Significance
Topoisomerases are a family of proteins that alter DNA topology to accommodate for the stresses imparted by processes such as transcription, replication, and mitotic chromosome condensation. Topoisomerase 2A (TOP2A) is a key member of this family that prominently stains along the axis of mitotic chromosomes. Assessing mitotic-specific functions of TOP2A has been thwarted by the many essential roles the protein plays across the cell cycle. Our study used an ultrafast protein depletion system in human cells to dissect the mitotic-specific function of TOP2A. We have shown that TOP2A is necessary for keeping the structure of chromatids together once they have formed (maintenance), in addition to being involved in the establishment of the mitotic chromosome.
Abstract
Topoisomerase IIα (TOP2A) is a core component of mitotic chromosomes and important for establishing mitotic chromosome condensation. The primary roles of TOP2A in mitosis have been difficult to decipher due to its multiple functions across the cell cycle. To more precisely understand the role of TOP2A in mitosis, we used the auxin-inducible degron (AID) system to rapidly degrade the protein at different stages of the human cell cycle. Removal of TOP2A prior to mitosis does not affect prophase timing or the initiation of chromosome condensation. Instead, it prevents chromatin condensation in prometaphase, extends the length of prometaphase, and ultimately causes cells to exit mitosis without chromosome segregation occurring. Surprisingly, we find that removal of TOP2A from cells arrested in prometaphase or metaphase cause dramatic loss of compacted mitotic chromosome structure and conclude that TOP2A is crucial for maintenance of mitotic chromosomes. Treatments with drugs used to poison/inhibit TOP2A function, such as etoposide and ICRF-193, do not phenocopy the effects on chromosome structure of TOP2A degradation by AID. Our data point to a role for TOP2A as a structural chromosome maintenance enzyme locking in condensation states once sufficient compaction is achieved.
The correct formation of condensed mitotic chromosomes is a crucial step in the cell division cycle and is absolutely required for faithful segregation of sister chromatids to daughter cells. Once considered a “black box,” recent developments and technologies have shed new light on the structural organization of condensed mitotic chromosomes (1) and the way in which mitotic chromatin is looped by condensins. Mitotic chromosome condensation requires the coordinated action of both histone and nonhistone proteins. Indeed, nonhistone proteins comprise 40% of the total protein mass of mitotic chromosomes (2), and most of these proteins are organized in the chromosome scaffold (3, 4).
The chromosome scaffold is a structural and organizational component of mitotic chromosomes. It comprises proteins that occupy the chromatid core and display a distinct axial localization along the chromosome (3, 5–7). Key scaffold proteins include condensins I and II, KIF4A, and topoisomerase IIα (TOP2A), with TOP2A being the most abundant by mass (2, 4, 8). TOP2A is a type II topoisomerase that functions as a dimer to resolve double stranded DNA (dsDNA) entanglements. Type II topoisomerases are enzymes that can create a transient break in one DNA duplex via transesterification of the phosphodiester bond, covalently linking the DNA ends to tyrosyl groups in each monomer. A second duplex is then passed through the break, and the break is sealed by reversing transesterification (9). This activity is essential for myriad cellular processes, including decatenation of sister chromatids prior to mitosis and relief of supercoiling that builds up during transcription and replication (10). In vertebrates, there are two isozymes of topoisomerase II (TOP2)—alpha (TOP2A) and beta (TOP2B)—that share similar N-terminal ATPase and core domains, but differ in their C-terminal domains. The isoforms also differ functionally, with TOP2A expressed at higher levels in G2 and mitosis and strongly localizing to mitotic chromatin (4, 11, 12). TOP2B is undetectable on early mitotic chromatin but starts accumulating on chromatin in late anaphase and telophase (13). TOP2B cannot substitute for the mitotic functions of TOP2A (14). Although depletion of TOP2B alone does not affect mitotic chromosome structure, removal of both together produces a stronger phenotype than TOP2A depletion alone (15).
A large body of literature has described the mitotic consequences of TOP2A disruption in vitro and in vivo. However, the pleiotropic nature of TOP2A has made it challenging to study its cell cycle-specific functions. Most studies of TOP2A in mitosis have relied on either systems that deplete protein over more than one cell cycle, making it difficult to discern primary from secondary phenotypes, or drugs targeting TOP2A. These drugs (e.g., etoposide and ICRF-193) form stable complexes with DNA (16, 17) and target both TOP2A and TOP2B. The drug treatments thus induce potential confounding effects not necessarily related to loss of TOP2A function. As a major proliferation-linked gene (18), TOP2A is targeted in the treatment of many cancers (19, 20), and it is therefore important to understand the effects of TOP2A-targeting drugs.
Landmark in vitro experiments showed that TOP2A, together with nucleosomes and condensins, constitute the essential core elements for formation of mitotic chromosomes (21). In vivo studies across a broad range of species showed that disruption of TOP2A causes defects in mitotic chromosome condensation and chromosome segregation (15, 22–26). In vertebrates, disruption of TOP2A leads to abnormally long and thin mitotic chromosomes (15, 26, 27). Collectively, these studies show that TOP2A is necessary for the establishment of mitotic chromosome structure. A previous in vitro study found evidence suggesting that TOP2A is not required for the maintenance of mitotic chromosome structure (28). Another study showed that chromosomes depleted of TOP2A with RNAi were able to recover compacted structure after unfolding, and the authors suggested that TOP2A is not important for structural integrity of mitotic chromosomes (26). However, with the advent of new efficient protein depletion technologies, we aimed to revisit this question.
In this study, we dissect the role of TOP2A in mitosis directly by combining rapid auxin-inducible degradation of TOP2A with high-resolution live cell imaging. We find that TOP2A is dispensable for prophase condensation, but is acutely required in prometaphase. Surprisingly, we reveal that removal of TOP2A from precondensed mitotic chromosomes causes loss of compacted mitotic chromosome structure and premature mitotic exit. Our results suggest that TOP2A is crucial for maintenance of mitotic chromosome structure.
Results
Auxin-Inducible Degron of TOP2A.
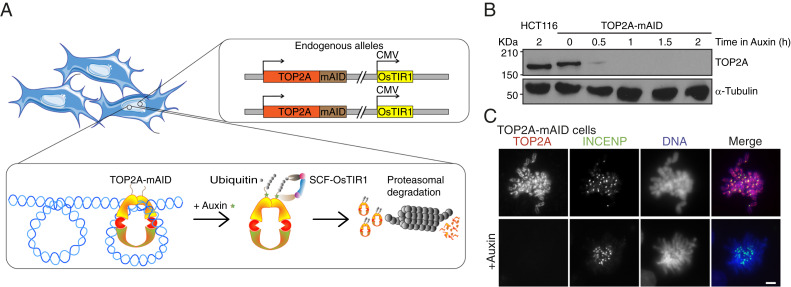
Due to the short duration of mitosis, it is difficult to study mitotic protein function when using methods that rely on transcriptional shut-off such as RNAi or Tet-OFF (29), which are limited by the rate of protein turnover. To our knowledge, no studies have depleted the TOP2A protein rapidly and specifically from cells already in mitosis. To achieve this, we constructed a conditional, auxin-inducible, TOP2A knockout in human HCT116 +CMV-OsTIR1 cells (30). In this system, a sequence encoding a 7.9-kDa protein tag, mAID, is inserted in frame with the target gene. The plant hormone auxin can bind mAID and direct the SCF ubiquitin ligase in complex with OsTIR1 to ubiquitinate the target protein, which is then rapidly degraded by the proteosome (31) (Fig. 1A). We used CRISPR-Cas9–directed cloning and introduced the mAID tag at the 3′ end of both alleles of the TOP2A gene, overwriting the stop codon (SI Appendix, Fig. S1A). Introduction of mAID and the two resistance genes was confirmed by genomic PCR, suggesting that the homology directed repair (HDR) was complete (SI Appendix, Fig. S1 B–E, 3). Expression of TOP2A-mAID was confirmed by immunoblotting, which showed a higher MW corresponding to the 7.9-kDa mAID tag (Fig. 1B). We used 500 μM of the synthetic auxin indole-3-acetic acid (IAA) (30) for all auxin treatments, which will be referred to henceforth as “auxin treatment.” TOP2A was barely detectable following 30 min of auxin treatment and was completely depleted after 1 h (Fig. 1B). Depletion could be achieved in both interphase (SI Appendix, Fig. S1 F, i) and mitosis (SI Appendix, Fig. S1 F, ii), and was confirmed using standard immunofluorescence conditions (SI Appendix, Fig. S1F) and by preextraction where the cytoplasm and nucleoplasm were precleared of soluble protein before fixation (SI Appendix, Fig. S1G). Chromosome spread experiments showed the characteristic axial and centromeric staining pattern of TOP2A in mitosis (Fig. 1C) was retained with the TOP2A-mAID protein (8, 32–35). In interphase, the pan-nucleoplasmic staining of TOP2A could be removed by preextraction to reveal a strong nucleolar localization of TOP2A (SI Appendix, Fig. S1 G, i) consistent with a previous study (35).
Fig. 1.
Auxin-inducible degron of TOP2A. (A) The TOP2A-mAID system. The mAID tag was inserted in-frame with both alleles of the TOP2A gene in HCT116 cells expressing OsTIR1 from CMV promoter. When auxin is added to the cells, it binds TOP2A-mAID, which is targeted for proteasomal degradation by SCF-OsTIR1–mediated ubiquitination. (B) TOP2A immunoblot of lysates from parental HCT116 CMV-OsTIR1 cells (labeled as HCT116) or HCT116 CMV-OsTIR1 TOP2A-mAID (labeled as TOP2A-mAID) cells treated with auxin for the indicated periods of time. A-tubulin was immunoblotted as loading control. (C) Immunofluorescent staining of TOP2A and INCENP and DAPI staining of DNA on chromosome spreads from TOP2A-mAID cells grown with or without auxin. (Scale bars: 5 μm.)
TOP2A Depletion during Prometaphase Causes Chromatin Entanglement.
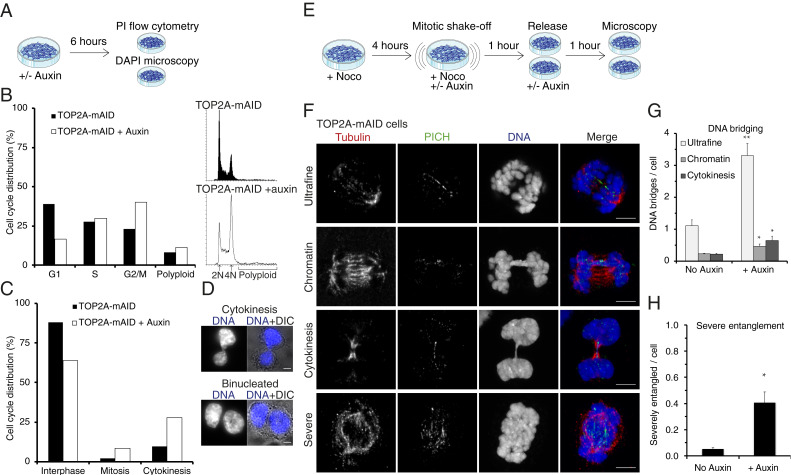
Inhibition of TOP2A by the bisdioxopiperazines ICRF-187 or ICRF-193 was shown previously to arrest cells at the G2/M border at the so-called decatenation checkpoint (36–40). This checkpoint was proposed to arrest cells until TOP2A has performed sufficient sister chromatid resolution/decatenation. In our study, TOP2A-mAID cells treated for 6 h with auxin (Fig. 2A) caused a >15% increase in the 4N population consistent with a cell cycle arrest in either G2 or mitosis in addition to a 3% increase in polyploid (>4N) cells (Fig. 2B). When analyzing corresponding DAPI-stained samples by microscopy (Fig. 2C), we observed that the increased 4N and >4N populations reflects a >5% increase in mitotic cells and a >15% increase in cells that are either undergoing cytokinesis (Fig. 2D, cytokinesis) or have aborted cytokinesis (Fig. 2D, binucleated). This argues that cells depleted of TOP2A do not arrest in G2. This is also supported by a study using doxycycline-inducible TOP2A knockdown where the authors found that the ICRF-193–induced G2 checkpoint requires the presence of TOP2A (25).
Fig. 2.
TOP2A depletion from prometaphase causes severe chromatin entanglement. (A) Experimental conditions for B–D. (B) Quantification of cell cycle profiles of TOP2A-mAID cells +/− auxin. 2N denotes the G1 peaks and 4N the G2/M peaks. Polyploid is defined as nonaggregates with more than G2/M DNA content. (C) TOP2A-mAID cells stained with DAPI and analyzed by fluorescent microscopy. Cells were quantified as either interphase, mitosis, or cytokinesis based on their DNA condensation level and conformation. (D) Representative images of cells undergoing cytokinesis or binucleated. (E) Experimental conditions for F–H. (F) Examples of DNA entanglements visualized by superresolution microscopy, quantified in G and H. PICH (green) and tubulin (red) were immunostained, and DNA stained with DAPI. (G) Quantification of chromatin and ultrafine DNA bridges per anaphase B, or cytokinesis bridges per cytokinesis, cell. (H) Quantification of severe entanglement. (G and H) Data points are averages of at least three independent experiments. Error bars denote SEM. Asterisks denote P values: *P < 0.05 and **P < 0.01. (Scale bars: D and F, 5 μm.)
TOP2A is required for decatenation of the centromeres (26) and to resolve chromatin bridges and ultrafine DNA bridges (UFBs) (26, 41, 42) in anaphase. We wanted to test with the AID system whether DNA bridging is still prominent in anaphase when TOP2A is depleted from cells in prometaphase. Therefore, we developed a synchronization protocol that ensured a highly enriched prometaphase population. TOP2A-mAID cells were arrested in prometaphase for 4 h by nocodazole treatment, isolated by mitotic shake-off, incubated with nocodazole for 1 h in the presence (+) or absence (−) of auxin, and then released from nocodazole into +/− auxin for 1 h (Fig. 2E). This, presumably, allowed time in prometaphase for the cells to resolve DNA entanglements and to condense chromatin before TOP2A depletion. Using this synchronization protocol, DNA bridging in anaphase was then analyzed by staining with DAPI and immunostaining of the UFB marker PICH (41, 42) (Fig. 2F). Quantification of IAA-treated versus control cells showed significant increases in UFBs (threefold) and chromatin bridges (twofold) in anaphase B and in cytokinetic bridges (threefold) in telophase (Fig. 2G). This indicates that a proportion of DNA bridges requires both active spindle forces and TOP2A to be resolved. During this analysis, we identified a subset of cells that had a single DNA mass even though tubulin and PICH staining suggested the cells were in anaphase (Fig. 2F, severe, and SI Appendix, Fig. S2). These severely entangled cells had high numbers of PICH threads inside the DNA mass (Fig. 2F, severe, and SI Appendix, Fig. S2B) and stretched centromere staining (SI Appendix, Fig. S2). Although the severely entangled nuclear masses might be interpreted as being from a disorganized prometaphase, staining with the centromere antibody CREST and tubulin revealed that many of these severely entangled cells were in anaphase (SI Appendix, Fig. S2) and that most of the PICH threads, but not all, originated from centromeres (SI Appendix, Fig. S2B). These cells could not be scored for DNA bridging since it was impossible to discern the number of individual bridges. Instead, they were scored individually as cells with severe entanglement, and this analysis showed an eightfold increase compared to control cells (Fig. 2H). This latter abnormality represents a more drastic phenotype than conventional DNA bridging, where typically only a single chromosome or subset is entangled, and hints at a more global defect in mitotic chromosome structure and segregation in the absence of TOP2A. We therefore went on to analyze gross chromosome architecture.
TOP2A Is Required for Correct Mitotic Chromosome Structure.
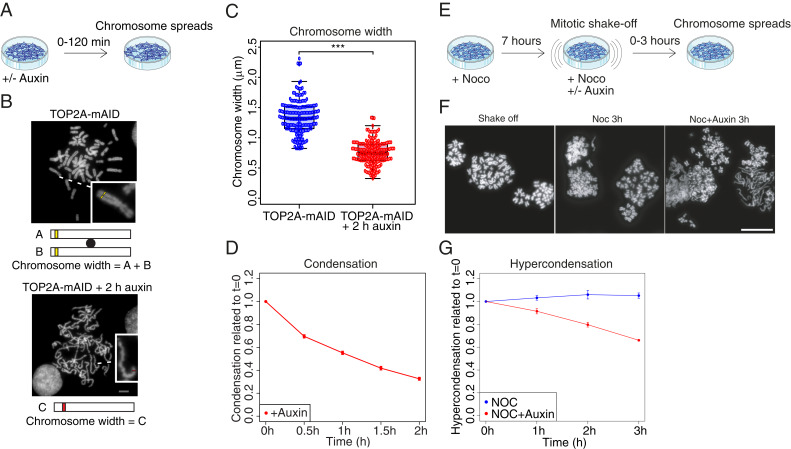
Knockdown of TOP2A or inhibition by ICRF-193 was shown previously to cause the formation of atypical, long and thin, mitotic chromosomes (15, 26, 27). We observed a similar phenotype when rapidly depleting TOP2A from asynchronous cultures of TOP2A-mAID cells (Fig. 3A) and analyzing chromosome width and general morphology on chromosome spreads. The width of chromosomes was decreased significantly (almost twofold; Fig. 3B), as was the interkinetochore distance, as measured by the distance between sister foci of CENP-C on chromosome spreads (SI Appendix, Fig. S3 A and B). This confirmed previous findings (26, 43, 44). These data do not necessarily suggest a defect in kinetochore or centromere structure per se, but could simply be explained by the fact that chromosomes are thinner and longer following TOP2A depletion. We classified chromosome morphology on chromosome spreads as either hypercondensed, intermediate, hypocondensed, or amorphous (SI Appendix, Fig. S3C). Asynchronous TOP2A-mAID cells grown in the presence of auxin showed a time-dependent increase in hypocondensed and amorphous morphologies compared to control chromosomes, which predominantly had a normal morphology (Fig. 3D and SI Appendix, Fig. S3D). These results are consistent with previous studies (15). Nevertheless, the short depletion time (30 to 60 min auxin treatment) required to produce such a severe phenotype was surprising. This led us to speculate that TOP2A might not only be involved in formation of mitotic chromosomes, but also in the continued maintenance of mitotic chromosome structure. To investigate this, we treated TOP2A-mAID cells with nocodazole for 7 h before further enriching for prometaphase cells by mitotic shake-off, and then added auxin as well as nocodazole, before analyzing chromosome spreads at 1-h intervals over a 3-h period (Fig. 3E). This protocol permitted cells to finish mitotic chromosome condensation and hypercondense chromosomes before TOP2A was removed. The mitotic index at this point was ∼84% and remained at similarly high levels throughout the experiment (SI Appendix, Fig. S3F). Strikingly, removal of TOP2A from these cells with precondensed chromosomes appeared to reverse condensation and produced pronounced hypocondensed and amorphous phenotypes (Fig. 3 F and G and SI Appendix, Fig. S3 C, E, and F). These results indicate that TOP2A is required for maintenance of mitotic chromosome structure, and we went on to confirm this surprising finding.
Fig. 3.
TOP2A is required to maintain mitotic chromosome morphology. (A) Diagram of experimental conditions for B–D. (B) Chromosome width measured on chromosome spreads. Representative chromosome spreads are shown. (Insets) Enlargements of individual chromosomes. Width was quantified as A+B when sister chromatids were distinguishable or C when they were not (predominantly auxin-treated). Error bars denote SEM. Asterisks denote P value: ***P < 0.001. (D) Quantification of chromosome condensation in asynchronous culture as the ratio of spreads with normal condensation compared to hypo 1, 2 and amorph categories (SI Appendix, Fig. S3 C and D). (E) Diagram of experimental conditions for F and G. (F) Examples of chromosome spreads quantified in G. (G) Quantification of hypercondensation of precondensed chromosomes following auxin treatment, presented as the ratio of spreads with hypercondensation compared to normal, hypo 1, 2, and amorph categories (SI Appendix, Fig. S3 C and E). (D and G) Results are relative to t = 0, which was set as 1. Data points are averages of three independent experiments. (Scale bars: B and F, 5 μm.)
TOP2A Is Not Required for Localization of Chromosome Condensation Proteins.
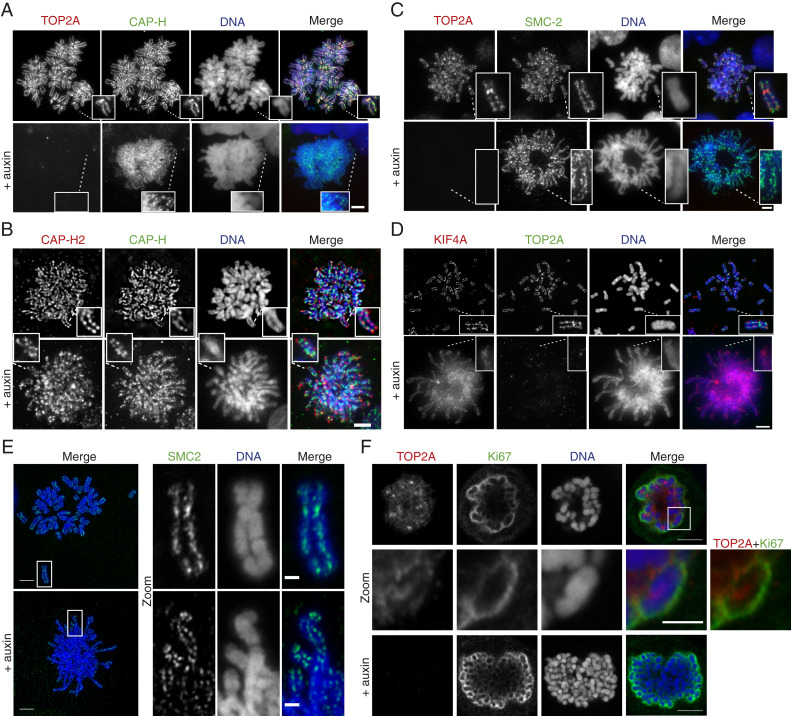
Unlike condensin I, condensin II, and KIF4A, TOP2A has not previously been attributed a role in the maintenance of mitotic chromosome structure (28). Therefore, a simple explanation for TOP2A’s role in mitotic chromosome maintenance might be that it affects the correct localization of other chromosome maintenance proteins. To investigate if the chromosome scaffold was disrupted following TOP2A degradation, we performed immunofluorescence staining of three scaffold proteins (condensins I and II and KIF4A) on chromosome spreads from asynchronous cultures of TOP2A-mAID cells grown in media +/− auxin for 2 h. TOP2A depletion did not appear to affect the scaffold localization of condensin I (CAP-H and SMC2; Fig. 4 A–C), condensin II (CAP-H2 and SMC-2; Fig. 4 B and C), or KIF4A (Fig. 4D). Condensin and KIF4A binding was evident and localized to the central region of each chromatid. This is consistent with previous studies using either siRNA-mediated depletion of TOP2A or inhibition of TOP2A by ICRF-193 (26, 27, 43). Condensin II was of particular interest, since it has not been analyzed specifically in TOP2A mutants and only indirectly in other studies by analysis of SMC2, which is shared by both condensins. Condensin II, whose depletion similarly causes long and thin mitotic chromosomes (and a defective prophase) (45, 46), was still localized to the scaffold region following rapid depletion of TOP2A. Nevertheless, although not disrupted, the scaffold appeared irregular and often twisted around its own axis (Fig. 4 A–D). This phenotype became even more apparent when analyzing chromosome structure using Airyscan superresolution microscopy (Fig. 4E).
Fig. 4.
TOP2A is not required for localization of chromosome condensation proteins. (A–E) TOP2A-mAID cells were grown in media +/− auxin for 2 h and harvested for chromosome spreads and coimmunofluorescent staining for TOP2A, CAP-H, CAP-H2, SMC2, or KIF4A, with DAPI staining of DNA. (Insets) Enlargements of individual chromosomes. (F) TOP2A-mAID cells were grown in media +/− auxin for 2 h and harvested for coimmunofluorescent staining for TOP2A and Ki67, with DAPI staining of DNA. (A–F) (Scale bars: regular images, 5 μm; Zoom, 2 μm.)
Another protein important for mitotic chromosome morphology is Ki67, which localizes to the perichromosomal compartment in mitosis (47, 48) and has been suggested to act as a surfactant on mitotic chromosomes (49). Superresolution microscopy analysis of the location of TOP2A and Ki67 showed that both have a perichromosomal staining pattern (Fig. 4F). Depletion of TOP2A by auxin treatment for 2 h of TOP2A-mAID cells did not, however, change the Ki67 staining pattern (Fig. 4F). Together, the data suggest that removal of TOP2A has a direct effect on mitotic chromosome topology rather than an indirect effect caused by mislocalization or perturbation of other key mitotic structure proteins.
TOP2A Depletion Prevents Prometaphase Condensation.
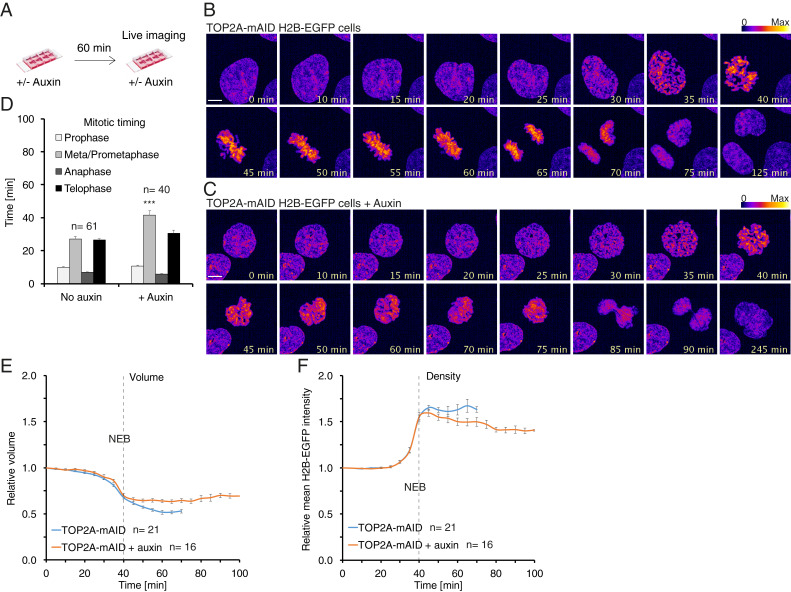
To analyze TOP2A depletion by live cell imaging, we established TOP2A-mAID + histone H2B-EGFP cells by tagging an endogenous locus of H2B using Alt-R CRISPR-Cas9 (Methods). The cells were then grown in media +/− auxin for 1 h (Fig. 5A) before being imaged by live cell spinning-disk confocal microscopy for 5 h (Fig. 5 B and C and Movies S1 and S2). The timing of the different mitotic phases was then quantified (Methods provides definitions of each phase; Fig. 5D). The timing of prophase was unaffected by auxin treatment (Fig. 5 B–D). Hence, TOP2A is not required for timely prophase progression. Moreover, the morphology and configuration of chromosomes in prophase appeared similar in the presence and absence of auxin (Fig. 5 B and C). In contrast, auxin treatment increased the average length of prometaphase and metaphase significantly from 27 min in control cells to 42 min in auxin-treated cells (Fig. 5D). Auxin treatment also had a profound effect on cell morphology in prometaphase, with the cells failing to show individualized chromosomes characteristic of prometaphase but rather forming an amorphous DNA mass (Fig. 5C). These cells rarely formed a metaphase plate or completed mitosis (Fig. 5C). Among the relatively few auxin-treated cells that did reach anaphase and telophase, the timing of these phases was similar to that of control cells (Fig. 5D). Taken together, these data indicate that TOP2A is not important for gross prophase chromosome structure, but is crucial for the reorganization of chromosomes that takes place in prometaphase.
Fig. 5.
TOP2A depletion prevents prometaphase condensation. (A) Diagram of experimental conditions for B–F. TOP2A-mAID +H2B-EGFP cells were seeded in chambered slides and grown in media +/− auxin for 60 min before being imaged over time by spinning-disk confocal microscopy. (B) Example of an untreated TOP2A-mAID cell going through mitosis. (C) Example of an auxin-treated TOP2A-mAID cell entering and exiting mitosis. (B and C) Fluorescence intensity is displayed as fire LUT. (Scale bars: 5 μm.) (D) Quantification of the duration of mitotic phases from live cell imaging experiments. Each data point is an average of the n number of cells. The data were collected from at least three independent experiments. Error bars denote SEM. Asterisks denote P values from parametric Student t tests: ***P < 0.001. (E and F) Three-dimensional modeling analysis of chromatin volume (E) and density (F) of nuclei entering mitosis. Values are normalized to the time point 40 min before nuclear envelope breakdown (NEB), which is set as 1. The dashed lines mark NEB. Each data point is an average of n number of nuclei analyzed. Error bars denote SEM.
Mitotic chromosome condensation is characterized by a reduced volume, but higher density, of chromatin. To investigate the role of TOP2A in mitotic chromosome condensation more directly, we quantified H2B-EGFP volume and mean fluorescence intensity, representing chromatin volume and density, respectively. To compare different cells in real time, we normalized measurements to the time point 40 min prior to NEB for each cell analyzed (set to a value of 1.0). As a proof of principle, 3D modeling of an untreated cell entering and traversing mitosis (SI Appendix, Fig. S4A and Movie S3) and quantification of its chromatin volume and density is shown (SI Appendix, Fig. S4B). From prophase through NEB (at 40 min) into prometaphase, metaphase, and anaphase, volume decreases while density increases. From early telophase through to nuclear envelope reformation (NER), volume increases and density decreases until values reach interphase levels (∼1.0). Using this method, we analyzed chromatin condensation profiles of asynchronous cells depleted of TOP2A. In prophase, from condensation to NEB, this analysis showed no difference in chromatin volume (Fig. 5E) or density (Fig. 5F) between control and auxin-treated cells. This result suggests that TOP2A is not required for global DNA condensation in prophase, although its depletion may affect local DNA topology or structure in a way not detectable by this method. Analysis of prometaphase chromatin in untreated cells from NEB onward showed a decreased volume (Fig. 5E) and increased density (Fig. 5F). This is consistent with increasing chromatin condensation during prometaphase. In auxin-treated cells, by contrast, chromatin volume was unchanged (Fig. 5E) and chromatin density decreased (Fig. 5F) before cells exited mitosis, generally, without chromosome segregation (Fig. 5C). This shows that TOP2A is principally required for condensation of mitotic chromosomes in prometaphase.
TOP2A Depletion, but Not Inhibition, Reverses Chromosome Condensation.
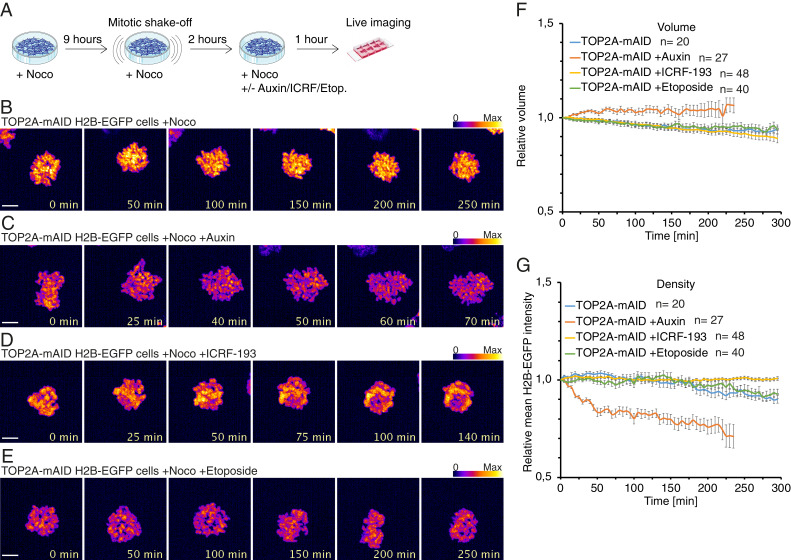
To analyze if the condensation state of mitotic chromosomes could be reversed following TOP2A depletion or inhibition, we enriched for cells with precondensed chromatin using a 9-h nocodazole treatment and mitotic shake-off followed by an additional 2-h nocodazole treatment (Fig. 6A). These precondensed prometaphase cells were then treated with auxin, ICRF-193, or etoposide. We imaged and quantified volume and density of mitotic chromatin over time in the manner described for Fig. 5 E and F, except that values were normalized to the starting timepoint. In control nocodazole-treated cells (Fig. 6B and Movie S4), the prometaphase chromatin appeared largely unchanged. In contrast, chromatin of cells grown in the presence of auxin (Fig. 6C and Movie S5) appeared to decondense and lose defined mitotic structure over time. Surprisingly, cells with precondensed chromosomes and grown with either 25 μM ICRF-193 (Fig. 6D and Movie S6) or etoposide (Fig. 6E and Movie S7) did not exhibit similar changes in chromatin structure. Correspondingly, quantification of chromatin volume (Fig. 6F) and density (Fig. 6G) showed that control and ICRF-193- and etoposide-treated cells had decreased volume and marginally increased density over time. By contrast, auxin-treated cells displayed a gradual increase in chromatin volume (Fig. 6F) and a decrease in density (Fig. 6G). As a control, we confirmed that the relative levels of chromatin volume or density was similar at the first time point (t = 0) for the different treatments (SI Appendix, Fig. S4C). These results show that depletion of TOP2A from precondensed chromosomes causes decondensation and loss of mitotic chromosome structure.
Fig. 6.
TOP2A depletion from prometaphase reverses chromosome condensation. (A) Diagram of experimental conditions. (B–E) Representative images from time-lapse live cell imaging of TOP2A-mAID cells arrested in prometaphase with nocodazole and treated with solvent (B), auxin (C), ICRF-193 (D), or etoposide (E). Fluorescence intensity is displayed in fire LUT. (Scale bars: 5 μM.) (F and G) Quantification of average chromatin volume (F) and chromatin density (G) over time before cells exit mitosis. (F and G) Error bars denote SEM. n values denote number of cells analyzed.
Treatment with auxin or ICRF-193 often overrode the nocodazole block, leading to mitotic exit without chromosome segregation. Hence, only timepoints before cells exited mitosis were quantified and displayed in the representative quantifications for auxin-treated (0 to 70 min) and ICRF-193–treated (0 to 140 min) cells (Fig. 6 C and D and SI Appendix, S4 D and E). This is because we were interested in the direct consequences of TOP2A impairment on chromatin structure. The drastic chromatin changes accompanying mitotic exit would otherwise skew the overall quantification. Importantly, these experiments indicate that the decondensation occurs before mitotic exit and not as a result of it. Complete chromatin volume and density quantifications corresponding to the representative examples in Fig. 6 B–E are shown in SI Appendix, Fig. S4 F–I. The mitotic exit phenotypes are also described (see Fig. 8).
Fig. 8.
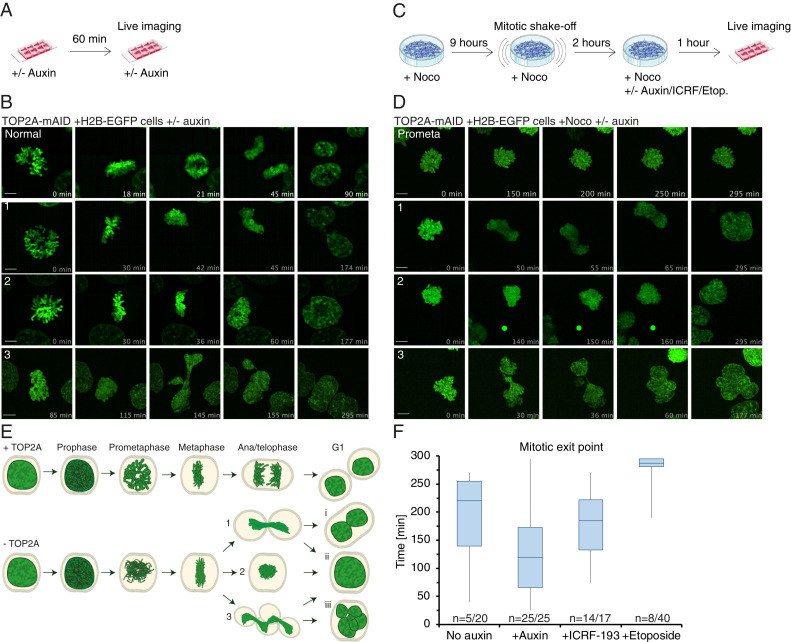
TOP2A depletion causes cytokinesis failure and premature mitotic exit. (A) Experimental conditions for B. (B) Representative time-lapse image series of TOP2A-mAID cells grown without (normal) or with auxin (1–3) from prometaphase to mitotic exit. (C) Experimental conditions for D and E. (D) Representative time-lapse image series of TOP2A-mAID cells arrested in prometaphase (prometa) or arrested and then grown with auxin with similar mitotic exit phenotypes as in B and C (1–3). (E) Schematic of mitosis with TOP2A (Top) and premature mitotic exit phenotypes (1–3) of cells traversing mitosis without TOP2A and their possible interphase outcomes, binucleation (i), giant mononucleation (ii), or multinucleation (iii). (F) Timing of mitotic exit following the indicated treatments, specified in C. n denotes number of cells exited out of the total number of cells analyzed.
In opposition to our results, an early study found that treatment with an etoposide-like TOP2A poison, VM-26, caused mitotic chromosomes to decompact (50). In that study, hamster cells were arrested in G1 by serum deprivation, released into 1 µg/mL aphidicolin for 24 h, released for 6 h, and then arrested with Colcemid before treatment with 5 μM VM-26. We cannot explain this difference, but perhaps speculate that aphidicolin treatment for 24 h would cause severe replication stress and several problems in the following mitosis and that they did not necessarily treat cells with fully condensed mitotic chromosomes.
To ensure that the ICRF-193 and etoposide treatments were efficient, we performed a series of control experiments (SI Appendix, Fig. S5). These experiments showed that ICRF-193 treatment of asynchronous cultures arrests the cells in G2 and prometaphase, whereas etoposide arrests cells in the S and G2 phases (SI Appendix, Fig. S5 A–F). Very few of the ICRF-193- or etoposide-treated mitotic cells shaken off in SI Appendix, Fig. S5 E and F were in anaphase, except for a few that appeared severely entangled, and hence we concluded that these treatments prevented cells from entering anaphase. Instead the ICRF-193–treated mitotic cells were in an abnormal prometaphase (SI Appendix, Fig. S5E). To better understand the mitotic context of the TOP2A targeting phenotypes, we also tested if our treatments caused DSBs in mitosis. To this end, we arrested cells in prometaphase with a 6-h nocodazole treatment followed by mitotic shake-off and a 2-h additional nocodazole treatment. We then added solvent, 25 μM ICRF-193, or 25 μM etoposide for 1 h before adding solvent or auxin for another 2 h (3 h ICRF-193/etoposide and 2 h of auxin at endpoint; SI Appendix, Fig. S5H). The cells were harvested for immunofluorescence staining of γH2AX and TOP2A, and relative total chromatin γH2AX intensity was quantified (SI Appendix, Fig. S5 H and I). We observed a mild increase in γH2AX staining upon auxin (twofold) and ICRF-193 (threefold) treatments compared to control treatments, consistent with a previous study (51). The addition of auxin to ICRF-193–treated mitotic cells increased γH2AX fluorescence intensity slightly. In contrast, etoposide treatment induced a drastic (27-fold) increase in γH2AX staining over control, which was slightly lower upon cotreatment with auxin (SI Appendix, Fig. S5 H and I). We took these results as confirmation that our treatments were effective.
Taken together, our results show that the removal of TOP2A from chromatin induces decondensation of precondensed chromosomes, whereas inhibition (ICRF-193) or poisoning (etoposide) of chromatin-associated TOP2A does not.
TOP2A Is Essential for Maintenance of Metaphase Chromosome Structure.
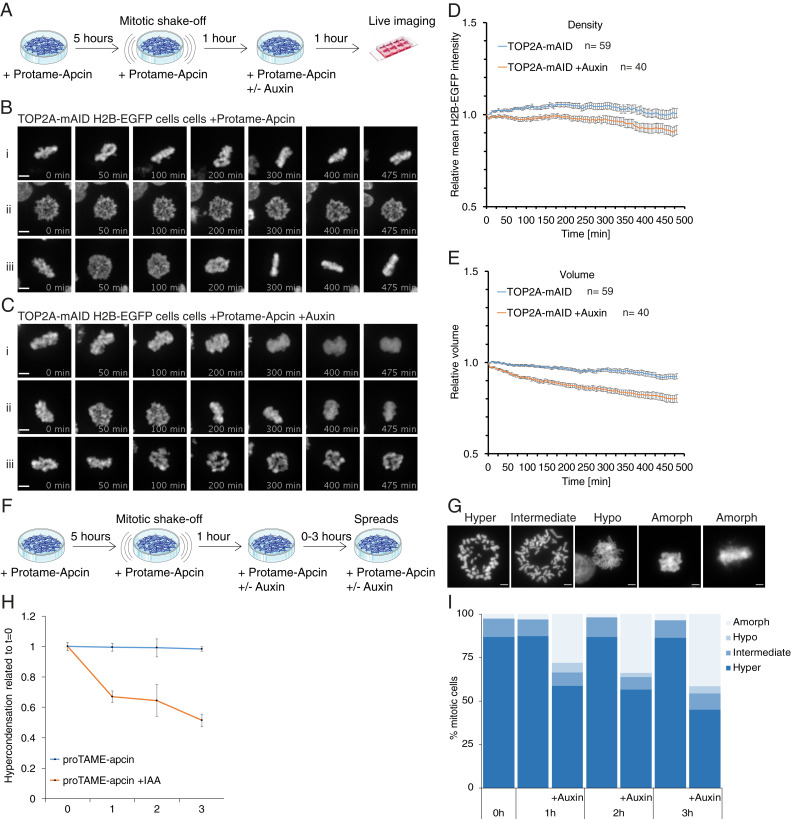
The results discussed earlier revealed that TOP2A is necessary for maintenance of mitotic chromosome structure in prometaphase in the absence of active spindle forces (nocodazole; Figs. 3 E–G and 6). Next, we wanted to analyze the role of TOP2A under conditions where the Anaphase Promoting Complex/Cyclosome (APC/C) is blocked, arresting cells in metaphase with spindles acting on the chromosomes. Blocking the APC/C does not activate the spindle assembly checkpoint (SAC), but it inhibits cyclin-B1 degradation, which renders cells unable to exit mitosis. Overexpression of cyclin-B1 has been shown to rescue the anaphase decondensation defects in SMC2-knockout cells (52) and also prevents anaphase chromosomes from decompacting in normal cells (53). In our experiments, we used the potent APC inhibitor combination proTAME-apcin (54) and found that 12.5 μM proTAME combined with 25 μM apcin was the lowest combination of drug doses to arrest HCT116 cells efficiently in mitosis (62% G2/M, 5 h treatment; SI Appendix, Fig. S6A). Mitotic cells were enriched by blocking with the drug combination and then performing mitotic shake-off before auxin was added (Fig. 7A), followed by live cell imaging. Five hours of treatment was chosen since prolonged arrest with proTAME-apcin can cause cohesion fatigue (54), which we also observed for some cells during our analysis (SI Appendix, Fig. S6B and Movie S8) and decided to quantify this. We scored cohesion fatigue as the first timepoint where cells had bipolar chromatin (H2B-EGFP) signals separated from the chromatin mass (e.g., SI Appendix, Fig. S6B, 160 min). At t = 0 when cells had been treated for 1 h with auxin, there was only a marginal difference in the number of cells with cohesion fatigue compared to controls (SI Appendix, Fig. S6C). At t = 480 min, however, there was a twofold reduction compared to controls (SI Appendix, Fig. S6D). This supports the idea that TOP2A depletion from precondensed chromosomes causes sister chromatid entanglement, supplying extra cohesion and preventing them from separating.
Fig. 7.
TOP2A depletion from metaphase causes loss of mitotic chromosome structure. (A) Experimental conditions for live imaging of cells in proTAME-apcin metaphase block (B–E). (B and C) Representative time-lapse images of TOP2A-mAID cells treated like in A without auxin (B) and with auxin (C). (D and E) Quantification of chromatin density (D) and volume (E) in cells treated like depicted in A. Error bars denote SEM. (F) Experimental conditions for chromosome spreads from proTAME-apcin metaphase block (G and H). (G) Examples of quantified categories of chromosome spreads in H and I. (H and I) Quantification of chromosome spreads from cells treated like in F.
The results of the morphological and structural analysis paralleled our nocodazole experiments, with cells grown without auxin maintaining compacted mitotic chromosome structure (Fig. 7B and Movies S9–S11), while chromosomes lost mitotic structure following auxin treatment (Fig. 7C and Movies S12–S14). Similarly to the effect seen during a nocodazole block, chromatin density declined over time in auxin-treated cells (Fig. 7D and SI Appendix, S6E). Nevertheless, following auxin treatment, the volume occupied by H2B-EGFP decreased significantly over time compared to controls (Fig. 7E and SI Appendix, S6F), the opposite of what we observed in the nocodazole-arrested cells.
To confirm the somewhat surprising results from live cell imaging, we performed mitotic chromosome spreads following the proTAME-apcin protocol (Fig. 7F). Overall, the results (Fig. 7 G–I) resembled nocodazole-blocked chromosome spreads (Fig. 3 F and G and SI Appendix, Fig. S3D), with a dramatic loss of hypercondensed chromosome configuration (Fig. 7H) and with increased hypo- and particularly amorphous chromosome configurations. The amorphous chromosome spread phenotypes (Fig. 7I) resemble the live cell imaging phenotype after auxin exposure (Fig. 7C), showing that the decreased chromatin volume observed in live cell imaging is not an artifact. A budding yeast study showed previously that spindle-attached plasmid DNA becomes positively supercoiled in vivo following top2 degradation from an APC/C inhibition block, but not from a nocodazole block (55). Interestingly, this activity was condensin-dependent. Perhaps a global spindle-dependent change in DNA topology occurs following TOP2A depletion from proTAME-apcin block, but not from nocodazole block. Supercoiling state determines the handedness of nucleosome assembly (56). It is therefore possible that a global reversal in supercoiling might, over time, cause nucleosome expulsion, thereby reducing overall H2B-EGFP staining. This is supported by studies showing that the spindle forces are strong enough to expel nucleosomes from dsDNA (57, 58). Combined with the fact that, in the proTAME-apcin block, chromosomes are held in close proximity by the spindles, it might explain the differing chromatin volume results from the two mitotic blocks. It is clear from both experiments, though, that TOP2A is essential for maintenance of mitotic chromosome structure in prometa- and metaphase.
TOP2A Depletion Causes Premature Mitotic Exit and Cytokinesis Failure.
Cells depleted of TOP2A from an asynchronous cell culture (Fig. 8A) cannot segregate their chromosomes and often exit mitosis prematurely without chromosome segregation (Fig. 8B and SI Appendix, Fig. S7A and Movies S15–S18). We defined three morphologically distinct premature mitotic exit phenotypes (Fig. 8 B and D, 1–3) with different outcomes in interphase. Exit phenotype 1 led to either binucleation (i) or giant mononucleation (ii), phenotype 2 led to mononucleation (ii), and phenotype 3 led to mononucleation (ii) or multinucleation (iii) (Fig. 8 B and D, summarized in E). To investigate if the phenotypes were due to a defect in anaphase or prometaphase, and if they were dependent on proper spindle attachment, we enriched for cells held in prometaphase using nocodazole and mitotic shake-off (Fig. 8C). Control cells generally stayed in the prometaphase block (Figs. 6B and 8 D and F, Movie S19, and SI Appendix, Fig. S7B); however, we observed all three phenotypes of premature mitotic exit following auxin or ICRF-193 treatment (Fig. 8 D and F, Movies S20–S22, and SI Appendix, Fig. S7B). All auxin-treated cells exited the prometaphase block and mitosis, and did so earlier than controls (Fig. 8F). Likewise, ICRF-193–treated cells often exited mitosis prematurely, although later than auxin-treated cells (Fig. 8F). Indeed, these mitotic exit phenotypes occur despite nocodazole-induced activation of the SAC. By contrast, cells treated with etoposide rarely left mitosis prematurely (Fig. 6E and SI Appendix, Fig. S7B) and generally stayed in prometaphase (Fig. 8E). The three phenotypes of premature mitotic exit are all characterized by membrane blebbing and cytokinetic ingressions (SI Appendix, Fig. S7C, 1–3), similar to regular cytokinesis (SI Appendix, Fig. S7C, normal).
One explanation for the mitotic exit from the nocodazole-induced arrest following TOP2A depletion could be that Aurora B function is affected. We tested this hypothesis by measuring total chromatin fluorescence intensity of the Aurora B substrate Ser-10 phosphor-H3 (PH3) in TOP2A-mAID cells arrested in prometaphase. However, there were no significant differences between TOP2A-depleted and control cells (SI Appendix, Fig. S8 A and B). In addition, there was no evidence for mislocalization of either INCENP, a member of the chromosome passenger complex (SI Appendix, Fig. S8C), or BUBR1 (found at sister kinetochores; SI Appendix, Fig. S8D). This suggests that the SAC was still active in nocodazole-arrested cells lacking TOP2A, and therefore that the premature mitotic exit phenotype following TOP2A depletion does not depend on SAC inactivation. Although the drastic missegregation and premature prometaphase exit phenotypes suggest that TOP2A is continuously required by cells during this mitotic stage, we did not observe premature mitotic exit from a proTAME-apcin block (Fig. 7 B and C). Even following auxin treatment and loss of defined mitotic chromosome structure, cells did not adopt interphase chromatin conformation (Fig. 7C and Movies S12–S14).
In conclusion, premature mitotic exit triggered by TOP2A deficiency is not dependent on spindle forces or the SAC since it occurs from both asynchronous culture and nocodazole block. It does not depend on TOP2A itself either because both depletion of TOP2A by auxin treatment and inhibition of TOP2A by ICRF-193 treatment induces premature mitotic exit. It is, however, dependent on APC/C activity and likely degradation of cyclin B1.
Discussion
Previous studies have argued for an active role of TOP2A in the establishment of mitotic chromosome structure (14, 15, 21, 22, 25, 27, 28, 59–61). In agreement with that, our data show that TOP2A is crucial for chromosome condensation after NEB. Importantly, we define another role for TOP2A in the maintenance of mitotic chromosome structure. Our results differ from earlier pioneering in vitro studies, using Xenopus egg extracts, which showed that TOP2A was responsible for establishment, but not ongoing maintenance, of mitotic chromosomes (28). Our results also provide clear delineation between TOP2A-targeting drugs and TOP2A depletion, and show that these drugs cannot be taken as a direct proxy of TOP2A depletion. Our studies with ICRF-193 differ from a recent study in Drosophila (61), which showed decompaction of mitotic chromosomes upon addition of ICRF-193. Comparison is difficult in this instance, as Drosophila seemingly has a different mechanism of chromosome condensation, with condensin II present at very low levels and not necessary for mitotic chromosome structure (62, 63). Furthermore, in our study, we quantified whole chromatin, used a lower concentration of ICRF-193, and imaged over a longer period: 25 μM ICRF-193 over 5 h compared to chromatids from Drosophila embryos that were treated with 280 μM ICRF-193 and followed for 15 min.
TOP2A is the main enzyme needed to resolve DNA bridges in anaphase and is essential for chromosome segregation. We show that depletion of TOP2A from cells held in prometaphase increases the number of DNA bridges in anaphase and telophase and drastically increases the number of cells with severely entangled chromatin (Fig. 2). This suggests either that TOP2A depletion in prometaphase causes reentanglement of DNA or that a portion of TOP2A-mediated DNA resolution cannot be completed in prometaphase and instead takes place in anaphase or telophase. Indeed, previous studies suggest that TOP2A is required for decatenation of the centromere in anaphase (64). Likely, the spindle forces are needed to supply directionality for the TOP2A enzyme to resolve interchromatid entanglements, as suggested previously (55, 65). The cell might even benefit from the extra “cohesion” from interchromatid entanglements to counterbalance cohesion fatigue (66) and prevent premature sister chromatid disjunction. Indeed, even in an unperturbed mitosis, UFBs connecting centromeres and rDNA persist into anaphase, where they are rapidly resolved (41, 67). However, some reentanglement likely occurs to give rise to the severe chromatin entanglement phenotype. These results demonstrate the key role the enzyme plays specifically in prometaphase.
Despite showing a strong localization to prophase chromosomes (68), we find that neither the timing of prophase nor the condensation of prophase chromatin are affected by TOP2A depletion (Fig. 5). This is different to that seen in condensin II-depleted cells (that also show long and thin chromosomes), which instead show a completely defective prophase (45, 46). Notably in the absence of TOP2A, condensin II still localized to mitotic chromosomes (Fig. 4). Our data are consistent with a study which showed prophase chromosome volume was unaffected by ICRF-193 treatment (60). The same study found that resolution of sister chromatid arm intertwining takes place predominantly in prophase and is dependent on TOP2A. Our results do not argue against a role for TOP2A in prophase resolution of arm chromatin. Instead, we show that TOP2A depletion from asynchronous cultures causes a dramatic prometaphase delay in mitosis and prevents chromosome condensation after NEB (Fig. 5). This suggests a role in facilitating proper condensin I-mediated condensation, since condensin I is predominantly absent from chromatin until NEB (69).
Precondensed mitotic chromatin loses its compact individualized structure following rapid removal of TOP2A by AID from cells held in prometaphase (Figs. 3 and 6 and SI Appendix, Fig. S3) or metaphase (Fig. 7). This shows that TOP2A is crucial for mitotic chromosome maintenance. Nevertheless, neither ICRF-193 nor etoposide treatment causes a similar effect (Fig. 6). Both drugs immobilize the enzyme on DNA and prevent catalytic strand passage activity by TOP2A. Therefore, TOP2A’s role in maintenance is likely linked to the enzyme’s ability to entrap two DNA strands and hold them in place. It appears that TOP2A is, in fact, a structural chromosome maintenance enzyme.
The interplay between TOP2A and condensin I appears to be crucial for the shaping of prometaphase chromosomes, a contention supported by several studies (55, 61, 65, 70). Condensin is able to positively supercoil DNA (55, 71), and condensin-induced positive supercoiling can efficiently stimulate TOP2A decatenation of catenated minichromosomes (55, 70). Therefore, the TOP2A reactions in maintenance of prometaphase chromatin are likely guided by condensin-mediated changes in topological state supplying directionality to TOP2A catenation or decatenation activity. Conversely, the loss of mitotic chromosome structure following rapid TOP2A depletion might be caused by the uncoupling of TOP2A from condensin I activity. This proposal implies that mitotic chromosome condensation must be a continuous process, which does not reach an endpoint. DNA in already formed mitotic chromosomes appears to be heavily self-entangled, which in itself might be needed to stabilize the mitotic chromosome (72). Indeed, a hypothesis has been put forward based on studies from the Oliveira lab (61) and the Aragon lab (73) that TOP2A can reentangle mitotic chromatin, and that this potentially problematic activity is counteracted by the continuous activity of condensin I.
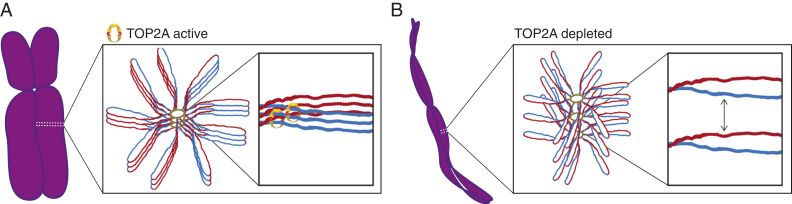
Our data point to a role of TOP2A in maintaining mitotic chromosome structure, but how might the enzyme achieve this? Such a proposal is somewhat paradoxical given the known activity of TOP2A. Since TOP2A has the ability to entrap DNA, it is possible that a pool of TOP2A might remain in a locked conformation to seal condensation states at the end of prometaphase. Alternatively, locking of condensation states might be dynamic and achieved by balancing the DNA-metabolizing activities of TOP2A and condensin I. Our data support a hypothesis where TOP2A plays a dual role in first resolving entanglements and relieving torsional stress during establishment of compact mitotic chromosomes and then locking in condensation states once sufficient compaction is achieved (Fig. 9).
Fig. 9.
TOP2A locks in condensation states. Diagram of how TOP2A might prevent axial lengthening of late mitotic chromosomes and thereby decondensation. (A) A portion of the TOP2A pool in prometaphase cells locks axial chromatin loops to maintain condensed chromosome structure. (B) With TOP2A absent from late prometaphase chromosomes, axial chromatin loops move apart, causing lengthening of chromosomes and decondensation.
In summary, we show that TOP2A is required for maintenance of mitotic chromosomes. The advent of new sophisticated single-molecule microscopy and genome-wide techniques will enable the molecular mechanism behind this activity to be illuminated.
Methods
Construction of Auxin-Inducible TOP2A Degron and H2B-EGFP Cells.
Guide DNAs (gDNAs) targeting the 3′ UTR of the TOP2A gene were designed using Zhang Lab design resources (https://zlab.bio/guide-design-resources; MIT 2013). gDNA oligos with BbsI/BpiI overhangs were annealed and ligated into pSpCas9(BB)-2A-GFP (PX458)*BpiI (no. 48138, Addgene; gift from Feng Zhang, Broad Institute, Cambridge, MA) (74). HDR-targeting constructs were assembled using Hifi Gibson Assembly (NEB). The 397-bp (5′) and 467-bp (3′) homology arms flanking the TOP2A 3′UTR and overwriting the stop codon with an 18-bp linker sequence were produced by PCR with Q5 Hot start high-fidelity polymerase (NEB) using the “TOP2A 5′ arm” or “TOP2A 3′ arm” “forward” and “reverse” primers (SI Appendix, Table S1). The arms had 35-bp overlaps bearing homology to both mAID-HYGRO or mAID-BSR pieces cut by BamHI from plasmids pMK287 and pMK288 [no. 72825 and no. 72826, Addgene (30), gifts from Masato Kanemaki, National Institute of Genetics (NIG), Shizuoka, Japan] and pSK_BS+*EcorI as backbone. Plasmids were transfected into HCT116 +CMV-OsTIR1 cells (gift from Masato Kanemaki) (30) using Lipofectamine 3000 (Thermo Fisher) according to manufacturer protocol. At 24 h following transfection, cells were reseeded and selected with 125 µg/mL hygromycin and 7.5 µg/mL blasticidin for >12 d before colony selection. Correct clones were confirmed by PCR, immunoblotting, and immunofluorescence microscopy (TOP2A antibody; sc-166934, Santa Cruz). The Alt-R CRISPR-Cas9 method (IDT) was used according to manufacturer instructions for endogenous tagging of histone H2B with EGFP. Cells were transfected using the Neon transfection system (Thermo Fisher Scientific) according to manufacturer instructions. Transfectants were single-cell flow sorted for EGFP fluorescence. Primer and gDNA sequences are listed in SI Appendix, Table S1.
Materials and Data Availability.
Cell lines, plasmids, and other reagents created in this study will be sent upon request. Data files were uploaded to the public repository Zenodo (https://doi.org/10.5281/zenodo.3724099) according to the principles of the Findable, Accessible, Interoperable, Reusable (FAIR) initiative.
Supplementary Material
Acknowledgments
The authors thank Dr. Matthew Burton for providing flow cytometry and microscopy technical support. This work was supported by National Health and Medical Research Council Project Grants GNT1127209 and GNT145188, by the Victorian Government’s Operational Infrastructure Support Program, The Danish National Research Foundation (DNRF115), and a Carlsberg Foundation Fellowship (CF16-0206). We thank members of the D.F.H., P.K., and Hickson laboratories for helpful discussions. Special thanks to Professor Ian D. Hickson, Professor Bill C. Earnshaw, Dr. Kumiko Samejima, and Dr. Kata Sarlos for critical feedback on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data files have been deposited in the public repository Zenodo (https://doi.org/10.5281/zenodo.3724099).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001760117/-/DCSupplemental.
References
- 1.Gibcus J. H., et al. , A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohta S., et al. , The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulson J. R., Laemmli U. K., The structure of histone-depleted metaphase chromosomes. Cell 12, 817–828 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Earnshaw W. C., Halligan B., Cooke C. A., Heck M. M., Liu L. F., Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100, 1706–1715 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeshima K., Laemmli U. K., A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell 4, 467–480 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Ono T., et al. , Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115, 109–121 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Zhang T., et al. , Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosome Res. 24, 243–269 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw W. C., Heck M. M., Localization of topoisomerase II in mitotic chromosomes. J. Cell Biol. 100, 1716–1725 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roca J., Wang J. C., DNA transport by a type II DNA topoisomerase: Evidence in favor of a two-gate mechanism. Cell 77, 609–616 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Nitiss J. L., DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer K. N., et al. , Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J. Cell Biol. 136, 775–788 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linka R. M., et al. , C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35, 3810–3822 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen M. O., et al. , Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 157, 31–44 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grue P., et al. , Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J. Biol. Chem. 273, 33660–33666 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Farr C. J., Antoniou-Kourounioti M., Mimmack M. L., Volkov A., Porter A. C., The α isoform of topoisomerase II is required for hypercompaction of mitotic chromosomes in human cells. Nucleic Acids Res. 42, 4414–4426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pommier Y., Leo E., Zhang H., Marchand C., DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roca J., Ishida R., Berger J. M., Andoh T., Wang J. C., Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl. Acad. Sci. U.S.A. 91, 1781–1785 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heck M. M., Earnshaw W. C., Topoisomerase II: A specific marker for cell proliferation. J. Cell Biol. 103, 2569–2581 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T., Sun Y., Ji P., Kopetz S., Zhang W., Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene 34, 4019–4031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortune J. M., Osheroff N., Topoisomerase II as a target for anticancer drugs: When enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 64, 221–253 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Shintomi K., Takahashi T. S., Hirano T., Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol. 17, 1014–1023 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Adachi Y., Luke M., Laemmli U. K., Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell 64, 137–148 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Chang L. K., Schmidt R. E., Johnson E. M. Jr, Alternating metabolic pathways in NGF-deprived sympathetic neurons affect caspase-independent death. J. Cell Biol. 162, 245–256 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez R. E., et al. , Effects of conditional depletion of topoisomerase II on cell cycle progression in mammalian cells. Cell Cycle 10, 3505–3514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson M., Phua H. H., Bennett S. C., Spence J. M., Farr C. J., Studying vertebrate topoisomerase 2 function using a conditional knockdown system in DT40 cells. Nucleic Acids Res. 37, e98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spence J. M., et al. , Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 120, 3952–3964 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samejima K., et al. , Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J. Cell Biol. 199, 755–770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano T., Mitchison T. J., Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 120, 601–612 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson D. F., Morrison C., Ruchaud S., Earnshaw W. C., Reverse genetics of essential genes in tissue-culture cells: ‘dead cells talking’. Trends Cell Biol. 12, 281–287 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Natsume T., Kiyomitsu T., Saga Y., Kanemaki M. T., Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Hudson D. F., Vagnarelli P., Gassmann R., Earnshaw W. C., Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell 5, 323–336 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Rattner J. B., Hendzel M. J., Furbee C. S., Muller M. T., Bazett-Jones D. P., Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 134, 1097–1107 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schueler M. G., Sullivan B. A., Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genomics Hum. Genet. 7, 301–313 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Tavormina P. A., et al. , Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158, 23–29 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bower J. J., et al. , Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene 29, 4787–4799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deming P. B., et al. , The human decatenation checkpoint. Proc. Natl. Acad. Sci. U.S.A. 98, 12044–12049 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deming P. B., Flores K. G., Downes C. S., Paules R. S., Kaufmann W. K., ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J. Biol. Chem. 277, 36832–36838 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Downes C. S., et al. , A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells/. Nature 372, 467–470 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Skoufias D. A., Lacroix F. B., Andreassen P. R., Wilson L., Margolis R. L., Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell 15, 977–990 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Chan K. L., North P. S., Hickson I. D., BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumann C., Körner R., Hofmann K., Nigg E. A., PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Coelho P. A., et al. , Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 6, e207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyoda Y., Yanagida M., Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell 17, 2287–2302 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green L. C., et al. , Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 125, 1591–1604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J. M., Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117, 6435–6445 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Booth D. G., et al. , Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife 3, e01641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takagi M., et al. , Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. J. Cell Sci. 131, jcs212092 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Cuylen S., et al. , Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535, 308–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberge M., Th’ng J., Hamaguchi J., Bradbury E. M., The topoisomerase II inhibitor VM-26 induces marked changes in histone H1 kinase activity, histones H1 and H3 phosphorylation, and chromosome condensation in G2 phase and mitotic BHK cells. J. Cell Biol. 111, 1753–1762 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang K. C., et al. , Topoisomerase II poisoning by ICRF-193. J. Biol. Chem. 276, 44488–44494 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Vagnarelli P., et al. , Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 8, 1133–1142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parry D. H., O’Farrell P. H., The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr. Biol. 11, 671–683 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sackton K. L., et al. , Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baxter J., et al. , Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331, 1328–1332 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Vlijm R., Kim S. H., De Zwart P. L., Dalal Y., Dekker C., The supercoiling state of DNA determines the handedness of both H3 and CENP-A nucleosomes. Nanoscale 9, 1862–1870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennink M. L., et al. , Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 8, 606–610 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Houchmandzadeh B., Marko J. F., Chatenay D., Libchaber A., Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol. 139, 1–12 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson H., Roberge M., Topoisomerase II inhibitors affect entry into mitosis and chromosome condensation in BHK cells. Cell Growth Differ. 7, 83–90 (1996). [PubMed] [Google Scholar]
- 60.Nagasaka K., Hossain M. J., Roberti M. J., Ellenberg J., Hirota T., Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nat. Cell Biol. 18, 692–699 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Piskadlo E., Tavares A., Oliveira R. A., Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. eLife 6, e26120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartl T. A., Sweeney S. J., Knepler P. J., Bosco G., Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 4, e1000228 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herzog S., et al. , Functional dissection of the Drosophila melanogaster condensin subunit Cap-G reveals its exclusive association with condensin I. PLoS Genet. 9, e1003463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L. H., Mayer B., Stemmann O., Nigg E. A., Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 123, 806–813 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Leonard J., et al. , Condensin relocalization from centromeres to chromosome arms promotes Top2 recruitment during anaphase. Cell Rep. 13, 2336–2344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daum J. R., et al. , Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21, 1018–1024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen C. F., Hickson I. D., PICH promotes mitotic chromosome segregation: Identification of a novel role in rDNA disjunction. Cell Cycle 15, 2704–2711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang Z., et al. , Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion cycles. Cell 161, 1124–1137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walther N., et al. , A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J. Cell Biol. 217, 2309–2328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charbin A., Bouchoux C., Uhlmann F., Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res. 42, 340–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kimura K., Hirano T., ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell 90, 625–634 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Kawamura R., et al. , Mitotic chromosomes are constrained by topoisomerase II-sensitive DNA entanglements. J. Cell Biol. 188, 653–663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sen N., et al. , Physical proximity of sister chromatids promotes top2-dependent intertwining. Mol. Cell 64, 134–147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.