Significance
Pancreatic cancer remains a challenge for modern oncology practice because of its dismal prognosis, 5-y survival rate of <10%, and lack of effective therapies. Here we show that the transcription factor KLF7 promotes pancreatic cancer growth and metastasis by regulating IFN-stimulated genes (ISGs) and Golgi complex integrity. Our results demonstrate a cell-intrinsic role for ISGs that is independent of innate immunity. We also demonstrate a previously unappreciated tumorigenic role of the Golgi complex as a cellular apparatus that promotes the secretion of cancer-promoting growth factors. Our results have implications for the understanding of pancreatic cancer as a disease and for the development of new therapeutic interventions for pancreatic cancer.
Keywords: KLF7, pancreatic cancer, metastasis, IFN-stimulated genes, Golgi complex
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer with a dismal prognosis. Currently, there is no effective therapy for PDAC, and a detailed molecular and functional evaluation of PDACs is needed to identify and develop better therapeutic strategies. Here we show that the transcription factor Krüppel-like factor 7 (KLF7) is overexpressed in PDACs, and that inhibition of KLF7 blocks PDAC tumor growth and metastasis in cell culture and in mice. KLF7 expression in PDACs can be up-regulated due to activation of a MAP kinase pathway or inactivation of the tumor suppressor p53, two alterations that occur in a large majority of PDACs. ShRNA-mediated knockdown of KLF7 inhibits the expression of IFN-stimulated genes (ISGs), which are necessary for KLF7-mediated PDAC tumor growth and metastasis. KLF7 knockdown also results in the down-regulation of Discs Large MAGUK Scaffold Protein 3 (DLG3), resulting in Golgi complex fragmentation, and reduced protein glycosylation, leading to reduced secretion of cancer-promoting growth factors, such as chemokines. Genetic or pharmacologic activation of Golgi complex fragmentation blocks PDAC growth and metastasis similar to KLF7 inhibition. Our results demonstrate a therapeutically amenable, KLF7-driven pathway that promotes PDAC growth and metastasis by activating ISGs and maintaining Golgi complex integrity.
Pancreatic ductal adenocarcinoma (PDAC) is the fourth-leading cause of cancer-related deaths in the United States and is expected to become the second-leading cause by 2020 (1, 2). The five-year disease-free survival rate for patients with PDAC is extremely low and has remained <10% for several decades (2, 3). Currently, there is no effective therapy for PDAC, and even immunotherapies that have worked effectively in patients with other cancer types have failed to provide meaningful clinical benefits in patients with PDAC (1). Therefore, further molecular and functional evaluation of PDAC is needed to identify and develop better therapeutic strategies.
Transcriptional deregulation is a major driver of cancer growth and metastasis that shapes the cancer transcriptome and consequently the cancer cell proteome (4–7). In fact, most cancers and their subtypes can be classified based on mRNA expression patterns alone (8, 9). Several key transcriptional regulators have been shown to play important roles in PDAC tumor growth and metastasis (10–12). Key deregulated pathways in PDAC include oncogenic mutations in the KRAS gene (found in >90% of PDACs) and mutations in the tumor-suppressor gene TP53 (found in up to two-thirds of PDACs), both of which mediate their cancer-promoting effects in part through their ability to regulate transcription (13–15). Overall, previous studies have shown that various transcription factors and transcriptional networks play major roles in facilitating PDAC tumor growth and metastasis.
Krüppel-like factor (KLF) proteins are transcriptional regulators that are conserved among mammals from human to mouse (16). Functionally, the KLF proteins can be classified into three major groups. Group 1 (including KLFs 3, 8, and 12) and group 3 (including KLFs 9, 10 11, 13, 14, 15, and 16) function largely as transcriptional repressors (16). The group 2 proteins, which include KLF7 in addition to KLFs 1, 2, 4, 5, and 6, function predominantly as transcriptional activators (16).
In this study, we show that KLF7 promotes PDAC growth and metastasis by up-regulating the expression of IFN-stimulated genes (ISGs) and by maintaining Golgi integrity. Our results suggest that the PDAC-promoting, KLF7-regulated transcriptional pathway is pharmacologically tractable for PDAC therapy.
Results
KLF7 Is Overexpressed in PDACs and Necessary for PDAC Tumor Growth and Metastasis.
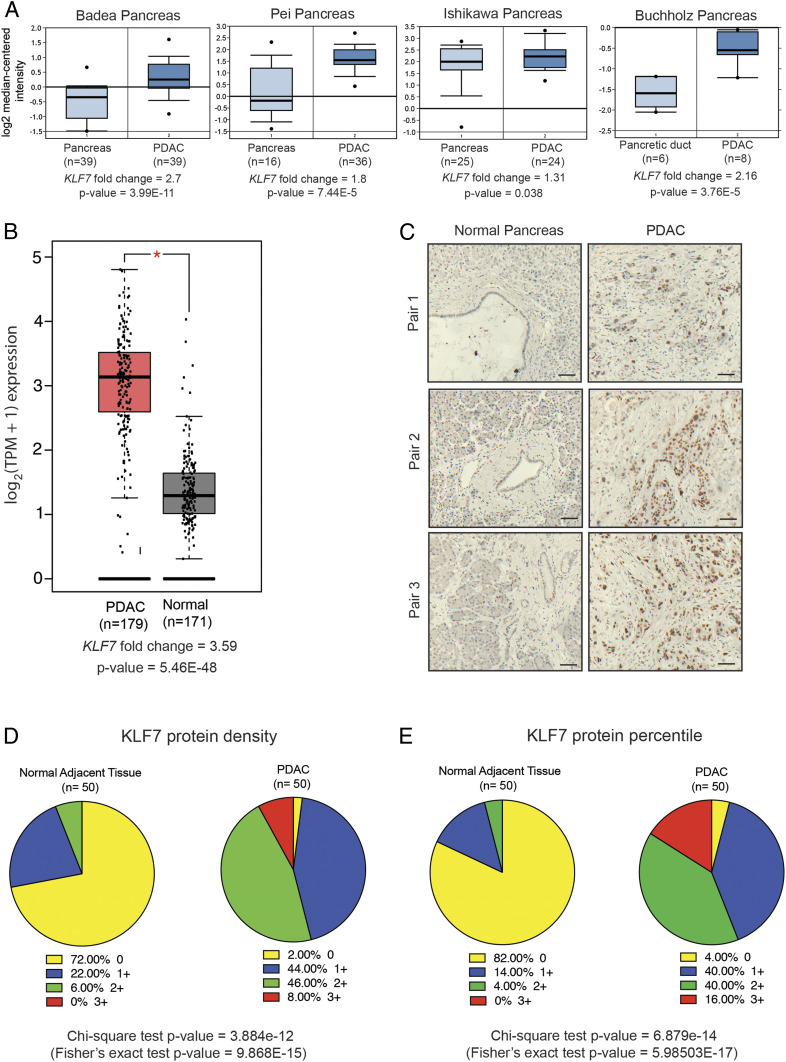
Analysis of previously published mRNA expression data from patient-derived PDAC samples revealed that KLF7 mRNA expression was significantly overexpressed in PDAC samples compared with normal pancreas samples (Fig. 1 A and B and SI Appendix, Fig. S1A) (17–22). To investigate these findings further, we analyzed KLF7 protein expression using immunohistochemistry and a tissue microarray of patient-derived PDAC samples (n = 50) and matched normal pancreas samples (n = 50). We found that KLF7 protein was significantly overexpressed in a large majority of the PDAC samples compared with the matched normal pancreas samples (Fig. 1 C–E and SI Appendix, Table S1). Taken together, the results of these analyses show that KLF7 is overexpressed in PDAC.
Fig. 1.
KLF7 is overexpressed in PDAC and is predictive of poor overall patient survival. (A) PDAC patient datasets were analyzed for KLF7 mRNA expression. The up-regulation of KLF7 mRNA in the PDAC samples relative to that in normal pancreas samples is shown. (B) KLF7 mRNA expression was significantly higher in TCGA PDAC samples compared with GTEx and TCGA normal samples combined. (C) Analysis of KLF7 protein expression in a tissue microarray (TMA) containing PDAC and matched normal adjacent pancreatic tissues (n = 50 each). Immunohistochemical staining for KLF7 in PDAC and matched normal adjacent pancreatic tissues at 200× magnification. Representative images are shown. (Scale bar: 50 μm.) (D) Analysis of immunohistochemical data from a TMA with PDAC and adjacent normal pancreatic tissues, with the average densities of KLF7 staining plotted. (E) Analysis of immunohistochemical data from a TMA with PDAC and adjacent normal pancreatic tissues, with the average percentiles of KLF7 staining plotted.
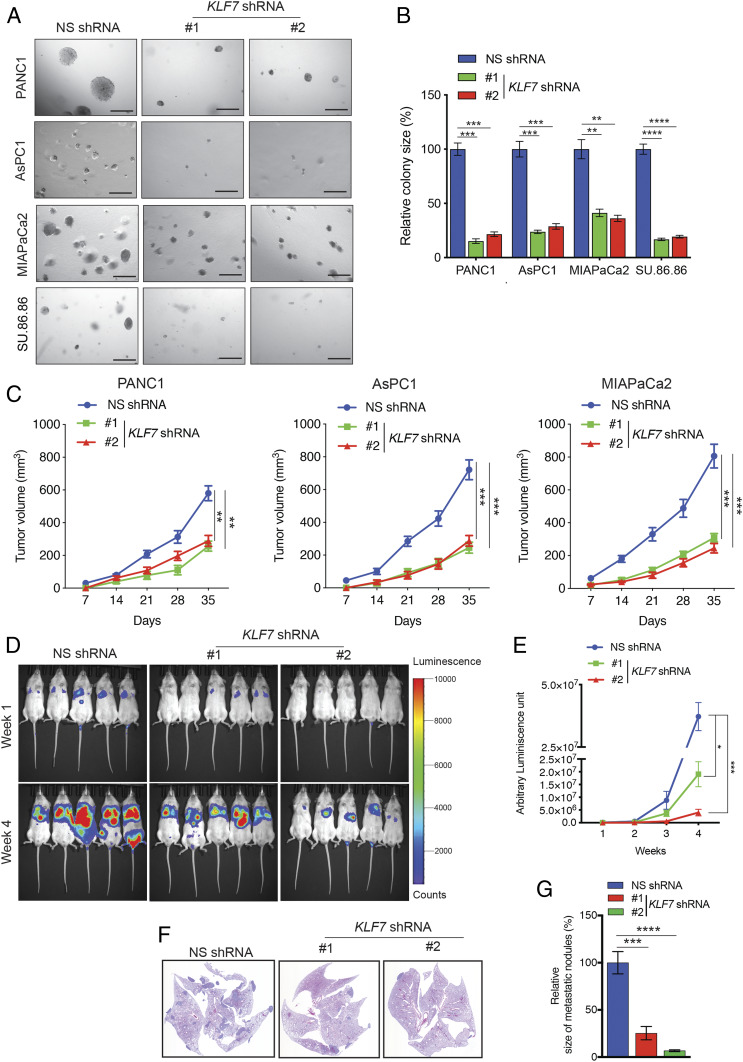
Due to the fact that KLF7 is overexpressed in PDAC led us to rationalize that KLF7 might be important for PDAC tumor growth and metastasis. To determine this possibility, we knocked down KLF7 expression in four different PDAC cell lines—PANC1, AsPC1, MIAPaCa2, and SU.86.86—using two sequence-independent short hairpin RNAs (shRNAs) (SI Appendix, Fig. S1 B and C). We then tested the outcome of the KLF7 knockdown on the ability of PDAC cells to form colonies in soft agar. We choose to perform the soft agar assay because measurement of anchorage-independent growth in soft agar serves as a reliable surrogate assay for estimating in vivo tumorigenesis (23, 24). KLF7 knockdown in PDAC cells resulted in a significant reduction in their ability to form colonies in soft agar (Fig. 2 A and B).
Fig. 2.
KLF7 loss inhibits PDAC tumor growth and metastasis. (A) Representative images of soft agar colonies of PDAC cell lines expressing either NS or KLF7 shRNAs. (Scale bar: 500 μm.) (B) Average colony size of the data shown in A. (C) PDAC cell lines expressing NS or KLF7 shRNAs were injected s.c. into athymic nude mice (n = 5) and analyzed for tumor formation. The average tumor volumes at the indicated time points are shown. (D) Firefly luciferase-labeled PANC1 cells expressing NS or KLF7 shRNAs were injected into mice via the tail vein (n = 5). Representative bioluminescence images taken 1 wk and 4 wk after injection are shown. (E) Quantitation of bioluminescence in the mice at the indicated time points. (F) Representative images of hematoxylin and eosin-stained lung sections from the week 4 groups shown in D. (G) Relative size of metastatic nodules in week 4 lungs with Firefly luciferase-labeled PANC1 cells expressing KLF7 shRNA compared with the NS shRNA-expressing metastatic nodules that were considered 100%. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next asked whether KLF7 knockdown could inhibit PDAC tumor growth in vivo. To test this, we subcutaneously injected PANC1, AsPC1, and MIAPaCa2 PDAC cells, expressing either KLF7-specific shRNAs or a nonspecific (NS) control shRNA, into the flanks of mice. We found that, similar to the results of the cell culture experiments, KLF7 knockdown inhibited PDAC tumor growth in the mice (Fig. 2C).
To explore whether KLF7 is necessary for PDAC metastasis, we first analyzed the effects of KLF7 knockdown on PDAC cell migration and invasion. We found that KLF7 knockdown significantly inhibited the invasiveness (SI Appendix, Fig. S2 A and B) and migration (SI Appendix, Fig. S2 C and D) of PDAC cells. To determine whether KLF7 expression plays a role in PDAC metastasis in vivo, we injected Firefly luciferase-labeled PANC1 (F-luc-PANC1) cells expressing NS or KLF7 shRNAs into the tail veins of mice to mimic lung metastasis. KLF7 knockdown significantly reduced the metastatic growth of PDAC cells in the mouse lungs (Fig. 2 D–G). Taken together, our findings demonstrate that genetic inhibition of KLF7 attenuates PDAC tumor growth and metastasis.
Activation of the MAP Kinase Pathway or Loss of p53 Results in Transcriptional Overexpression of KLF7 in PDAC Cells.
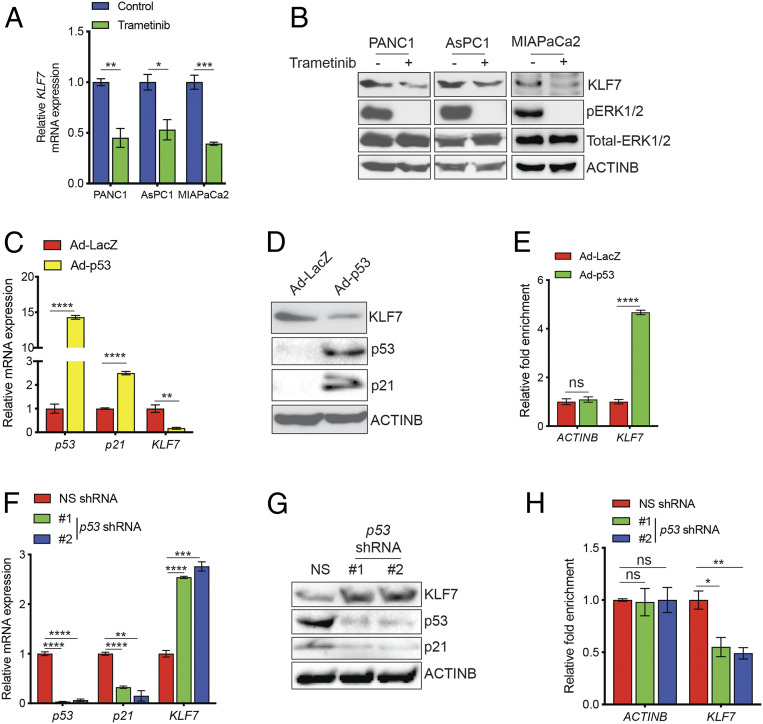
Our analyses of previously published PDAC gene expression data from patient-derived PDAC samples revealed increased KLF7 at the mRNA level, leading us to hypothesize that the elevated expression was the result of altered transcriptional regulation. Oncogenic KRAS mutation and inactivation of the tumor suppressor p53, due to either deletion or mutation, occur in a large majority of PDACs (14). Therefore, we asked whether inhibition of KRAS signaling or p53 function would affect KLF7 expression. We first determined whether inhibition of key oncogenic pathways downstream of KRAS, MAP kinase (MAPK) and PI3K (25), altered KLF7 expression. We inhibited each of these pathways by treating PDAC cells with the MEK inhibitor trametinib (26) or the PI3K inhibitor wortmannin (27) and then measured the mRNA and protein expression of KLF7. We found that trametinib treatment resulted in reduced KLF7 mRNA and protein levels (Fig. 3 A and B), whereas wortmannin treatment had no effect on KLF7 expression (SI Appendix, Fig. S3A).
Fig. 3.
KLF7 transcription is activated by the MAPK pathway and repressed by transcription factor p53. (A and B) PDAC cell lines were treated with dimethyl sulfoxide (DMSO) or trametinib (250 nM) for 24 h. KLF7 mRNA expression was analyzed by RT-qPCR. KLF7 mRNA expression is shown relative to that in the DMSO-treated cells (A). The indicated proteins were detected by immunoblotting (B). (C and D) AsPC1 cells were infected with adenovirus expressing either p53 (Ad-p53) or β-galactosidase as a control (Ad-LacZ). The mRNA expression levels of KLF7 and p21 (C) and immunoblots for the indicated proteins (D) are shown. (E) AsPC1 cells expressing Ad-p53 or Ad-LacZ were analyzed by ChIP assay to evaluate p53 binding to the KLF7 promoter or the ACTINB promoter as a control. The relative enrichment of p53 on the promoters is shown. (F) HPNE-hTERT cells expressing either NS or p53 shRNAs were analyzed for mRNA expression of p53, p21, and KLF7. (G) HPNE-hTERT cells expressing either NS or p53 shRNAs were analyzed by immunoblotting for the indicated proteins. (H) HPNE-hTERT cells expressing either NS or p53 shRNAs were analyzed for p53 recruitment on the KLF7 and ACTINB promoters by ChIP assay. Relative p53 enrichment on the promoters in HPNE-hTERT cells expressing either NS or p53 shRNAs is shown. Data are shown as mean ± SEM. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next investigated the role of p53 in the regulation of KLF7 transcription. We infected human pancreatic adenocarcinoma AsPC1 cells, which lack p53 due to the genetic deletion of p53, with a recombinant adenovirus expressing either p53 (Ad-p53) or, as a control, β-galactosidase (Ad-LacZ). Ectopic expression of p53, but not of LacZ, resulted in down-regulation of KLF7 at the mRNA and protein levels (Fig. 3 C and D). To determine whether p53 directly regulates KLF7 transcription, we first analyzed the KLF7 promoter sequence using the in silico program rVista2.0, which predicts potential DNA-binding sites for transcription factors (28). The results identified a DNA-binding site for p53 in the KLF7 promoter (SI Appendix, Fig. S3B). On the basis of these findings, we performed chromatin immunoprecipitation (ChIP) to confirm the association of p53 with the endogenous KLF7 promoter in AsPC1 cells expressing either Ad-p53 or Ad-LacZ. The ChIP results showed that p53 directly binds to the KLF7 promoter in vivo (Fig. 3E).
To further substantiate the role of p53 as a transcriptional repressor of KLF7, we knocked down p53 expression in hTERT-immortalized primary human pancreatic ductal epithelial (HPNE-hTERT) cells and analyzed KLF7 expression. We found that p53 knockdown increased the levels of KLF7 mRNA and protein in the cells (Fig. 3 F and G). In addition, we confirmed that the p53 knockdown resulted in reduced p53 recruitment to the KLF7 promoter (Fig. 3H). Collectively, our results demonstrate that activation of the MAPK pathway or inactivation of p53 are sufficient for the transcriptional overexpression of KLF7 in PDAC cells.
Loss of KLF7 Results in the Repression of ISGs and Consequential Inhibition of PDAC Growth and Metastasis.
We next aimed to identify the mechanism by which KLF7 inhibition attenuates PDAC tumor growth and metastasis. Because KLF7 is a transcription factor, we hypothesized that KLF7 mediates its effects on PDAC tumor growth and metastasis by regulating gene expression. To test this idea, we performed transcriptome-wide mRNA sequencing (RNA-seq) analysis of PDAC cell lines expressing either NS or KLF7 shRNAs (29). The loss of KLF7 expression resulted in a significant decrease in the expression of various ISGs (SI Appendix, Fig. S3C), which are known to promote cancer cell growth via cell-extrinsic and cell-intrinsic mechanisms (30–33).
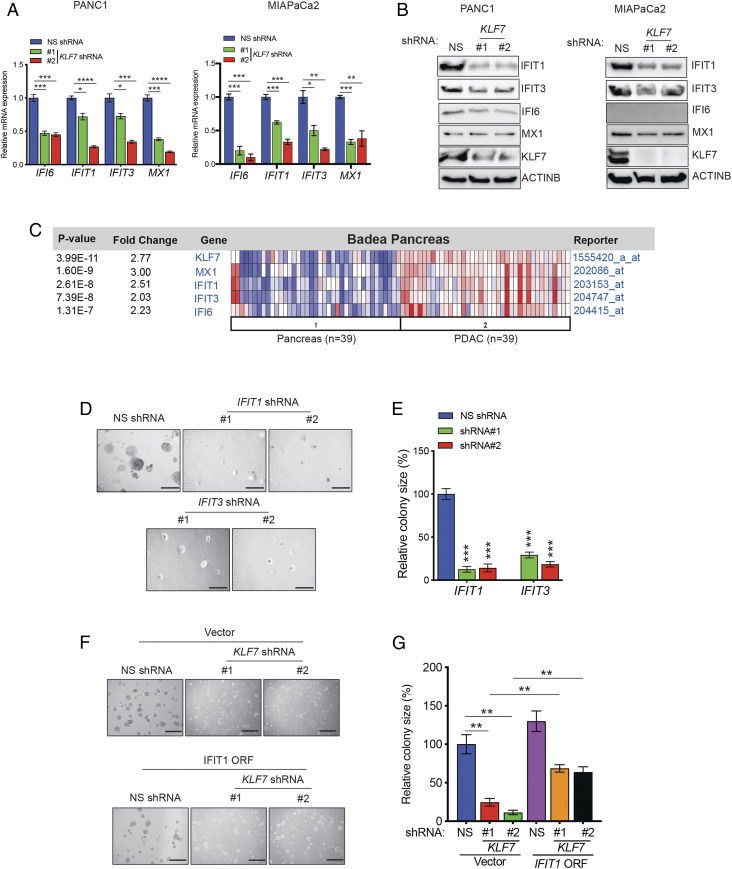
We validated the results of the RNA-seq analysis and confirmed that KLF7 knockdown inhibited the expression of ISGs in PDAC cell lines at both the mRNA and protein levels (Fig. 4 A and B). However, we observed variation in ISG expression and the extent of their down-regulation among different PDAC cell lines on KLF7 knockdown (Fig. 4B). To determine whether the regulation of ISGs by KLF7 is clinically relevant, we asked whether, similar to KLF7, ISGs are overexpressed in PDAC patient samples. Consistent with the RNA-seq results, the expression of ISGs was significantly higher in patient-derived PDAC samples than in matched normal pancreas samples (Fig. 4C and SI Appendix, Fig. S3D).
Fig. 4.
Loss of KLF7 inhibits the expression of ISGs and PDAC tumor growth and metastasis. (A) Relative gene expression data for ISGs in PANC1 and MIAPaCa2 cells expressing NS or KLF7 shRNAs were analyzed. ISG expression levels are plotted relative to those in NS shRNA-expressing cells. (B) Immunoblot analyses of the indicated proteins in PANC1 and MIAPaCa2 cells expressing KLF7 or NS shRNAs. ACTINB was used as a loading control. (C) The Badea dataset containing normal pancreas and PDAC patient samples was analyzed for mRNA expression of KLF7 and ISGs using the Oncomine database. Fold changes in expression and P values for the indicated genes are shown. (D) Anchorage-independent growth was measured by soft agar assay in PANC1 cells expressing NS or ISG-specific shRNAs. Representative images are shown. (Scale bar: 500 μm.) (E) Plot showing the relative colony sizes from D. (F) Anchorage-independent growth was measured by soft agar assay in PANC1 cells expressing KLF7 or NS shRNAs along with the V5-tagged IFIT1 ORF or empty vector (pLX304) control. Representative images are shown. (Scale bar: 500 μm.) (G) Plot showing the relative colony sizes from F. Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To further substantiate the importance of ISGs in KLF7-mediated PDAC growth, we knocked down the expression of IFIT1 and IFIT3, two ISGs whose expression was consistently down-regulated at the mRNA and protein levels in PDAC cell lines (Fig. 4 A and B), and measured the effect of the loss of these ISGs on PDAC growth in a soft agar assay. We found that the knockdown of IFIT1 or IFIT3 was sufficient to inhibit the growth of PDAC cells in soft agar (SI Appendix, Fig. S3E and Fig. 4 D and E), indicating potentially important roles downstream of KLF7.
Finally, to directly confirm the role of the ISGs downstream of KLF7, we asked whether ectopic expression of IFIT1 could rescue the inhibition of PDAC tumor growth induced by shRNA-mediated knockdown of KLF7. We found that ectopic expression of IFIT1 partly rescued the inhibition of PDAC growth induced by the loss of KLF7 (SI Appendix, Fig. S3F and Fig. 4 F and G).
Previous studies have shown that ISGs have antiapoptotic roles (34, 35). Therefore, we asked whether the reduced ISG expression caused by the loss of KLF7 results in increased apoptosis and, furthermore, whether ectopic expression of ISGs in KLF7-knockdown cells could rescue tumor growth by inhibiting apoptosis. We performed a caspase-3 activity assay and found that KLF7 knockdown resulted in increased apoptosis, which was partially rescued by the ectopic expression of IFIT1 (SI Appendix, Fig. S3G). Collectively, our results demonstrate that KLF7 facilitates PDAC tumor growth by stimulating the expression of ISGs, such as IFIT1.
KLF7 Maintains Golgi Integrity and PDAC Tumor Growth and Metastasis by Promoting DLG3 Expression.
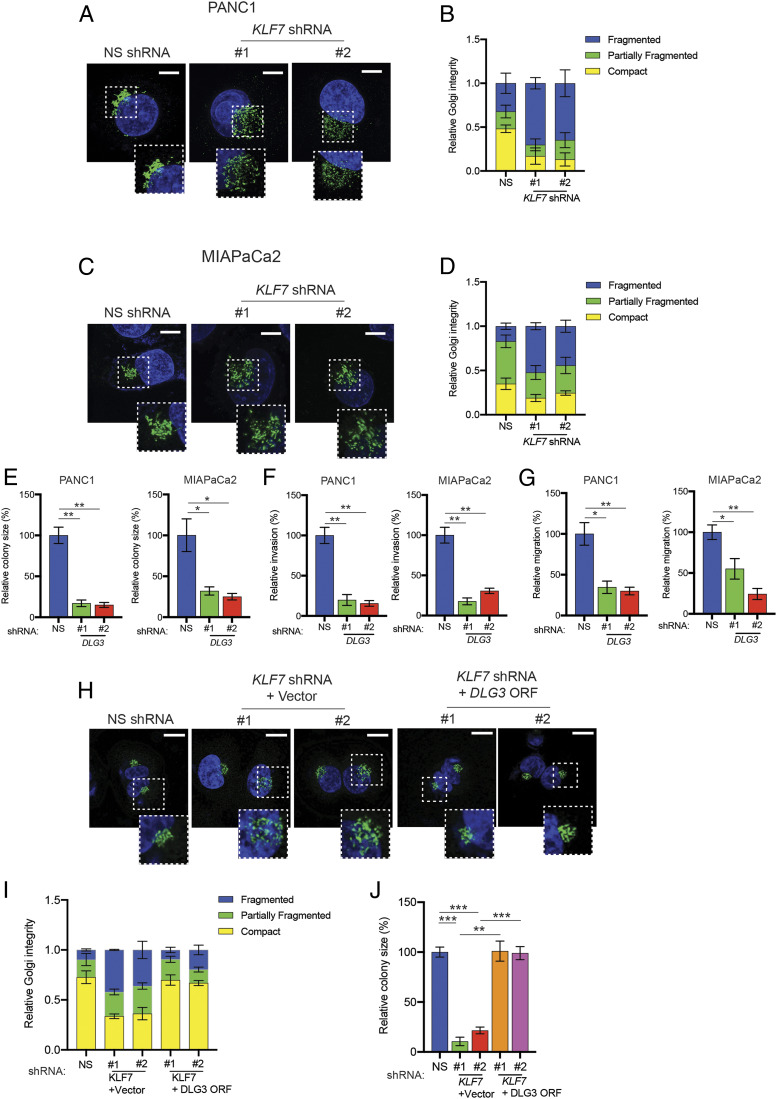
In addition to the down-regulation of ISGs in KLF7-knockdown PDAC cells in our RNA-sequencing results, we also observed that the loss of KLF7 expression resulted in a significant decrease in the expression of DLG3 (SI Appendix, Fig. S4 A and B). DLG3 has been shown to play an important role in maintaining the integrity of the Golgi complex (36), a membrane-bound organelle that functions in the posttranslational modification, sorting, and packaging of proteins for secretion (37–40). Our observation that KLF7 knockdown resulted in reduced DLG3 expression suggested that KLF7 knockdown cells may have reduced Golgi integrity. To test this idea, we performed immunofluorescence staining using GM130, a cis-Golgi matrix marker protein used to monitor Golgi integrity (41). We found that PDAC cells expressing KLF7 shRNAs showed significantly dispersed GM130 staining, indicative of higher Golgi fragmentation, compared with PDAC cells expressing NS shRNA (Fig. 5 A–D). Similarly, PDAC cells expressing DLG3 shRNAs showed increased Golgi fragmentation compared with PDAC cells expressing NS shRNA (SI Appendix, Fig. S4 C–F).
Fig. 5.
KLF7 loss causes Golgi fragmentation and inhibition of tumor growth and metastasis by regulating DLG3 transcription. (A) PANC1 cells expressing either NS or KLF7 shRNAs were analyzed for Golgi fragmentation by immunofluorescence staining with the Golgi marker GM130. Representative confocal images are shown. (Scale bar: 5 μm.) (B) Quantitation of cells with compact, partly fragmented, or fragmented Golgi based on the data shown in A. (C) MIAPaCa2 cells expressing either NS or KLF7 shRNAs were analyzed for Golgi fragmentation by immunofluorescence staining with GM130. Representative confocal images are shown. (Scale bar: 5 μm.) (D) Quantitation of cells with compact, partly fragmented, or fragmented Golgi based on the data shown in C. (E) PANC1 and MIAPaCa2 cells expressing DLG3 shRNAs were analyzed for anchorage-independent growth by soft agar assay. Relative colony sizes of cells expressing the indicated shRNAs are shown. (F) PANC1 and MIAPaCa2 cells expressing DLG3 shRNAs were analyzed by a Matrigel invasion assay. Relative invasion by cells expressing the indicated shRNAs is shown. (G) Migration of PANC1 and MIAPaCa2 cells expressing DLG3 shRNAs was analyzed in a wound-healing assay. Relative migration of cells expressing the indicated shRNAs is shown. (H) PANC1 cells expressing NS shRNA alone or KLF7 shRNAs along with empty vector or in combination with the DLG3 ORF were analyzed for Golgi fragmentation by immunofluorescence with GM130. Representative confocal images are shown. (Scale bar: 5 μm.) (I) Quantitation of cells with compact, partly fragmented, or fragmented Golgi based on the data shown in H. (J) PANC1 cells expressing NS shRNA alone or KLF7 shRNAs along with empty vector or in combination with the DLG3 ORF were analyzed for anchorage-independent growth by soft agar assay. Relative colony sizes of the PANC1 cells under the indicated conditions are shown. Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
We next asked whether, similar to KLF7 loss, DLG3 loss inhibits PDAC tumor growth and metastasis. We analyzed the ability of PDAC cells expressing NS or DLG3 shRNAs to form colonies in soft agar and also to migrate and invade. The results showed that DLG3 knockdown inhibits PDAC growth in soft agar (Fig. 5E and SI Appendix, Fig. S4G), invasion (Fig. 5F and SI Appendix, Fig. S4H), and migration (Fig. 5G and SI Appendix, Fig. S4I). To determine whether the reduction in Golgi complex integrity and inhibition of PDAC tumor growth on KLF7 knockdown was due to decreased expression of DLG3, we ectopically expressed DLG3 in PDAC cells expressing the KLF7 shRNAs (SI Appendix, Fig. S5A) and measured the growth of these cells. We found that ectopic expression of DLG3 rescued Golgi integrity (Fig. 5 H and I) and tumor-growth phenotypes (Fig. 5J and SI Appendix, Fig. S5B). Collectively, our results demonstrate that the loss of KLF7 in PDAC cells results in Golgi fragmentation due to transcriptional inhibition of DLG3, thereby blocking PDAC tumor growth and metastatic attributes.
Targeting of Golgi Integrity Maintenance Factor STX5 and Pharmacologic Activation of Golgi Fragmentation Inhibit PDAC Tumor Growth and Metastatic Attributes.
To conclusively determine whether the loss of Golgi integrity causes reduced PDAC tumor growth and metastasis, we performed two lines of experiments. We first knocked down Syntaxin 5 (STX5), a factor required for the maintenance of Golgi integrity (42), in PDAC cells. We found that STX5 knockdown increased Golgi fragmentation (SI Appendix, Fig. S5 C–E), strongly inhibited PDAC cell growth in soft agar (SI Appendix, Fig. S5 F and G), and reduced the ability of PDAC cells to invade and migrate (SI Appendix, Fig. S6 A–D).
We next asked whether chemical agents that activate Golgi fragmentation could inhibit PDAC tumor growth and metastatic attributes. We induced Golgi fragmentation using two agents: brefeldin A, which reversibly disassembles the Golgi complex by accentuating tubule formation between the trans-Golgi network and endosomes while preventing tubule detachment, and golgicide A, which causes reversible dissociation and dispersal of the Golgi and trans-Golgi network into small vesicles that subsequently disseminate throughout the cell (43). We found that treatment with either drug resulted in Golgi fragmentation in PDAC cells (SI Appendix, Fig. S6 E and F).
Finally, we asked whether the induction of Golgi fragmentation by drug treatment attenuated PDAC cell growth and metastatic attributes. Treatment of PDAC cells with brefeldin A or golgicide A strongly inhibited the growth of cells in soft agar (SI Appendix, Fig. S7 A and B) and also inhibited cell invasion and migration (SI Appendix, Fig. S7 C–F). Collectively, our results demonstrate that drugs that induce Golgi fragmentation exert tumor-suppressive effects and suppress the metastatic attributes of PDAC.
Golgi Fragmentation as a Result of KLF7 Loss Is Tumor Suppressive because of Reduced Protein Glycosylation and Consequent Reduced Secretion of Cancer-Promoting Growth Factors.
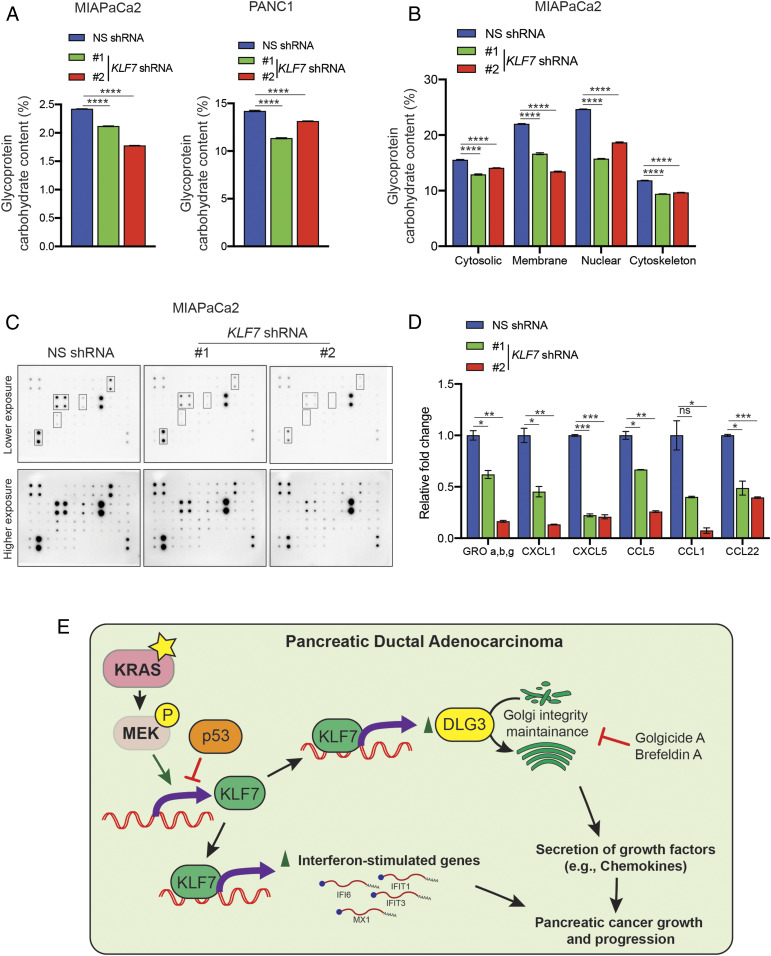
We next determined the mechanistic basis by which Golgi fragmentation is tumor suppressive. The Golgi body plays an important role in the posttranslational modification of proteins, particularly glycosylation, which in turn is necessary for the secretion of many growth-promoting factors, such as chemokines (44–46). Therefore, we hypothesized that KLF7 knockdown causes Golgi fragmentation that in turn reduces protein glycosylation and the secretion of cancer-promoting factors. To test this hypothesis, we analyzed the overall glycosylation of proteins in PDAC cell lines expressing NS or KLF7 shRNAs. Protein glycosylation is a measure of addition of a carbohydrate moiety to the protein molecule and is shown to play an important role in cancer progression (47). In agreement with our hypothesis, we observed a significant reduction in protein glycosylation in PDAC cells expressing KLF7 shRNAs (Fig. 6A).
Fig. 6.
Golgi fragmentation resulted in reduced global protein glycosylation and reduced secretion of growth-promoting chemokines. (A) MIAPaCa2 and PANC1 cells expressing NS or KLF7 shRNAs were analyzed for total glycosylated protein. The amounts of glycosylated proteins relative to those in NS shRNA-expressing cells are shown. (B) MIAPaCa2 cells expressing NS or KLF7 shRNAs were analyzed for total glycosylated protein in various cellular fractions. The amounts of glycosylated proteins in cytosolic, membrane, nuclear, and cytoskeleton in KLF7 shRNA-expressing cells relative to those in NS shRNA-expressing cells is shown. (C) Expression of 38 human chemokines was measured in MIAPaCa2 cells expressing NS or KLF7 shRNAs using human chemokine arrays. Representative images of the array membranes are shown. (D) Quantitation of the membranes shown in C, Left. (E) Model summarizing the role of KLF7 in promoting the expression of ISGs and the maintenance of Golgi integrity, which is required for KLF7 to retain its ability to promote PDAC tumor growth and metastatic attributes. Data are presented as mean ± SEM. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To further delineate this effect on a subcellular level, we fractionated PDAC cells expressing either KLF7 shRNA or control NS shRNA into cytoplasmic, membrane, nuclear, and cytoskeletal fractions and analyzed the protein glycosylation levels in each of these fractions. In this case also, we saw a significant reduction of protein glycosylation in various cellular fractions in PDAC cells expressing KLF7 shRNAs (Fig. 6B and SI Appendix, Fig. S8A).
We next asked whether KLF7 knockdown in PDAC cells reduces the secretion of factors that can promote tumor growth in an autocrine and paracrine manner. We performed immunoblotting on an antibody array that allows the detection of changes in the expression of 38 different human chemokines. We found that the secretion of a number of chemokines was reduced in PDAC cells expressing KLF7 shRNAs (Fig. 6 C and D and SI Appendix, Fig. S8 B and C). Collectively, our results support the hypothesis that increased Golgi fragmentation induced by the loss of KLF7 causes reduced protein glycosylation, which may contribute to reduced secretion of cancer growth-promoting factors.
Discussion
Effective treatment of PDAC has remained a significant clinical challenge. A better understanding of the functional landscape of PDAC is important for the development of new and effective treatments. In this study, we found that the transcription factor KLF7 is overexpressed in PDAC. We also found that KLF7 drives a transcriptional program that promotes PDAC tumor growth and metastasis. Our results are summarized in Fig. 6E and are described in detail below.
KLF7 Promotes PDAC Tumor Growth and Metastasis.
KLF proteins have important roles in the regulation of a wide variety of biological processes as well as human diseases, such as cardiovascular disease and cancer (48). KLF7 is required for neuronal morphogenesis (49) and for TrkA gene expression and the development of nociceptive sensory neurons (50). KLF7 is overexpressed in human oral squamous cell carcinoma (51) and was also shown to mediate the effects of miR-185 and the STAT3-induced long noncoding RNA LINC00668 on the growth and progression of non–small-cell lung cancer (52, 53). Those previous studies were largely correlative in nature but did not establish a direct role of KLF7 in facilitating tumor growth and/or progression. We showed that KLF7 is overexpressed in cells harboring either MAPK pathway activation or tumor suppressor p53 inactivation, two aberrations that frequently occur in PDACs (13–15). We also showed that the loss of KLF7 results in attenuated PDAC tumor growth and metastasis both in cell culture and in mice. The MAPK pathway has been shown to regulate a large number of transcription factors (e.g., MYC, AP-1, NF-κB) by regulating either their expression or their activity (54–56). Therefore, it is possible that one or more of these transcription factors downstream of the MAPK pathway might regulate the expression of KLF7 in PDAC cells. In addition, it is also possible that MAPK and p53 may synergize to regulate KLF7 expression in PDAC cells (57). Collectively, our findings demonstrate that KLF7 is overexpressed in PDAC and is necessary for PDAC tumor growth and metastasis.
Regulation of ISGs by KLF7 and Implications for PDAC Tumor Growth and Metastasis.
ISGs compose a group of genes that derive their names from the presence of IFN-responsive elements in their promoters (58). Functionally, ISGs exert diverse biological functions ranging from the regulation of viral replication to innate immune regulation via paracrine mechanisms to the regulation of cell proliferation and apoptosis via autocrine mechanisms (31, 58). A previous study showed that oncogenic NRAS transcriptionally activates ISGs, and that the ISG IFI6 is necessary for NRAS-driven tumor initiation and progression (31). Another study showed that in breast cancer, ISGs are down-regulated by the progesterone receptor, potentially leading to tumor evasion, although that hypothesis was not experimentally tested (59). In addition, ISGs are involved in the regulation of apoptosis and drug responses (34, 35). For example, IFI6 inhibits apoptosis via a mitochondria-dependent pathway in vascular endothelial cells infected with dengue virus 2 (60). Similarly, ISGs have been shown to be involved in cross-resistance to radiotherapy in tamoxifen-resistant breast cancer (61). We found that KLF7 loss results in the down-regulation of several ISGs. Moreover, we demonstrated that ISGs are necessary for PDAC tumor growth. Finally, we showed that ectopic expression of at least one of the ISGs, IFIT1, partly rescued the inhibition of PDAC growth induced by the loss of KLF7, which may be due to the ability of KLF7 to negatively regulate apoptosis induction in PDAC cells via activating ISGs. Taken together, our results show that KLF7, by activating ISGs, facilitates PDAC growth.
KLF7, DLG3, and the Tumor-Suppressive and Metastasis-Suppressive Effects of Golgi Fragmentation in PDAC.
We found that KLF7 is necessary for DLG3 expression in PDAC cells and that, similar to KLF7 knockdown, DLG3 knockdown resulted in increased Golgi fragmentation. Ectopic expression of DLG3 was able to reverse the effects of DLG3 loss, further supporting the role of DLG3 as an essential component downstream of KLF7 that promotes PDAC tumor growth and metastasis. The knockdown of other genes that maintain Golgi integrity inhibited PDAC tumor growth and metastatic attributes similar to the knockdown of KLF7 or DLG3. Moreover, pharmacologic agents that induce Golgi fragmentation (brefeldin A and golgicide A) had similar effects. These results indicate that Golgi fragmentation suppresses tumor growth and metastatic attributes in PDAC cells.
To determine the mechanism by which Golgi fragmentation suppresses PDAC tumor growth and metastatic attributes, we analyzed two key functions of the Golgi body: protein glycosylation and regulation of protein secretion. We found that KLF7 knockdown cells with higher amounts of Golgi fragmentation displayed reduced protein glycosylation and significantly reduced secreted levels of several cancer-promoting chemokines. Chemokines promote cancer growth in both an autocrine and a paracrine manner (45, 62). Taken together, our results suggest that the reduction in PDAC tumor growth and metastatic attributes resulting from the loss of KLF7 is mediated by reduced secretion of chemokines, and possibly other growth factors, due to Golgi fragmentation.
Materials and Methods
Cell Culture.
The PANC1, AsPC1, MIAPaCa2, SU.86.86, and hTERT-HPNE cell lines were obtained from American Type Culture Collection and maintained as recommended. The cells were maintained in a 5% CO2 atmosphere at 37 °C in Dulbecco’s modified Eagle’s medium (Life Technologies) or RPMI-1640 medium (Life Technologies), each supplemented with 10% fetal bovine serum (Life Technologies) and 1% penicillin/streptomycin (Life Technologies) (63).
Plasmids and Cloning.
V5-tagged IFIT1 and V5-tagged DLG3 lentiviral expression constructs cloned into the pLX304-Blast-V5 plasmid were purchased from Horizon Discovery and are listed in SI Appendix, Table S2. Adenoviruses expressing GFP or p53 were purchased from Vector Biolabs.
Statistical Analysis.
All experiments were conducted with at least three biological replicates. The results of individual experiments are expressed as mean ± SEM. The statistical significance for KLF7 immunohistochemistry data was tested using the Pearson χ2 test with Yates continuity correction in R version 3.6.1. Because the χ2 approximation may be incorrect due to the presence of small numbers in some of the cells, Fisher’s exact test was also performed in R version 3.6.1. For measurements of tumor progression in mice and MTT assays, the area under the curve of the relevant data were analyzed using GraphPad Prism version 7.0 for Macintosh (https://www.graphpad.com/). For the remaining experiments, P values were calculated using the two-tailed unpaired Student’s t test with GraphPad Prism version 7.0, for Macintosh. *P < 0.05; **P <0.01; ***P < 0.001; ****P < 0.0001.
Data Availability Statement.
The RNA-seq data of this study have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE107184). All additional data discussed in the paper are available in the main text and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the following grants from the NIH: R01 CA218008 (to M.R.G. and N.W.), R01 CA196566 (to N.W.), R01 CA200919 (to N.W.), R21 CA229927 (to N.W. and R.G.), and R03 CA230815 (to R.G.). N.W. is also supported by a Research Scholar Grant from the American Cancer Society (128347-RSG-15-212-01-TBG).
Footnotes
The authors declare no competing interest.
Data deposition: The RNA-seq data of this study have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE107184).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005156117/-/DCSupplemental.
References
- 1.Garrido-Laguna I., Hidalgo M., Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 12, 319–334 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J., et al. , Pancreatic cancer. Nat. Rev. Dis. Primers 2, 16022 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Bradner J. E., Hnisz D., Young R. A., Transcriptional addiction in cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan K. D., Galbraith M. D., Andrysik Z., Espinosa J. M., Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan M. A., Shilatifard A., Chromatin signatures of cancer. Genes Dev. 29, 238–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang P., Freedman M. L., Liu J. S., Liu X. S., Inference of transcriptional regulation in cancers. Proc. Natl. Acad. Sci. U.S.A. 112, 7731–7736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou C. M., et al. , Molecular portraits of human breast tumours. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Hoshida Y., et al. , Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittle M. C., et al. , RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 161, 1345–1360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krebs A. M., et al. , The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017). [DOI] [PubMed] [Google Scholar]
- 12.DeNicola G. M., et al. , Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpa A., et al. , Pancreatic adenocarcinomas frequently show p53 gene mutations. Am. J. Pathol. 142, 1534–1543 (1993). [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey P. et al.; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 16.McConnell B. B., Yang V. W., Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badea L., Herlea V., Dima S. O., Dumitrascu T., Popescu I., Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 55, 2016–2027 (2008). [PubMed] [Google Scholar]
- 18.Buchholz M., et al. , Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 24, 6626–6636 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Pei H., et al. , FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16, 259–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa M., et al. , Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci. 96, 387–393 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grützmann R., et al. , Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 6, 611–622 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iacobuzio-Donahue C. A., et al. , Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 162, 1151–1162 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westbrook T. F., et al. , A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Lin L., et al. , A large-scale RNAi-based mouse tumorigenesis screen identifies new lung cancer tumor suppressors that repress FGFR signaling. Cancer Discov. 4, 1168–1181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones S., et al. , Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun C., et al. , Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Ui M., Okada T., Hazeki K., Hazeki O., Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem. Sci. 20, 303–307 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Loots G. G., Ovcharenko I., rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32, W217–W221 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta R., Malvi P., Wajapeyee N., The role of KLF7 in pancreatic adenocarcinoma (PDAC) progression. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107184. Deposited 20 November 2017.
- 30.Benci J. L., et al. , Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta R., et al. , Interferon alpha-inducible protein 6 regulates NRASQ61K-induced melanomagenesis and growth. eLife 5, e16432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cañadas I., et al. , Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 24, 1143–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han H. G., Moon H. W., Jeon Y. J., ISG15 in cancer: Beyond ubiquitin-like protein. Cancer Lett. 438, 52–62 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Wagner S., et al. , Suppression of interferon gene expression overcomes resistance to MEK inhibition in KRAS-mutant colorectal cancer. Oncogene 38, 1717–1733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo C., et al. , MicroRNA-138 enhances TRAIL-induced apoptosis through interferon-stimulated gene 15 downregulation in hepatocellular carcinoma cells. Tumour Biol. 39, 1010428317710410 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Anitei M., et al. , A high-throughput siRNA screen identifies genes that regulate mannose 6-phosphate receptor trafficking. J. Cell Sci. 127, 5079–5092 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Huang S., Wang Y., Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000 Res. 6, 2050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emr S., et al. , Journeys through the Golgi—Taking stock in a new era. J. Cell Biol. 187, 449–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickner W., Schekman R., Protein translocation across biological membranes. Science 310, 1452–1456 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Marsh B. J., Howell K. E., The mammalian Golgi—Complex debates. Nat. Rev. Mol. Cell Biol. 3, 789–795 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Nakamura N., et al. , Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715–1726 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudlyk T., Willett R., Pokrovskaya I. D., Lupashin V., COG6 interacts with a subset of the Golgi SNAREs and is important for the Golgi complex integrity. Traffic 14, 194–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sáenz J. B., et al. , Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5, 157–165 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witsch E., Sela M., Yarden Y., Roles for growth factors in cancer progression. Physiology (Bethesda) 25, 85–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sporn M. B., Roberts A. B., Autocrine growth factors and cancer. Nature 313, 745–747 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Balkwill F., Cancer and the chemokine network. Nat. Rev. Cancer 4, 540–550 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Stowell S. R., Ju T., Cummings R. D., Protein glycosylation in cancer. Annu. Rev. Pathol. 10, 473–510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bialkowska A. B., Yang V. W., Mallipattu S. K., Krüppel-like factors in mammalian stem cells and development. Development 144, 737–754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laub F., et al. , Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol. Cell. Biol. 25, 5699–5711 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei L., et al. , The zinc finger transcription factor Klf7 is required for TrkA gene expression and development of nociceptive sensory neurons. Genes Dev. 19, 1354–1364 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding X., et al. , KLF7 overexpression in human oral squamous cell carcinoma promotes migration and epithelial-mesenchymal transition. Oncol. Lett. 13, 2281–2289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L., et al. , miR-185 inhibits the proliferation and invasion of non-small cell lung cancer by targeting KLF7. Oncol. Res. 27, 1015–1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.An Y. X., et al. , STAT3-induced long noncoding RNA LINC00668 promotes migration and invasion of non-small cell lung cancer via the miR-193a/KLF7 axis. Biomed. Pharmacother. 116, 109023 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Zhang W., Liu H. T., MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Chang F., et al. , Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: Potential targeting for therapeutic intervention. Leukemia 17, 1263–1293 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Carlson S. M., et al. , Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci. Signal. 4, rs11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurray H. R., et al. , Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 453, 1112–1116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter K. R., et al. , Interferon-stimulated genes are transcriptionally repressed by PR in breast cancer. Mol. Cancer Res. 15, 1331–1340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi Y., et al. , Correction: IFI6 inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2-infected vascular endothelial cells. PLoS One 10, e0138896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Post A. E. M., et al. , Interferon-stimulated genes are involved in cross-resistance to radiotherapy in tamoxifen-resistant breast cancer. Clin. Cancer Res. 24, 3397–3408 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Hattermann K., et al. , Transmembrane chemokines act as receptors in a novel mechanism termed inverse signaling. eLife 5, e10820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malvi P., Janostiak R., Nagarajan A., Cai G., Wajapeyee N., Loss of thymidine kinase 1 inhibits lung cancer growth and metastatic attributes by reducing GDF15 expression. PLoS Genet. 15, e1008439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data of this study have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE107184). All additional data discussed in the paper are available in the main text and SI Appendix.