Significance
RAS proteins, critical regulators of cell growth and differentiation, are the most frequently mutated oncogenes in humans. RAS functions as dimers/coclusters on cell membranes. We developed an improved split luciferase complementation assay coupled to a powerful genetic system to show that colocalization within the same membrane domain enables formation of RAS dimers/coclusters with itself and other membrane-associated proteins. Membrane association-facilitated interactions (MAFIs) are not sufficient for RBD-mediated Ras inhibition, which additionally requires high-affinity domain-mediated interactions. Notably, we show that MAFI augments the impact of domain-mediated interactions to elicit autophagy/lysosome-mediated elimination of nonfunctional RAS complexes. This broadly applicable strategy enables discovery of low-affinity protein interactions mediated by membrane tethering and analysis of their impact on biologic function.
Keywords: KRAS, NRAS, HRAS, protein–protein interaction, split-luciferase complementation
Abstract
HRAS, NRAS, and KRAS4A/KRAS4B comprise the RAS family of small GTPases that regulate signaling pathways controlling cell proliferation, differentiation, and survival. RAS pathway abnormalities cause developmental disorders and cancers. We found that KRAS4B colocalizes on the cell membrane with other RAS isoforms and a subset of prenylated small GTPase family members using a live-cell quantitative split luciferase complementation assay. RAS protein coclustering is mainly mediated by membrane association-facilitated interactions (MAFIs). Using the RAS–RBD (CRAF RAS binding domain) interaction as a model system, we showed that MAFI alone is not sufficient to induce RBD-mediated RAS inhibition. Surprisingly, we discovered that high-affinity membrane-targeted RAS binding proteins inhibit RAS activity and deplete RAS proteins through an autophagosome–lysosome-mediated degradation pathway. Our results provide a mechanism for regulating RAS activity and protein levels, a more detailed understanding of which should lead to therapeutic strategies for inhibiting and depleting oncogenic RAS proteins.
Cellular signal transduction pathways are executed through a variety of transient and dynamic protein–protein interactions (PPIs). The PPI binding affinities (or dissociation constants) are usually in the micromolar to millimolar range (1). The PPIs in the RAS–MAPK pathway, one of the most mutated oncogenic pathways in human cancer, illustrate the importance of transient and weak protein interactions for network regulation. The small GTPase RAS family consists of four isoforms, KRAS4A/KRAS4B (splice variants), HRAS, and NRAS, which share 90% similarity in the N-terminal catalytic G domain (2). RAS proteins can form homodimer or nanoclusters on the inner surface of the plasma membrane (3, 4). Computational and biophysical studies suggest that the dimerization affinity of KRAS ranges from millimolar to micromolar (5, 6). However, the cellular concentration of RAS proteins is estimated to be 0.033 to ∼0.5 µM (7). This is significantly less than the RAS protein concentration required to spontaneously dimerize. Hence, cells must have a mechanism to bring RAS proteins into proximity to increase their local concentrations and restrict random protein tumbling (8) to enable RAS dimer/nanocluster formation.
One potential mechanism to promote protein proximity and increase the effective molarity of low-abundance proteins involves targeting to, and localization in, the two-dimensional plane of a membrane (9). Importantly, protein prenylation on the RAS C-terminal hypervariable–CAAX motif (HVR–CAAX) is both necessary and sufficient to promote RAS membrane attachment and effector interaction to activate downstream signal transduction (10, 11). More recently, the RAS HVR–CAAX motif was shown to mediate membrane attachment and enable dimerization of an appended fluorescent protein (4). However, it remains to be determined whether the weak intrinsic affinity between independent RAS protomers is sufficient to promote and sustain RAS dimer formation (12). Furthermore, although a dimerization domain (DD) fused to KRAS can boost downstream signaling in the presence of a chemical dimerizer (4), whether RAS functions are regulated by weak interactions between RAS proteins in dimers/nanoclusters in the cell membrane remains a significant gap in our understanding of this critical oncogenic pathway.
RAS proteins also interact transiently and reversibly with a variety of downstream effector proteins that bind with micromolar to nanomolar affinities (13). Among them, the interaction between activated RAS–GTP and the RAS binding domain (RBD) of RAF proteins plays a central role in regulating the ERK–MAPK signaling pathway (14). The crucial amino acids that provide bonding energy to stabilize the interaction in the RAS–RBD complex have been well-defined, and mutations that reduce dimerization affinity and destabilize the RAS–RBD complex have been described (15, 16). Here, we improved a split luciferase-based system to enable detection of the weak and transient protein interactions in the RAS–MAPK pathway when the proteins are expressed at physiologic levels and the interactions evaluated in living cells in real time. We also determine whether the intrinsic dimerization affinity measured by in vitro computational and biophysical methodologies can contribute to RAS dimer/nanocluster formation in vivo. This approach also enabled us to ascertain the extent to which cell membrane localization is required for RAS protein dimerization/clustering in living cells. Finally, we took advantage of the well-studied RAS–RBD interaction as a model system to investigate whether weak intrinsic affinities between RAS–RAS complexes on the cell membrane impact RAS downstream signaling. Our results disclose a previously unanticipated autophagosome–lysosome-mediated mechanism by which cells can eliminate nonproductive complexes formed by interaction of RAS and tight binding RBD or engineered RAS binding proteins. These studies have implications for the generation of a class of pan-RAS antagonists.
Results
Quantitative ReBiL2.0 Assay.
Protein-fragment complementation assays such as split luciferase (1/2luc) complementation (or bimolecular luciferase complementation [BiLC]) (17–20) enable analysis of PPIs in diverse biological processes. However, concerns have been raised about whether the residual binding energy between 1/2luc fragments could cause or contribute to the reconstitution of the 1/2luc fragments without requirement for the fused interacting proteins whose interaction is intended to be analyzed. Given that the 1/2luc fusion construct quantitatively recovers the dissociation constant (Kd) for known protein interactors, the free energy (ΔG) is very low (17). Based upon error analysis of the assay, we can put a lower limit on the binding affinity for the 1/2luc pair ≧ 600 mM (SI Appendix, Supplemental Note). Since the binding affinities of most cellular PPIs typically range from micromolar and nanomolar (1), the extremely low affinity of the 1/2luc fragments is not likely to contribute significantly to self-assembly. We verified this experimentally in control studies using 1/2luc fragments alone as negative controls (see below). Thus, reconstituted luminescent signals must be driven by the fused candidate proteins or other cellular events such as colocalization of candidate proteins where their effective molarity can be increased (9).
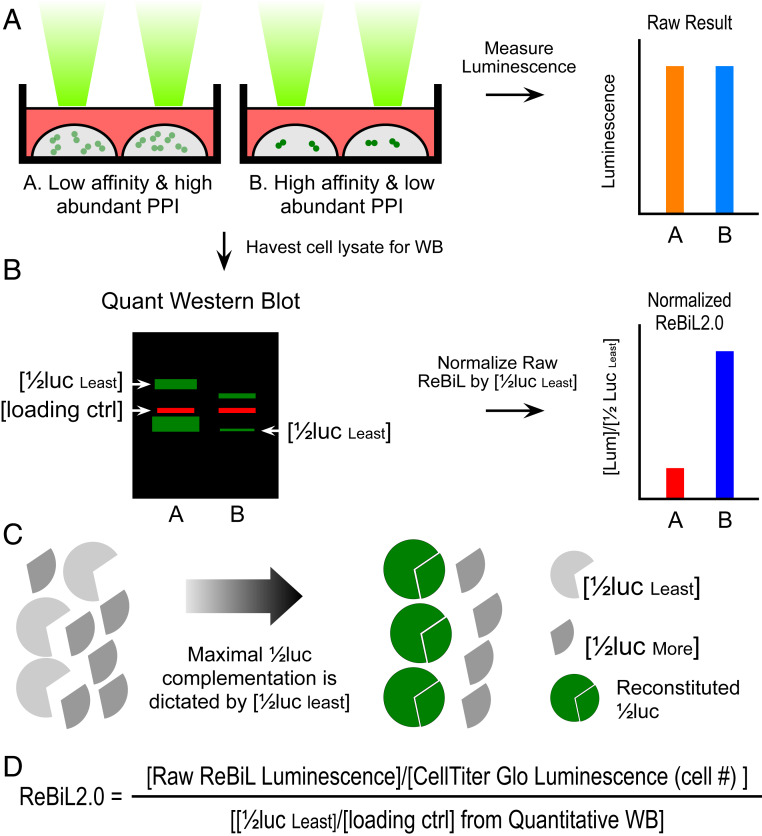
The luminescent signal output in BiLC assays depends not only on the binding affinity of candidate PPI pairs fused to 1/2luc fragments, but also on the levels at which they are expressed. Hence, accurate comparison of BiLC signals between different PPI pairs can be affected by the different levels at which the 1/2luc fusion proteins are expressed (Fig. 1A). We overcame this potential pitfall by normalizing the luminescent signal to the amount of expressed 1/2luc fusion proteins that generate the luminescent signal (Fig.1 B and C). We refer to this improvement in the original ReBiL (recombinase-enhanced BiLC) approach (20) as ReBiL2.0 (Fig. 1D; see Materials and Methods for detailed explanation of ReBiL2.0).
Fig. 1.
Development of ReBiL2.0 assay. (A) The raw luminescent signal reported by the original ReBiL assay does not necessarily reflect the affinity of a pair of PPIs since ReBiL signals depend on both the binding affinity of the interacting proteins and the levels at which they are expressed. That is, signals generated by low-affinity, high-abundance protein interaction (represented by light green dot pairs) may generate luminescent signals similar to those resulting from high-affinity and low-abundance protein interactions (represented by dark green dot pairs). (B) ReBiL2.0 corrects this potential pitfall by normalizing the raw luminescent signal to the expressed level of each 1/2luc fusion protein. The amount of each 1/2luc fusion protein and loading control (actin) were quantified by probing Western blots with anti-HA and anti-actin antibodies simultaneously using the LI-COR Odyssey two-channel system (Left). (C) Although controlled by the same TRE (tetracycline response element) bidirectional promoter, 1/2luc fusion proteins are not always expressed in a 1:1 stoichiometric ratio. Thus, the maximal luminescent output generated by reconstituted split-luciferase is determined by the least abundant 1/2luc fusion [1/2luc least] when the 1/2luc fusions are not expressed in 1:1 stoichiometry. Since each 1/2luc fusion protein contains a single copy of an HA-epitope tag in the linker region (Fig. 2A), quantitative Western blotting with an anti-HA antibody enables accurate determination of their relative expression levels as shown in B. (D) ReBiL 2.0 formula. The raw ReBiL signal [Raw ReBiL Luminescence] is measured by a luminometer. Then, the relative viable cell number in each sample is measured using CellTiter Glo [CellTiter Glo Luminescence (Cell #)] upon termination of the ReBiL2.0 assay. The [1/2lucleast] and [loading ctrl] were determined by the corresponding band intensities in Western blots using the LI-COR Odyssey system (SI Appendix, Fig. S2I and Table S3).
We employed the ReBiL2.0 assay to study interactions between RAS and related proteins in living cells. Since RAS is a membrane-bound protein, the cell membrane cannot be omitted when studying its interactions with other cellular proteins. Unlike most in vitro biochemical and biophysical studies, the cell-based ReBiL2.0 assay enables quantitative studies of protein interactions between RAS and other cellular proteins in a physiological environment where the RAS activity can also be measured. Importantly, ReBiL2.0 offers a platform for studying such complex interactions in real time, in living cells.
Membrane Association Facilitates Colocalization of KRAS with NRAS, HRAS, and Other Prenylated Proteins.
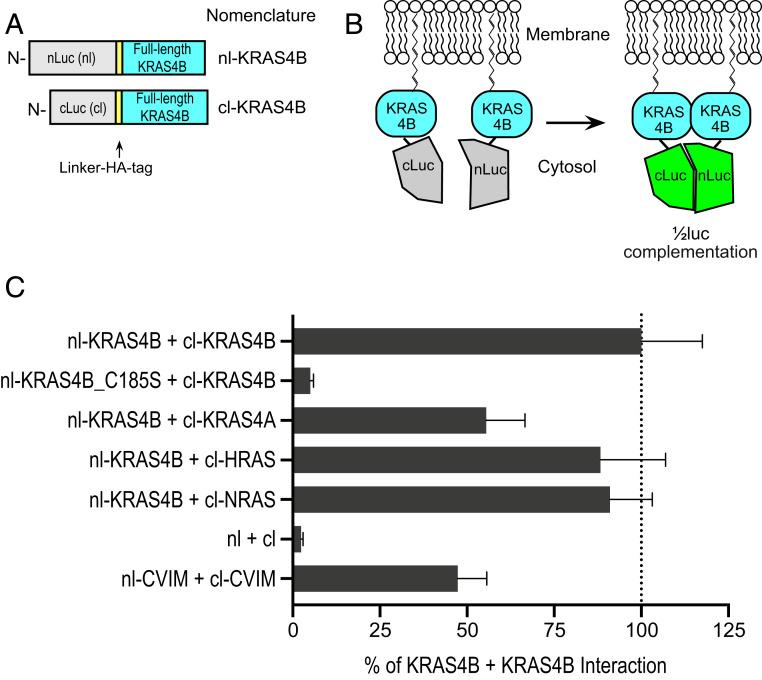
KRAS4B can form homodimer or nanoclusters on cell membranes (3, 4). We first validated the ReBiL2.0 approach for the RAS system by assessing its ability to capture KRAS4B dimers resulting from interaction of N-terminal split luciferase (nl) and C-terminal split luciferase (cl) appended to the N terminus of KRAS4B (designated nl–KRAS4B and cl–KRAS4B, respectively; Fig. 2 A and B). This fusion configuration preserves the intact C-terminal HVR–CAAX motif required for RAS prenylation and retains KRAS4B activity as determined by NIH/3T3 mouse fibroblast growth transformation assays (SI Appendix, Fig. S1A). When expressed at physiologically relevant levels (SI Appendix, Fig. S1B), nl–KRAS4B and cl–KRAS4B generated robust 1/2luc complementation signals (Fig. 2C and SI Appendix, Fig. S2A). Since functional reconstitution of 1/2luc fragments requires a minimum 1:1 ratio of nl–KRAS4B and cl–KRAS4B (Fig. 1C), we interpret these results to indicate that KRAS4B minimally forms homodimers, consistent with prior observations made using an orthogonal approach (4). However, formation of higher-order KRAS coclusters cannot be excluded (3).
Fig. 2.
Validation of ReBiL2.0 for analyzing protein interactions with RAS isoforms. (A) The diagram depicts the configuration and nomenclature of a subset of the split-luciferase (1/2luc) KRAS4B fusion proteins analyzed. The N-terminal 1/2luc (nl) or C-terminal 1/2luc (cl) followed by a linker and HA-epitope tag were fused to the N terminus of KRAS to preserve the integrity and function of the C-terminal HVR–CAAX domain. (B) Membrane association facilitates interaction of the nl–KRAS4B and cl–KRAS4B that reconstitutes functional luciferase to generate luminescent signals. (C) ReBiL2.0 assay shows the quantitative homo/heteromer formations between KRAS4B and all four isoforms of RAS family members (KRAS4B, KRAS4A, HRAS, and NRAS). The bar chart reports quantitative ReBiL2.0 scores normalized to the KRAS4B + KRAS4B interaction (set to 100%). The 1/2luc alone lacking fusion partners (nl + cl) served as the negative control showing the background reading for the ReBiL2.0 assay applied to RAS. CVIM represents the last 20 amino acids from KRAS (KMSKDGKKKKKKSKTKCVIM). Data show mean ± SEM (n = 4).
Previous studies showed that a single Cys-to-Ser mutation (KRAS4B_C185S) in the CAAX farnesylation motif abolishes RAS protein membrane association and inactivates cellular transformation activity (21, 22). Consistent with this, a single cysteine mutation in nl–KRAS4B CAAX motif (nl–KRAS4B_C185S) prevented 1/2luc complementation (Fig. 2C and SI Appendix, Fig. S2B). Western blotting analysis showed that the absence of luminescent signal was not due to lack of nl–KRAS4B_C185S protein expression (SI Appendix, Fig. S2I). This result confirms the importance of membrane association for KRAS4B dimer/cocluster formation on cell membranes in living cells. We infer that the weak intrinsic affinity of KRAS4B proteins measured by in vitro assays (5, 6) is insufficient for 1/2luc complementation and, by extension, dimer/cocluster formation. Importantly, the 1/2luc fragments (nl and cl) alone failed to generate a luminescent signal above background, confirming their extremely weak intrinsic affinity (∼600 mM) (Fig. 2C and SI Appendix, Fig. S2F and Supplemental Note). However, appending each 1/2luc fragment to the last 20 amino acids from KRAS that determine KRAS4B membrane tethering (designated as nl–CVIM and cl–CVIM), generated robust luminescent signals (Fig. 2C and SI Appendix, Fig. S2G). Similar results were observed by photoactivated localization microscopy analysis of fluorescent protein mCherry fused with the HVR and CAAX domains of KRAS4B (4). Together, these results demonstrate that the very weak intrinsic affinity between KRAS4B monomers measured by in vitro biochemical and biophysical assays is insufficient to promote RAS dimer/cocluster formation in vivo. Rather, we interpret our studies to indicate that, under physiologic conditions, KRAS4B protomers form closely colocalized protein complexes driven by membrane association. We suggest that this two-dimensional protein concentration facilitated by membrane localization provides sufficient energy to enable detectable reconstitution of enzymatic luciferase reaction. We thus define such proximity-dependent and intrinsic affinity-independent protein interactions as membrane association-facilitated interactions (or MAFIs).
RAS isoforms (KRAS4A/KRAS4B, HRAS, and NRAS) share 90% sequence identity in their N-terminal G domains, but the C-terminal membrane targeting hypervariable regions (HVRs) are highly divergent (8% sequence identity) (2). We asked whether these different HVRs can direct RAS isoforms to the proximal membrane domains where MAFI-dependent 1/2luc complementation can occur (refer to SI Appendix, Fig. S2 for analysis details and raw data). Surprisingly, all three cl–KRAS4A, cl–HRAS, and cl–NRAS proteins efficiently reconstituted 1/2luc activity when paired with nl–KRAS4B (Fig. 2C and SI Appendix, Fig. S2 C–E). Additionally, we fused the C-terminal HVR–CAAX motif from each RAS isoform to nl–TagBFP fusion protein to determine whether the HVR–CAAX alone is sufficient to facilitate their interaction. Indeed, each pair tested generated robust 1/2luc complementation (SI Appendix, Fig. S3A). The slightly lower 1/2luc complementation signals from nl–TagBFP–HVR–CAAX may result from a less favorable protein configuration of the nl–TagBFP interaction partner for 1/2luc complementation. Alternatively, the higher 1/2luc complementation signals from the full-length RAS isoforms may reflect a contribution of the weak binding affinity between RAS G domains once they are tethered and concentrated in the plasma membrane. These results indicate that different RAS isoforms localize sufficiently closely on membrane domains in cells to enable MAFI to occur. It is important to note that the induced expression levels of the RAS proteins in the ReBiL system were adjusted to be as close to endogenous as possible (e.g., SI Appendix, Fig. S1B). Although our results measured under physiologically relevant conditions in living cells might appear inconsistent with earlier reports that RAS isoforms reside on distinct membrane microdomains (3, 23, 24), a more recent analysis showed that KRAS and HRAS can cocluster through lipid-mediated spatial cross talk (25). Thus, another interpretation is that the different RAS proteins can occupy both distinct and common membrane localization domains in vivo, and that their proximity offers opportunities for MAFI.
A significant subset of small GTPases of the RAS and RHO (RAS homologous) families terminate with CAAX farnesylation or geranylgeranylation signal motifs that are essential for promoting membrane association and subcellular localization (26). Based on the results presented above, we determined whether KRAS4B might interact with other prenylated proteins via MAFI. ReBiL2.0 readily detected interactions between KRAS4B and the farnesylated and plasma membrane-associated DIRAS3, a genetically defined KRAS suppressor (27) (SI Appendix, Fig. S3B), as well as two plasma membrane-associated RHO family small GTPases, geranylgeranyl-modified RAC1, and RAC2 (SI Appendix, Fig. S3C). All interactions required prenylation since a cysteine-to-serine mutation in the CAAX motif abolished 1/2luc complementation (SI Appendix, Fig. S3). This is consistent with these interactions occurring via MAFI-dependent protein complexes. Interestingly, the hot-spot cancer-associated driver point mutations KRAS4B_G12D and RAC1_P29S (28) showed higher 1/2luc complementation scores with RAC1 and KRAS4B, respectively, suggesting they might cocluster with higher probability, although the underlying mechanisms remain unclear (SI Appendix, Fig. S3C). Of significance, KRAS4B did not exhibit efficient interaction with the RAS family small GTPase RHEB (SI Appendix, Fig. S3B), a farnesylated protein that localizes to endomembranes (29). Furthermore, KRAS4B did not colocalize significantly with the farnesylated RHO small GTPase RND3 (SI Appendix, Fig. S3B), which also localizes to the plasma membrane (29). We infer that either KRAS4B and RND3 reside in nonoverlapping membrane compartments, or that the strength of their lipid interactions with membranes is not sufficient for MAFI. Taken together, our results demonstrate that KRAS4B resides in close proximity to itself, KRAS4A, HRAS, NRAS, and a subset of prenylated proteins on the plasma membrane where they are sufficiently close to allow MAFI-dependent 1/2luc complementation.
Using RAS–RBD Interactions to Model PPI and MAFI.
Our observations raise a critical question: does MAFI-dependent colocalization of a protein complex such as RAS–RAS contribute to RAS regulation or biological functions in vivo, or is their ability to interact just a reflection of the sensitivity of the ReBiL2.0 assay? To address this important question, we took advantage of the well-defined RAS–RBD (RAS binding domain of CRAF) interaction (16) as a model system to investigate whether conventional PPI and MAFI-driven protein complexes can affect RAS pathway regulation, downstream signaling, and phenotypic outcome.
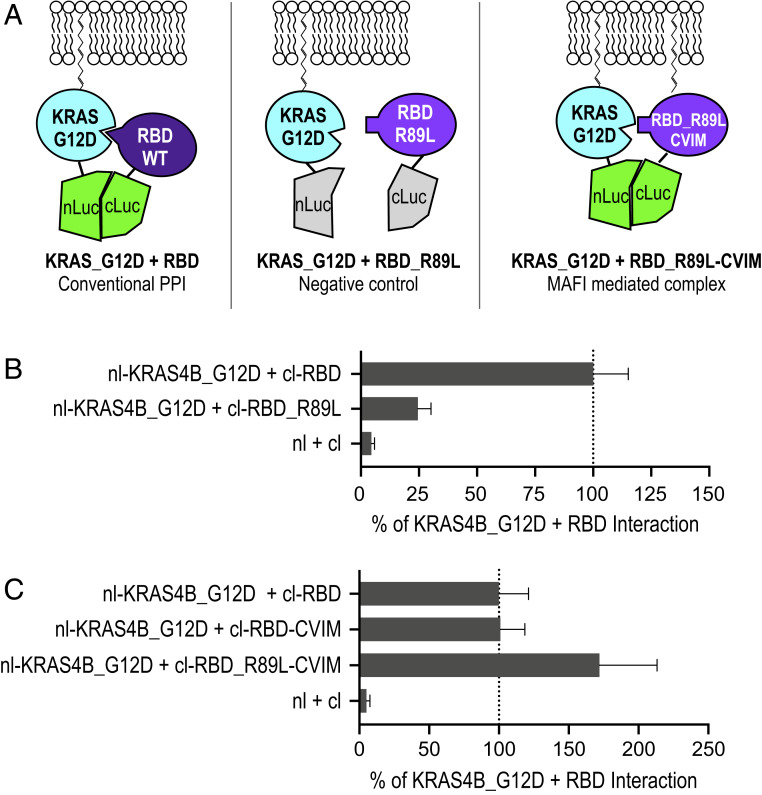
We used the wild-type RAS–RBD interaction to model the effects of a conventional domain-based PPI (Fig. 3 A, Left). The wild-type RBD (amino acids 51 to 131 of CRAF) binds tightly (Kd, 0.13 µM in physiological ionic strength) to RAS–GTP (16), whereas the arginine-89-to-leucine mutation (RBD_R89L) decreases the binding affinity to RAS–GTP to the micromolar range (15, 16), which is similar to the affinity range of the RAS–RAS interaction (5, 6). If MAFI can contribute significantly to RAS–RBD interactions, we predict that the HVR–CAAX domain from KRAS appended to the C terminus of cl–RBD_R89L (designated cl–RBD_R89L–CVIM) should interact with RAS–GTP (Fig. 3 A, Right). We first confirmed the PPI between KRAS4B and RBD using a ReBiL2.0 reporter cell line. Consistent with the literature, we observed robust 1/2luc complementation of nl–KRAS4B_G12D (representing RAS–GTP) and cl–RBD (cl fused to the amino acids 51 to 220 of CRAF) (30) (Fig. 3B). The negative control cl–RBD_R89L showed significantly reduced 1/2luc complementation when paired with nl–KRAS4B_G12D (Fig. 3B), confirming the loss of intrinsic dimerization ability. By contrast, the cl–RBD_R89L–CVIM efficiently restored 1/2luc complementation with nl–KRAS4B_G12D (Fig. 3C). Thus, the interaction between KRAS4B_G12D and cl–RBD_R89L–CVIM fulfills the prediction that they would interact via MAFI. We suggest that the higher luminescent signal generated by this pair is due to decreased rigidity of the RAS–RBD complex in which lower intrinsic affinity due to the R89L mutation enables more freedom to better accommodate 1/2luc fragments in this pair.
Fig. 3.
Model PPI and MAFI by RAS–RBD interaction. (A) Diagrams depict the experimental design. The interaction of nl–KRAS4B_G12D and wild-type cl–RBD represents the conventional PPI. cl–RBD_R89L mutant serves as a negative control. The weak intrinsic affinity between nl–KRAS4B_G12D and membrane-associated cl–RBD_R89L–CVIM should mimic membrane association-facilitated interaction (MAFI). (B) ReBiL2.0 assay shows that KRAS4B_G12D interacts with RBD, but not RBD_R89L. The bar chart reports quantitative ReBiL2.0 scores normalized to the KRAS4B_G12D + RBD interaction (set to 100%). The 1/2luc alone lacking fusion partners (nl + cl) served as the negative control. Data show mean ± SEM (n = 4). (C) ReBiL2.0 assay shows that RBD_R89L–CVIM restores interaction with KRAS4B_G12D. The bar chart reports quantitative ReBiL2.0 scores normalized to the KRAS4B_G12D + RBD interaction (set to 100%). The 1/2luc alone lacking fusion partners (nl + cl) served as the negative control. Data show mean ± SEM (n = 4).
RAS Inhibition Requires Conventional RAS–RBD Interaction.
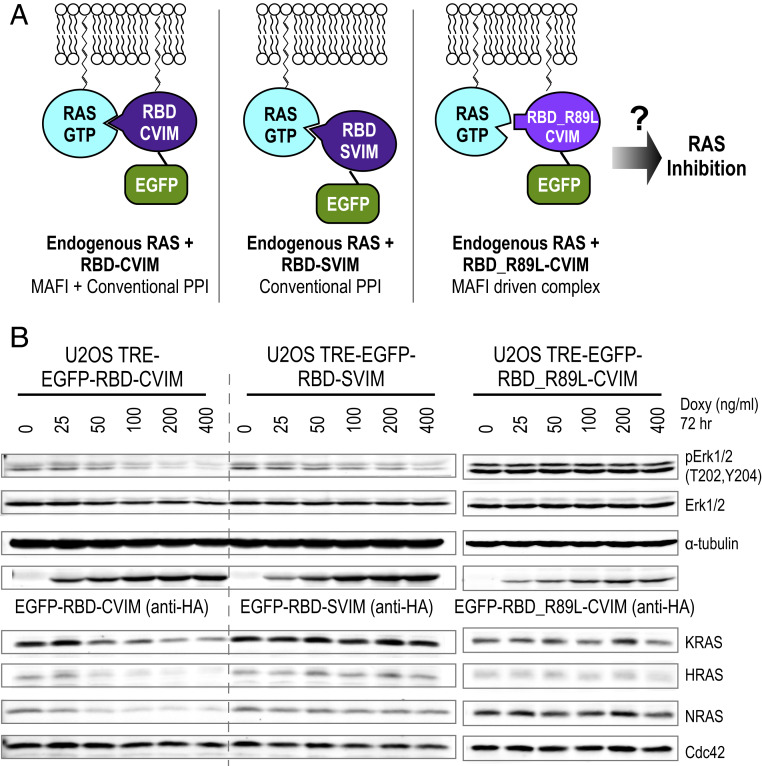
We next asked whether the weak affinity between MAFI-mediated RBD_R89L–CVIM and the RAS complex is sufficient to reduce RAS function. We used ERK phosphorylation (pERK) as a phenotypic readout for RAS inhibition since overexpressed RBD can act as a dominant-negative mutant to inhibit RAS–RAF-mediated ERK phosphorylation (30, 31). To test whether the RBD_R89L–CVIM inhibits RAS, we engineered doxycycline-inducible U2OS cell lines that stably encode the wild-type RBD and RBD_R89L transgenes. As the RBD is a small protein (∼19.3 kDa), we appended an EGFP (∼27 kDa) on its N terminus to increase its size to enable it to be distinguished from endogenous RAS proteins (∼21 kDa) using Western blotting. We generated a cysteine-to-serine mutation (RBD–SVIM) to prevent protein prenylation and membrane localization to enable assessment of the effects of nonmembrane-associated RBD. Thus, RBD–SVIM and RBD_R89L–CVIM differ by only two amino acids (R89L and C185S) that both prevent domain-based interaction and MAFI-enabled proximity localization (Fig. 4A). We used RMCE (recombinase-mediated cassette exchange) (20, 32) to integrate a single-copy RBD transgene to the preselected chromosomal locus where transgene expression is tightly controlled by a doxycycline-inducible promoter. This allows transcription of the RBD transgene to be minimized during cell line engineering and expansion, which is important since RBD expression inhibits pERK formation and impedes cell growth. Consistent with the literature, the positive control, EGFP–RBD–CVIM and EGFP–RBD–SVIM caused a doxycycline dose-dependent reduction of endogenous pERK (Fig. 4B), indicating the endogenous RAS activity was inhibited by both RBD–CVIM and RBD–SVIM. Importantly, RBD_R89L–CVIM did not inhibit RAS activity (Fig. 4 B, Right) despite its robust 1/2luc complementation with KRAS_G12D (Fig. 3C). These results clearly demonstrate that the MAFI-mediated low-affinity interaction between RBD_R89L and RAS is not sufficient to inhibit RAS activity. Rather, high-affinity classic PPI between RAS and RBD is required to inhibit ERK phosphorylation. However, as shown below, combining classic PPI and MAFI engenders an unpredicted mode of RAS protein regulation.
Fig. 4.
Conventional PPI causes RAS inhibition, while conventional PPI combined with membrane association engenders both RAS inhibition and depletion. (A) Diagram depicts the experimental design. The interaction between wild-type RBD and endogenous RAS–GTP represents an interaction mediated by conventional PPI. By contrast, RBD_R89L–CVIM forms a MAFI-driven complex with endogenous RAS proteins. (B) Western blot analyses show that expression of EGFP–RBD–CVIM inhibits ERK phosphorylation and depletes RAS isoforms including KRAS, HRAS, and NRAS in a doxycycline dose-dependent manner. CDC42 that does not bind RBD has no effect on protein level. EGFP–RBD–SVIM interacts with RAS via conventional PPI, and inhibits ERK phosphorylation, but has no effect on pan-RAS protein levels. EGFP–RBD_R89L–CVIM does not interact with RAS via conventional PPI and, while it does localize to the plasma membrane, does not inhibit RAS activity measured ERK phosphorylation.
A Mechanism for RAS Protein Depletion Mediated by Membrane-Targeted High-Affinity RAS Binders via a Lysosome-Mediated Process.
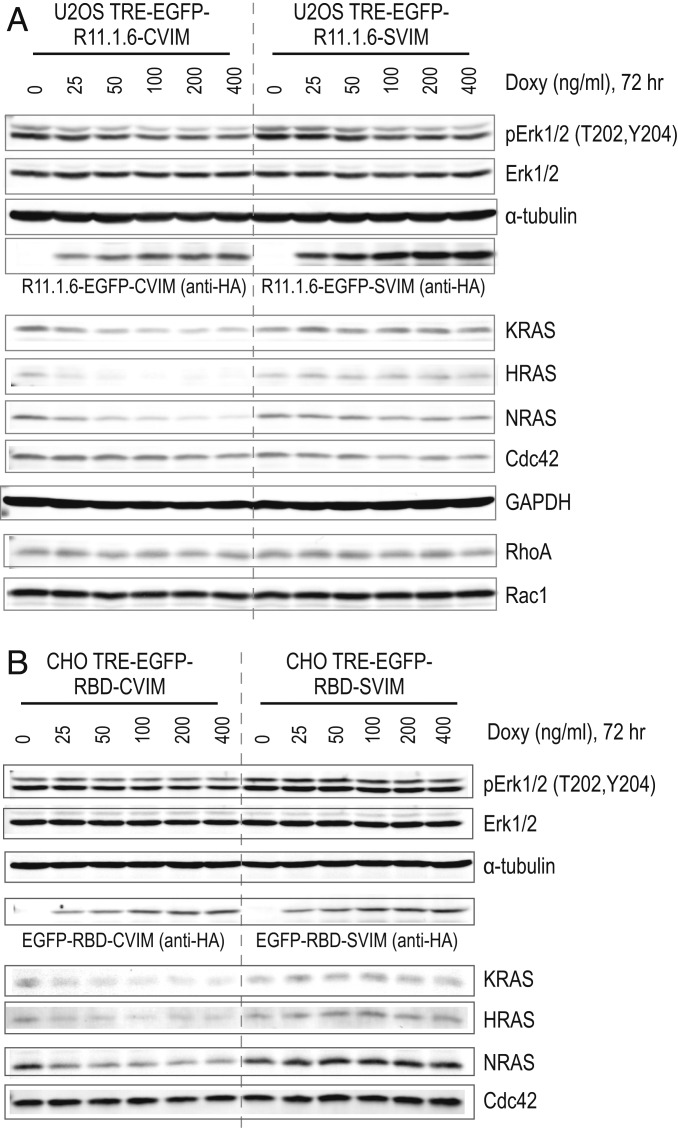
Although RBD_R89L–CVIM failed to inhibit RAS activity, the membrane-targeted RBD–CVIM (a combination of conventional PPI and MAFI; Fig. 4A) exhibited stronger inhibitory effects in both ERK phosphorylation (Fig. 4B) and cell growth than RBD–SVIM (SI Appendix, Fig. S4). A similar phenotype was reported previously using a membrane-associated intrabody (iDab#6-memb) that was shown to exhibit superior RAS inhibition activity (33). Therefore, we explored the difference between RBD–CVIM and RBD–SVIM in greater detail. We were surprised to find that expression of RBD–CVIM not only inhibited RAS activity but also substantially reduced the abundance of endogenous HRAS, KRAS, and NRAS proteins (Fig. 4B). The membrane-associated RHO-family small GTPases (34) such as CDC42 (Figs. 4B and 5 A and B), RHOA, and RAC1 (Fig. 5A) were not decreased since they do not bind to RBD, indicating that the effect does not generally affect other membrane-associated proteins. Additionally, doxycycline-induced expression of EGFP–RBD–CVIM in Chinese hamster ovary (CHO) cells also caused similar depletion of RAS proteins (Fig. 5B), indicating that this is a general phenomenon that can occur in both human and rodent cells.
Fig. 5.
Membrane-targeted tight RAS binders deplete pan-RAS proteins. (A) Western blot shows that expression of EGFP–R11.1.6–CVIM in U2OS cells inhibits ERK phosphorylation and depletes KRAS, HRAS, and NRAS proteins. Expression of EGFP–R11.1.6–SVIM inhibits ERK phosphorylation but has no effect on RAS protein level. The RHO-family small GTPase CDC42, RHOA, and RAC1 serve as the negative control. α-Tubulin and GAPDH serve as the loading control. (B) Western blot shows that expression of EGFP–RBD–CVIM in CHO cells inhibits ERK phosphorylation and depletes KRAS, HRAS, and NRAS proteins. Expression of EGFP–RBD–SVIM does not do so.
The ability of RBD–CVIM, but not RBD_R89L–CVIM, to cause RAS depletion suggests that high-affinity RAS binding proteins (e.g., RBD–CVIM) might be required to trigger RAS protein depletion in the plasma membrane, whereas weaker binders (e.g., RBD_R89L–CVIM) lack such potential. If this hypothesis is correct, then other high-affinity RAS binding proteins such as R11.1.6, an engineered RAS binding protein (35) based on the small protein Sso7D (∼7 kDa, 63 amino acids) scaffold (36) (in vitro KRAS Kd = 2.2 ∼50.3 nM) (35) should also inhibit and deplete RAS proteins. We used the RMCE strategy to stably integrate either EGFP–R11.1.6–CVIM or EGFP–R11.1.6–SVIM transgenes into the U2OS genome. Consistent with the results presented above, doxycycline-induced expression of either transgene inhibited RAS activity as demonstrated by decreased ERK phosphorylation (Fig. 5A). Notably, only the membrane-associated EGFP–R11.1.6–CVIM depleted K, N, and H-RAS proteins (Fig. 5A). Our hypothesis also predicts that a weak RAS binder should neither inhibit nor deplete RAS proteins. As expected, the RAS association domain (37) (RA) (amino acids 202–361) from RASSF5, a weaker RAS binder (Kd, 0.2 ∼0.4 µM) (13) neither inhibited RAS activity nor decreased RAS protein levels (SI Appendix, Fig. S5). Taken together, these results support our notion that high-affinity RAS binding proteins can significantly impact RAS activity and protein homeostasis when targeted to the proximity of RAS in the cell membrane in living cells. We attribute the RAS inhibition by RBD and R11.1.6 to a dominant-negative mechanism that competes with the binding of RAS with downstream effector RAF proteins.
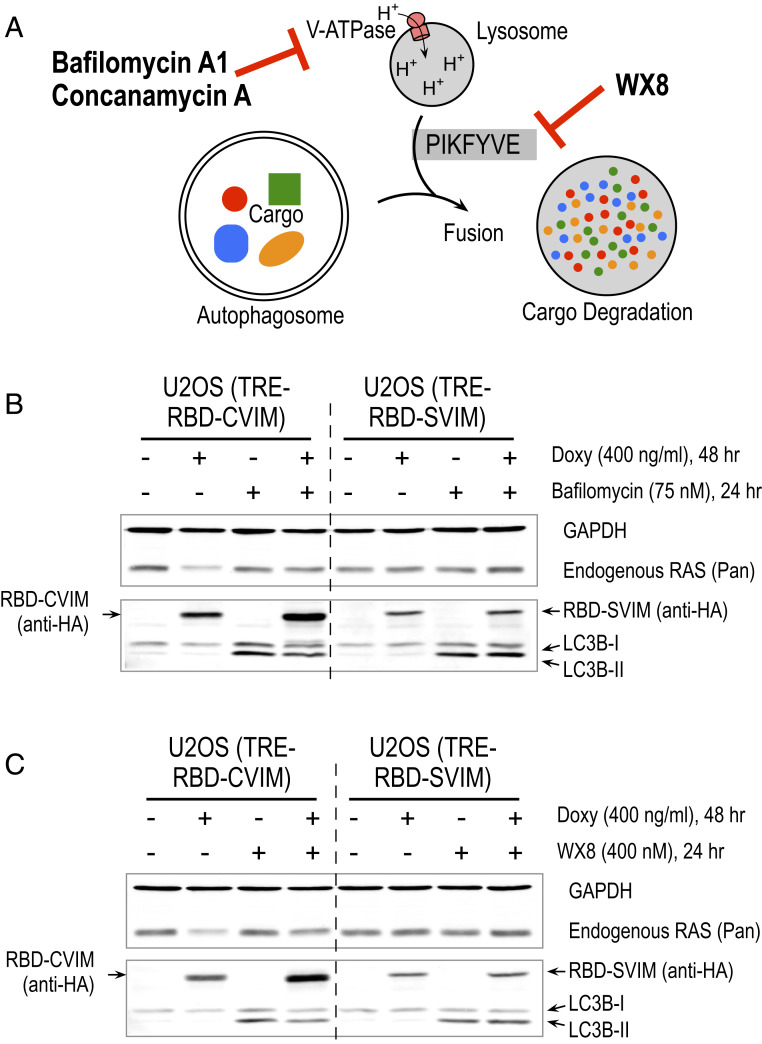
The above observations led us to investigate the mechanism(s) responsible for RAS protein depletion by membrane-targeted high-affinity RAS binding proteins. Analyses of RAS mRNA levels by quantitative RT-PCR showed that the mRNAs from the three RAS isoforms were not changed when EGFP–RBD–CVIM and EGFP–RBD–SVIM were induced by doxycycline (SI Appendix, Fig. S6 A and B), indicating that decreased RAS protein levels were not due to changes in RAS mRNA expression. We then focused on mechanisms of cellular protein homeostasis such as proteasomal or lysosomal protein degradation. Attempts to block RAS protein depletion using proteasome inhibitors (e.g., MG132) were unsuccessful because such treatment induced cell death before RAS protein depletion occurred in U2OS reporter cell lines. Inhibition of the NEDD8-activating enzyme with the antagonist MLN4924 failed to prevent RAS protein depletion by membrane-targeted RBD–CVIM (SI Appendix, Fig. S7). We next explored whether disruption of an autophagosome–lysosome-mediated protein degradation pathway can prevent RBD–CVIM-induced RAS protein depletion. We used three inhibitors to either block the V-ATPase proton pump or the PIKFYVE enzymatic function to test this idea (Fig. 6A). The V-ATPase proton pump inhibitors such as bafilomycin A1 and concanamycin A can prevent autophagosome and lysosome fusion and disrupt lysosomal degradation (38). Treating cells with both bafilomycin A1 and concanamycin A blocked RBD–CVIM-induced RAS protein depletion (Fig. 6B and SI Appendix, Fig. S8). Unlike the V-ATPase proton pump inhibitor, the newly identified small-molecule inhibitor WX8 impairs lysosome function by preventing fusion between autophagosomes and lysosomes by inhibiting PIKFYVE enzymatic function (39). WX8 treatment also blocked RBD–CVIM-induced RAS protein depletion (Fig. 6C). These results clearly implicate an autophagosome–lysosome-mediated degradation pathway in RBD–CVIM-mediated RAS protein depletion.
Fig. 6.
The membrane-targeted RBD-induced pan-RAS depletion is mediated by an autophagosome–lysosome pathway. (A) Diagram depicts the targets of bafilomycin A1, concanamycin A, and WX8 in the autophagosome–lysosome pathway. (B and C) Western blot shows that RBD–CVIM-dependent RAS protein depletion was blocked by bafilomycin A1 (B) and WX8 (C) treatment. Cells were treated by doxycycline to induce RBD–CVIM or RBD–SVIM for 24 h followed by bafilomycin A1 or WX8 treatment for another 24 h. GAPDH serves as a loading control and LC3B-II, which increases in response to bafilomycin A1 treatment (38).
Discussion
We used a live-cell, real-time strategy and found that KRAS4B residence in the cell membrane occurs in close proximity to other RAS isoforms. This enables RAS isoforms to interact, and KRAS4B to interact with a subset of other membrane proteins via MAFIs. Our results are consistent with previous image-based studies showing that KRAS forms dimers or nanoclusters on the cell membrane (3, 4). However, the inability of KRAS4B_C185S (Fig. 2C and SI Appendix, Fig. S2B) to form dimers reveals that the weak intrinsic affinity between RAS proteins is not sufficient for dimer formation in the absence of membrane tethering. This is reminiscent of a recent biophysical study showing that KRAS4B lacks intrinsic dimerization capability and remains monomeric even on supported lipid bilayers (12). We thus define such weak protein interactions between protomers in close association in a membrane as MAFI. Importantly, the ReBiL2.0 assay is able to detect both conventional PPIs (17, 20) and MAFI-mediated protein complexes, and can distinguish between them by comparing isogenic cell lines encoding appropriate mutations in classic protein interaction domains, or in the membrane targeting domain. Without using such genetic approaches, and expression at physiologic levels, it is difficult to distinguish MAFI from conventional dimerization domain-mediated interactions. This limitation is likely shared by similar proximity-based assays such as proximity ligation (40) and fluorescence resonance energy transfer (41).
Using the RAS–RBD interaction as a model system, we showed that MAFI efficiently enables colocalization of RAS and RBD_R89L–CVIM. However, no inhibitory effect was observed (Fig. 4B) due to the apparent negligible intrinsic dimerization ability of these two proteins. Similarly, RAF protein dimerization is required to activate the downstream kinase cascade. However, dimer interface mutations such as arginine 481 (FRAF1_DROME equivalent to arginine 509 in BRAF_HUMAN) and arginine 401 (CRAF_HUMAN) failed to efficiently activate downstream kinase when they were artificially forced to form “dimers” with wild-type RAF protein (42, 43). These results provide evidence to support our notion that colocalization of proteins without intrinsic interaction affinity does not necessarily lead to a functional outcome. It is worth noting that our results do not exclude the possibility that RAS protein clusters on membranes could attract RAF proteins to localize at sufficient concentrations and also restrict random tumbling of RAF proteins (8) through RAS–RBD interactions to promote RAF dimerization (44). Several genetic studies were interpreted as evidence that wild-type RAS can function as a tumor suppressor to inhibit oncogenic RAS mutants (45–47). One model to explain the inhibitory effect of wild-type RAS is that it forms a nonproductive dimer with mutant RAS through direct interactions. However, the binding affinities of both RAS–RAS and RAS–RBD_R89L are in the millimolar range. As we found that RBD_R89L–CVIM does not detectably inhibit RAS activity, we infer that such weak intrinsic affinity between wild-type RAS and mutant RAS might be inadequate to sustain an inhibitory effect. If direct interaction alone were sufficient, then one would expect to observe inhibitory effects between the different RAS isoforms since our data imply that KRAS4B interacts with HRAS and NRAS in the ReBiL assay (Fig. 2C). However, the inhibitory effects of wild-type RAS are only seen in the context of mutation of the cognate RAS isoform (48). Taken together, the data indicate that wild-type RAS inhibition of mutant RAS through direct dimerization likely represents an oversimplification of what is occurring in the context of the required membrane association needed for RAS function. Although many possible models have been proposed and discussed (49), precisely whether and how wild-type RAS functions as a tumor suppressor have not been definitively established.
It is puzzling that the RAS binding small protein R11.1.6, which has nanomolar affinity, failed to inhibit RAS activity in a panel of human cancer cell lines harboring wild-type KRAS and mutant KRAS including G12D, G12C, and G12V oncogenic mutations (50). We reason that this could be partly due to technical issues related to how these cell lines were generated or because the R11.1.6 protein was introduced by lentiviral infection. Establishing cell lines to stably express RBD or other inhibitory RAS binding proteins is impractical due to their strong negative effect on the RAS/MAPK pathway activity needed for growth of many commonly used cell lines. Applying strong drug selection to these cells often enriches for cells escaping expression of transgene-encoded inhibitory proteins. We overcame this limitation by inserting the inhibitory transgene into a single preselected chromosomal locus by RMCE and then preventing transcription to enable cell expansion. Importantly, the doxycycline-inducible system employed encodes both the transcriptional repressor TetR-KRAB and the activator rtTA2S-M2. This enables suppression of transcription in the absence of doxycycline, thereby reducing transcriptional leakage to undetectable levels. The combination of TetR-KRAB and rtTA2S-M2 achieved 6,000- to ∼70,000-fold increases in luciferase activity (32). The extremely low background in this dual system enables very high fold inducibility and signal-to-noise ratios. This is much higher than most commercially available doxycycline-inducible cell lines, likely due to the high leakage background in systems only encoding the transactivator rtTA. We infer that the extremely low background expression in our system greatly facilitated the establishment of cell lines encoding growth-inhibitory transgenes. The robustness of this system enabled us to analyze in depth the functional consequences of expressing growth-inhibitory proteins in a physiologically relevant cellular environment.
We were surprised to find that membrane-targeted strong RAS binding proteins deplete K, N, and H-RAS proteins. We envision that such strong binders act as decoys that mimic strong RAF binding without activation of downstream signaling pathways due to their lack of a kinase domain. We discovered that cells thus eliminate such nonproductive protein complexes through an autophagosome–lysosome pathway (Fig. 6). These results may have clinical relevance if KRAS_G12C cancer cells handle the adduct of KRAS_G12C and its covalent inhibitors (51–53) through a similar autophagosome–lysosome-dependent pathway for eliminating such nonproductive adducts. Further investigation is needed to uncover the underlying molecular mechanism(s) of this type of quality control system able to distinguish between active versus nonproductive RAS–RBD protein complexes and subsequently activate an autophagosome–lysosome degradation pathway. Understanding these mechanisms may help to identify therapeutic avenues to promote targeted depletion of RAS proteins. Such improvements could greatly benefit the large number of patients with pancreatic and other cancers that depend on oncogenic activation of RAS proteins.
Materials and Methods
Construction of ReBiL and RMCE Targeting Plasmids and Cell Lines.
The methods for constructing ReBiL and RMCE plasmids and cell lines have been previously described (20, 32). ReBiL cell lines were maintained in DMEM (Corning; 10-013-CV) with 10% (vol/vol) FBS, 10 µg/mL ciprofloxacin (Corning; 61-277-RG), 200 to ∼400 µg/mL G418 (Corning; 61-234-RG), 1 ng/mL doxycycline (Sigma; D9891), and 4 µg/mL blasticidin (Thermo Fisher; R21001). All ReBiL and RMCE targeting plasmids and cell lines used in this report are listed in SI Appendix, Tables S1 and S2.
Antibodies.
Antibodies used here are anti-RAS (Pan, mouse monoclonal; Millipore 05-516; lot #3072322), anti-KRAS4B (mouse monoclonal; Sigma; WH0003845M1), anti-HRAS (rabbit polyclonal; Proteintech; 18295-1-AP; lot #00022367), anti-NRAS (mouse monoclonal; Santa Cruz; sc-31; lot #L1115), anti-CDC42 (rabbit monoclonal; Cell Signaling; #2466), anti-RHOA (rabbit monoclonal, Cell Signaling; #2117), anti-RAC1 (mouse monoclonal; Millipore; 05-389; lot #2727207), anti-β-tubulin (rabbit polyclonal; LI-COR; P/N 926-42211), anti-HA-tag (rabbit monoclonal; Cell Signaling; #3724), anti-β-actin (mouse monoclonal; LI-COR; P/N 926-42212), anti-phospho-ERK1/2 (rabbit monoclonal; Cell Signaling; #4370), anti-ERK1/2 (mouse monoclonal; Cell Signaling; #4696), anti–α-tubulin (mouse monoclonal; Sigma), anti-GAPDH (rabbit monoclonal; Cell Signaling; #5174; or mouse monoclonal; Proteintech; 60004-1), anti-LC3B (rabbit polyclonal; Cell Signaling; #2775), anti-p21 (mouse monoclonal; BD; 610233), goat anti-rabbit secondary antibody (Alexa Fluor 680; Thermo Fisher; A-21109), and goat anti-mouse secondary antibody (IRDye 800CW; LI-COR; P/N 926-32210; and DyLight 800 4× PEG; Thermo Fisher; SA5-35521).
ReBiL2.0 Assay.
This assay consists of 1) real-time BiLC assay that has been described previously (SI Appendix, Fig. S2) (20) and 2) quantitative Western blotting to determine the amount of each 1/2luc fusion protein (SI Appendix, Fig. S2I and Table S3). The assay medium (I) consists of DMEM/F12 (Thermo Fisher; phenol-red free), 10% (vol/vol) FBS, and 10 µg/mL ciprofloxacin. The assay medium (II) is based on medium (I) with freshly added 40 ng/mL doxycycline, and 400 to 600 µM d-luciferin (Biosynth; L-8200 or L-8220). The 384-well (Corning; 3570; white, flat-bottom, tissue-culture treated) and 6-well plates were first loaded with 20 µL and 1.6 mL of medium (II) for each well, respectively. ReBiL cells were trypsinized, and cell numbers were determined by Cellometer Auto T4. The required number of cells were collected, centrifuged (200 rcf, 5 min at room temperature) to remove supernatant, and resuspended to 250 cells per µL with medium (I). Then, either 20 µL or 1.6 mL of ReBiL cells were seeded into each well of a 384-well plate (5,000 cells per well) or each well of a 6-well plate (400,000 cells per well), respectively. The final concentration of each component was 1× medium DMEM/F12 (phenol-red free), 10% (vol/vol) FBS, 10 µg/mL ciprofloxacin, 20 ng/mL doxycycline, and 200 to 300 µM d-luciferin. The 384-well plate was sealed with MicroAmp Optical Adhesive Film (Thermo Fisher), and luminescence was read in a Tecan M200 microplate reader (integration time, 2 s; 15 min per cycle for a total of 24 h at 37 °C). The six-well plate was incubated at 37 °C incubator with 7% CO2. Every ReBiL experiment contains a negative control (nl + cl). Upon termination of the ReBiL assay (24 h), the Optical Adhesive Film was removed, and viable cell number was assessed using CellTiter-Glo (Promega; 1:5 diluted with PBS, 35 µL per 384-well) (SI Appendix, Fig. S2H). To quantify the 1/2luc fusion proteins, ReBiL cells from six-well plates were harvested with RIPA lysis buffer (50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 0.25% [wt/vol] deoxycholic acid, 1% [vol/vol] IGEPAL CA-630, 1 mM EDTA, 2 mM Na3VO4, 20 mM NaF, and 1× cOmplete Protease Inhibitor mixture; Roche) after 24 h. The clear lysate was mixed with LDS sample buffer (Thermo Fisher) with 10% (vol/vol) β-mercaptoethanol and denatured at 70 °C for 5 min. The denatured lysates were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore; no. IPFL00010) as described previously (20). The PVDF membrane was then probed with the anti-HA and anti-Actin antibodies in 0.5× Odyssey PBS Blocking Buffer (LI-COR; part no. 927-40000; 1:1 diluted with PBS or 0.5× Odyssey TBS blocking buffer; LI-COR; part no. 927-50000; 1:1 diluted with TBS) with 0.05% Tween 20 at 4 °C overnight. After incubation with secondary antibodies that are conjugated with Alexa Flour 680 (Thermo Fisher) and IRDye 800 (LI-COR), the membrane was scanned in the LI-COR Odyssey Imaging System (Odyssey Application Software 3.0) using the following parameters: resolution, 84 µm; quality, high; focus offset, 0.0 mm; intensity, 700 5.0 and 800 5.0. The relative band intensities of [1/2luc Least], and the actin loading control [loading ctrl] were determined (SI Appendix, Fig. S2I) and exported to Microsoft Excel using ImageStudio Software (version 2.1.10; LI-COR) and the [1/2luc Least]/[Actin] was then calculated (SI Appendix, Table S3). The raw luminescent data collected by the Tecan luminometer were imported to Prism 8 (GraphPad). More cells generated higher luminescent signals. This cell number effect was first corrected by calculating the percentage of [Raw ReBiL Luminescence]/[CellTiter Glo Luminescence]. The [Raw ReBiL Luminescence] was the luminescent reading at the 24-h time point (SI Appendix, Fig. S2 A–G) and the [Cell # by CellTiter Glo] was the luminescent reading from CellTiter-Glo cell viability assay performed at the end of ReBiL kinetic assay (SI Appendix, Fig. S2H). Then the ReBiL2.0 score was calculated by dividing the value of ([Raw ReBiL Luminescence]/[CellTiter Glo Luminescence]) by the corresponding ([1/2luc Least]/[Actin]) (Fig. 1D). The results are reported as bar charts with the x axis representing the percent of the positive control (set to 100%) interaction (SI Appendix, Fig. S2J).
Syto60 Cell Growth Assay.
Syto60 is a cell-permeant red-fluorescence dye that stains nucleic acid. The intensity of Syto60 fluorescence stain is thus proportional to the cell numbers. U2OS cells carrying TRE–EGFP–RBD–CVIM and TRE–EGFP–RBD–SVIM were seeded in a 96-well plate (1,000 cells and 200 µL of medium [DMEM, 10% FBS, 10 µg/mL ciprofloxacin, 200 µg/mL G418] per well) with indicated concentration of doxycycline at day 0. At the times indicated, cells were fixed with 100 µL of 10% buffered formalin (protocol 67-56-1) for 10 min at room temperature, washed twice with PBS, and once with TBS (20 mM Tris, 150 mM NaCl, pH 7.4). Cells were then stained with 1 µM Syto60 dye (Thermo Fisher; S11342 prepared in TBS) for 10 min in the dark. Cells were washed three times with TBS. The mean intensity of Syto60 fluorescence was quantified with the LI-COR Odyssey system and plotted by Prism 8 (GraphPad).
RT-qPCR Analysis.
RNA was isolated using RNeasy Micro Plus and Mini kits (Qiagen; 74034) and converted to cDNA using iScript RT Supermix (Bio-Rad; 1708840). Quantitative real-time PCR was performed using a QuantStudio 5 Real-Time PCR System (Thermo Fisher) by mixing cDNAs, SYBR Green PCR Master Mix (Thermo Fisher; 4309155), and gene-specific primers. Primer sequences are available in SI Appendix, Table S4. All RT-qPCR data were normalized to GAPDH.
Inhibitor Treatment.
Cells in six-well plates (DMEM, 10% FBS, 10 µg/mL ciprofloxacin, 200 µg/mL G418) were treated with or without doxycycline (400 ng/mL) for 24 h, followed by adding the inhibitor or vehicle (DMSO or acetonitrile) to the cell culture medium for another 24 h. Cells were washed with ice-cold PBS twice and harvested by RIPA lysis buffer. Inhibitors used in this study were MLN4924 (Selleckchem; S7109; Batch No. 02; resuspended in DMSO), bafilomycin A1 (BioViotica; catalog #BVT-0252; resuspended in DMSO), concanamycin A (Cayman; item 11050; batch 0564072-5; 100 µg in 1 mL of acetonitrile), and WX8 (resuspended in DMSO; a generous gift from the laboratory of M. DePamphilis, NIH, Bethesda, MD).
Data Availability.
All protocols are described in Materials and Methods or in the references therein. All plasmids and cell lines used in this study are listed in SI Appendix, and they are available upon request from the corresponding author G.M.W.
Supplementary Material
Acknowledgments
We thank E. Stites, M. Philips, N. Rosen, and Z. Yao for constructive comments. We also thank M. DePamphilis and A. Buchwalter for sharing reagents. Work in the laboratory of G.M.W. was supported, in part, by Cancer Center Core Grant CA014195, the Susan G. Komen Foundation (Grant SAC110036), NIH/National Cancer Institute (Grant R35 CA197687), the Leona M. and Harry B. Helmsley Charitable Trust (Grant 2012-PG-MED002), the Freeberg Foundation, the Greenfields, Sorrento Biosciences, and Genentech.
Footnotes
Competing interest statement: F.M. is a consultant for the following companies: Aduro Biotech, Amgen, Daiichi Ltd., Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc., PellePharm, Pfizer, Inc., PMV Pharma, Portola Pharmaceuticals, and Quanta Therapeutics. F.M. has received research grants from Daiichi Ltd. and from Gilead Sciences. F.M. is a consultant and cofounder for the following companies (with ownership interest including stock options): BridgeBio, DNAtrix, Inc., Olema Pharmaceuticals, Inc., and Quartz. F.M. is scientific director of the NCI Ras Initiative at Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. The rest of authors declare no competing financial interests.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000848117/-/DCSupplemental.
References
- 1.Perkins J. R., Diboun I., Dessailly B. H., Lees J. G., Orengo C., Transient protein-protein interactions: Structural, functional, and network properties. Structure 18, 1233–1243 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Cox A. D., Der C. J., Philips M. R., Targeting RAS membrane association: Back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21, 1819–1827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior I. A., Muncke C., Parton R. G., Hancock J. F., Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nan X., et al. , Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muratcioglu S., et al. , GTP-dependent K-Ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash P., et al. , Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep. 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujioka A., et al. , Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 281, 8917–8926 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Grasberger B., Minton A. P., DeLisi C., Metzger H., Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. U.S.A. 83, 6258–6262 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuriyan J., Eisenberg D., The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Willumsen B. M., Christensen A., Hubbert N. L., Papageorge A. G., Lowy D. R., The p21 ras C-terminus is required for transformation and membrane association. Nature 310, 583–586 (1984). [DOI] [PubMed] [Google Scholar]
- 11.Buss J. E., Solski P. A., Schaeffer J. P., MacDonald M. J., Der C. J., Activation of the cellular proto-oncogene product p21Ras by addition of a myristylation signal. Science 243, 1600–1603 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Chung J. K., et al. , K-Ras4B remains monomeric on membranes over a wide range of surface densities and lipid compositions. Biophys. J. 114, 137–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakhaeizadeh H., Amin E., Nakhaei-Rad S., Dvorsky R., Ahmadian M. R., The RAS-effector interface: Isoform-specific differences in the effector binding regions. PLoS One 11, e0167145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavoie H., Therrien M., Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Fabian J. R., Vojtek A. B., Cooper J. A., Morrison D. K., A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc. Natl. Acad. Sci. U.S.A. 91, 5982–5986 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Block C., Janknecht R., Herrmann C., Nassar N., Wittinghofer A., Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 3, 244–251 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Luker K. E., et al. , Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U.S.A. 101, 12288–12293 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villalobos V., et al. , Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem. Biol. 17, 1018–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald-Obermann J. L., Piwnica-Worms D., Pike L. J., Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc. Natl. Acad. Sci. U.S.A. 109, 137–142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y. C., et al. , A versatile platform to analyze low-affinity and transient protein-protein interactions in living cells in real time. Cell Rep. 9, 1946–1958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson J. H., et al. , Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proc. Natl. Acad. Sci. U.S.A. 87, 3042–3046 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock J. F., Paterson H., Marshall C. J., A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–139 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Chiu V. K., et al. , Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4, 343–350 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Ahearn I. M., Haigis K., Bar-Sagi D., Philips M. R., Regulating the regulator: Post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39–51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Hancock J. F., Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Vigil D., Cherfils J., Rossman K. L., Der C. J., Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton M. N., et al. , DIRAS3 (ARHI) blocks RAS/MAPK signaling by binding directly to RAS and disrupting RAS clusters. Cell Rep. 29, 3448–3459.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodis E., et al. , A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanker A. B., et al. , Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene 29, 380–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondeva T., Balla A., Várnai P., Balla T., Structural determinants of Ras-Raf interaction analyzed in live cells. Mol. Biol. Cell 13, 2323–2333 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brtva T. R., et al. , Two distinct Raf domains mediate interaction with Ras. J. Biol. Chem. 270, 9809–9812 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Wong E. T., et al. , Reproducible doxycycline-inducible transgene expression at specific loci generated by Cre-recombinase mediated cassette exchange. Nucleic Acids Res. 33, e147 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka T., Williams R. L., Rabbitts T. H., Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 26, 3250–3259 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaelson D., et al. , Differential localization of Rho GTPases in live cells: Regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111–126 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauke M. J., et al. , An engineered protein antagonist of K-Ras/B-Raf interaction. Sci. Rep. 7, 5831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gera N., Hussain M., Wright R. C., Rao B. M., Highly stable binding proteins derived from the hyperthermophilic Sso7d scaffold. J. Mol. Biol. 409, 601–616 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Stieglitz B., et al. , Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 27, 1995–2005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma G., et al. , A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy 15, 1694–1718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Söderberg O., et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Rajakulendran T., Sahmi M., Lefrançois M., Sicheri F., Therrien M., A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Nan X., et al. , Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 18519–18524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philips M. R., Der C. J., Seeing is believing: Ras dimers observed in live cells. Proc. Natl. Acad. Sci. U.S.A. 112, 9793–9794 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bremner R., Balmain A., Genetic changes in skin tumor progression: Correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 61, 407–417 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z., et al. , Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 29, 25–33 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ambrogio C., et al. , KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 48.To M. D., Rosario R. D., Westcott P. M., Banta K. L., Balmain A., Interactions between wild-type and mutant Ras genes in lung and skin carcinogenesis. Oncogene 32, 4028–4033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenney C., Stites E. C., Analysis of RAS as a tumor suppressor. bioRxiv:10.1101/153692 (21 June 2017).
- 50.Kauke M. J., et al. , A Raf-competitive K-ras binder can fail to functionally antagonize signaling. Mol. Cancer Ther. 17, 1773–1780 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zeng M., et al. , Potent and selective covalent quinazoline inhibitors of KRAS G12C. Cell Chem. Biol. 24, 1005–1016.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Janes M. R., et al. , Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Canon J., et al. , The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All protocols are described in Materials and Methods or in the references therein. All plasmids and cell lines used in this study are listed in SI Appendix, and they are available upon request from the corresponding author G.M.W.