Significance
Interorganelle communication occurs at membrane contact sites via dynamic trafficking of diverse biomolecules between the two organelles. The mitochondria-associated membrane (MAM) is the most critical cellular contact site of mitochondria and endoplasmic reticulum. However, a proteomics method to profile in vivo components at the contact sites is still unavailable. Here, we introduce Contact-ID, a split-BioID method, which specifically biotinylates the proteins localized at MAM and the biotinylated sites are identified by mass spectrometry, thereby definitively disclosing the MAM proteome in live cells. As a result, we report 115 MAM-specific proteins with diverse functions (e.g., cholesterol metabolism and antiapoptosis process). We expect this method to be utilized to profile the local proteome at any organelle–membrane contact site in live cells.
Keywords: mitochondria-associated membrane (MAM), membrane contact site, proximity labeling, membrane protein topology, FKBP8
Abstract
The mitochondria-associated membrane (MAM) has emerged as a cellular signaling hub regulating various cellular processes. However, its molecular components remain unclear owing to lack of reliable methods to purify the intact MAM proteome in a physiological context. Here, we introduce Contact-ID, a split-pair system of BioID with strong activity, for identification of the MAM proteome in live cells. Contact-ID specifically labeled proteins proximal to the contact sites of the endoplasmic reticulum (ER) and mitochondria, and thereby identified 115 MAM-specific proteins. The identified MAM proteins were largely annotated with the outer mitochondrial membrane (OMM) and ER membrane proteins with MAM-related functions: e.g., FKBP8, an OMM protein, facilitated MAM formation and local calcium transport at the MAM. Furthermore, the definitive identification of biotinylation sites revealed membrane topologies of 85 integral membrane proteins. Contact-ID revealed regulatory proteins for MAM formation and could be reliably utilized to profile the proteome at any organelle–membrane contact sites in live cells.
Recent advances in biological imaging techniques have enabled the identification of membrane contact sites used for communication among various cellular organelles (1, 2) and their functional significance for regulation of cellular homeostasis (3). For example, the mitochondrial-associated membrane (MAM) of the endoplasmic reticulum (ER) is reported to regulate many physiological processes such as calcium homeostasis (4), lipid transport (5), organelle biogenesis (6), organelle stress regulation (7–9), and cell cycle regulation (10, 11). Additionally, the MAM is closely related to a wide spectrum of metabolic diseases, such as type 2 diabetes (12) and neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (13), and Wolfram syndrome (14). This accumulating evidence suggests that MAM functions as a molecular sensory hub and conducts cellular signaling in a variety of physiological and pathological processes, including viral infection, pharmacological stresses, and aging (8, 9, 15–17). Conventionally, the MAM proteome has been isolated by sequential centrifugation-based fractionation (18, 19). Although >1,000 MAM proteins have been identified in cell lines or in the mouse brain, to date, with this method (20–22), only 40% of these proteins were reproducibly identified between replicates, and a substantial number of irrelevant organelle proteins (e.g., nucleus or microsome) were included (21, 23). These anomalies are attributed to the technical limitations in the pure isolation of the MAM fraction (19, 22). Furthermore, the conventional method cannot identify the temporal resident proteins at the MAM, which can be easily missed during the serial centrifugation steps (19, 22).
To overcome these limitations of local proteome identification, a proximity-based labeling method using enzymatic biotinylating reactions was recently developed based on peroxidase (APEX) (24), horseradish peroxidase (HRP) (25, 26), and promiscuous biotin ligase (pBirA) (27). These enzymes generate reactive biotin species with short half-lives in aqueous solution and can label proximal proteins <10 to 20 nm from their points of origin. Given the advantages of these enzymatic-labeling methods in revealing protein networks in live cells, which cannot be achieved by conventional in vitro approaches, they have been actively utilized in the proteome mapping of various compartments (28–30). However, the MAM-localized proteome has not been directly mapped by a proximity-labeling approach to date, largely due to the lack of MAM-specific marker proteins.
Here, we report Contact-ID, a proximity-labeling method designed for organelle membrane contact site proteome identification, which was based on the biotinylation activity of two split-BioID components after proximity-dependent reconstitution. Using this method, we identified 115 MAM-localized proteins in live cells. Among these proteins, we revealed that the function of FKBP8 is closely related to MAM formation and calcium transport from the ER to the mitochondria at the MAM.
Results
Development of a Split Promiscuous Biotin Ligase Pair.
To construct a split-proximity labeling system for the in situ labeling of the proteome at the contact sites of two organellar membranes, the following two criteria have to be met: 1) the reconstituted biotinylation activity of the split-BioID system is solely proximity dependent and validated with a strictly proximity-controllable biological system, such as rapamycin-induced protein–protein interactions (31, 32); and 2) the intrinsic affinity between the two split fragments should be negligible to avoid artificial MAM formation and consequent false-positive identification. An artificially generated MAM increases the local calcium concentration in mitochondria (33, 34), which can compromise the mitochondrial oxidative capacity and thus augment the oxidative stress level, thereby significantly impairing cellular physiology.
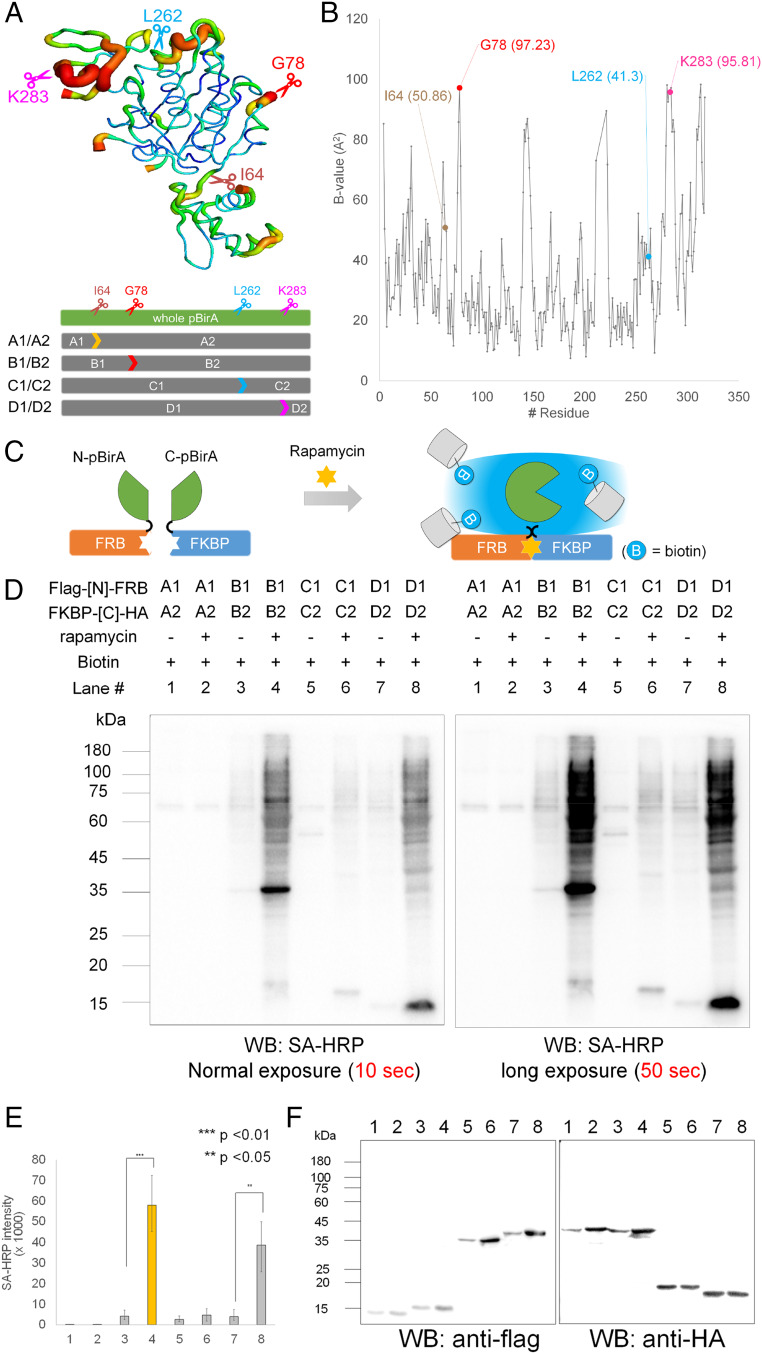
Currently, proteome mapping of the membrane contact site has not yet been attempted with the available split-proximity labeling systems, including the split-HRP (35), split-APEX2 (36, 37), and split-BioID systems (38, 39). Since the MAM is a hydrogen peroxide-sensitive compartment (40), we excluded peroxidase-based split systems and selected the split-BioID system for mapping the MAM proteome because biotin ligase (pBirA) does not require hydrogen peroxide for labeling. Although split-BioID systems using domainwise splitting approaches were previously reported (38, 39), we attempted to develop a split-BioID system by splitting enzymes based on temperature factors (B factors), which has not been attempted yet. Since the B factor of a specific residue indicates its atomic flexibility in a crystalline state (41, 42) (Fig. 1A), it is desirable to select a site with a high B factor as a splitting site on the flexible loop to preserve the structural integrity and biotinylation activity of the BioID when reconstituted (43). Using the crystal structure of wild-type biotin ligase (PDB ID: 1HXD), we selected two sites, G78 and K283, with considerably high B factors, 97.23 and 95.81, respectively, as the splitting site for our split-BioID system (Fig. 1 A and B). As control systems, we selected I64 and L262 with low B factors of 50.86 and 41.3, respectively. Thus, the following four split constructs of BioID were generated, including control pairs: A1(N-I64)/A2(Q65-C) (control), B1(N-G78)/B2(G79-C), C1(N-L262)/C2(S263-C) (control), and D1(N-K283)/D2(E284-C) (Fig. 1 A and B).
Fig. 1.
Design of Contact-ID based on the structural flexible region of Escherichia coli biotin ligase (BirA). (A) Crystal structure of wild-type E. coli biotin ligase (BirA PDB: 1HXD) and selected four split sites. (B) Plot of B factor (y axis) along the amino acid chain of the biotin ligase (x axis). (C) Schematic view of the restoration of biotinylating activity of split-BioIDs using the FRB–FKBP system in the presence of rapamycin. (D) Streptavidin-horseradish peroxidase (SA-HRP) Western blot result of biotinylated proteins from four split candidate pairs of pBirA in the FRB–FKBP system with or without rapamycin. Biotin (50 µM) was treated for 16 h. (E) SA-HRP signal intensity of triplicate experiments of D; ***P < 0.01, **P < 0.05. (F) Anti-HA and anti-FLAG Western blot results of the same samples of D.
To verify the proximity-dependent reconstitution of the biotinylation activity for these split-BioID pairs, we adopted the FRB–FKBP interaction system because this interaction is controllable with the addition of rapamycin (Fig. 1C) (23, 31, 32, 35, 44, 45). For this assay, the N-terminal and C-terminal fragments were genetically conjugated with FKBP12 and FRB in a mammalian expression vector, respectively, for all four split pairs. As shown in Fig. 1 D and E, the turn-on biotinylation activity of split pairs B1/B2 and D1/D2 was increased three- to fivefold with rapamycin treatment, while the control split pairs A1/A2 and C1/C2 exhibited weak activities even with rapamycin (streptavidin-HRP Western blot). Furthermore, in the same FKBP–FRB system, the biotinylation activity of the B1/B2 pair showed substantially higher turn-on biotinylation activity compared to that of the most recently reported split-BioID pairs (E256/G257) (38) in the presence of rapamycin (SI Appendix, Fig. S1 A and B). This result supports that our B factor-based determination of a splitting site might be a reasonable approach. Even with long exposure, pair B1/B2 showed negligible biotinylation activity without rapamycin, while pair D1/D2 generated a subtle level of biotinylation activity without rapamycin (Fig. 1E). We also confirmed that neither fragment (B1 or B2) had any biotinylating activity at all with or without treatment of rapamycin (SI Appendix, Fig. S1 D and E). These results demonstrated that the reconstituted biotinylation activity of pair B1/B2, i.e., Contact-ID, was strictly controlled by the enforced proximity condition, suggesting that the intrinsic affinity between the pair should be very low.
Contact-ID Is Actively Biotinylated to Reliably Identify the Localized Proteome at the ER–Mitochondrial Contact Site in Live Cells.
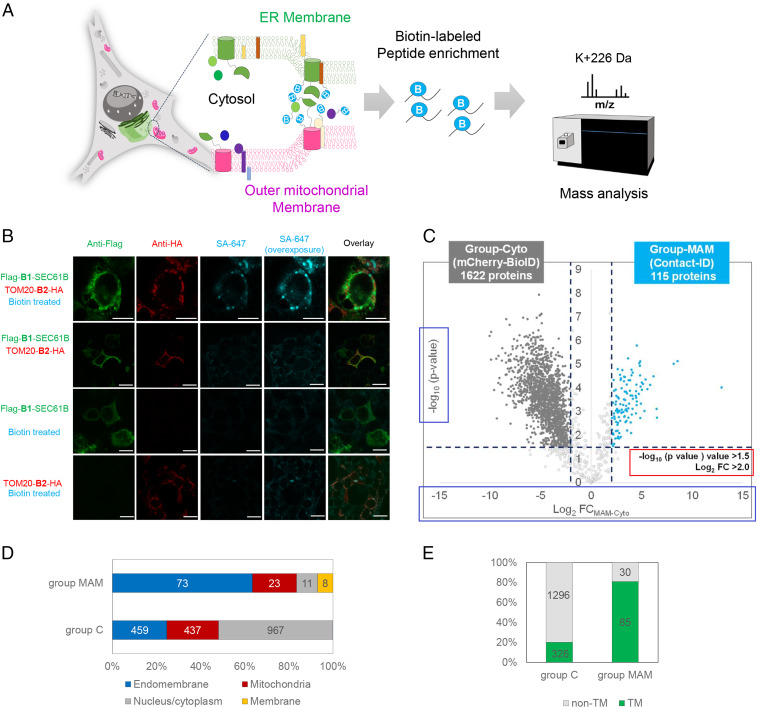
To map the proteome of the ER–mitochondria contact site in live cells, we developed a Contact-ID system consisting of the N-G78 (B1) and G79-C (B2) fragments of pBirA, which were genetically conjugated with the cytosol-facing N terminus of SEC61B (B1-SEC61B) at the ER membrane (ERM) and the C terminus of TOM20 (TOM20-B2) at the outer mitochondrial membrane (OMM), respectively. Because SEC61B (>80,000 copies per cell) (46) and TOM20 (>1,000 copies per cell) (46) are abundantly expressed at the ERM and OMM, respectively, we expected that the two split-BioID fragments fused with SEC61B or TOM20 could be colocalized and, consequently, reconstitute the biotinylating activity at the contact site of both membranes (Fig. 2 A and B). A unique biotinylation pattern stained by streptavidin-Alexa Fluor 647 was observed only at the junction of B1-SEC61B and TOM20-B2 expression in HEK293T cells (Fig. 2B) and other mammalian cells (HEK293T, U2-OS, and HeLa; SI Appendix, Fig. S2 B–E), which was expected since the biotinylation activity would be MAM formation dependent in our Contact-ID system. Electron microscopy (EM) imaging showed that most of the proteins biotinylated by our MAM-Contact-ID system (see detailed procedures in SI Appendix) were localized at the junction of the OMM with ERM or nuclear envelope (NE), which contains a part of the ERM (red arrows in SI Appendix, Fig. S2A). At the MAM, Contact-ID showed higher biotinylation activity (over twofold) than the previously reported split-BioID pair E256/G257 (38) (SI Appendix, Fig. S3). Thus, our Contact-ID appears to solely satisfy the criteria for a split proximity-labeling system for membrane contact site mapping: strictly proximity-dependent reconstitution of biotinylation activity, demonstrating its reliability for MAM proteome mapping experiments. To identify the MAM proteome in the normal state by Contact-ID, HEK293 cells coexpressing B1-SEC61B and TOM20-B2 were treated with 50 μM biotin in full FBS medium for 16 h. The cytosolic control cell line stably expressing mCherry-BioID was prepared as a control sample because biotinylation of the MAM proteome by Contact-ID should occur in the cytosolic space of the membrane. All experiments were performed reproducibly in biological triplicates for a given biological condition (R2 > 0.85, SI Appendix, Fig. S4A).
Fig. 2.
ER–mitochondrial contact site mapping using Contact-ID. (A) Overview of the MAM proteome mapping workflow by Contact-ID. (B) Confocal microscopy imaging of MAM biotinylation by Contact-ID in HEK293T cells. Flag-B1-SEC61B was visualized by anti-Flag antibody (AF488-conjugated, green fluorescence channel) and TOM20-B2-HA was visualized by anti-HA antibody (AF568-conjguated, red fluorescence channel). Biotinylated proteins were visualized by AF647-conjugated streptavidin (Cy5 fluorescence channel). Control samples (i.e., no biotin treatment, Flag-B1-SEC61B expression only, and TOM20-B2-HA expression only) showed no significant biotinylation. (Scale bars, 20 µm.) (C) Volcano plots showing statistically significant enrichment of biotinylated proteins (group-MAM) by Contact-ID over the cytosolic biotinylated proteins (group C) by EGFP-BioID. Biotin (50 µM) treatment, 16 h. See Dataset S1 for details. (D) Subcellular distribution of proteins in group-MAM and in group-Cyto with prior annotated localization information in Uniprot. See Dataset S1 table for details. (E) Number of transmembrane proteins in group C and group-MAM.
From triplicate samples, 3,462 biotinylated peptide-spectrum matches (PSMs), corresponding to 36.5% of the total PSMs, and 920 nonredundant biotinylated peptides, were reproducibly identified in at least two replicates under a 1% false discovery rate at the biotinylated protein level via MaxQuant analysis. It is noteworthy that the majority of biotinylated peptides from Contact-ID could be only observed in the postenriched sample of the streptavidin enrichment (SI Appendix, Fig. S4B). Additionally, cytosolic proteins were profiled (mCherry-BioID) as a negative control for precise identification of MAM proteins. Conventionally, ratiometric criteria for a labeling-treated sample against a negative control sample are applied in mass spectrometric analysis to quantitatively cut off the nonbiotinylated proteins and determine the biotinylated proteins, generating a certain level of uncertainty for the identified biotinylated proteins owing to its indirect identification (19). However, we previously developed the Spot-BioID method (47, 48), allowing for the biotinylated lysine residue (K + 226 Da) in biotinylated peptides or proteins to be unambiguously identified via tandem mass spectrometric analysis. Using this Spot-BioID workflow, the confidence level of the proximity-labeled proteins in our Contact-ID could be significantly boosted compared to that of the conventional approach.
Via label-free quantification (LFQ) and statistical analysis with the biotinylated proteins only, we determined 115 biotinylated proteins as MAM proteins (group-MAM [group M]) and 1,622 proteins as cytosolic proteins (group-Cyto [group C]) (Fig. 2C). The gene names of group-MAM are shown in SI Appendix, Fig. S5 and Dataset S1. Gene ontology analysis of documented group-MAM proteins for subcellular localization showed significant enrichment in the endomembrane system (73 proteins, 63.4%) and mitochondria (23 proteins, 20%) (Fig. 2D). This is a reasonable result given that the majority of MAM proteins should originate from either the mitochondria or ER (Fig. 3A and SI Appendix, Fig. S6A).
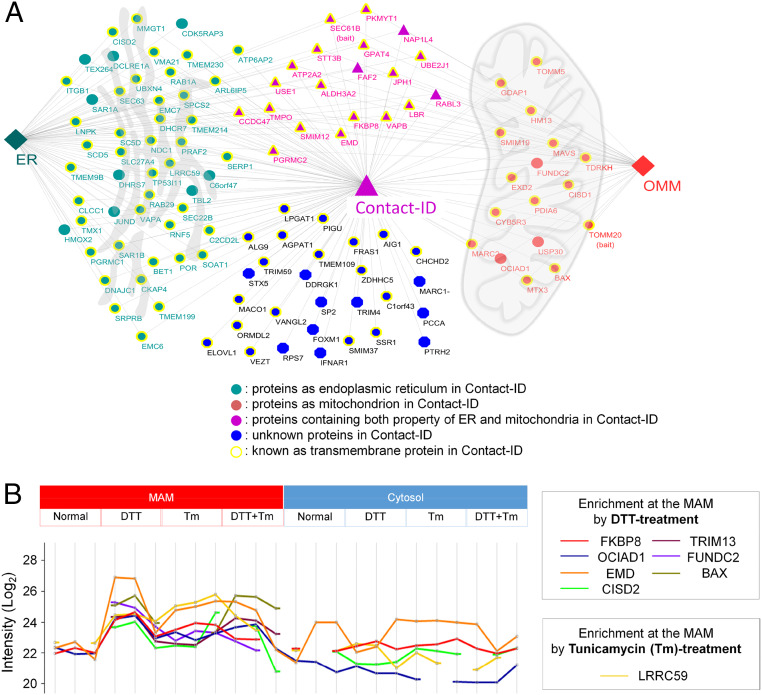
Fig. 3.
Clusters of group-MAM proteins. (A) Organellar distribution of group-MAM proteins over the mitochondria, ER, and cytosol. (B) Enriched proteins at the MAM under the normal, DTT, tunicamycin (Tm), and DTT + tunicamycin-treated condition. Y value represents Log value of LFQ mass signal intensity of the biotinylated peptides from each labeled protein by Contact-ID (MAM) or by mCherry-BioID (cytosol) under the different conditions. see Dataset S5 for details.
Notably, the majority of group-MAM proteins (85 of 115, 74%) are membrane proteins that contain a single or multiple transmembrane domain(s) (Fig. 2E). This proportion of membrane proteins was significantly higher than that of the cytosolic proteins (group-Cyto: 326 of 1,622, 20%) or the general abundance of transmembrane proteins in the whole human proteome (5,526 of 15,498, 36%) (49). Twelve of the group-MAM proteins are well-characterized MAM proteins (e.g., VAPB, TMX1, CISD2, MAVS, EMC6, BAX, RNF5, STX5, SAR1A, CLCC1, TDRKH, and TBL2), and the other 103 proteins in our list could be regarded as MAM proteins (SI Appendix, Fig. S7 and Dataset S1).
Enriched Functional Protein Clusters at the MAM.
Since the MAM is a contact site of the ER and mitochondria, it is reasonable that many of the identified proteins originated from the ERM and OMM (Fig. 3A). Interestingly, 28% (32/115) of group-MAM proteins showed dual subcellular annotation (SI Appendix, Fig. S8A). Among these proteins, BAX (50), VAPB (51), CISD2 (52), and FKBP8 (53) have been characterized to show dual localization in the mitochondria and ER by previous imaging or biochemical assays (SI Appendix, Fig. S8C); thus, these dual-localized proteins could be regarded as mobile MAM proteins (SI Appendix, Fig. S8). Moreover, 24% (28/115) of group-MAM proteins have no previous annotation to the ERM or OMM (Fig. 3A), suggesting their in situ recruitment to the MAM from the cytosol. Detailed information of the Contact-ID-labeled proteome with origin subcellular organelle information is shown in Dataset S1. Based on a literature search of the documented functions of the 115 MAM proteins, we found diverse cellular functions enriched at the MAM (Dataset S2). Among them, the group related to lipid and steroid metabolism was the largest functionally enriched group with 18 proteins of group-MAM (16%, SI Appendix, Fig. S6B). Among these proteins, TMPO is known to be related to cholesterol transfer from the outer to inner mitochondrial membrane during steroidogenic processes (54), and our result implies that TMPO can function at the MAM, which is known as a cholesterol-enriched region (55, 56). In addition, some other proteins (e.g., SOAT1, SCD5, PGRMC1, and CYB5R3) with functions related to steroidogenesis were included in our MAM protein list.
Moreover, many proteins related to typical MAM-related functions were included in the list, such as autophagy control (e.g., TEX264, FUNDC2, and FKBP8), vesicle transport (e.g., BET1 and STX5), cell cycle and apoptosis control (e.g., BAX and EMC6), and antiviral response (e.g., MAVS, IFNAR1, and TRIM59) (SI Appendix, Fig. S6B). Several nucleic acid-binding proteins (e.g., EXD2, PIGU, and TDRKH) were also included in the list, which might be related to the actions of local mRNAs at the MAM for translation at the ERM- and OMM-attached ribosomes (24, 57). Several proteins related to redox metabolism (e.g., CHCHD2, CISD1, HMOX2, and MARC2) on our list might be related to the redox homeostasis process occurring at the MAM (8, 40). Thus, our findings reflect that these characterized or yet-to-be characterized physiological processes can occur at the MAM.
We also checked whether our Contact-ID can capture dynamic change of MAM proteome composition under pharmacologically stressed conditions using tunicamycin and/or reducing agents that have been reported to modulate MAM physiology (8, 9). For this experiment, we have constructed a dually stable cell line of Contact-ID constructs in Flp-in HEK293T-Rex cells to minimize cellular stress from the transfection agents (e.g., lipofectamine) and conducted mass analysis of the biotinylated peptidome under the different stress conditions of dithiothreitol (DTT) (1 mM for 2 h) and/or tunicamycin treatment (0.5 μg/mL for 4 h, see SI Appendix for details). As shown in Fig. 3B and SI Appendix, Fig. S9, a substantial number of proteins were highly enriched at the MAM upon pharmacological stresses: FKBP8 at the DTT treatment and LRRC59 at the tunicamycin treatment. We could also observe that FAM184B and TMEM57 were exclusively enriched at the MAM upon DTT treatment while SUN1 and FAF2 are enriched upon tunicamycin treatment (SI Appendix, Fig. S9 and see Dataset S5 for details). This result supports that our Contact-ID can be used to identify the subtle quantitative changes of MAM proteome upon various cellular conditions.
Contact-ID Reveals Membrane Topological Information of MAM Proteins.
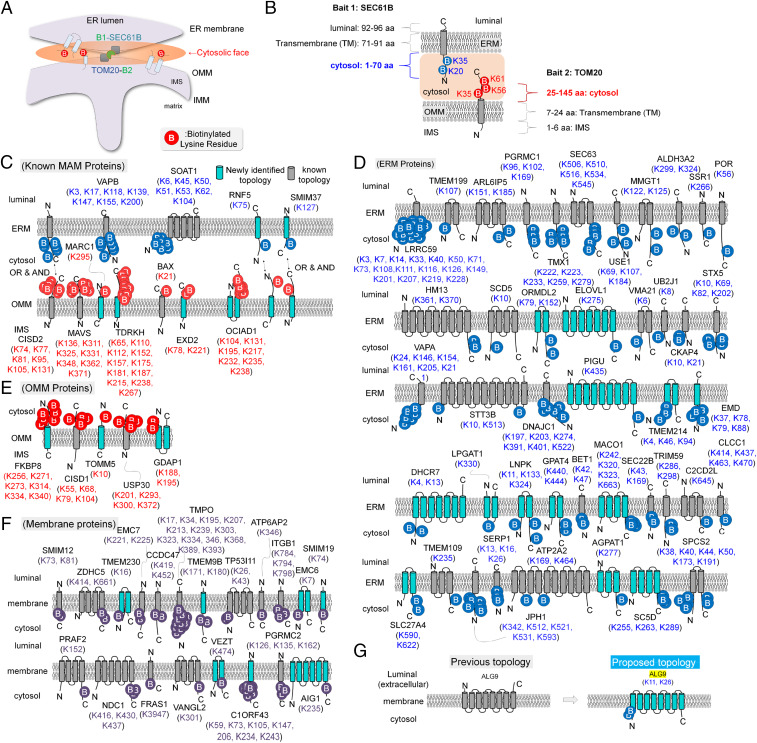
As described above, we unambiguously identified 327 biotinylation sites from a total of 352 nonredundant biotinylated peptides of the 115 MAM proteins at a high confidence level by virtue of Contact-ID. All of these peptides have at least one biotinylation site (226-Da addition on the lysine residue) in their sequences (see Dataset S3 for detailed information). Among these biotinylated peptides, 272 biotinylation sites belong to the 85 MAM proteins with a transmembrane domain (TM). Since the biotinylating reaction of Contact-ID occurs at the cytosolic face of the MAM, the biotinylated sites on the proximity-labeled transmembrane proteins are likely positioned at the cytosolic side of the membrane (Fig. 4A). For example, the biotinylated sites of the two bait TM proteins of Contact-ID, SEC61B and TOM20 (e.g., K20 and K35 of SEC61B, and K35, K56, and K61 of TOM20) are well matched to their previously characterized cytosolic-exposed domains (Fig. 4B). Furthermore, over 50 TM proteins showed good matches with previously characterized membrane topologies and our biotinylation site information by Contact-ID (Fig. 4 C–F).
Fig. 4.
Membrane protein topology identification of group-MAM proteins. (A) Overview of the Contact-ID generation of biotinylated sites at the cytosolic faces of organelle membranes. Since all of our MS-detected biotinylated sites by Contact-ID can be considered as sites on the cytosolic domain, we propose the membrane topology of all identified proteins in our current study. See Dataset S3 for details. (B) Representative results of biotin-labeled sites on the cytosolic domain of bait proteins (SEC61B and TOM20) of Contact-ID. (C) Observed membrane topologies of previously characterized MAM proteins in group-MAM. Newly identified membrane topologies in the current study are colored in light blue. (D) Observed membrane topologies of ERM-originated membrane proteins in group-MAM. (E) Observed membrane topologies of OMM-originated membrane proteins in group-MAM. (F) Observed membrane topologies of other endomembrane proteins in group-MAM. (G) Proposed membrane topology of ALG9 by our labeled site results. Previously annotated topologies of these proteins are shown on the Left, and our proposed topologies are shown on the Right.
In addition, we can suggest the membrane topologies of 32 proteins at their origin membranes, ERM or OMM, based on our biotinylation site information of Contact-ID (Fig. 4 C–G). Among these findings, there is conflict between previously characterized and our biotinylation site-based membrane topologies for one transmembrane protein, such as ALG9 (Fig. 4G). For example, ALG9 has a multitransmembrane helix domain and the N-terminal domain was previously characterized to be headed in the luminal space of the ER (58), whereas the lysine residues of the N-terminal domains were clearly biotinylated in our Contact-ID dataset, implying their cytosolic face localization. Moreover, the N-terminal peptides of ALG9 were not predicted as ER luminal-targeting peptides or signal peptides by SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/). Collectively, we propose revised membrane topologies based on the biotinylation sites on these proteins determined by Contact-ID (Fig. 4G).
FKBP8 Regulates Formation of the Mitochondrial–ER Interface and Calcium Transport at the MAM.
FKBP8 (also known as FKBP38) was one of the heavily biotin-labeled MAM proteins determined by Contact-ID with multiple biotinylated lysine residues, similar to other MAM proteins (e.g., CISD2, VAPB, and MAVS) (Fig. 4E). FKBP8 is a proline peptide isomerase protein with its C-terminal tail transmembrane domain anchored at the OMM. Its molecular functions have been characterized as an antiapoptotic player under the stress conditions (53, 59–61), Sonic hedgehog signaling (61), and mitophagy (62), but there are no studies to date that have been related to MAM physiology; therefore, we decided to conduct further investigation of FKBP8 related to maintenance of the MAM.
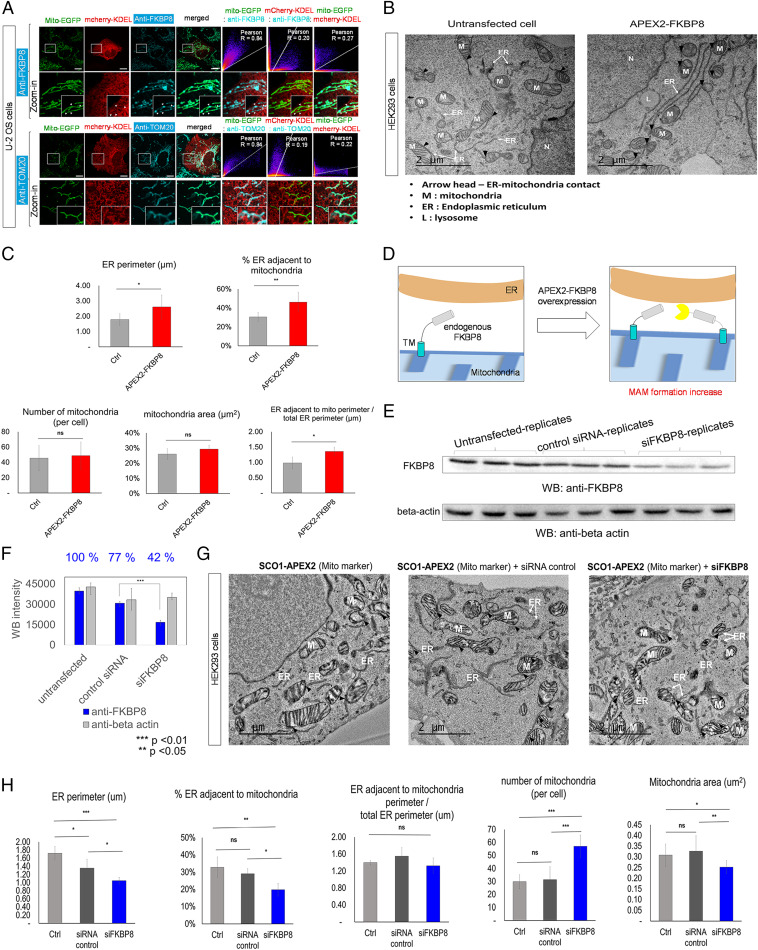
We investigated its primary localization by fluorescence microscopy. As shown in Fig. 5A, the imaging pattern of endogenous FKBP8 using anti-FKBP8 clearly overlapped with that of mitochondria marker proteins (Mito-EGFP, Pearson correlation r = 0.84) in U-2 OS cell lines, while there was lower but still significant overlap with ER marker proteins (mCherry-KDEL, r = 0.27). As a control experiment, anti-TOM20 showed slightly lower overlap with mCherry-KDEL (r = 0.22) than that of anti-FKBP8 (Fig. 5A) in U-2 OS cell lines. In the electronically magnified confocal microscope image (Fig. 5A) and the Airy Scan microscope image (SI Appendix, Fig. S15C), we could see that anti-FKBP8 immunofluorescence (Cy5, cyan) is mainly localized at the mitochondrial tubule (Mito-GFP, green); however, it also showed a punctate pattern at the mitochondria contact site with the ER tubule (mCherry-KDEL) (SI Appendix, Fig. S15).
Fig. 5.
Overexpression or knockdown of FKBP8 leads to mitochondrial–ER contact formation or perturbation. (A) Confocal microscopy imaging of endogenous FKBP8 and the outer mitochondrial marker TOM20 in U-2 OS cells. Endogenous FKBP8 and endogenous TOM20 were visualized by AF647-conjugated anti-FKBP8 and anti-TOM20, respectively (Cy5 channel). The green channel was used for imaging the mitochondrial marker (Mito-EGFP), and the red channel was used for imaging the ER marker (mCherry-KDEL). (Scale bars, 10 µm.) Pearson correlation results between fluorescence signals at each channel are shown on the Right. The punctate localizations of FKBP8 at the interfaces of mitochondria and ER tubules are marked by arrows in the digitally magnified (zoom-in) images. (B) TEM imaging of untransfected HEK293 cells (Left) and APEX2-FKBP8 transfected HEK293 cells (Right). APEX2-FKBP8 overexpressed cells showed increased mitochondria–ER contact sites and morphological changes of the mitochondria and ER. M, mitochondria; arrowhead, contact between the mitochondria and ER. (Scale bars, 2 μm.) (C) Statistical comparison of ER perimeter (micrometers), number and area (square micrometers) of mitochondria, number of contacts between the mitochondria and ER, and normalized ER length adjacent to mitochondria by total ER length in each group; *P < 0.01, **P < 0.005; ns, not significant. (D) Scheme of increased MAM formation by FKBP8 overexpression. (E) Inhibition of FKBP8 protein expression in HEK293 cells by siFKBP8 determined by Western blotting using anti-FKBP8 antibody. (F) Statistical analysis of FKBP8 knockdown by siFKBP8 from triplicate experiments; ***P < 0.001. (G) TEM imaging results of ER and mitochondrial morphological changes by FKBP8 knockdown. SCO1-APEX2 stable cell line (HEK293) was used for DAB/OsO4 staining in the mitochondrial cristae region (78). The siFKBP8-treated sample is shown on the Right. Untransfected (Left) and control siRNA-treated (Right) samples are shown as controls. (Scale bars, 2 μm.) M, mitochondria; arrowhead, contact between the mitochondria and ER. The morphometric analyses of the TEM images (total 250 μm2) were conducted from an average of 16 montage images per experimental group (from five different cells per group). (H) Statistical comparison of ER perimeter (micrometers), number and area (square micrometers) of mitochondria, number of contacts between the mitochondria and ER, and normalized ER length adjacent to mitochondria by total ER length in each group. *P < 0.05, **P < 0.005, ***P < 0.001.
To test the hypothesis that FKBP8 overexpression (FKBP8-OE) facilitates MAM formation (Fig. 5D), we prepared a V5-APEX2-FKBP8 construct for transmission electron microscope (TEM) imaging using an APEX-EM staining method (24). Interestingly, in FKBP8-OE cells, both increased ER and mitochondrial contact sites (25 to 55%) and an increased overall ER perimeter size (1.7 μm to 2.8 μm) were observed (Fig. 5 B and C). Notably, the normalized ER perimeter adjacent to the mitochondria relative to the total ER perimeter also significantly increased in FKBP8-OE cells (Fig. 5C). Although the total number of mitochondria and area were largely unchanged, most of the mitochondria in FKBP8-OE cells was in close contact with the ER. Overall, these data support that FKBP8 plays an important role in regulating MAM formation and controls mitochondrial and ER morphology (Fig. 5C and SI Appendix, Fig. S10).
We further examined whether a decreased level of FKBP8 could affect MAM formation. FKBP8 expression was knocked down with specific small interfering RNA (siRNA), which significantly decreased the endogenous FKBP8 level down to 42% compared to that of cells transfected with the negative control (Fig. 5 E and F). As shown in Fig. 5G, substantial morphological changes of the mitochondria, ER, and their connections were observed after siFKBP8 treatment. Quantitative analysis showed that ER perimeter and the proportion of the ER in close contact with mitochondria was significantly decreased in the FKBP8-knockdown (FKBP8-KD) cells than those in controls (Fig. 5H). The number of mitochondria approximately doubled in FKBP8-KD cells and the mitochondrial area significantly decreased compared with those of the control group (Fig. 5H).
In addition, the number of contact sites was steadily maintained in FKBP8-overexpressed cells compared to the wild-type cells under either stress condition of DTT or tunicamycin treatment while MAM formation was significantly perturbed under the DTT treatment in FKBP8-knocked down cells (SI Appendix, Fig. S11). Together with our mass proteomics result of FKBP8 enrichment at the MAM under the stress condition of DTT treatment (SI Appendix, Fig. S9), these EM imaging results indicate that FKBP8 is an essential player in MAM formation under the stress condition. This result can provide another model to explain how FKBP8 confers cells with greater resistance under stress which has been shown in previous studies (53, 59–61).
FKBP8 Expression Level Controls Calcium Ion Transport from ER to the Mitochondria.
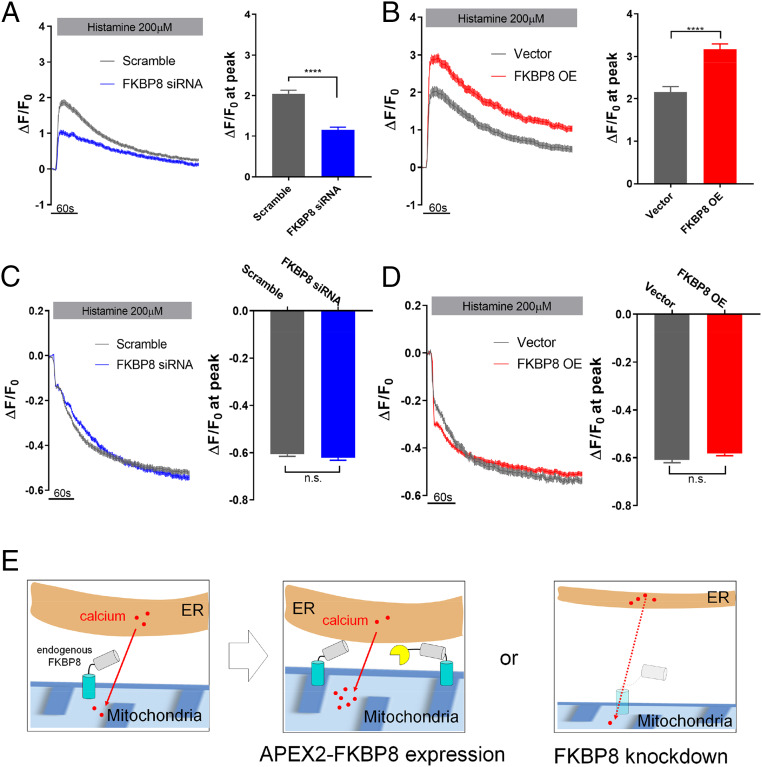
Next, we assessed whether the MAM formation regulated by FKBP8 can affect local calcium transfer from ER to mitochondria. We further expected that the FKBP8 expression level would also affect the calcium levels of the mitochondria and ER (Fig. 6E). To confirm this hypothesis, we employed two genetically encoded calcium sensors: GCaMP6mt (63), a mitochondrial matrix-targeted calcium sensor with green fluorescence, and R-CEPIAer (64), an ER lumen-targeted calcium sensor with red fluorescence. Using these two constructs, we conducted live cell experiments to measure mitochondria and ER calcium levels under FKBP8-KD or FKBP8-OE conditions in the same cells simultaneously. FKBP8-KD cells exhibited a significantly lower level of Ca2+ transfer from the ER to mitochondria relative to that of control cells (Fig. 6A, SI Appendix, Fig. S12, and Movie S1 A and B). Conversely, FKBP8-OE cells displayed higher mitochondrial Ca2+ uptake upon histamine treatment (Fig. 6B). No significant changes in Ca2+ release from the ER store were observed in either FKBP8-KD and FKBP8-OE conditions (Fig. 6 C and D, SI Appendix, Fig. S10, and Movie S1), suggesting that FKBP8 function is likely associated with mitochondrial calcium uptake at the MAM. We checked that the mitochondrial calcium uniporter (MCU) complex subunits and the mitochondrial membrane potentials were not significantly altered by FKBP8 silencing (SI Appendix, Figs. S12 and S13). We also confirmed that the defects in ER–mitochondrial calcium transfer in FKBP8-KD cells were rescued by an artificial increase of ER–mitochondrial contact using a rapamycin-inducible bridge-forming module (RiBFM) (33) (SI Appendix, Fig. S14). All of these findings support the notion that FKBP8 regulates MAM formation, thereby controlling Ca2+ crosstalk between these two organelles.
Fig. 6.
FKBP8 regulates Ca2+ transfer from ER to mitochondria. (A and C) Plasmids for GCaMP6mt and R-CEPIA1er were cotransfected with scramble siRNA (gray), and FKBP8 siRNA (blue), respectively; 200 μM histamine was utilized to stimulate ER Ca2+ release and changes of GCaMP6mt, and R-CEPIA1er fluorescence was recorded simultaneously and normalized to the basal signals (F0). Bar graphs represent the peak amplitude of ΔF/F0 in mitochondria and ER. The data were assembled and analyzed from three independent sets of experiments. (B and D) An equivalent experimental setting used in A and C and applied to FKBP8 overexpression (red) and control (gray). Lines and bars represent mean ± SE n.s., not significant; *P < 0.05; ****P < 0.0001 determined by unpaired t test (cell numbers = 256 for scrambled siRNA, FKBP8 siRNA, FKBP8-OE, and 140 for vector control). (E) Schematic figure of FKBP expression level related to calcium transport between the ER and mitochondria.
Proximity-Labeling Experiment with BioID-FKBP8 Reveals FKBP8’s Interaction Partners.
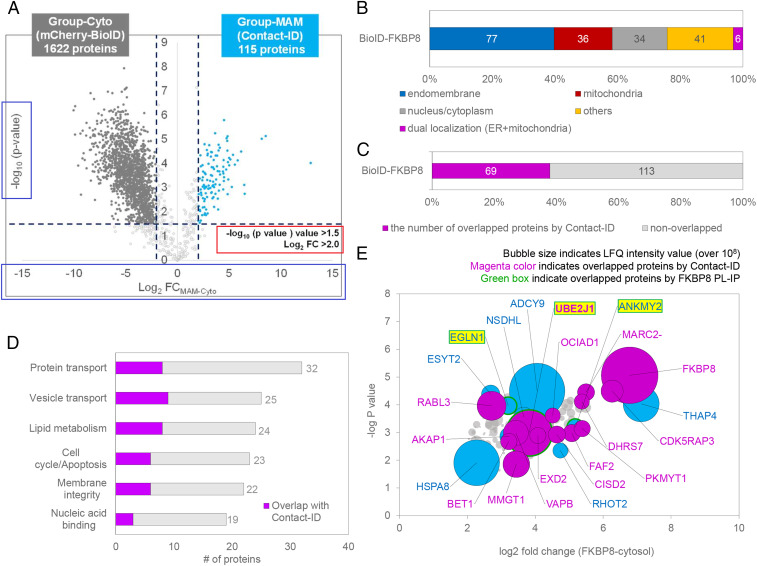
Finally, we sought to resolve the remaining question as to how FKBP8 expression can induce MAM formation. Since endogenous FKBP8 is characterized as an OMM protein (65), we assumed that it might have an interacting protein at the ERM for MAM formation. Thus, we conducted another proximity-labeling experiment with mCherry-BioID-HA-FKBP8 (BioID-FKBP8) in which the BioID is genetically conjugated at the N terminus of FKBP8 (SI Appendix, Fig. S15A). Triplicate mass analysis identified 182 proteins that were exclusively labeled in BioID-FKBP8 compared to mCherry-BioID, the cytosolic control (Fig. 7A). Nearly half of these labeled proteins (42%) originate from the endomembrane system (Fig. 7B), and a considerable number of proteins (69, 38%) showed strong overlap with the 115 MAM proteins identified by Contact-ID (Fig. 7 C and D). These results suggested that FKBP8 is localized at the MAM. Several ERM proteins, including THAP4, ADCY9, UBE2J1, and VAPB, were strongly labeled by BioID-FKBP8 based on the biotinylated mass signal intensity (Fig. 7E), suggesting ERM proteins as possible interacting partners with FKBP8 for MAM formation. Notably, VAPB was also identified in our Contact-ID analysis, which has been previously recognized as a MAM marker protein (66, 67).
Fig. 7.
BioID-FKBP8 reveals the FKBP8 interactome. (A) Volcano plots showing statistically significant enrichment of biotinylated proteins (182 proteins) by BioID-FKBP8 over the cytosolic biotinylated proteins by mCherry-BioID. See Dataset S4 for details. (B) Subcellular distribution of proteins in the selective biotinylated proteins (182 proteins) by BioID-FKBP8 with prior annotated localization information in Uniprot. (C) Number of overlapped proteins between group-MAM and BioID-FKBP8 proteins. (D) Functional clustering of BioID-FKBP8 proteins according to the annotated function in Uniprot. See Dataset S4 for details. (E) Representative biotinylated proteins by BioID-FKBP8. The bubble size indicates the mass signal intensity of the biotinylated peptide by BioID-FKBP8. Protein names for biotinylated peptides with a mass signal intensity over 108 au are shown in the volcano plot of A. Group-MAM proteins in this plot are shown in magenta and proteins found in coimmunoprecipitation after proximity-labeling experiment (PL-IP) are outlined in green (“green box”) (see Dataset S4 for details).
To specify the physical interacting proteins with FKBP8 among the biotinylated protein by BioID-FKBP8, we performed proteomics profiling of the streptavidin-enriched peptides from the elution samples via anti-HA immunoprecipitation (PL-IP) experiment (SI Appendix, Fig. S16). From this analysis, we could see that ANKMY2, EGLN1, and UBE2J1 were significantly enriched in the anti-HA elution sample compared to the control elution sample (Fig. 7E and see Dataset S4 for details). ANKMY2 and EGLN1 are the known interaction partners of FKBP8 (61, 68, 69) and UBE2J1 is an E2 ubiquitin-conjugating ER membrane protein related to ER-associated degradation (70) under the ER stress condition. Thus, it might be intriguing to further investigate how MAM formation and calcium transport can be regulated by the protein network of FKBP8 under the various conditions.
Discussion
We designed a split version of BioID, Contact-ID, for local proteome mapping at the contact sites of the ER and mitochondria. We unequivocally identified 115 proteins that were reproducibly enriched in the Contact-ID-labeled datasets over controls. Membrane proteins were highly enriched in our labeled proteome, including many cholesterol metabolism-related proteins. This is a reasonable result given that the MAM has been characterized as a cholesterol-enriched intracellular lipid raft domain (55, 56), and our result implies that steroidogenesis might be one of the important functions of the MAM. Since a steroidogenic process has not been extensively discussed as one of the MAM functions to date, our results provide strong evidence for the possibility of a role of steroidogenesis in MAM formation in steroidogenic tissues.
Although we identified various interesting MAM proteins with the present tool, our method failed to cover some well-known MAM proteins such as GRP75 (71–73), which has been long known as a MAM protein at the cytosolic side (74, 75). However, GRP75 contains very strong mitochondrial matrix-targeting peptides at its N terminus (SI Appendix, Fig. S17A). We performed TEM imaging with GRP75-APEX2 cells and observed its clear mitochondrial matrix localization (SI Appendix, Fig. S17B). Our TEM imaging result is in agreement with previous findings of GRP75 in the mitochondrial matrix fraction (76) and in mitochondrial matrix proteome lists (77, 78). Thus, it is reasonable that GRP75 was not included in our MAM protein list of Contact-ID.
In our labeling experiment, we did not incorporate an additional linker between the split enzyme and anchored proteins. Thus, our current Contact-ID system can preferentially function at the tight-MAM junction where lipid transport occurs (78, 79). This might explain the relatively high enrichment of membrane proteins related to lipid metabolism in our list. Thus, it would be interesting to test whether Contact-ID can cover other functional MAM proteins related to calcium transport (79, 80) if additional linkers are introduced to the constructs.
Overall, we demonstrated that our Contact-ID method can successfully map the local proteome at membrane contact sites. Thus, we expect that this method could be generally utilized to map the local proteome at any membrane contact site (e.g., ER–plasma membrane, ER–lysosome, and mitochondria–lipid droplet) in various cell types under the various stress conditions as demonstrated in this study with DTT and tunicamycin (SI Appendix, Fig. S9). We also expect that our Contact-ID method can resolve the proteomic architecture of an abnormally increased MAM in type 2 diabetes (81), obesity (34), and neurodegenerative diseases (10, 13, 14, 82).
Materials and Methods
Molecular Biology and Biochemistry.
Details on plasmids, siRNAs, and antibodies are in SI Appendix.
Mass Sampling and LC-MS/MS Analysis.
Details on proteome digestion, enrichment of biotinylated peptides, LC-MS/MS analysis of the enriched peptidome, and MS data processing are in SI Appendix.
EM Imaging.
Details on correlative light and electron microscopy imaging and EM imaging of mitochondria and ER are in SI Appendix.
Calcium Measurement.
Details on measurement of calcium concentration of ER and mitochondria are in SI Appendix.
Proteomic Data Availability.
Detailed information of identified proteins in this study is shown in Datasets S1–S5. The raw proteomics analysis data file is deposited in the MassIVE database (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) (accession no. PXD015534) (FTP download link: ftp://massive.ucsd.edu/MSV000084362/).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF-2019R1A2C3008463) and the Organelle Network Research Center (NRF-2017R1A5A1015366). S.S. and J.-S.K. are supported by IBS-R008-D1 of the Institute for Basic Science from the Ministry of Science and ICT of Korea. J.Y.M. and M.J. are supported by KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (Information & Communication Technology) (20-BR-01-09). EM data were acquired at Brain Research Core Facilities in KBRI. S.K.P. is supported by the Mid-Career Researcher Program (2017R1A2B2009031) funded by the National Research Foundation of Korea. S.K.P. and H.-W.R. were supported by the Samsung Science and Technology Foundation (SSTF-BA1601-14 to S.K.P. and SSTF-BA1401-11 to H.-W.R.). The MCU antibody was a gift from Prof. Soo Hyun Eom, Gwangju Institute of Science and Technology (GIST), Korea.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw LC-MS data generated from this study have been deposited on ProteomeXchange Consortium (accession no. PDX015534) via the Mass Spectrometry Interactive Virtual Environment repository (MassIVE ID MSV000084362).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916584117/-/DCSupplemental.
References
- 1.Murley A., Nunnari J., The emerging network of mitochondria-organelle contacts. Mol. Cell 61, 648–653 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbaz Y., Schuldiner M., Staying in touch: The molecular era of organelle contact sites. Trends Biochem. Sci. 36, 616–623 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Phillips M. J., Voeltz G. K., Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17, 69–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzuto R., et al. , Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Vance J. E., Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 (1990). [PubMed] [Google Scholar]
- 6.Friedman J. R., et al. , ER tubules mark sites of mitochondrial division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamasaki M., et al. , Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Gilady S. Y., et al. , Ero1alpha requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperones 15, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo R., et al. , Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 124, 2143–2152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillusson S., et al. , There’s something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 39, 146–157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmen T., Lynes E. M., Gesson K., Thomas G., Oxidative protein folding in the endoplasmic reticulum: Tight links to the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 1798, 1465–1473 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tubbs E., et al. , Disruption of mitochondria-associated endoplasmic reticulum membrane (MAM) integrity contributes to muscle insulin resistance in mice and humans. Diabetes 67, 636–650 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Schon E. A., Przedborski S., Mitochondria: The next (neurode)generation. Neuron 70, 1033–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delprat B., Maurice T., Delettre C., Wolfram syndrome: MAMs’ connection? Cell Death Dis. 9, 364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janikiewicz J., et al. , Mitochondria-associated membranes in aging and senescence: Structure, function, and dynamics. Cell Death Dis. 9, 332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter L., Hajnóczky G., Mitochondria and endoplasmic reticulum: The lethal interorganelle cross-talk. J. Bioenerg. Biomembr. 37, 191–206 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Theurey P., Rieusset J., Mitochondria-associated membranes response to nutrient availability and role in metabolic diseases. Trends Endocrinol. Metab. 28, 32–45 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Wieckowski M. R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P., Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Poston C. N., Krishnan S. C., Bazemore-Walker C. R., In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteomics 79, 219–230 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Ma J. H., et al. , Comparative proteomic analysis of the mitochondria-associated ER membrane (MAM) in a long-term type 2 diabetic rodent model. Sci. Rep. 7, 2062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horner S. M., Wilkins C., Badil S., Iskarpatyoti J., Gale M. Jr, Proteomic analysis of mitochondrial-associated ER membranes (MAM) during RNA virus infection reveals dynamic changes in protein and organelle trafficking. PLoS One 10, e0117963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis A., Tsai S.-Y., Su T.-P., “Detection of isolated mitochondria-associated ER membranes using the Sigma-1 receptor” in Lipid Signaling Protocols, Waugh M. G., York S. N., York N., Eds. (Springer, New York, NY, 2016), pp. 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J., et al. , mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 9, 96–102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martell J. D., et al. , Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143–1148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X.-W., et al. , New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J. Biol. Chem. 289, 14434–14447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees J. S., Li X.-W., Perrett S., Lilley K. S., Jackson A. P., Selective proteomic proximity labeling assay using tyramide (SPPLAT): A quantitative method for the proteomic analysis of localized membrane-bound protein clusters. Curr. Protoc. Protein Sci. 80, 19.27.11–19.27.18 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loh K. H., et al. , Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166, 1295–1307.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glasgow H. L., et al. , Laminin targeting of a peripheral nerve-highlighting peptide enables degenerated nerve visualization. Proc. Natl. Acad. Sci. U.S.A. 113, 12774–12779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinke A. W., Balla K. M., Bennett E. J., Troemel E. R., Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nat. Commun. 8, 14023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi J., Chen J., Schreiber S. L., Clardy J., Structure of the FKBP12-Rapamycin complex interacting with binding domain of human FRAP. Science 273, 239–242 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Komatsu T., et al. , Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods 7, 206–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csordás G., et al. , Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell 39, 121–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arruda A. P., et al. , Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 20, 1427–1435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martell J. D., et al. , A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Y., et al. , Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue M., et al. , Optimizing the fragment complementation of APEX2 for detection of specific protein-protein interactions in live cells. Sci. Rep. 7, 12039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schopp I. M., et al. , Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8, 15690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Munter S., et al. , Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 591, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Yoboue E. D., Sitia R., Simmen T., Redox crosstalk at endoplasmic reticulum (ER) membrane contact sites (MCS) uses toxic waste to deliver messages. Cell Death Dis. 9, 331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carugo O., How large B-factors can be in protein crystal structures. BMC Bioinformatics 19, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Z., Zhao J., Wang Z.-X., Flexibility analysis of enzyme active sites by crystallographic temperature factors. Protein Eng. 16, 109–114 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Chernov K. G., Neuvonen M., Brock I., Ikonen E., Verkhusha V. V., Introducing inducible fluorescent split cholesterol oxidase to mammalian cells. J. Biol. Chem. 292, 8811–8822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabantous S., Waldo G. S., In vivo and in vitro protein solubility assays using split GFP. Nat. Methods 3, 845–854 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Kamiyama D., et al. , Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck M., et al. , The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S.-Y., Seo J. K., Rhee H.-W., “Direct identification of biotinylated proteins from proximity labeling (Spot-BioID)” in Proximity Labeling: Methods and Protocols, Sunbul M., Jäschke A., York S. N., York N., Eds. (Springer, New York, NY, 2019), pp. 97–105. [DOI] [PubMed] [Google Scholar]
- 48.Lee S.-Y., et al. , Proximity-directed labeling reveals a new rapamycin-induced heterodimer of FKBP25 and FRB in live cells. ACS Cent. Sci. 2, 506–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thul P. J., et al. , A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Nutt L. K., et al. , Bax and Bak promote apoptosis by modulating endoplasmic reticular and mitochondrial Ca2+ stores. J. Biol. Chem. 277, 9219–9225 (2002). [DOI] [PubMed] [Google Scholar]
- 51.De Vos K. J., et al. , VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21, 1299–1311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang N. C., Nguyen M., Germain M., Shore G. C., Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 29, 606–618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhujabal Z., et al. , FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18, 947–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rone M. B., et al. , Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol. Endocrinol. 26, 1868–1882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Area-Gomez E., et al. , Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106–4123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi T., Fujimoto M., Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 77, 517–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fazal F. M., et al. , Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank C. G., et al. , Identification and functional analysis of a defect in the human ALG9 gene: Definition of congenital disorder of glycosylation type IL. Am. J. Hum. Genet. 75, 146–150 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Misaka T., et al. , FKBP8 protects the heart from hemodynamic stress by preventing the accumulation of misfolded proteins and endoplasmic reticulum-associated apoptosis in mice. J. Mol. Cell. Cardiol. 114, 93–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirane M., Nakayama K. I., Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 5, 28–37 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Saita S., Shirane M., Ishitani T., Shimizu N., Nakayama K. I., Role of the ANKMY2-FKBP38 axis in regulation of the Sonic hedgehog (Shh) signaling pathway. J. Biol. Chem. 289, 25639–25654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saita S., Shirane M., Nakayama K. I., Selective escape of proteins from the mitochondria during mitophagy. Nat. Commun. 4, 1410 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Park S. J., et al. , DISC1 modulates neuronal stress responses by gate-keeping ER-mitochondria Ca2+ transfer through the MAM. Cell Rep. 21, 2748–2759 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Suzuki J., et al. , Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 5, 4153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhujabal Z., et al. , FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18, 947–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cho I.-T., et al. , Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J. Biol. Chem. 292, 16382–16392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoica R., et al. , ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 5, 3996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huttlin E. L., et al. , Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barth S., et al. , Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 284, 23046–23058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burr M. L., et al. , HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 2034–2039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Honrath B., et al. , Glucose-regulated protein 75 determines ER-mitochondrial coupling and sensitivity to oxidative stress in neuronal cells. Cell Death Discov. 3, 17076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D’Eletto M., et al. , Transglutaminase type 2 regulates ER-mitochondria contact sites by interacting with GRP75. Cell Rep. 25, 3573–3581.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Theurey P., et al. , Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J. Mol. Cell Biol. 8, 129–143 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Szabadkai G., et al. , Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–911 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rizzuto R., et al. , Ca2+ transfer from the ER to mitochondria: When, how and why. Biochim. Biophys. Acta 1787, 1342–1351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Z., et al. , Mitochondria chaperone GRP75 moonlighting as a cell cycle controller to derail endocytosis provides an opportunity for nanomicrosphere intracellular delivery. Oncotarget 8, 58536.– (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S.-Y., et al. , Architecture mapping of the inner mitochondrial membrane proteome by chemical tools in live cells. J. Am. Chem. Soc. 139, 3651–3662 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Flis V. V., Daum G., Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. 5, a013235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giacomello M., Pellegrini L., The coming of age of the mitochondria-ER contact: A matter of thickness. Cell Death Differ. 23, 1417–1427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patergnani S., et al. , Calcium signaling around mitochondria associated membranes (MAMs). Cell Commun. Signal. 9, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rieusset J., “Role of endoplasmic reticulum-mitochondria communication in type 2 diabetes” in Organelle Contact Sites: From Molecular Mechanism to Disease, Tagaya M., Simmen T., Eds. (Springer, Singapore, 2017), pp. 171–186. [DOI] [PubMed] [Google Scholar]
- 82.Area-Gomez E., Schon E. A., Mitochondria-associated ER membranes and Alzheimer disease. Curr. Opin. Genet. Dev. 38, 90–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.