynthetic precursor syn-pimara-7,15-dien-19-oic acid [3] (Fig. 3A). When the Pichia yeast cells expressing CpCYP964A1 were fed with syn-pimara-7,15-diene, a product was detected in the windows of m/z 288 selected ion scanning, whereas the negative control did not show any significant peak. The mass spectrum of the product was identical to that of authentic 3β-hydroxy-9β-pimara-7,15-diene [4] reported previously, suggesting that CpCYP964A1 is responsible for catalyzing hydroxylation at the C3 position of syn-pimara-7,15-diene [2] (Fig. 3D) (26, 27).
To provide further validation of the C. plumiforme momilactone A biosynthetic gene function, we also examined their function by transiently expressing them in Nicotiana benthamiana. The CpDTC1/HpDTC1, CpCYP970A14, and CpCYP964A1 genes were cloned into a pEAQ-HT expression vector (28) and then introduced into N. benthamiana leaves using the Agroinfiltration method. As shown in Fig. 3E, it was possible to detect the in planta production of syn-pimaradiene following transient expression of CpDTC1/HpDTC1 [2]. When CpCYP970A14 was coexpressed with CpDTC1/HpDTC1 in N. benthamiana, it was possible to detect the in planta conversion of syn-pimaradiene [2] to syn-pimara-7,15-dien-19-oic acid [3] (Fig. 3F). This was based on direct comparison to the spectrum of the syn-pimara-7,15-dien-19-oic acid (as methyl ester derivative) successfully produced by rice CYP99A3 in N. benthamiana (SI Appendix, Fig. S6). Similarly, CpCYP964A1 was also found to catalyze the conversion from syn-pimaradiene [2] to 3β-hydroxy-9β-pimara-7,15-diene [4] in N. benthamiana (Fig. 3F). These enzymatic activities were identical to what was observed in our yeast expression system (Fig. 3D).
When both CpCYP970A14 and CpCYP964A1 were coexpressed, neither syn-pimaradienoic acid [3] nor 3β-hydroxy-9β-pimara-7,15-diene [4] produced by CpCYP970A14 or CpCYP964A1 could be detected (Fig. 3F), presumably due to a further metabolism into a hypothetical intermediate 3OH-syn-pimaradienoic acid (Fig. 3A, compound X). However, there was no relevant signal corresponding to the expected molecular ion (m/z 332 as methyl ester derivative) of compound X in GC-MS analysis (Fig. 3F). On the other hand, synthesis of 3OH-syn-pimaradienolide [5] was detected using LC-MS/MS (Fig. 3G, peak 2). Furthermore, accumulation of momilactone A [6] was more clearly defined in the N. benthamiana leaves expressing CpDTC1/HpDTC1, CpCYP970A14, and CpCYP964A1 (Fig. 3G, peak 1). These results imply that CpCYP970A14 and CpCYP964A1 would possibly be able to catalyze two oxidation steps in conversion from syn-pimaradienoic acid [3] and 3β-hydroxy-9β-pimara-7,15-diene [4] to 3OH-syn-pimaradienolide [5] via the hypothetical intermediate 3,6OH-syn-pimaradienoic acid (Fig. 3A, compound Y). We also found that the potential enzymatic activity of momilactone A [6] synthesis from 3OH-syn-pimaradienolide [5] existed endogenously in N. benthamiana leaves, based on in vitro assays with crude protein and in planta feeding assays (SI Appendix, Fig. S7). Eventually, coexpression of CpMAS with other clustered genes increased the production of momilactone A (Fig. 3H), and in planta conversion of 3OH-syn-pimaradienolide to momilactone A was apparently enhanced by CpMAS expression in N. benthamiana as compared to that in the vector control plant (SI Appendix, Fig. S8).
Taken together, these results indicate that the two CYP genes adjacent to the syn-pimara-7,15-diene synthase gene CpDTC1/HpDTC1 and the momilactone A synthase gene CpMAS are functionally involved in momilactone A formation, supporting our conclusion that this is in fact a BGC for momilactone biosynthesis in the moss. These results are also a demonstration of the successful reconstitution of the momilactone biosynthetic pathway by heterologous gene expression in momilactone nonproducing plants.
Evolution of Momilactone Gene Clusters in Plants.
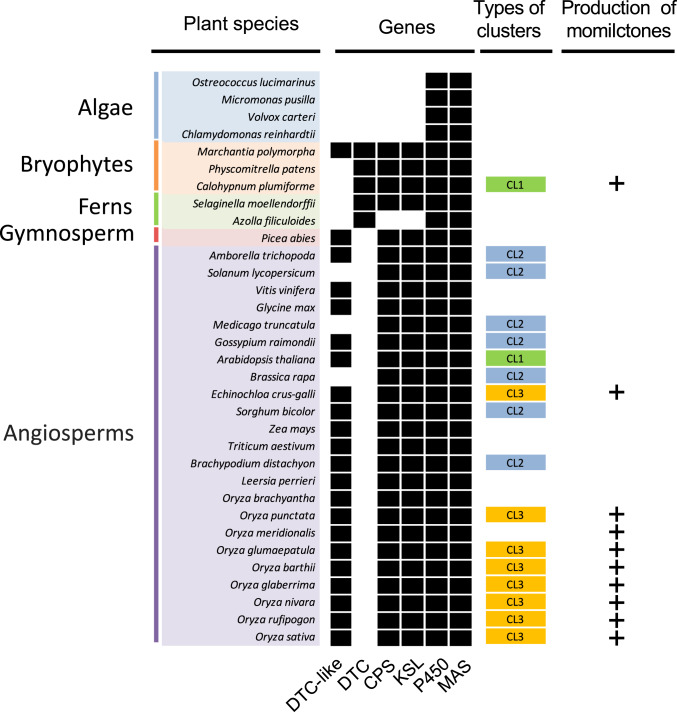
To investigate the evolutionary trajectory of the momilactone BGCs, we broadly surveyed existence of any other possible momilactone gene clusters in the plant kingdom. To this end, we collected available public genomic sequence and annotation information of 107 plants and conducted a data-mining exercise on this genomic information. We first classified all genes into four types based on the Pfam domain annotation of momilactone genes in O. sativa, namely CYP genes (Pfam ID PF00067), terpene synthase (TPS) genes (Pfam ID PF01397/PF03936), short-chain dehydrogenase/reductase (SDR), MAS genes (Pfam ID PF13561/PF00106/PF08659), and all others (SI Appendix, Fig. S9). Next, we scanned all 107 plant genomes within a window of 100 kb using in-house scripts to select candidate regions where TPS, P450, and SDR genes coexisted, and the adjacent windows holding these three types of genes would be merged. As a result, in addition to the known momilactone gene clusters in Oryza species and barnyard grass (8, 16), 43 putative TPS gene clusters in 35 plant species were identified in seed plants, whereas no such candidate gene clusters were found in bryophytes other than in C. plumiforme, fern, and gymnosperm (Fig. 4 and SI Appendix, Table S6). No apparent syntenic relationship in gene arrangement was observed between the known momilactone gene cluster and the 43 candidate clusters.
Fig. 4.
Genome-wide identification of the clustered genes for momilactone biosynthesis in plants. Black boxes represent existence of the genes in the corresponding plant genome. Types of gene clustering: CL1, DTC, or DTC-like type of terpene synthase gene + P450 + MAS; CL2, one type of terpene synthase gene (CPS or KS or DTC or DTC-like) + P450 + MAS; CL3, two types of terpene synthase genes (CPS and KSL) + P450 + MAS. Momilactones were only detected in the species with CL1 or CL3 gene clusters.
Homologs of the bifunctional DTC gene CpDTC1/HpDTC1, a key pathway gene responsible for producing the core diterpene substrate, could be identified in other bryophytes, such as P. patens and fern species, but were not found in flowering plants (Table 2). We identified many terpene synthases in flowering plants with the two D-rich motifs (i.e., “DXDD” and “DDXXD” motif); however, we termed them as “DTC-like” because their DXDD motif was not located at its typical position (SI Appendix, Fig. S10A) and they lacked the third motif (“SXYDTAW”). Interestingly, we could not find any terpene cyclases in the algal species examined. Phylogenetic analysis of the terpene biosynthesis-related genes in the momilactone gene clusters of the two grasses indicated that the CPS and KSL genes in the BGCs did not appear to have evolved from the clustered terpene cyclase gene (CpDTC1/HpDTC1) in the moss (SI Appendix, Figs. S10B and S11).
Table 2.
Summary of the TPS family in plant genomes
| Species | DTC type | DTC-like type | CPS type | KSL type | Other type |
| M. polymorpha | 2 | 1 | 2 | 3 | 12 |
| P. patens | 1 | 0 | 1 | 2 | 2 |
| C. plumiforme | 5 | 0 | 1 | 1 | 0 |
| S. moellendorffii | 3 | 0 | 4 | 13 | 5 |
| A. filiculoides | 1 | 0 | 0 | 0 | 0 |
| P. abies | 0 | 1 | 2 | 35 | 18 |
| E. crus-galli | 0 | 2 | 2 | 42 | 29 |
| O. sativa | 0 | 2 | 3 | 38 | 10 |
| A. thaliana | 0 | 5 | 1 | 24 | 4 |
DTC-type has three motifs (SXYDTAW, DXDD, and DDXXD). DTC-like type has two D-rich motifs (DXDD and DDXXD). CPS-type and KSL-type have either of one D-rich motif. Other type does not have D-rich motif (more details can be found in SI Appendix, Table S7).
Up to now, only two types of gene clusters (CL1 and CL3) have been shown to contain genes involved in the biosynthesis of momilactones (Fig. 4). Intriguingly, we found that A. thaliana also has a potential CL1-type gene cluster that contains a DTC-like gene and CYP and MAS gene homologs. However, the DTC-like gene has been reported to be TPS10 [coding for (3R)-linalool synthase] and it is coexpressed with the CYP genes, CYP71B31 and CYP76C3, which are located outside the cluster region that further metabolizes (3R)-linalool into hydroxylinalool (29). Therefore, the only gene cluster harboring a DTC-type bifunctional terpene cyclase gene for momilactone formation found in our survey is that present in C. plumiforme.
While our results suggest that the 43 candidate clusters are not likely to be responsible for the production of momilactones, they are potentially involved in the formation of other terpenoid metabolites. The lack of synteny and phylogenetic distance supports the conclusion that the momilactone BGCs in O. sativa, E. crus-galli, and C. plumiforme arose independently with acquisition of conserved core component enzymes of syn-pimaradiene synthase, CYPs, and SDR. Notably, the momilactone BGC in C. plumiforme seems to be a unique case of clustering of biosynthetic genes responsible for a specialized secondary metabolite for plant defense and allelopathy in nonseed plants.
Discussion
In this study, we have generated a high-quality genome assembly of the moss, C. plumiforme, and shown that it contains a BGC responsible for the production of the defense and allelopathic metabolite momilactone. This is the only evidence of gene clustering for the biosynthesis of specialized defensive metabolites identified in a bryophyte lineage and is distinct from those previously identified in the grasses, which includes O. sativa, wild Oryza species, and the paddy weed barnyard grass E. crus-galli. This observation suggests that the evolutionary process of BGC formation in all plants is widely distributed. Our evidence indicates that there is little syntenic relationship among the momilactone BGCs in C. plumiforme and the grasses. Therefore, momilactone biosynthesis gene clustering in the plant genome was likely the product of convergent evolution leading to the formation of the same distinctive secondary metabolite (momilactone) in plants from very different families (mosses and grasses). How and why this occurred is unknown, but it is likely tied to the necessity of maintaining a capability to regulate the formation of this important defense and allelopathic molecule.
The four genes in the C. plumiforme momilactone BGC (i.e., CpMAS, CpCYP970A14, CpDTC1/HpDTC1, and CpCYP964A1) are coordinately induced under stress treatment (Fig. 2B). Among the four genes, CpDTC1/HpDTC1 had a very low basal level of expression (based on its fragments per kilobase of transcript per million mapped reads value of 9.23 in untreated samples in the RNA-seq analysis) and consequently the highest level of induction (85-fold with chitosan treatment and 540-fold with CuCl2 treatment as determined by qRT-PCR). In contrast, the other three genes in the cluster had higher basal expression levels than CpDTC1/HpDTC1 and were more up-regulated with the chitosan (by 4- to 24-fold) and CuCl2 (by 8- to 150-fold) treatments than that in untreated samples. Since C. plumiforme constitutively produces momilactones under natural conditions (9), the induction of CpDTC1/HpDTC1 expression seems to be the major regulatory step controlling momilactone production in response to stress. The low-level induction of the CpMAS, CpCYP970A14, and CpCYP964A1 genes involved in momilactone biosynthesis in moss is in contrast to that observed in the orthologous genes of the momilactone BGC in O. sativa. Here it has been observed that all of the rice genes in the cluster are highly induced in response to environmental stress (30). Clearly, additional studies are needed to determine the regulatory elements and transcription factors important in determining the differential levels of gene expression of the momilactone biosynthetic genes in the C. plumiforme cluster. Studies toward understanding the control mechanisms of the gene regulatory process, compared to those involved in regulation of the genes in the momilactone BGCs in various grasses, are also warranted. The availability of the C. plumiforme genome sequence and gene annotation data will certainly assist in these future studies.
Several BGC prediction pipelines have been developed for the plant genomes, including plantiSMASH (31), PlantClusterFinder (32), and PhytoClust (33). However, there are still limitations in the use of prediction methods with bryophyte genomes, which have a long divergence distance to flowering plants. In our genome survey, we added domain searching in our analysis pipeline for P450 and ADH domains. It is also known that biochemical and chemical analyses are indispensable to fully delineate newly predicted pathways associated with candidate BGCs (34). Hence, evidence from gene expression and functional experiments were additionally used to support the prediction of the momilactone BGC in C. plumiforme. As a result, we found that four genes existed as a cluster in one contig (Scaffold38) in our BGC prediction. This potential gene cluster is composed of CpDTC1/HpDTC1 (syn-pimaradiene synthase), two P450 genes (CpCYP970A14 and CpCYP964A1, and a dehydrogenase gene (CpMAS). CpMAS is highly similar to OsMAS, a rice momilactone A synthase at the amino acid level, and our functional analysis of E. coli-produced recombinant CpMAS protein showed the activity for momilactone A synthase. On the other hand, although the two P450s in the cluster, CpCYP970A14 and CpCYP964A1, do not share high levels of identity to the two known P450s, CYP99A2 and CYP99A3, involved in the momilactone A biosynthesis in rice, our functional verification of CpCYP970A14 and CpCYP964A1 using two different approaches (i.e., expression in yeast and transient expression in tobacco) demonstrated their direct involvement in momilactone A biosynthesis. Moreover, the successful reconstitution of the momilactone biosynthetic pathway in N. benthamina suggests that only two P450s (CpCYP970A14 and CpCYP964A1) are required for the formation of the momilactone A precursor 3OH-syn-pimaradienolide [5] in the moss. This is in contrast with rice, which appears to require at least three P450s (CYP99A2/A3, CYP707A8, and CYP76M8) in the oxidation step. The CpCYP964A1 gene is responsible for the conversion of syn-pimaradiene to 3OH-syn-pimaradiene and the CpCYP970A14 gene is involved in the synthesis of syn-pimaradienoic acid. Our results suggest that either, or both, are capable of catalyzing two consecutive oxidation steps at the C19 and C6 positions to yield 3OH-syn-pimaradienolide [5] through a spontaneous lactonization of a putative intermediate 3β, 6β-dihydroxy-9β-pimara-7,15-dien-19-oic acid. However, we currently do not know if such a putative intermediate is produced by the consecutive reactions catalyzed by CpCYP970A14 and CpCYP964A1. Further studies will clearly be necessary to establish the whole pathway of the momilactone A and B synthesis and its precise biosynthetic machinery in the moss.
A distribution of TPS and potential gene clusters in distinctive plant families were evidenced by our genomic survey in the plant kingdom (Fig. 4). It appears that many vascular plants have CL2-type gene clusters comprising one type of terpene cyclase gene (CPS or KSL) with CYP and MAS homologs. However, these clusters are not likely to be responsible for the momilactone synthesis, and further investigation will be required to understand if they are functional metabolic gene clusters for specialized metabolite production in each plant. Based on previous studies of terpenoid biosynthesis in bryophytes, DTC in mosses are shown to be bifunctional enzymes possessing both CPS (class I/type B, DXDD motif only) and KSL (class II/type A, DDXXD motif only) activities (9). This is distinct from the situation in vascular plants where separate CPS- and KSL-type genes are present in the genome (8). Intriguingly, three bryophytes also appear to possess potential homologs of CPS and KSL, and DTC-like gene homologs are present in most vascular plants surveyed in this study (Fig. 4). However, the exact types of terpenoids produced by these TPSs remain to be determined.
Our phylogenetic analysis of TPS genes in 107 plant genomes also clearly shows that only nonseed plants have the bifunctional TPS gene, such as CpDTC1/HpDTC1, including four bryophytes (M. polymorpha, C. plumiforme, P. patens, and Sphagnum fallax) and two ferns (S. moellendorffii and Azolla filiculoides) (Table 2). Among these lower plants harboring bifunctional TPS, only C. plumiforme has the ability to produce momilactone (Fig. 4). We found five homologs of CpDTC1/HpDTC1 in the C. plumiforme genome, whereas P. patens only had one copy of the homologous gene, annotated as ent-kaurene synthase (KS) involved in gibberellin-like ancient plant hormone synthesis (Table 2 and SI Appendix, Table S7) (35). This observation supports our hypothesis that gene duplication and neofunctionalization likely occurred in C. plumiforme, leading to the evolution of KS variants with the ability to synthesize syn-pimaradiene, a distinct precursor of momilactone. Additionally, our phylogenetic analysis clearly shows that CpDTC1/HpDTC1 is derived from a bifunctional CPS/KS but not from monofunctional KS (SI Appendix, Figs. S10B and S11). The CYP genes in the momilactone BGCs in moss and grasses were found to have different phylogenetic affiliations, with the two CYPs in C. plumiforme (CpCYP970A14 and CpCYP964A1) belonging to the CYP71A (85 clan) and CYP707A (71 clan) subfamilies on one branch, whereas the CYPs in the momilactone BGC in O. sativa and E. crus-galli (CYP99A and CYP76M, respectively) are located on a different branch (SI Appendix, Fig. S12). These data further support the hypothesis that the molecular evolution of the relevant CYPs responsible for the momilactone biosynthesis occurred independently in these plant species. The origin of the final step of momilactone synthesis catalyzed by MAS is less clear. It appears that many SDR have a potential dehydrogenation activity capable of converting 3OH-syn-pimaradienolide to produce momilactone A, likely because of their low substrate specificity. In fact, N. benthamiana and fission yeast have been found to have the endogenous metabolic capacity to catalyze the conversion of 3OH-syn-pimaradienolide to momilactone A (SI Appendix, Figs. S7 and S13).
Functional gene clustering and coordinated expression leading to the production of a specialized metabolite clearly provides a distinctive advantage in response to environmental perturbations, such as abiotic and biotic stress and for dealing with competition with other organisms for space and resources in an uncertain ecology. The observation that bryophytes like C. plumiforme independently evolved a momilactone BGC, such as that found in grasses, underscores the value of this molecule as an important, broad-spectrum agent useful in defense and allelopathy, and indicates the power of gene duplication and neofunctionalization in plant evolution. The defense and allelopathic function could have been part of the positive selection pressure for assembling the BGC in C. plumiforme, as previously suggested (36). In addition, it has also been suggested that negative selection pressure could play a role in BGC assembly if inheritance of single genes or incomplete BGCs leads to the build-up of detrimental pathway intermediates (4, 36). It is currently not known whether any intermediates in the momilactone biosynthetic pathway have deleterious effects on plant metabolism or growth. This is certainly an area that could be tested in the future using gene-editing technologies and would contribute to the understanding of the contributions of positive and negative selective pressures in momilactone BGC evolution.
The convergent evolution of distinctive pathways leading to the formation of momilactones indicated the profligate nature of TPSs and their ability to provide new combinations of actions to extend plant biochemistry and the production of need-driven specialized plant metabolites. These results set the stage for future work aimed at examining a broader group of bryophytes to determine whether they produce momilactones and have similar gene clusters, as well as determining the extent to which gene clusters for specialized metabolites occur broadly in ancient plants.
Materials and Methods
Plant Material and Genome Sequencing.
The previously reported wild-type strain of C. plumiforme was cultured on BCDATG agar medium (37) under continuous white light at 24 °C and was used throughout this study. Genomic DNA was extracted from the C. plumiforme gametophore using DNeasy Plant Mini Kit (Qiagen). The C. plumiforme genome was sequenced using both the Illumina HiSEq. 4000 and PacBio RS II sequencing platforms. In total, 88 Gb of Illumina HiSEq. 4000 sequencing data (203× genome coverage) was generated, including two paired-end libraries with insertion sizes of 360 bp and 350 bp and two mate-pair libraries with insertion sizes of 10 kb and 20 kb (SI Appendix, Table S1). A total of 1.54 Gb of Pacbio sequencing data (3.5× genome coverage) with an average subreads length of 5.26 kb was generated with the PacBio RS II platform, including one SMRT cell from P5C3 chemistry.
De Novo Genome Assembly, Genome Annotation, and Identification of Gene Clusters.
All Illumina and Pacbio data were used to perform hybrid assembly and genome annotation and gene-clustering identification were conducted with methods described in SI Appendix, SI Materials and Methods.
Phylogenetic Tree and Divergence Time Estimation.
Based on the annotated protein sequences from C. plumiforme and the other 10 plants (P. patens, M. polymorpha, Picea abies, Ginkgo biloba, Amborella trichopoda, A. thaliana, O. sativa, O. punctata, E. crus-galli, and Chara vulgaris); orthogroups or gene families were identified using Orthofinder with default setting (38). After the Gblocks alignment optimization (39), the alignment of single-copy genes by MAFFT (40) was used to estimate species divergence times by an uncorrelated relaxed clock in BEAST v1.10.4, with C. vulgaris as an outgroup (41). See SI Appendix, SI Materials and Methods for the details.
Functional Analysis of Clustered Genes.
For functional analysis of clustered genes, see SI Appendix, SI Materials and Methods.
Data Availability.
The genome assembly and full-length cDNAs of C. plumiforme by this study have been deposited in the Genome Sequence Archive (GSA) under accession no. PRJCA001833 and DNA Data Bank of Japan under accession nos. LC494432 (CpMAS), LC494433 (CpCYP970A14), and LC494434 (CpCYP964A1), respectively.
Supplementary Material
Acknowledgments
We thank Prof. Toshiyuki Ohnishi (Shizuoka University) for providing the plasmid pRI-ATR1, and Prof. George Lomonossoff (John Innes Centre) for N. benthamiana expression vector pEAQ-HT. We also thank Professor David R. Nelson (University of Tennessee, Memphis, TN) for giving the gene names of C. plumiforme P450s. This work was supported by grants from the National Natural Science Foundation (9143511), Zhejiang Natural Science Foundation (LZ17C130001), Jiangsu Collaborative Innovation Center for Modern Crop Production and 111 Project B17039 (to L.F.), and Grants-in-Aid for Scientific Research - KAKENHI 17H03811 (to K.O.) and 19H02894 (to H. Kawaide).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.O. is a guest editor invited by the Editorial Board.
Data deposition: The genome assembly and full-length cDNAs of Calohypnum plumaeforme by this study have been deposited in the Genome Sequence Archive (GSA) (accession no. PRJCA001833) and the DNA Data Bank of Japan (accession nos. LC494432 [CpMAS], LC494433 [CpCYP970A14], and LC494434 [CpCYP964A1]). RNA-seq data were deposited in DDBJ Sequence Read Archive (DRA), https://www.ddbj.nig.ac.jp/dra/submission-e.html (accession no. DRA010138).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914373117/-/DCSupplemental.
References
- 1.Kato T., et al. , Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett. 39, 3861–3864 (1973). [Google Scholar]
- 2.Nozaki H., et al. , Momilactone A and B as allelochemicals from moss Hypnum plumaeforme: First occurrence in bryophytes. Biosci. Biotechnol. Biochem. 71, 3127–3130 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Kato-Noguchi H., Hasegawa M., Ino T., Ota K., Kujime H., Contribution of momilactone A and B to rice allelopathy. J. Plant Physiol. 167, 787–791 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Xu M., et al. , Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 193, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyomasu T., et al. , Reverse-genetic approach to verify physiological roles of rice phytoalexins: Characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol. Plant. 150, 55–62 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Lu X., et al. , Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 30, 1119–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S. J., Park H. R., Park E., Lee S. C., Cytotoxic and antitumor activity of momilactone B from rice hulls. J. Agric. Food Chem. 55, 1702–1706 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto K., et al. , Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 87, 293–304 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Okada K., et al. , HpDTC1, a stress-inducible bifunctional diterpene cyclase involved in momilactone biosynthesis, functions in chemical defence in the moss Hypnum plumaeforme. Sci. Rep. 6, 25316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye Z., et al. , Biochemical synthesis of uniformly 13C-labeled diterpene hydrocarbons and their bioconversion to diterpenoid phytoalexins in planta. Biosci. Biotechnol. Biochem. 81, 1176–1184 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Frey M., et al. , Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Qi X., et al. , A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilderman P. R., Xu M., Jin Y., Coates R. M., Peters R. J., Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135, 2098–2105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanno Y., et al. , Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci. Biotechnol. Biochem. 70, 1702–1710 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Shimura K., et al. , Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Guo L., et al. , Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8, 1031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rensing S. A., et al. , The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Newton A. E., Wikstrom N., Shaw A. J., “Mosses (Bryophyta)” in The Timetree of Life, Hedges S. B., Kumar S., Eds. (Oxford University Press, 2009), pp. 138–145. [Google Scholar]
- 19.Liu Y., et al. , Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat. Commun. 10, 1485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nützmann H. W., Scazzocchio C., Osbourn A., Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Haas B. J., et al. , Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman J. L., et al. , Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Nelson D. R., The cytochrome p450 homepage. Hum. Genomics 4, 59–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q., Hillwig M. L., Peters R. J., CYP99A3: Functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 65, 87–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitaoka N., Wu Y., Xu M., Peters R. J., Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl. Microbiol. Biotechnol. 99, 7549–7558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yajima A., et al. , Stereocontrolled total synthesis of (±)-3β-hydroxy-9β-pimara-7,15-diene, a putative biosynthetic intermediate of momilactones. Tetrahedron Lett. 52, 3212–3215 (2011). [Google Scholar]
- 28.Sainsbury F., Thuenemann E. C., Lomonossoff G. P., pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Ginglinger J. F., et al. , Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25, 4640–4657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada A., et al. , Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 65, 177–187 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kautsar S. A., Suarez Duran H. G., Blin K., Osbourn A., Medema M. H., plantiSMASH: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 45, W55–W63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schläpfer P., et al. , Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Töpfer N., Fuchs L. M., Aharoni A., The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res. 45, 7049–7063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nützmann H. W., Osbourn A., Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26, 91–99 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki S., et al. , An ancestral gibberellin in a moss Physcomitrella patens. Mol. Plant 11, 1097–1100 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Chu H. Y., Wegel E., Osbourn A., From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 66, 66–79 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Nishiyama T., Hiwatashi Y., Sakakibara I., Kato M., Hasebe M., Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7, 9–17 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Emms D. M., Kelly S., OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly and full-length cDNAs of C. plumiforme by this study have been deposited in the Genome Sequence Archive (GSA) under accession no. PRJCA001833 and DNA Data Bank of Japan under accession nos. LC494432 (CpMAS), LC494433 (CpCYP970A14), and LC494434 (CpCYP964A1), respectively.