Significance
The androgen receptor (AR) antagonist enzalutamide is one of the principal treatments for men with metastatic castration-resistant prostate cancer. However, not all patients respond, and de novo resistance mechanisms are largely unknown. To clarify mechanisms that contribute to enzalutamide resistance, we conducted a single-arm enzalutamide clinical trial. Metastatic tissue biopsies were required prior to study entry so we could attempt to identify molecular features associated with de novo resistance. Transcriptional profiling identified specific gene sets—including those linked to reduced AR transcriptional activity and a stemness program—that were activated in nonresponders. Our results suggest that patients whose tumors harbor this program should be considered for clinical trials testing rational agents to overcome de novo enzalutamide resistance.
Keywords: enzalutamide, resistance, androgen receptor, stemness
Abstract
The androgen receptor (AR) antagonist enzalutamide is one of the principal treatments for men with castration-resistant prostate cancer (CRPC). However, not all patients respond, and resistance mechanisms are largely unknown. We hypothesized that genomic and transcriptional features from metastatic CRPC biopsies prior to treatment would be predictive of de novo treatment resistance. To this end, we conducted a phase II trial of enzalutamide treatment (160 mg/d) in 36 men with metastatic CRPC. Thirty-four patients were evaluable for the primary end point of a prostate-specific antigen (PSA)50 response (PSA decline ≥50% at 12 wk vs. baseline). Nine patients were classified as nonresponders (PSA decline <50%), and 25 patients were classified as responders (PSA decline ≥50%). Failure to achieve a PSA50 was associated with shorter progression-free survival, time on treatment, and overall survival, demonstrating PSA50’s utility. Targeted DNA-sequencing was performed on 26 of 36 biopsies, and RNA-sequencing was performed on 25 of 36 biopsies that contained sufficient material. Using computational methods, we measured AR transcriptional function and performed gene set enrichment analysis (GSEA) to identify pathways whose activity state correlated with de novo resistance. TP53 gene alterations were more common in nonresponders, although this did not reach statistical significance (P = 0.055). AR gene alterations and AR expression were similar between groups. Importantly, however, transcriptional measurements demonstrated that specific gene sets—including those linked to low AR transcriptional activity and a stemness program—were activated in nonresponders. Our results suggest that patients whose tumors harbor this program should be considered for clinical trials testing rational agents to overcome de novo enzalutamide resistance.
We now know that persistent intratumoral androgens that activate the androgen receptor (AR) are commonly found in castration-resistant prostate cancer (CRPC) tumors despite androgen deprivation therapy (ADT). The main sites of androgen production in men with CRPC include the adrenal glands and tumor cells themselves. Because of this knowledge, new and more potent inhibitors of androgen activation of the AR have been developed in recent years (1–4). Importantly, treatment with the androgen synthesis inhibitor abiraterone acetate or the AR antagonist enzalutamide improves progression-free survival (PFS) and overall survival (OS) in patients with metastatic CRPC (5–8). Furthermore, enzalutamide, apalutamide, and the newer AR antagonist darolutamide all improve metastasis-free survival in men with nonmetastatic CRPC (9–11).
Enzalutamide is commonly used in the first-line treatment of men with CRPC. While the majority of patients with metastatic CRPC benefit from enzalutamide treatment, nearly one-quarter to one-half do not (5, 6). A few studies to date have prospectively examined samples from men with enzalutamide-naïve CRPC to identify determinants of response or resistance. However, these studies have largely been restricted to mutational profiling, were small in size, or focused on acquired—rather than de novo—resistance (12–14). Thus, predictors and determinants of de novo enzalutamide resistance in CRPC remain largely unknown.
We hypothesized that a more detailed characterization of the genomic landscape of baseline CRPC samples in patients beginning enzalutamide treatment would clarify determinants of de novo resistance. To test this hypothesis, we initiated a multiinstitutional, prospective enzalutamide clinical trial in men with metastatic CRPC who had a metastatic lesion amenable to a pretreatment biopsy. In this report, we describe baseline genomic and transcriptional features that differed between those patients whose tumors either responded or failed to respond to enzalutamide treatment.
Results
Patient Characteristics.
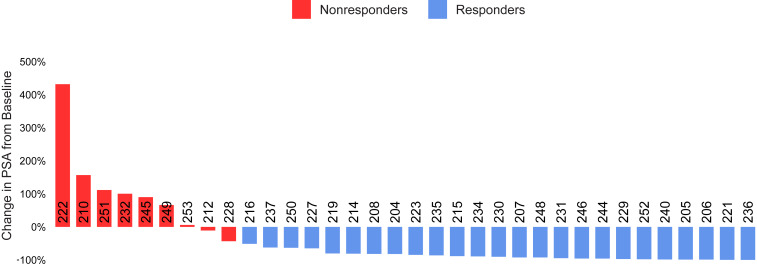
The clinical trial Genetic and Molecular Mechanisms in Assessing Response in Patients with Prostate Cancer Receiving Enzalutamide Therapy enrolled 36 patients with metastatic CRPC who had not previously received enzalutamide. Patients with prior use of docetaxel or abiraterone were ineligible for this study. Study enrollment, follow-up, and analyses are depicted in SI Appendix, Fig. S1. The demographic characteristics for all 36 patients enrolled on the study are shown in SI Appendix, Table S1. Thirty-four patients were evaluable for the primary end point of ≥50% PSA decline at 12 wk, and their demographic features are shown in Table 1. There were 9 nonresponders (<50% PSA decline at 12 wk) and 25 responders (≥50% PSA decline at 12 wk) (Fig. 1).
Table 1.
Baseline patient characteristics
| Characteristic | All evaluable patients (n = 34) |
| Age (y) | |
| Median | 72.3 |
| Range | 56.6–88.7 |
| Total Gleason score, n (%) | |
| 6 | 2 (5.9) |
| 7 | 13 (38.2) |
| 8 | 3 (8.8) |
| 9 | 10 (29.4) |
| Unavailable | 6 (17.6) |
| Metastatic sites at time of biopsy, n (%) | |
| Bone | 25 (73.5) |
| Lung | 2 (5.9) |
| Liver | 0 (0.0) |
| Visceral (other than lung and liver)* | 1 (2.9) |
| Lymph nodes | 6 (17.6) |
| ECOG performance status score, n (%) | |
| 0 | 20 (58.8) |
| 1 | 14 (41.2) |
| 2–4 | 0 (0.0) |
| PSA | |
| Median | 36.6 |
| Range | 2.3–2,137.3 |
Patient demographics for evaluable patients. Demographic information for the 34 evaluable patients is shown. ECOG, Eastern Cooperative Oncology Group.
Ischioanal fossa mass.
Fig. 1.
PSA waterfall plot. PSA change from baseline for patients by response group (9 nonresponders and 25 responders). Each bar represents one patient with patient identification indicated along zero axis. PSA response was determined based on change at 12 wk vs. the baseline value.
Clinical Outcomes.
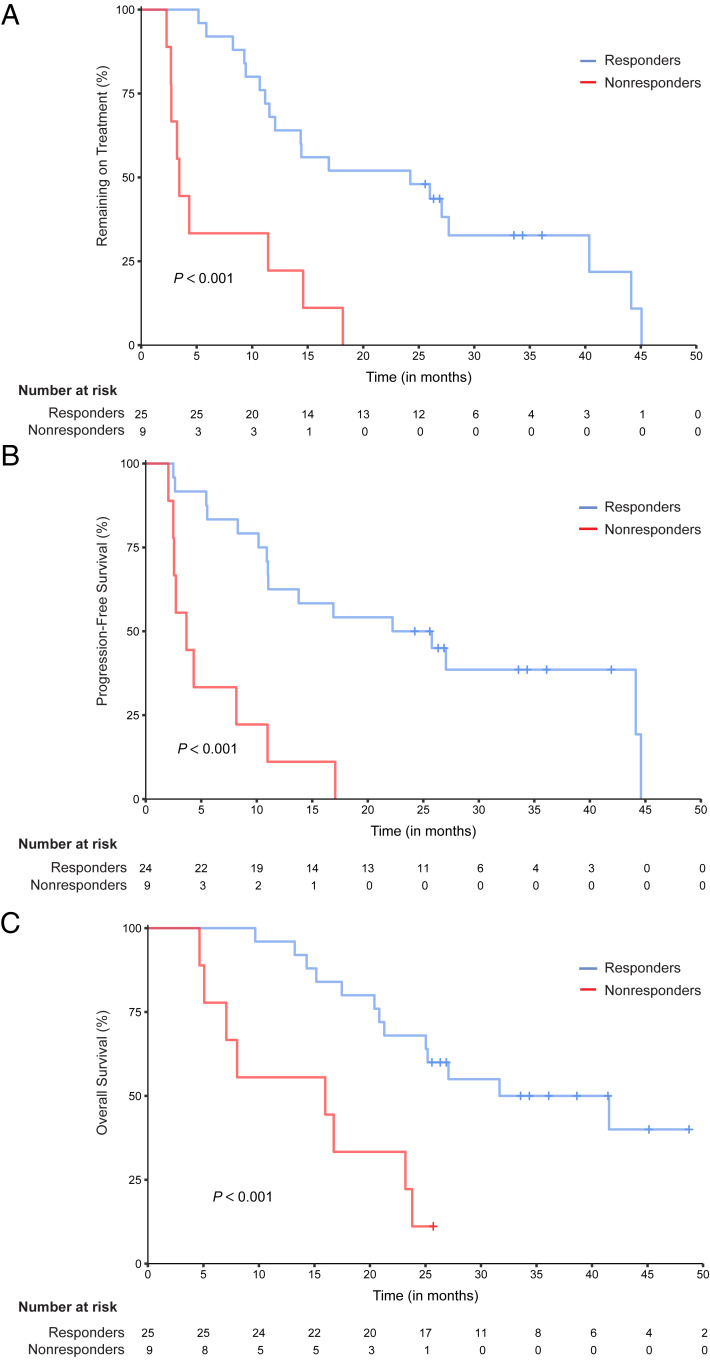
We next examined clinical outcomes for the entirety of the trial participants and for the nonresponders vs. responders. Thirty-eight percent (13 of 34) of evaluable patients had radiographic responses by response evaluation criteria in solid tumors (RECIST), and all radiographic responses were seen in those with a PSA50 response. Overall median time on treatment (TOT) was 14.4 mo. Overall median PFS was 11.03 mo, and OS was 25.11 mo. Compared with responders, nonresponders had a statistically significant shorter median TOT (3.4 vs. 24.2 mo, P < 0.001, hazard ratio (HR) = 4.90 [2.03 to 11.82]), PFS (3.67 vs. 24 mo, P < 0.001, HR = 5.51 [2.2 to 13.81]), and OS (15.97 vs. 36.6 mo, P < 0.001, HR = 4.41 [1.71 to 11.44]) (Fig. 2). Thus, PSA50 response was a strong predictor of clinical benefit.
Fig. 2.
Kaplan–Meier curves stratified by PSA response. Tick marks indicate censoring events. P values were determined using the log-rank test to compare outcome measures between nonresponders and responders. (A) TOT. Probability that patients remain on treatment during follow-up in months. (B) PFS. Probability that patients are alive and without evidence of disease progression during follow-up in months. One subject was excluded due to missing progression information. (C) OS. Probability that patients are still alive during follow-up in months.
Targeted DNA-Sequencing.
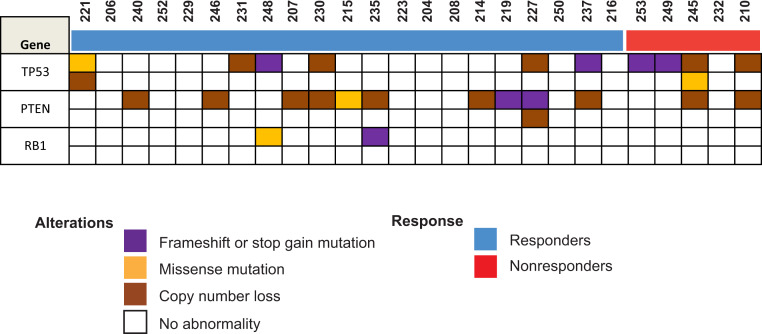
For 26 of 36 patients, there was sufficient DNA to perform targeted DNA-sequencing (DNA-seq). Five of these patients were nonresponders, and 21 were responders. We focused on TP53, RB1, and PTEN as those genes have been linked previously to poor outcomes for men with CRPC or resistance to AR-targeting therapies in preclinical or clinical studies (12, 15–18). No statistically significant differences were detected between the two groups for these genes (Fig. 3) or other genes examined (SI Appendix, Fig. S2). However, there was a strong trend toward greater likelihood of TP53 alterations in nonresponders (Fisher’s exact test P = 0.055).
Fig. 3.
DNA-seq results for TP53, PTEN, and RB1. An OncoPrint reflecting the copy number alteration and mutation status for the indicated genes in responders and nonresponders is shown.
Assessments of AR Genomic Aberrations and Expression.
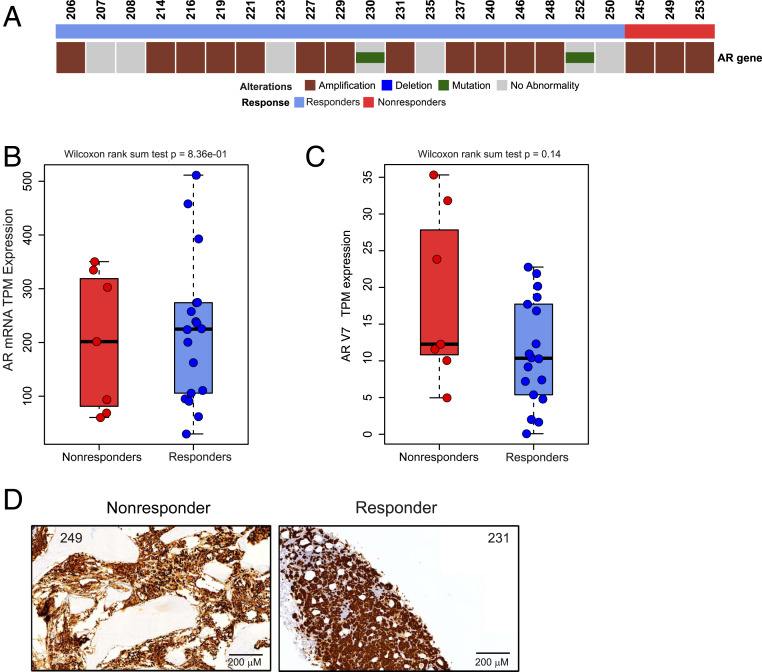
Since AR is the target of enzalutamide, we next determined if there were genomic differences in the AR between groups using AR DNA-seq. Of the 22 unique patients who had AR DNA-seq data, there were 3 nonresponders and 19 responders. No statistically significant differences in AR genomic alterations were found between these two groups (Fig. 4A). We next quantified AR mRNA expression in the 25 patients whose tumors underwent RNA-sequencing (RNA-seq): 7 nonresponders and 18 responders. AR messenger ribonuleic acid (mRNA) expression was similar between both groups (Fig. 4B). Furthermore, expression levels of the transcript variant AR-V7 were low and were similar between both groups (Fig. 4C). AR immunohistochemistry was available in 22 patients. Strong, nuclear AR staining was present in tumors from all 6 nonresponder samples and in all 15 responder samples that were tested (SI Appendix, Table S2). Representative examples are shown in Fig. 4D. Thus, neither alterations in the AR gene nor AR expression correlated with de novo enzalutamide resistance.
Fig. 4.
AR DNA-seq results and expression status. (A) An OncoPrint reflecting the copy number alteration and mutation status of the AR in responders and nonresponders is shown. (B and C) Boxplots of the mRNA expression of the AR (B) and AR-V7 (C) in nonresponders and responders are shown. Wilcoxon rank sum test was used to test the difference between the two groups. (D) Representative AR immunohistochemistry staining is shown for two patients.
Pathway Analysis.
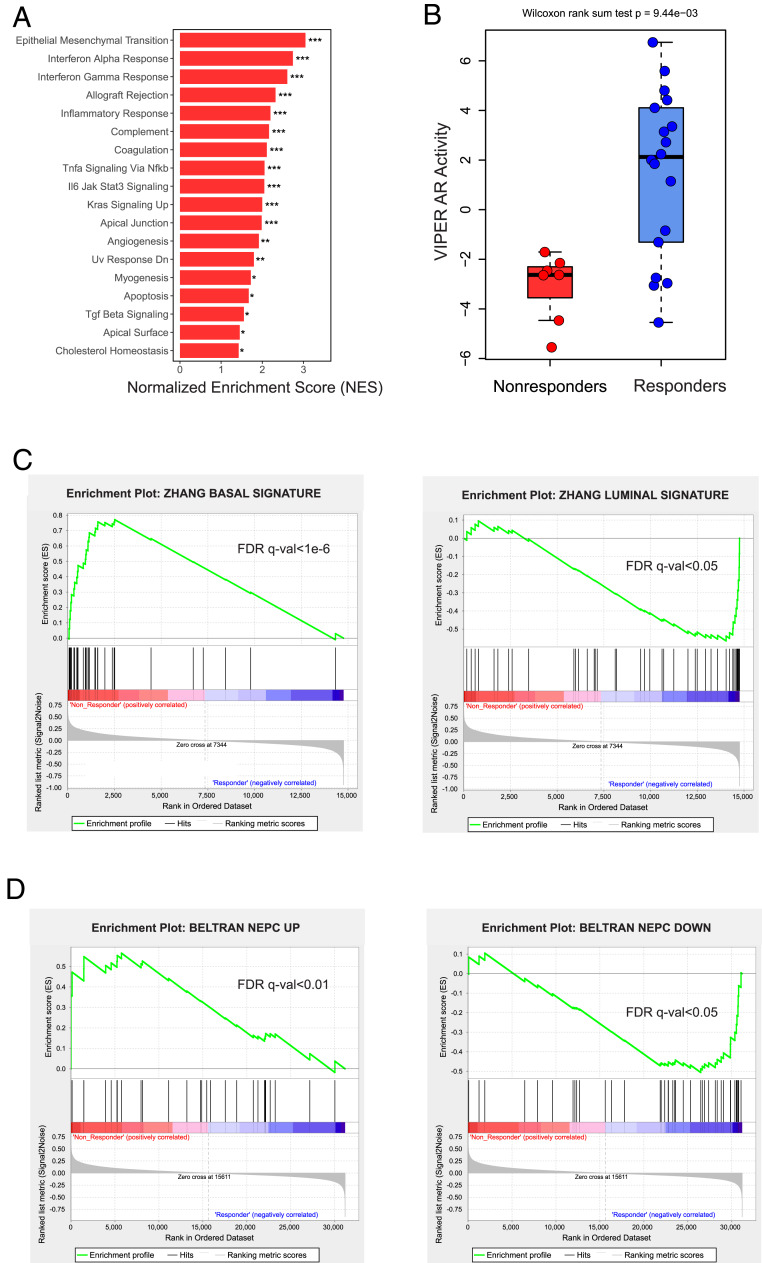
Next, we sought to determine if there were differentially activated pathways in nonresponders vs. responders in the RNA-seq data. To identify these pathways, we performed gene set enrichment analysis (GSEA) using all candidate gene sets within the Hallmark database. This analysis determined that baseline tumors from nonresponders vs. responders had enrichment of 18 statistically significant pathways using a false discovery rate (FDR) q-value cutoff of 0.05 (Fig. 5A and SI Appendix, Fig. S3). Epithelial mesenchymal transition (EMT) was the top gene set activated in nonresponders, and other pathways linked to EMT, including IL-6/Jak/Stat3 signaling, TGF-β signaling, and TNFα signaling via NFΚB, were also activated. Furthermore, immune hallmark pathways were also activated in nonresponders, including IFN-α, IFN-γ, allograft rejection, inflammatory response, and complement.
Fig. 5.
Transcriptional measurements implicate an AR activity-low, stemness program in nonresponders. (A) Results from the GSEA pathway analysis using the Broad Hallmark pathways are shown. The 18 hallmark pathways activated in the nonresponders with FDR q value < 0.05 are shown. *FDR q value < 0.05; **FDR q value < 1e-3; ***FDR q value < 1e-6. (B) Boxplot indicates the AR activity differences between nonresponders and responders. Significance of the difference between nonresponders and responders was calculated using the Wilcoxon rank sum test. (C) Basal and luminal signature enrichment plots for nonresponders and responders with FDR q values are shown. (D) Beltran NEPC up-regulated and down-regulated gene set enrichment plots for nonresponders and responders with FDR q values are shown.
AR Activity Measurements.
AR signaling was not one of the top gene sets differentially activated between nonresponders and responders. However, there were many gene sets activated in enzalutamide nonresponders in our trial—including those linked to EMT, stemness, and immune hallmarks—that were also activated in subsets of localized, hormone-naïve prostate cancer that had low AR activity in a recent report from our group (19). Importantly, low AR activity in that prior report was associated with shorter metastasis-free survival or time to biochemical recurrence, demonstrating the clinical significance of AR activity-low prostate cancers and the utility of this signature (the Spratt signature) (19). Therefore, we next measured AR function in RNA-seq data from our enzalutamide trial specimens using the Spratt signature. There was a strong trend toward reduced AR activity in nonresponders vs. responders using this signature (Wilcoxon rank sum test P = 0.06) (SI Appendix, Fig. S4). We also quantified AR activity using the Virtual Inference of Protein activity by Enriched Regulon analysis (VIPER) algorithm (20). VIPER analysis also demonstrated that enzalutamide nonresponders had lower AR activity (Fig. 5B). Overall, these results suggest that low AR transcriptional activity—rather than AR genomic changes or altered AR expression—may contribute to de novo enzalutamide resistance.
Stemness and Lineage Signature Measurements.
Low AR activity has been linked to stemness and lineage plasticity, or change in differentiation state, that are now recognized as a common cause of acquired resistance to AR-targeting therapies (21–25). Importantly, all of the baseline tumors from our study were adenocarcinomas, and most prostate adenocarcinoma tumors express a luminal lineage program (22). However, a basal lineage, neurogenic/stemness program is present in a subset of patients and is linked with prostate cancer aggressiveness (26, 27). Indeed, prior work demonstrated that localized prostate tumors with a basal lineage program are much less likely to respond to ADT vs. patients whose tumors harbor a luminal lineage program (22). Therefore, we next sought to measure signatures of these lineages (26) and determine if they were differentially activated between nonresponders and responders. Our analysis demonstrates that a basal lineage, neurogenic/stemness program is activated in nonresponders to enzalutamide treatment, while a luminal lineage program is more activated in responders (Fig. 5C), corroborating the prior work in prostate cancer examining the effect of basal lineage on promoting resistance to ADT (22).
To more fully examine the role of neurogenic gene expression, we determined if a set of genes used previously to distinguish neuroendocrine prostate cancer (NEPC) vs. adenocarcinoma patient tumors (28) was differentially expressed in nonresponders vs. responders. Importantly, genes up-regulated in NEPC patient tumors from that prior report were activated in nonresponders vs. responders; genes down-regulated in NEPC patient tumors from that prior report—many linked to AR activity—were deactivated in nonresponders from our trial (Fig. 5D). These results further suggest that nonresponder tumors have features of stemness and corroborate our findings suggesting that AR activity is lower in nonresponders. In summary, results from this clinical trial suggest that an AR activity-low, stemness program may contribute to de novo enzalutamide resistance.
Discussion
Two large, randomized, phase III trials demonstrate that enzalutamide treatment improves PFS and OS for men with metastatic CRPC (5, 6). Enzalutamide also improves PFS for men with nonmetastatic CRPC and OS and/or PFS for men with hormone-naïve metastatic prostate cancer in two additional clinical trials (10, 29, 30). However, approximately one-third of patients do not respond to enzalutamide treatment, and very little is known about determinants of de novo enzalutamide resistance in metastatic CRPC (5, 6). The Genetic and Molecular Mechanisms in Assessing Response in Patients with Prostate Cancer Receiving Enzalutamide Therapy clinical trial was designed to address this deficit. To identify molecular predictors of enzalutamide response, we performed both targeted DNA-seq and RNA-seq of tumors from prospectively collected pretreatment metastatic biopsies. Correlating the molecular features from these assays with the primary end point of ≥50% PSA decline 12 wk after starting enzalutamide treatment—that we confirmed to be a strong predictor of patient outcome (Fig. 2)—enabled us to identify factors associated with de novo resistance.
It is noteworthy that the patients enrolled on our trial had a lower PSA response rate, TOT, PFS, and OS than patients with metastatic CRPC enrolled on the PREVAIL trial who had not previously received enzalutamide, abiraterone, or docetaxel (5). There are several key differences between our trial and PREVAIL that may help explain these results. First, our trial—unlike PREVAIL—allowed enrollment of patients with symptomatic metastases. Second, our trial mandated that all eligible patients have a metastatic lesion that was amenable to biopsy. Thus, it is quite likely that the patients enrolled on our trial had more aggressive or advanced disease than those enrolled on PREVAIL, thus explaining the differences in outcomes.
Our institutional targeted DNA-seq panel did not detect any statistically significant differences in mutations found between nonresponders and responders. However, we did observe a strong trend toward statistical significance for TP53 mutations that were more common in nonresponders (4 of 5 vs. 6 of 21, Fisher’s exact test P = 0.055) (Fig. 3). This is consistent with previous findings by Annala et al. (12) who used cell-free DNA-seq of enzalutamide- or abiraterone-naïve patients and determined that alterations in TP53 were associated with poor outcomes. Furthermore, loss of TP53 has been linked to increased cellular reprogramming efficiency (31) or stemness/lineage plasticity (16, 17)—programs that we find to be enriched in nonresponders (Fig. 5 C and D). This suggest that TP53 alterations may contribute to the gene expression changes we observed in nonresponders vs. responders.
Because AR is the target of enzalutamide, we also examined alterations in the AR gene itself as well as measurements of AR transcriptional function. We did not observe statistically significant differences in AR gene alterations between the two groups (Fig. 4A), although this may be due to sample size limitations and the fact that not every patient had AR DNA-seq data available. Importantly, we did measure AR mRNA and protein expression. Levels of AR mRNA and protein expression were also similar between nonresponders and responders (Fig. 4B and SI Appendix, Table S2), suggesting that AR genomic changes or AR expression changes are not good predictors of enzalutamide response. Furthermore, expression levels of AR-V7 were low in both nonresponders and responders (Fig. 4C), which may be attributable to the fact that none of the patients had been treated previously with novel, potent AR inhibitors like enzalutamide or abiraterone. Conversely, we determined that lower AR transcriptional activity was associated with enzalutamide resistance using two independent measurements of AR function (Fig. 5B and SI Appendix, Fig. S4). Because AR is the target of enzalutamide, intuitively, this makes sense and strongly suggests that tumors with lower AR function are less AR signaling dependent and thus, less likely to respond to enzalutamide treatment. Understanding factors that lead to reduced AR activity despite persistent AR expression—inactivation of essential cofactors or chromatin conformation not conducive to AR activity—will be important to develop strategies to resensitize these tumors to drugs like enzalutamide.
Low AR activity has been linked to lineage plasticity, EMT, or stemness (19, 23–26). Importantly, GSEA of differentially expressed genes identified 18 pathways (P < 0.05) that were activated in tumors from patients with de novo enzalutamide resistance—most notably pathways that are often linked to stemness, lineage plasticity, or EMT (32). None of the histologic sections from our biopsies demonstrated a mesenchymal morphology or phenotype, so we believe that the GSEA results may indicate a partial EMT, stem-like state that has been previously linked to self-renewal and treatment resistance (33, 34). This interpretation is in keeping with the enrichment of a stemness signature (26) in nonresponder patients (Fig. 5 C and D). Indeed, a previously described basal lineage, neurogenic, stemness signature (26) was activated in nonresponders from our trial (Fig. 5C). Furthermore, we found that a gene set used previously to distinguish NEPC vs. adenocarcinoma patient tumors linked to lower AR activity (26) was differentially expressed in nonresponders vs. responders (Fig. 5D). The sum of these results suggests that reduced AR activity may promote a stemness program or that activation of this stemness program leads to reduced AR activity. While it is uncertain which event occurs first, it is clear that the presence of low AR activity and a stemness program are linked to de novo enzalutamide resistance.
Considerable prior work has shown that stemness-associated pathways are associated with acquired resistance to AR pathway inhibition (32). Indeed, previous work demonstrates that the IL-6–JAK-STAT3 signaling pathway is activated in tumors resistant to AR pathway inhibition and that IL-6–JAK-STAT3 promotes stem-like properties, EMT, and lineage plasticity (35). GSEA also implicated activation of the TGF-β signaling pathway that is upstream of SMAD3 in nonresponders. SMAD3 up-regulation was previously shown by Pal et al. (14) to mediate TGF-β as well as cyclin D1 signaling in circulating tumor cell samples from patients with acquired resistance to enzalutamide. However, our work demonstrates enrichment of IL-6–JAK-STAT3 and TGF-β pathways in CRPC patient tumors even prior to enzalutamide treatment and suggests that activation of these pathways may contribute to de novo enzalutamide resistance. Finally, that there are drugs to block key members of these pathways [e.g., IL-6 inhibitor siltuximab (36), JAK inhibitor ruxolitinib (37), STAT3 degraders (38), TGF-β receptor inhibitor galunisertib (39), and CDK4/6 inhibitors (40)] that target cyclin D1 suggests that it may be possible in the near term to determine if these drugs may overcome de novo enzalutamide resistance. One way to enrich these trials with appropriate subjects most likely to have de novo enzalutamide resistance is by using RNA-seq like in our trial. Importantly, Clinical Laboratory Improvement Amendments (CLIA) RNA-seq is now available commercially and at centers such as ours (University of Michigan), and results are often available within a 4-wk timeframe.
Low AR activity has also been linked to immune pathways (19). Indeed, IFN-γ, IFN-α, allograft rejection, and inflammatory response were also among the top gene sets activated in nonresponders (Fig. 5A). This suggests that differences in immune activation may contribute to enzalutamide response. Our samples underwent laser-capture microdissection to enrich for tumor cells prior to RNA-seq. Thus, it is not possible to reliably measure transcriptional signatures linked to nonepithelial cell types and deconvolute the tumor microenvironment. However, despite the approval of the autologous DNA vaccine sipuleucel-T (41) in 2011, phase III trials of anti-CTLA4 checkpoint inhibition have been disappointing in CRPC (42, 43), and anti-PD1 trials in men with metastatic CRPC have not demonstrated high response rates or durability of tumor control (44). In the future, it will be important to determine if immunotherapeutic agents may be useful for overcoming de novo enzalutamide resistance and to determine if differences in the microenvironmental components of tumors contribute to resistance to immunotherapy and drugs such as enzalutamide.
Despite the importance of our findings, there are several limitations of our study. First, fewer patients than anticipated had sufficient material for all of the molecular analyses. This is not unique to this study and is attributable to the fact that many of our patients underwent biopsies of metastatic bone lesions from which it can be challenging to obtain high-quality material for sequencing. Nonetheless, our study is the largest prospective clinical trial examining molecular predictors of enzalutamide response from metastatic tumor biopsies, and we look forward to the results of other studies with novel AR-targeting agents that may help to validate our results. Second, another limitation of our study is that mutational profiling primarily consisted of targeted DNA-seq rather than whole–exome-sequencing or whole–genome-sequencing (whole–genome-seq), limiting the number of genes examined across all patients. However, our panel did include key genes previously linked to enzalutamide resistance or EMT/lineage plasticity [TP53, RB1, and PTEN (12, 15–18)], and significant differences were not found. We cannot rule out that loss of function of these genes or others through epigenetic or posttranscriptional mechanisms could have also contributed to de novo enzalutamide resistance.
A further limitation of our study is that only a single metastatic site was biopsied prior to treatment, although it remains challenging to collect biopsies from multiple sites at the same time point. However, despite only obtaining a biopsy from a single site, we did determine that the site sampled had molecular features that strongly correlated with de novo enzalutamide resistance. This suggests that a single-site biopsy—if it contains adequate material for sequencing—may be sufficient to identify markers of enzalutamide response using the approach we describe herein. Indeed, our comprehensive transcriptome-based analysis with GSEA identified pathways and lineage signatures not previously linked to de novo clinical enzalutamide resistance.
In summary, this is the largest prospective enzalutamide clinical trial to perform mutational and transcriptional profiling of enzalutamide-naïve, metastatic CRPC biopsy samples. Importantly, using transcriptional profiling and GSEA, we identified several targetable pathways that may contribute to an AR activity-low, stemness program linked to de novo enzalutamide resistance. In the future, it will be important to determine whether mechanisms identified herein are also relevant to men who present with ADT- and enzalutamide-naïve prostate cancer because of the recent approval of enzalutamide in that setting, along with abiraterone and apalutamide. Moreover, it will be important to determine which specific factors that regulate the phenotype described herein are most critical for de novo enzalutamide resistance. Such experiments may provide the rationale to target these factors alone or in combination with enzalutamide in patients like those identified herein who are least likely to respond to enzalutamide monotherapy.
Materials and Methods
Study Design and Patients.
The clinical trial Genetic and Molecular Mechanisms in Assessing Response in Patients with Prostate Cancer Receiving Enzalutamide Therapy (principal investigator: J.J.A.) was registered on clinicaltrials.gov (NCT02099864). This was a multicenter, single-arm, phase II study evaluating baseline predictors of response to enzalutamide therapy in men with metastatic CRPC who had not received prior enzalutamide, abiraterone, or docetaxel. Institutional review board (IRB) approval was obtained at the participating centers (Oregon Health & Science University and the University of California, San Francisco) prior to enrollment of the first patient at each center. Written informed consent was obtained from all patients prior to participation in the study.
All eligible patients had a metastatic lesion amenable to image-guided biopsy. Prior to initiation of enzalutamide treatment, subjects underwent an image-guided biopsy of a metastatic lesion to obtain tumor tissue for baseline genomic analyses. Subjects received enzalutamide for the duration of the study and were assessed for clinical or radiographic progression every 12 wk. Study treatment was continued until withdrawal of consent, unacceptable toxicity, disease progression, or death.
Traditional power calculations cannot be used to determine the number of samples (i.e., patients) required because the assumptions of independence and normality do not hold (45). An alternative approach is to model this problem as a learning curve in which the classification error is characterized as an inverse power law (46): error = an−α + b, where error is the expected error rate given n training samples, a is the learning rate, α is the decay rate, and b is the Bayes error, or minimum error achievable. This approach has been applied to several cancer classification problems, and estimates for the variables can be found in the literature (46). For our purposes, we used the average values from seven cancer datasets to estimate a, α, and b. With a = 0.736, α = 0.65, and b = 0.001, we can estimate the error for various sample sizes. Prior literature demonstrates an ∼70% PSA response rate with enzalutamide in a similar patient population (5). Therefore, we estimated that we would need to enroll 36 patients, allowing for a 20% biopsy failure rate, to yield 30 patients with evaluable tumors (20 responders and 10 nonresponders) and that 30 patients would achieve an error rate of ∼0.08.
Per the Prostate Cancer Clinical Trials Working Group-2 criteria, disease progression was defined as radiographic progression (based on RECIST 1.1 criteria for soft tissue disease measured by computerized tomography (CT) scan or the appearance of two or more unequivocal bone lesions on bone scan) or clinical progression; PSA progression alone did not meet criteria for discontinuation of study treatment (47). Patients who discontinued treatment despite not meeting study-defined progression continued to be followed until they met progression criteria. Following discontinuation of study treatment, all subjects were followed for OS.
For the primary analysis, we assessed correlations between baseline molecular features and PSA50 response defined as a PSA decline of ≥50% at 12 wk compared with baseline. Secondary clinical end points included TOT, PFS, and OS, all measured from the date of enzalutamide initiation.
Thirty-six men with enzalutamide-, abiraterone-, and docetaxel-naïve metastatic CRPC progressing on ADT and who were eligible to receive treatment with enzalutamide were enrolled (SI Appendix, Fig. S1). Two subjects were not evaluable for analysis of the primary end point (≥50% PSA decline at 12 wk). One subject was excluded because of incomplete PSA data. The second patient was excluded because his PSA fell on treatment in the setting of rapid progression of disease, concerning for conversion to neuroendocrine prostate cancer; thus, it was determined that this subject’s PSA decline was not consistent with response. Therefore, a total of 34 patients were evaluable for analysis of the primary end point. Their clinical trial identification numbers are shown. SI Appendix, Fig. S1 includes information on the number of patients who underwent each molecular assay.
Statistical Analysis.
Descriptive statistics (median and interquartile range for continuous variables; frequencies and percentages for categorical variables) were used to summarize the demographic and clinical data by response group. Time-to-event analysis was performed using Kaplan–Meier curves and log-rank tests to evaluate the association between response group and TOT, PFS, and OS. Cox regression models were used to calculate hazard ratios by response group with 95% CIs. All statistical analyses were performed using R: A Language and Environment for Statistical Computing. P < 0.05 was considered statistically significant.
Analysis of Baseline Biopsy Samples and DNA-Seq.
At the time of consenting for the Genetic and Molecular Mechanisms in Assessing Response in Patients with Prostate Cancer Receiving Enzalutamide Therapy trial, patients also consented to the IRB-approved West Coast Prostate Cancer Dream Team tissue collection protocol through which their tumor samples were sequenced. Results in Fig. 3 and SI Appendix, Fig. S2 are based on a 37-gene targeted DNA-seq panel described previously (48). AR DNA-seq results in Fig. 4A are based on an alternate targeted DNA-seq panel that included the AR or whole–genome-seq that has been described previously (21, 49).
Characterization of AR Protein Expression.
AR protein expression was analyzed using immunohistochemical analysis. The AR [C6F11] XP rabbit monoclonal antibody was used as previously described, and AR nuclear staining was quantified as described previously: 0 (absent), 1+ (low), 2+ (intermediate), and 3+ (strong) (21).
Transcriptional Profiling.
Tumor cores were frozen in optimized cutting temperature (OCT) medium and underwent laser capture microdissection using methods described previously (49). The raw read counts and transcripts per kilobase million (TPM) of the gene expression measurements were derived by RSEM (50) based on the GRCh37 GENCODE version 19 (Ensembl 74) gene annotation. To filter out low-expressed genes, we required that the genes have TPM greater than one in at least three samples for further analysis. The TPM gene expression data quantified by RSEM from the RNA-seq may be found in Dataset S1; sample identifications highlighted in blue indicate responders, while those highlighted in red indicate nonresponders.
GSEA Pathway Analysis.
GSEA version 3.0 (51) was used to identify gene sets that were significantly activated in enzalutamide nonresponders compared with responders from the Hallmark database (52). The expression data normalized by variance stabilizing transformation in DESeq2 (53) were used as the input of GSEA, and the default metric Signal2Noise in GSEA was applied to calculate the differential expression with respect to the responders and nonresponders. The gene sets were considered to be activated if their FDR q value was less than 0.05.
AR Activity Analysis.
The Spratt AR activity scores were calculated based on nine genes described previously (19). To measure AR activity, we also used the VIPER R package (20). The input for VIPER is the log2 transformed TPM gene expression matrix of all samples. The regulon used for the AR activity used in VIPER analysis was curated from four databases as previously described (54).
Basal and Luminal Lineage Signature Analysis.
The Zhang Basal and Luminal gene signatures have been described previously (26). First, GSEA was used to determine whether the two signatures showed significant, concordant differences between the nonresponder and responder groups. Second, to quantify the activities of the two signatures in each sample, we used the single-sample gene set enrichment analysis (ssGSEA) (55) implemented in the GSVA (56) R package. ssGSEA is a rank-based comparison to access the expression levels of genes in the gene set against all other genes based on the expression profile of one sample. FDR q values were used to determine statistical significance.
NEPC Signature Analysis.
We used the previously described Beltran NEPC gene signature (28) to build gene sets. Specifically, 29 genes up-regulated in NEPC vs. adenocarcinoma patient tumors or 41 genes down-regulated in NEPC vs. adenocarcinoma patient tumors were used to develop two gene sets. We then used GSEA to determine whether each gene set showed enrichment differences between nonresponders and responders.
Supplementary Material
Acknowledgments
Support includes Stand Up to Cancer-Prostate Cancer Foundation (PCF) Prostate Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409. This research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. Other support includes The Pacific Northwest Prostate Cancer Specialized Programs of Research Excellence (SPORE)/National Cancer Institute (NCI) Grant P50 CA097186; the Michigan Prostate SPORE/NCI Grant P50 CA186786; the Knight Cancer Institute Biostatistics Shared Resource at Oregon Health & Science University Grant P30 CA069533-16 and P30 CA051008-16; a Rogel Scholar Award through the University of Michigan Rogel Cancer Center; and the Wayne D. Kuni and Joan E. Kuni Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI, the University of Michigan Rogel Cancer Center, or the Wayne D. Kuni and Joan E. Kuni Foundation. Oregon Health & Science University received research funding from Astellas/Pfizer to conduct this investigator-initiated trial. We thank Eva Rodansky and Joel Yates for help with manuscript preparation.
Footnotes
Competing interest statement: J.J.A. has received consulting income from Janssen Biotech and Merck, and honoraria for speaker’s fees from Astellas. T.M.B. has received research funding from Alliance Foundation Trials, Boehringer Ingelheim, Corcept Therapeutics, Endocyte Inc., Janssen Research & Development, Medivation, Inc./Astellas, OncoGenex, Sotio, and Theraclone Sciences/OncoResponse; consulting fees from AbbVie, AstraZeneca, Astellas Pharma, Bayer, Boehringer Ingelheim, Clovis Oncology, GlaxoSmithKline, Janssen Biotech, Janssen Japan, Merck, Novartis, and Pfizer; and stock ownership in Salarius Pharmaceuticals, and Arvinas Inc. J.H. serves as a consultant for Morehealth, OptraScan, Genetron, Gencode, York Biotechnology, Kingmed Diagnostics, and Gem Flower Healthcare and is a founder of Sisu Pharma. M.R. received research funding from Novartis, Johnson & Johnson, Merck, Astellas, and Medivation; is a member of a speakers’ bureau for Johnson & Johnson and Bayer; and served as a consultant for Constellation Pharmaceuticals. F.Y.F. has received consulting income from Astellas, Bayer, Celgene, Janssen, Sanofi, Genentech, and EMD Serono and has ownership interest (including patents) in PFS Genomics and Nutcracker Therapeutics. E.J.S. has received honoraria for speaker’s fees from Janssen; consulting income from Janssen, Fortis Therapeutics, Beigene, and Tolero; and stock ownership in Fortis Therapeutics and Harpoon Therapeutics. O.N.W. currently has consulting, equity, and/or board relationships with Trethera Corporation, Kronos Biosciences, Sofie Biosciences, Breakthrough Properties, and Allogene Therapeutics. None of these companies contributed to or directed any of the research reported in this article. T.M.B. and M.C. are coauthors on a 2019 article. D.E.S. and M.L. are coauthors on a 2019 article. None of the other authors have any relevant disclosures to declare.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922207117/-/DCSupplemental.
References
- 1.Attard G., et al. , Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J. Clin. Oncol. 26, 4563–4571 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Clegg N. J., et al. , ARN-509: A novel antiandrogen for prostate cancer treatment. Cancer Res. 72, 1494–1503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher H. I. et al.; Prostate Cancer Foundation/Department of Defense Prostate Cancer Clinical Trials Consortium , Antitumour activity of MDV3100 in castration-resistant prostate cancer: A phase 1-2 study. Lancet 375, 1437–1446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer T. M. et al.; PREVAIL Investigators , Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 371, 424–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher H. I. et al.; AFFIRM Investigators , Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 7.de Bono J. S. et al.; COU-AA-301 Investigators , Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan C. J. et al.; COU-AA-302 Investigators , Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fizazi K. et al.; ARAMIS Investigators , Darolutamide in nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 380, 1235–1246 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Hussain M., et al. , Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 378, 2465–2474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith M. R. et al.; SPARTAN Investigators , Apalutamide treatment and metastasis-free survival in prostate cancer. N. Engl. J. Med. 378, 1408–1418 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Annala M., et al. , Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 8, 444–457 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto D. T., et al. , RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal S. K., et al. , Identification of mechanisms of resistance to treatment with abiraterone acetate or enzalutamide in patients with castration-resistant prostate cancer (CRPC). Cancer 124, 1216–1224 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Chen W. S. et al.; West Coast Prostate Cancer Dream Team , Genomic drivers of poor prognosis and enzalutamide resistance in metastatic castration-resistant prostate cancer. Eur. Urol. 76, 562–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ku S. Y., et al. , Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu P., et al. , SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abida W., et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spratt D. E., et al. , Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment naïve primary prostate cancer. Clin. Cancer Res. 25, 6721–6730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez M. J., et al. , Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal R., et al. , Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao S. G., et al. , Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 3, 1663–1672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltran H., et al. , The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 25, 6916–6924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop J. L., et al. , The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 7, 54–71 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Handle F., et al. , Drivers of AR indifferent anti-androgen resistance in prostate cancer cells. Sci. Rep. 9, 13786 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D., et al. , Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat. Commun. 7, 10798 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith B. A., et al. , A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc. Natl. Acad. Sci. U.S.A. 112, E6544–E6552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltran H., et al. , Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong A. J., et al. , ARCHES: A randomized, phase III study of androgen deprivation therapy with enzalutamide or placebo in men with metastatic hormone-sensitive prostate cancer. J. Clin. Oncol. 37, 2974–2986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis I. D. et al.; ENZAMET Trial Investigators and the Australian and New Zealand Urogenital and Prostate Cancer Trials Group , Enzalutamide with standard first-line therapy in metastatic prostate cancer. N. Engl. J. Med. 381, 121–131 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Kawamura T., et al. , Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies A. H., Beltran H., Zoubeidi A., Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 15, 271–286 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Bae K. M., et al. , Expression of pluripotent stem cell reprogramming factors by prostate tumor initiating cells. J. Urol. 183, 2045–2053 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celià-Terrassa T., et al. , Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J. Clin. Invest. 122, 1849–1868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder A., et al. , Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res. 74, 1227–1237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudes G., et al. , A phase 1 study of a chimeric monoclonal antibody against interleukin-6, siltuximab, combined with docetaxel in patients with metastatic castration-resistant prostate cancer. Invest. New Drugs 31, 669–676 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Harrison C., et al. , JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med. 366, 787–798 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Bai L., et al. , A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 36, 498–511.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paller C., et al. , TGF-β receptor I inhibitor enhances response to enzalutamide in a pre-clinical model of advanced prostate cancer. Prostate 79, 31–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner N. C. et al.; PALOMA3 Study Group , Palbociclib in hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 373, 209–219 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Kantoff P. W. et al.; IMPACT Study Investigators , Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Beer T. M., et al. , Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J. Clin. Oncol. 35, 40–47 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Kwon E. D. et al.; CA184-043 Investigators , Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): A multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 15, 700–712 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antonarakis E. S., et al. , Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: Multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 38, 395–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pusztai L., Hess K. R., Clinical trial design for microarray predictive marker discovery and assessment. Ann. Oncol. 15, 1731–1737 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Mukherjee S., et al. , Estimating dataset size requirements for classifying DNA microarray data. J. Comput. Biol. 10, 119–142 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Scher H. I. et al.; Prostate Cancer Clinical Trials Working Group , Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the prostate cancer clinical trials working group. J. Clin. Oncol. 26, 1148–1159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao D. L., et al. , Molecular testing in patients with castration-resistant prostate cancer and its Impact on clinical decision making. JCO Precis. Oncol. 1, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quigley D. A., et al. , Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 175, 889 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A., et al. , The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson A. G., et al. , Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbie D. A., et al. , Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.