Significance
Inferring the precise timing of the origin of the HIV/AIDS pandemic is of great importance because it offers insights into which factors did—or did not—facilitate the emergence of the causal virus. Previous estimates have implicated rapid development during the early 20th century in Central Africa, which wove once-isolated populations into a more continuous fabric. We recovered the first HIV-1 genome from the 1960s, and it provides direct evidence that HIV-1 molecular clock estimates spanning the last half-century are remarkably reliable. And, because this genome itself was sampled only about a half century after the estimated origin of the pandemic, it empirically anchors this crucial inference with high confidence.
Keywords: HIV-1, evolution, virus, phylogeny
Abstract
With very little direct biological data of HIV-1 from before the 1980s, far-reaching evolutionary and epidemiological inferences regarding the long prediscovery phase of this pandemic are based on extrapolations by phylodynamic models of HIV-1 genomic sequences gathered mostly over recent decades. Here, using a very sensitive multiplex RT-PCR assay, we screened 1,645 formalin-fixed paraffin-embedded tissue specimens collected for pathology diagnostics in Central Africa between 1958 and 1966. We report the near-complete viral genome in one HIV-1 positive specimen from Kinshasa, Democratic Republic of Congo (DRC), from 1966 (“DRC66”)—a nonrecombinant sister lineage to subtype C that constitutes the oldest HIV-1 near full-length genome recovered to date. Root-to-tip plots showed the DRC66 sequence is not an outlier as would be expected if dating estimates from more recent genomes were systematically biased; and inclusion of the DRC66 sequence in tip-dated BEAST analyses did not significantly alter root and internal node age estimates based on post-1978 HIV-1 sequences. There was larger variation in divergence time estimates among datasets that were subsamples of the available HIV-1 genomes from 1978 to 2014, showing the inherent phylogenetic stochasticity across subsets of the real HIV-1 diversity. Our phylogenetic analyses date the origin of the pandemic lineage of HIV-1 to a time period around the turn of the 20th century (1881 to 1918). In conclusion, this unique archival HIV-1 sequence provides direct genomic insight into HIV-1 in 1960s DRC, and, as an ancient-DNA calibrator, it validates our understanding of HIV-1 evolutionary history.
The HIV pandemic is one of the most devastating in all human history: more than 70 million people have so far been infected with the virus and about 32 million have died (1). Though AIDS and its main causative agent HIV-1 were discovered almost four decades ago, the virus has likely been circulating for about a century (2). In the time before its discovery, the virus had already diversified into many of the lineages we would later classify as subtypes and their circulating recombinant forms (CRFs), and several strains had already spread from Central Africa to far corners of the world. These strains belong to HIV-1 group M (2), a phylogenetic clade that is set apart from the other HIV groups—HIV-1 groups N, O, P and the eight HIV-2 groups, each group having a distinct cross-species origin—by its pandemic scale and much higher prevalence, accounting for >95% of all cases (1).
As a retrovirus with an RNA genome, HIV-1 evolves so rapidly that sequence data collected at various time points can calibrate molecular clock models used to infer the timing of historical, unsampled events in the virus’ past. However, rates of molecular evolution appear to vary depending on the time frame considered, so rates estimated over a recent time frame cannot necessarily be extrapolated to a deeper time frame (3, 4). For example, molecular clock estimates based on the time that endogenous lentiviruses integrated in hosts’ genomes are several orders of magnitude slower than clock rates estimated from time-stamped extant samples (5). Clock rates measured with recent HIV and simian immunodeficiency virus (SIV) samples are also not reconcilable with the limited divergence between SIV strains in monkeys on an island established >10,000 y ago and SIV strains in the mainland relatives (6).
While time-dependent bias appears to grow in a continuous fashion with larger time frames (3), different time frame lengths reflect distinct biological processes (4). For RNA viruses, these include differences in host-induced selection pressures (7) and differences in population dynamics, for example when comparing intrahost to interhost dynamics (8) or newly emerged outbreaks versus endemically circulating viruses (9). As such, it should be possible to demarcate time frames with consistent epidemic behavior and therefore likely also consistent average molecular clock rates.
A crucial aspect in estimating molecular clock rates across such longer time frames is calibration by known events in the past (10). While ancient genomic DNA or RNA sequenced from archival samples or hosts’ remains are only available for relatively recent evolutionary times, they represent powerful calibrators as they offer direct evidence of an organism’s presence and identity at a point in the past. Short HIV-1 RNA fragments belonging to two different subtypes isolated from archival tissue specimens from 1959 and 1960 demonstrated that HIV-1 group M was at least that old (11, 12).
The wealth of genetic sequences of HIV-1 sampled throughout the decades since its discovery plus the few prediscovery archival sequences led to the phylogenetic inference of the time of the most recent common ancestor (TMRCA) of HIV-1 group M around 1920 in Central Africa (11, 13–15). The original spillover event of HIV-1 group M from central chimpanzees to humans must have occurred in rural southeast Cameroon (16) and, assuming the estimates of the TMRCA are correct, it must have occurred at or before this point in time. Early divergence events that resulted in the diversity of HIV-1 group M we see today likely took place in the major—and at that time massively growing—cities on the banks of the Congo River in what is now Republic of Congo (ROC) and Democratic Republic of Congo (DRC) (11, 13). This is also apparent from the much higher diversity of HIV-1 group M in DRC, ROC, and Cameroon, with divergent lineages and CRFs still being described from that region (17–20).
Genetic sequences from before HIV-1’s discovery in 1983 are scarce because 1) few archival specimens exist from that time period; 2) for those specimens that exist, the odds of retrospectively finding a patient’s samples with an unspecific syndrome are very small; 3) while screening by PCR amplification may be the most sensitive technique, sensitive existing primer sets might miss unknown divergent strains; 4) the further back in time one looks, the lower the expected prevalence of the virus; and 5) genetic molecules, especially viral RNA, tend to degrade during long-term storage or degrade during the original preservation of the specimen. Indeed, most long-term stored specimens are initially intended for histology, and while afterward remaining fairly stable at room temperature, the original formalin-fixation process degrades RNA and DNA tremendously. However, short viral DNA or RNA sequences around 50 to 200 bases in length are still recoverable from such formalin-fixed paraffin-embedded (FFPE) samples when using dedicated ultrasensitive amplification techniques (11, 21–24).
The M.W. laboratory recently described a particularly sensitive procedure, the “jackhammer RT-PCR” to detect and obtain high-quality, potentially divergent viral genomic sequences, yielding eight of the nine oldest HIV-1 group M genomes characterized to date (25).
Here, we screened 1,645 FFPE samples from Central Africa using a jackhammer RT-PCR procedure modified to accommodate the small size of RNA fragments present in such specimens. We determined the near full-length genome of an HIV-1 positive sample from Kinshasa, DRC, from 1966, the earliest HIV-1 genome assembled to date. We further investigated the effects of including or excluding this genome as a molecular clock calibration point from deep within the prediscovery phase of HIV/AIDS on the emergence and divergence time estimates of the HIV pandemic.
Methods
See SI Appendix for a more detailed description of the methods.
We screened a total of 1,645 FFPE samples from various tissues, originating from the DRC (n = 1628) and Rwanda (n = 17) from 1958 to 1966. Data recorded were unlinked to individual identifiers and the work was approved by the Human Subjects Protection Program at the University of Arizona.
Paraffin was dissolved with xylene and total RNA extracted using the High Pure FFPE RNA Micro kit (Roche) or Qiagen miRNeasy FFPE kit. The eight HIV-1 screening primer pairs listed in SI Appendix, Table S1, target regions 72 to 112 nucleotides (nt) in length of the gag, pol, and env genes plus a human β2-microglobulin positive control. These were applied in a jackhammer RT-PCR procedure (25). Reverse transcription was carried out for each RNA sample in an eightfold multiplex with a pool of the reverse primers and for each resulting cDNA sample a “preamplification” step, so that extremely rare template molecules are not lost during aliquoting (25), was then carried out in an eightfold multiplex PCR with a pool of the forward primers. In the final amplification step, each of the reactions was carried out as a singleplex PCR with each primer pair. Products were cloned and then Sanger sequenced at the University of Arizona Genetics Core facility.
FFPE tissue from a patient sampled in 1966 was HIV-1 positive. We subjected this HIV-1 sample, henceforth called “DRC66,” to jackhammer PCR attempts to sequence its coding genome. We designed 124 pan-HIV-1 group M primer pairs. Of these, 29 yielded HIV sequences, which were indicative of a subtype C-like genome. We then designed 219 primer pairs for subtype C-like genomes, of which 116 yielded HIV sequences (for 32 pairs after slight modification of either or both forward and reverse primers). We closed most remaining gaps using a primer-walking approach for which a total of 446 primers were designed. All PCRs were organized in a jackhammer approach. Sequences of all primers used are available in SI Appendix, Table S4. All DRC66 amplicon sequences were assembled and a consensus sequence is deposited in GenBank with accession number MN082768.
We searched for genetic signatures of preexisting drug resistance in DRC66 using the HIV Drug Resistance Database Program (26).
To compare our jackhammer PCR approach with a deep-sequencing approach, paired-end sequencing was carried out on an Illumina MiSeq. None of the reads mapped to HIV-1 but several bacterial, viral, and fungal reads were recovered.
We searched extensively for both partial and full genomic sequences deposited in GenBank that had close similarity to the DRC66 genomic sequence. We built an alignment for the pol region between 2,485 and 4,274 (HXB2 notation) based on four retrieved sequences together with our subsampled dataset A (see below) and “divergent C-like lineages” summarized in ref. 17. We investigated in recombination detection program 4 (RDP4) (27) that there had been no significant recombination among lineages within this alignment.
We then downloaded all near-complete (>7,000 nt) HIV-1 group M genomes sampled in Africa from the Los Alamos National Laboratories (LANL) database on November 19, 2018 (n = 2,342) and all HIV-1 group M genomes from other parts of the world that were sampled before 1985 (n = 78). We filtered out sequences that were designated as intersubtype recombinants, multiple sequences per patient, sequences with missing annotations, and hypermutated sequences as explained in SI Appendix. The final dataset contained 830 sequences (including the DRC66 sequence), which were codon aligned based on the translated gene sequences by the LANL HIVAlign tool (28) using the hidden Markov model (29). Intrasubtype recombination was investigated for sequences from each subtype separately in RDP4 (27). Sequence regions larger than 300 nt identified by at least six methods as (possibly) involved in recombination were masked from the alignment. This alignment was further down-sampled in five distinct subalignments (named subsampled datasets A to E), for the purpose of efficient computation, as well as to assess the inherent uncertainty through phylogenetic variation among random samples of actual HIV-1 diversity. For those HIV-1 genomes sampled after 1990, a random sample of 150 genomes was drawn five times without replacement. Because of the scarcity of nonsubtype B genomes sampled before 1990, all of these were retained in each subsampled dataset. For subtype B genomes sampled before 1990, seven or six were randomly sampled without replacement. Subsampled datasets A–C each contained 177 genomes; datasets D and E contained 176 genomes.
Maximum-likelihood (ML) trees of the complete dataset and subsampled datasets were estimated with RAxML v8.2 and, to explore clock-like signal, root-to-tip distance was plotted against sampling time with Tempest and R’s stats packages (30, 31). Tip-date calibrated phylogenetic trees were estimated using BEAST v.1.10.4 (32) for each subsampled dataset A–E. Convergence and effective samplesize (ESS) values of all parameters were evaluated in Tracer v1.6. The first 20% of the Markov chain Monte Carlo runs were removed as burn-in and the two to three parameter log and tree log output files for each dataset were combined in R v3.4.2 and Logcombiner (32). These analyses were repeated but with 1) the DRC66 sequence removed from the alignment and 2) without providing the sampling year of DRC66 but instead having its sampling date estimated from the rest of the data using BEAST’s “tip date sampling” option and a uniform prior bound between 0 and 1,000 (in years since youngest sampling date) (33). To explore how well tip dates of other samples could be estimated, we repeated the BEAST analyses of one of the subsampled alignments (A) for each of five different sequences in the dataset using BEAST’s tip date sampling option, where again for each analysis a uniform prior bound between 0 and 1,000 (in years since youngest sampling date) for the unspecified tip date was used.
Data Availability.
The DRC66 genome sequence is deposited in GenBank with accession number MN082768. Alignments and BEAST xml files are available on https://github.com/sophiegryseels/DRC66.
Results
HIV-1 Genomic Sequence from 1966 Characterized via Jackhammer PCR Is the Earliest Known Near-Complete HIV Genome.
Out of 1,645 archival FFPE specimens from DRC dated between 1958 and 1966, only two lymph nodes biopsied in 1966 in Kinshasa were found to be HIV positive. These two samples, taken about 1 mo apart, were both from a 38-y-old male and proved to have virtually identical sequences; hence we assume these are from the same person. Only data from the earlier sample were further used here. No pathological annotations were associated with the specimens. We henceforth call this sample DRC66.
A large part of the genomic sequence was determined via 54- to 106-nt PCR products yielding overlapping stretches of genomic RNA, for which the RT and a preamplification step were efficiently carried out in pools of no more than eight reactions while a final amplification step was carried out individually for each primer pair (collectively called the jackhammer PCR procedure). All final amplification products were cloned and multiple clones were Sanger sequenced. A final stretch covering 8,360 bases of the genome (HXB2 positions 816 to 9,175) was characterized, containing ∼1,957 undetermined sites (by comparison with HXB2 genome length), leaving a total of 6,390 characterized nucleotide sites.
DRC66 Is a Sister Lineage to the Subtype C Clade.
The maximum-likelihood tree based on an alignment with 829 complete HIV-1 genomes downloaded from GenBank (798 from Africa sampled between 1983 and 2014 and 31 from Europe and America sampled between 1978 and 1985) reveals that DRC66 represents a sister lineage to the clade conventionally denoted as subtype C (Fig. 1A). Analyses in RDP4 (27) showed no evidence for recombination with other HIV-1 group M subtypes or clades. Extensive BLAST searches and searches via neighbor-joining trees of downloaded HIV-1 sequences revealed that DRC66 is the only reported near-complete genome from this lineage. However, the subtype C-like portions of the genomes of three recently described circulating recombinant forms (designated CRF93_CPX) from Kinshasa and Mbuji-Mayi, DRC, sampled in 2008 (17), formed a monophyletic group with DRC66, as presented in a maximum-likelihood tree of a partial pol alignment (Fig. 1C). An ∼550-nt subtype C-like portion of a recombinant partial pol sequence sampled in Sweden in 2000, also clustered with this group. We further included partial pol sequences of other so-called divergent C lineages as summarized in Villabona-Arenas et al. (17), including those from CRF92_C2U, in this tree. The DRC66-CRF93_CPX clade is well supported, and as shown in Fig. 1C, constitutes a sister clade to the rest of subtype C-like sequences. The clade containing CRF92_C2U is also well supported. Some of the subtype C sequences from our subsample alignment A group with other divergent C lineages; though this clade and the relationships with the CRF92_C2U clade, other divergent C lineages, and the conventional subtype C cannot be recovered with confidence in this tree of partial pol sequences (Fig. 1C).
Fig. 1.
(A) Unrooted maximum-likelihood tree of the complete dataset of 830 HIV-1 group M genomes. (B) Evolutionary distances between the root and all tips of a rooted version of the tree shown in A plotted against the year the sequence was sampled. The root location was detemined in Tempest (30), minimizing the sum of the squared residuals from this exploratory regression. Root-to-tip plots based on ML trees of subsampled datasets are displayed in SI Appendix, Fig. S1. (C) Midpoint rooted ML tree of a 1,799-nt pol alignment that includes subsampled dataset A, sequences that cluster with the DRC66 lineage, plus multiple divergent subtype C-like sequences as summarized in ref. 17, some of which are derived from intersubtype recombinant genomes (e.g., “CU”) or of which only partial sequence for this alignment is available. Tips of the subtype C-related sequences, including DRC66, are labeled by subtype (marked with * if determined based on partial sequence only, e.g., “C*”), sampling year, sampling country, and GenBank accession number. For sampling country, COD = Democratic Republic of the Congo, BWA = Botwana, SWE = Sweden, ZAF = South Africa. In all three figures subtypes are color coded according to the color legend. The DRC66 sequence is indicated with a red star.
Phenotypic Characteristics.
A comparison of V3 amino acid sequences between DRC66 and subtype C samples with known coreceptor usage (34) suggests that DRC66 would have been an R5 virus utilizing the CCR5 coreceptor at the time of sampling.
Three residues present in the integrase of DRC66 have been previously determined to confer resistance to integrase inhibitor drugs: 51Y, 66I, and 145S. While H51Y and T66I mutations moderately reduce elvitegravir (EVG) susceptibility in patients, P145S induces high-level resistance to EVG in vitro, though it is rarely selected for in patients (26, 35, 36). The specific nucleotides that coded for the 51Y and 66I residues were present in 2/2 and 3/3 of successfully sequenced clones for those sites, but 145S was only present in 1/2 sequenced clones. A similar analysis of 565 integrase subtype C sequences sampled in Africa did not indicate any other strain harboring these three specific residues. No residues that confer resistance to protease or reverse transcriptase inhibitor drugs were detected in the DRC66 sequence of those corresponding genes.
Deep Illumina Sequencing Unsuccessful in Uncovering HIV-1 Sequence Data.
Illumina sequencing of this same sample yielded 68,847,162 paired reads, none of which mapped to HIV-1’s HXB2 genome, a subtype C genome (U46016) nor to the DRC66 consensus sequence generated by Sanger sequencing.
De novo assembly of reads into contigs >200 nt led to identification of mostly fungi and bacteria (see details in SI Appendix, Table S2). These were most likely environmental organisms that we speculate entered the FFPE lymph node tissue sample during sample preparation in 1966 or during its long storage time since. Nevertheless, organisms that often represent opportunistic infections in AIDS patients were also detected, such as Bartonella, Mycoplasma, and Candida (SI Appendix, Table S2), though it cannot be excluded that these were derived from later-invading environmental species of these genera rather than being present in the patient’s original biopsy material. Identification of a fungus-borne virus (related to the known yeast virus Saccharomyces cerevisiae virus l-A) provides evidence that our deep-sequencing approach was able to detect RNA viruses, although this virus contains a double-stranded RNA genome instead of the less stable ssRNA genomes of lentiviruses.
The Root-to-DRC66-Tip Distance in the ML Tree Is Consistent with Its Sampling Time.
There is a good correlation between sampling years and root-to-tip distances in the ML tree of the complete, full genome dataset (R2 = 0.389) (Fig. 1B). Although the R2 values of these regressions are difficult to interpret statistically because of nonindependence of the data points, they are indicative of the information held by the sampling dates on evolutionary rates. The root-to-tip distance of DRC66 is the shortest of all samples, as expected from the oldest sample in the dataset. The residual of −0.0395 of the DRC66 sample in this regression is small. Importantly, it is almost exactly the same as its residual (−0.0412) when calculating the regression without using the DRC66 data point but using all other root-to-tip data from the same ML tree (which yields a similar R2 of 0.374, SI Appendix, Fig. S1). The DRC66 sequence is thus not an outlier in the root-to-tip regression plots. This indicates that clock-like signal from more-recently sampled HIV-1 genomes can reliably estimate dates of events from decades earlier.
Correlations between root-to-tip divergences and sampling times were very comparable between the full dataset of 830 genomes and the subsampled datasets of 176 or 177 genomes, or even improved in the subsampled datasets, indicating that the temporal signal is maintained when subsampling the dataset (SI Appendix, Fig. S1). R2 values were also very similar when estimating root-to-tip vs. time correlations for phylogenies built from alignments that either included or excluded the DRC66 sequence (SI Appendix, Fig. S1).
Inclusion of the DRC66 Genome in BEAST Time-Stamped Phylogenies Indicates Robust Clock Inferences Using Recent Genomes.
The five subsampled datasets were each subjected to three types of time-stamped analysis in BEAST (see Methods for details): 1) subsampled dataset including DRC66 and its sampling time; 2) subsampled dataset including DRC66 but leaving its sampling time unknown and to be estimated; and 3) subsampled dataset excluding DRC66 sequence. Fig. 2 shows a time-scaled phylogenetic BEAST tree of subsampled dataset A that includes DRC66 and its date; the time-scaled trees of subsamples B–E are displayed in SI Appendix, Fig. S2. Mean evolutionary rates and dating estimates are summarized in Fig. 3 and SI Appendix, Table S3.
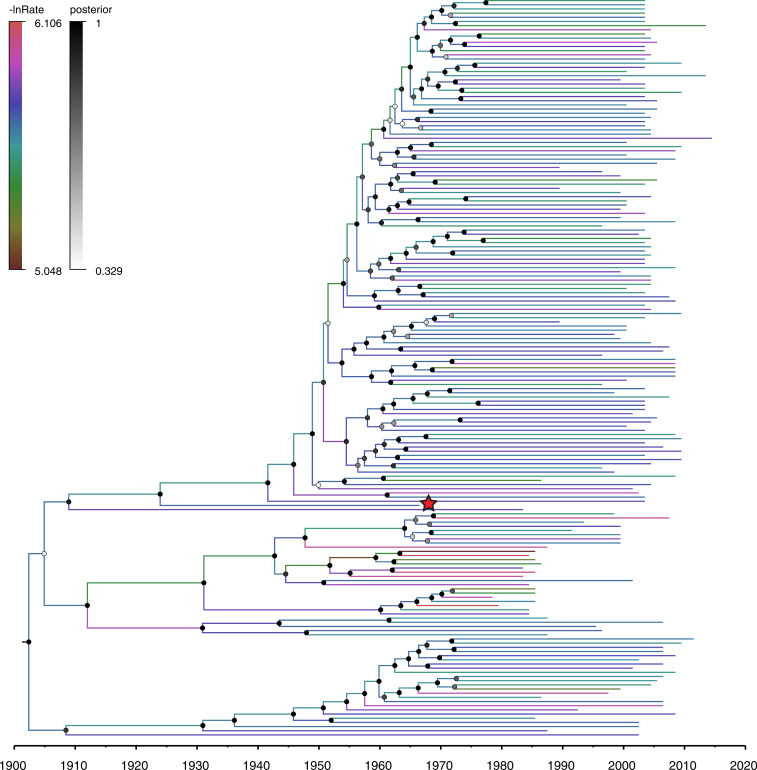
Fig. 2.
Time-scaled phylogenetic BEAST tree of subsampled dataset A estimated under a model that includes the sampling date of DRC66. Branches are color coded by the log of the estimated evolutionary rate for that branch (-lnRate), drawn from a log-normal distribution using the uncorrelated relaxed clock model (48). Node labels are coded on a gray scale by the posterior probability of clade support values. The DRC66 sample is marked with a red star. Trees with tip labels for all five subsampled datasets are displayed in SI Appendix, Fig. S2.
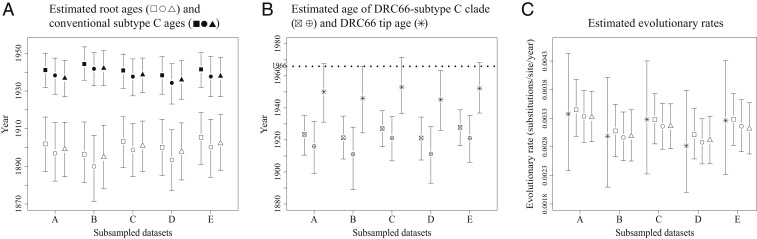
Fig. 3.
Mean node age and mean evolutionary rate estimates and their 95% HPD intervals for time-scaled phylogenies of the five different subsampled datasets (A–E), which were each analyzed in BEAST in three different ways: including DRC66 and its tip age (estimates represented by squares), including DRC66 but with its sampling date unknown and to be estimated (estimates represented by circles), and excluding the DRC66 sequence (estimates represented by triangles). See also SI Appendix, Table S1. (A) Age estimates of the root of the tree (open characters) and of the node representing the common ancestor of conventional subtype C (filled characters) for each of the three BEAST analyses. (B) Age estimates of the clade that encompasses both conventional subtype C and DRC66 (squares and circles with crosses) for the two BEAST analyses that included DRC66 and the estimated sample ages of DRC66 for those analyses in which this was left to be estimated (stars). (C) Estimates of the evolutionary rate along the terminal branch leading to DRC66 in BEAST analyses that included DRC66’s sampling date (stars) and mean evolutionary rates across entire phylogenies for each of the three BEAST analyses.
The posterior mean TMRCA estimates for the five different subsampled datasets A–E of HIV-1 group M genomes (that included DRC66 sequence and its age) ranged between 1896 (95% highest probability distribution [HPD] 1881 to 1914) and 1905 (95% HPD 1891 to 1918) (Fig. 3A and SI Appendix, Table S3). The 95% HPD intervals overlapped in all pairwise comparisons between subsampled datasets (Fig. 3A and SI Appendix, Table S3). While for each subsampled dataset the posterior mean estimates for the TMRCA were always slightly older when the DRC66 date was not specified and left estimated (between 4 and 7 y older) and when the DRC66 sequence was not included in the BEAST analyses (between 1 and 3 y older), this variation was smaller than the variation among some of the different subsampled datasets (Fig. 3A and SI Appendix, Table S3).
A similar pattern of little difference in age estimates between datasets including or excluding the DRC66 sample was observed for the node that represents the MRCA of conventional subtype C (the subtype C clade that does not include DRC66) (Fig. 3A and SI Appendix, Table S3). TMRCA estimates of the clade that incorporates both conventional subtype C and DRC66 varied among datasets as well as among analyses including or not the DRC66 tip date (Fig. 3B and SI Appendix, Table S3). In these BEAST analyses that included the DRC66 sample but left its sampling date to be estimated, the posterior mean estimates for the DRC66 age were between 13 and 21 y older than the actual sampling year of 1966, and for one of five subsampled datasets the upper 95% HPD interval just excluded the year 1966 (Fig. 3B and SI Appendix, Table S3). In subsequent BEAST analyses in which we estimated tip dates for five other HIV-1 sequences downloaded from GenBank, the posterior means for the tip date estimates were between 7 y younger and 29 y older than the real sampling dates (SI Appendix, Table S3). The latter was for a divergent sequence of an until-recently undetermined subtype (U) from 1983 (now designated subtype L; ref. 37), with an estimated tip date of 1954 (95% HPD 1934 to 1983), which indicates that dates of the tips of long external branches are particularly difficult to estimate under a relaxed clock model.
Estimated evolutionary rates averaged over the phylogenetic trees were very similar between datasets including or excluding the DRC66 sample or leaving its tip date unspecified (Fig. 3C and SI Appendix, Table S3). These mean evolutionary rates were also very similar to the rates estimated along the terminal branch leading to the time-stamped DRC66 tip (Fig. 3C).
Discussion
Here we present what is currently the oldest near-complete HIV genome, from 1966 in Kinshasa, DRC. This DRC66 sample is 10 y older than the previously earliest characterized full genome, an 01A1G strain that was isolated from blood in 1976, also in DRC, but which underwent cell culture passages before sequencing (38). There are only nine other HIV-1 genomes available from the prediscovery phase of AIDS (1978 to 1982), all subtype B from the United States (25). The oldest HIV-1 genomic fragments are derived from plasma and FFPE samples from 1959 and 1960, again both from Kinshasa, DRC (11, 12). While these provided undisputable evidence of the presence and major diversification of HIV-1 group M two decades before its discovery, the short sequences that were recovered do not allow complete characterization of the HIV-1 strain involved and contain only a fraction of the phylogenetic information that is present in complete genomes.
To achieve sequence coverage across the DRC66 archival genome, labor-intensive amplification of overlapping short fragments between 54 nt and 106 nt in a highly sensitive jackhammer PCR procedure proved necessary. In comparison, none of the >65 million reads of an Illumina MiSeq run without prior amplification on the same sample contained HIV-1 sequence data. The latter approach had provided a full genome at 3,000× coverage of an influenza A H1N1 strain in an FFPE sample from 1918, however (24). Perhaps the difference in success resulted from different storage conditions in a humid tropical versus a temperate region, as evidenced by the majority of our reads being derived from environmental organisms that could have invaded the sample during preparation or storage, or, more likely, from a comparatively low viral titer in the FFPE lymph node specimen.
Globally, more HIV-1 group M cases are caused by strains that belong to the subtype C clade than any other clade, largely because southern Africa holds the highest HIV-1 burden and subtype C predominates there (39). Estimated to have originated in southeastern DRC, phylodynamic analyses indicated subtype C strains have spread from there to southern Africa via connections between mining cities (13). At the LANL HIV sequence database, currently about 19% of HIV-1 sequences from DRC are classified as subtype C (mostly documented from partial gene sequences). The DRC66 sequence represents a sister lineage to the subtype C clade, and quite divergent: we estimate it shared a common ancestor with subtype C some 20 y before the time of the common ancestor of conventional subtype C. Parts of gag and pol from three recently described intersubtype recombinant genomes from Kinshasa and Mbuji-Mayi sampled in 2008 (17), and part of a partial pol sequence sampled in Sweden in 2000 (40), appear to be the only reported contemporary sequences that also belong to this lineage in part of their genomes, although we cannot be certain we did not miss any short sequence stretches of, e.g., complex recombinant forms that would also cluster with this clade. Villabona-Arenas et al. (17) and Rodgers et al. (19) describe additional so-called divergent C lineages sampled between 1997 and 2012 in DRC that are monophyletic with conventional C with respect to the DRC66 lineage, yet form distinct sister lineages to subtype C. Similarly, for most other HIV-1 subtypes, more divergent lineages can be found in DRC (in particular Kinshasa) and other central African countries than in other regions where the more restricted within-subtype diversity arose in a relatively short time after founder events. The DRC66 genome provides a unique insight into the subtype C-like diversity that would have been present in DRC in the 1960s. The fact that particular residues of the translated integrase protein of DRC66 are known to induce resistance to integrase inhibitor drugs, which were obviously developed long after DRC66 was sampled, highlights that the natural 1960s diversity already harbored some genetic basis for anti-HIV therapy failure.
We further investigated whether the phylogenetic information in the suite of HIV-1 genomes sampled across the past decades, almost all after the discovery of HIV-1, reliably captures HIV-1’s evolutionary rates over the longer time frame that includes HIV-1’s long prediscovery phase in humans. Few calibration points from direct biological observations are typically available to test such conclusions for real-world analyses, especially for such a medically important pathogen. Crucially, such ancient DNA calibration points can lead to dramatic changes in evolutionary histories once thought to be definitively established. For example, recently reported hepatitis B virus sequences from the Bronze age and Neolithic suggested a 100-fold slower evolutionary rate for this double-stranded DNA virus than previously thought (41–43), and such data are prompting updates to evolutionary clock models to better accommodate time-dependent rate variation (10). Because it is impossible to completely rule out such biases without complete genomic information from an early evolutionary time point, we believe it is important to attempt to recover such information from surviving HIV-1 specimens.
Reassuringly, in the context of HIV-1 group M, we do not observe that an “ancient” HIV-1 genome significantly changes evolutionary inferences based on phylogenies built from more-recent genomes. Indeed, there is remarkably little difference in key estimates—including the overall age of the pandemic lineage of HIV—when this sequence is included in phylogenomic analyses. Given that it is more than 50 y older than currently circulating HIV-1 strains, this sequence provides direct evidence for the reliability of dating estimates over the last half-century of HIV-1 circulation. This stands in contrast to the disconnection between short-term rates observed in SIVs and the rates at which SIV strains evolve when averaged across centuries or millennia of evolution in natural populations of different primate species, where molecular clock dating theory has difficulties accommodating the rate differences (6).
Interestingly, our analysis highlights an often-overlooked source of uncertainty in evolutionary divergence dating based on any sample of genomes. The suite of HIV-1 genomes sampled from patients and available in public databases is inevitably a very limited subsample of the true diversity of HIV-1 group M. To investigate the degree of variation such an unavoidable sampling process induces, we subsampled the available GenBank sample of nonintersubtype recombinant HIV-1 group M genomes from Africa, only retaining a small set of genome samples before 1990 in each sample. While credible intervals of all dating and rate estimates overlapped substantially, the overall variation between subsamples was larger than that induced in each subsample when DRC66 was either included or excluded. Besides variation in the underlying evolutionary models used in different studies, usage of different HIV-1 genome dataset samples could also explain why our HIV-1 group M TMRCA estimates are somewhat older here than previously reported: 1920 (95% HPD 1909 to 1930) (13), 1930 (1911 to 1945) (44), 1932 (1905 to 1954) (15), 1920 (1902 to 1939) (14), and 1908 (1884 to 1924) (11). Across our five investigated subsamples, HIV-1 group M TMRCA confidence intervals ranged from 1881 to 1918. We did not further explore the sensitivity of TMRCA estimates to various evolutionary model specifications, though it has been shown for example that the choice of coalescent tree prior may influence TMRCA estimates of HIV-1 for Bayesian inferences (11, 45). While a skygrid coalescent model should be appropriate (46), a recent study that was also based on complete HIV-1 genomes but that used a combination of an exponential and logistic growth model as tree prior (47) estimated 1915 to 1925 as the HIV-1 group M TMRCA. Taken together, while most estimates of the origin of the pandemic lineage of HIV-1 indeed converge to around the turn of the 20th century, phylogenetic uncertainty, evolutionary model specifications, and natural variation among samples of HIV-1’s genomic diversity prevent narrowing down the age estimate to less than a few decades.
In conclusion, using a highly sensitive amplification protocol for degraded archival samples, we here present the oldest HIV-1 near-complete genome available to date. While we are careful not to extrapolate to other pathogen–host systems and much deeper time scales evident in SIV, our study indicates that evolutionary rates calibrated from HIV-1 group M sequences sampled across the decades after its discovery can be used reliably to infer the timing of events that occurred during the prediscovery era. We note that in addition to evolutionary model specifications, the inherent stochasticity associated with a sample of the true viral diversity in nature inevitably introduces uncertainty to phylogenetic dating estimates, which is addressable by purposely subsampling datasets.
Supplementary Material
Acknowledgments
We thank Tatenda Mangurenje and Ryan Ruboyianes for their excellent technical help. This work was supported by NIH/National Institute of Allergy and Infectious Diseases (NIAID) grant R01AI084691 and the David and Lucile Packard Foundation (M.W.). S.G. was supported by a European Molecular Biology Organization (EMBO) long-term postdoctoral fellowship (ALTF-328) and an OUTGOING [Pegasus]2 Marie Skłodowska-Curie Fellowship of the Research Foundation–Flanders (12T1117N) during this work. P.L. acknowledges funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement 725422-ReservoirDOCS) and the Research Foundation–Flanders (grants G066215N, G0D5117N, and G0B9317N).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The DRC66 genome sequence is deposited in GenBank with accession number MN082768. Alignments and BEAST xml files are available at https://github.com/sophiegryseels/DRC66.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913682117/-/DCSupplemental.
References
- 1.World Health Organization , The 2018 Update, Global Health Workforce Statistics (World Health Organization, Geneva, 2018). [Google Scholar]
- 2.Sharp P. M., Hahn B. H., Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiewsakun P., Katzourakis A., Time-dependent rate phenomenon in viruses. J. Virol. 90, 7184–7195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho S. Y. W., et al. , Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Gifford R. J., Viral evolution in deep time: Lentiviruses and mammals. Trends Genet. 28, 89–100 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Worobey M., et al. , Island biogeography reveals the deep history of SIV. Science 329, 1487 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Wertheim J. O., Kosakovsky Pond S. L., Purifying selection can obscure the ancient age of viral lineages. Mol. Biol. Evol. 28, 3355–3365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemey P., Rambaut A., Pybus O. G., HIV evolutionary dynamics within and among hosts. AIDS Rev. 8, 125–140 (2006). [PubMed] [Google Scholar]
- 9.Scholle S. O., Ypma R. J. F., Lloyd A. L., Koelle K., Viral substitution rate variation can arise from the interplay between within-host and epidemiological dynamics. Am. Nat. 182, 494–513 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Membrebe J. V., Suchard M. A., Rambaut A., Baele G., Lemey P., Bayesian inference of evolutionary histories under time-dependent substitution rates. Mol. Biol. Evol. 36, 1793–1803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worobey M., et al. , Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu T., et al. , An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391, 594–597 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Faria N. R., et al. , HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salemi M., et al. , Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15, 276–278 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Yusim K., et al. , Using human immunodeficiency virus type 1 sequences to infer historical features of the acquired immune deficiency syndrome epidemic and human immunodeficiency virus evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 855–866 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keele B. F., et al. , Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villabona-Arenas C. J., et al. , Divergent HIV-1 strains (CRF92_C2U and CRF93_cpx) co-circulating in the Democratic Republic of the Congo: Phylogenetic insights on the early evolutionary history of subtype C. Virus Evol. 3, vex032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal N., et al. , Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74, 10498–10507 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodgers M., et al. , Sensitive next generation sequencing method reveals deep genetic diversity of HIV-1 in the Democratic Republic of the Congo. J. Virol. 91, e01841-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers M. A., et al. , Identification of rare HIV-1 Group N, HBV AE, and HTLV-3 strains in rural South Cameroon. Virology 504, 141–151 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Gilbert M. T. P., et al. , The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. U.S.A. 104, 18566–18570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert M. T., et al. , Multiplex PCR with minisequencing as an effective high-throughput SNP typing method for formalin-fixed tissue. Electrophoresis 28, 2361–2367 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Gilbert M. T., et al. , The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One 2, e537 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Y. L., et al. , High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J. Pathol. 229, 535–545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worobey M., et al. , 1970s and ‘Patient 0’ HIV-1 genomes illuminate early HIV/AIDS history in North America. Nature 539, 98–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T. F., Shafer R. W., Web resources for HIV type 1 genotypic-resistance test interpretation. Clin. Infect. Dis. 42, 1608–1618 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin D. P., Murrell B., Golden M., Khoosal A., Muhire B., RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, vev003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaschen B., Kuiken C., Korber B., Foley B., Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics 17, 415–418 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Eddy S. R., Profile hidden Markov models. Bioinformatics 14, 755–763 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Rambaut A., Lam T. T., Max Carvalho L., Pybus O. G., Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Development Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
- 32.Suchard M. A., et al. , Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro B., et al. , A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 28, 879–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cilliers T., et al. , The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77, 4449–4456 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee S.-Y., et al. , Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31, 298–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafer R. W., Rationale and uses of a public HIV drug‐resistance database. J. Infect. Dis. 194 (suppl. 1), S51–S58 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi J., et al. , Complete genome sequence of CG-0018a-01 establishes HIV-1 subtype L. J. Acquired Immune Defic. Syndr. (1999) 83, 319–322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi D. J., et al. , HIV type 1 isolate Z321, the strain used to make a therapeutic HIV type 1 immunogen, is intersubtype recombinant. AIDS Res. Hum. Retroviruses 13, 357–361 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Tebit D. M., Arts E. J., Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 11, 45–56 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Maljkovic I., Wilbe K., Sölver E., Alaeus A., Leitner T., Limited transmission of drug-resistant HIV type 1 in 100 Swedish newly detected and drug-naive patients infected with subtypes A, B, C, D, G, U, and CRF01_AE. AIDS Res. Hum. Retroviruses 19, 989–997 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Simmonds P., Aiewsakun P., Katzourakis A., Prisoners of war–host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 17, 321–328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mühlemann B., et al. , Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature 557, 418–423 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Krause-Kyora B., et al. , Neolithic and medieval virus genomes reveal complex evolution of hepatitis B. eLife 7, e36666 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korber B., et al. , Timing the ancestor of the HIV-1 pandemic strains. Science 288, 1789–1796 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Baele G., et al. , Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29, 2157–2167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gill M. S., et al. , Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 30, 713–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bletsa M., et al. , Divergence dating using mixed effects clock modelling: An application to HIV-1. Virus Evol. 5, vez036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A., Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DRC66 genome sequence is deposited in GenBank with accession number MN082768. Alignments and BEAST xml files are available on https://github.com/sophiegryseels/DRC66.