Significance
The continued high rates of using antibiotics in healthcare and livestock, without sufficient new compounds reaching the market, has led to a dramatic increase in antimicrobial resistance, with multidrug-resistant bacteria emerging as a major public health threat worldwide. Because the vast majority of antiinfectives are natural products or have originated from them, we assessed the predictive power of plant molecular phylogenies and species distribution modeling in the search for clades and areas which promise to provide a higher probability of delivering new antiinfective compound leads. Our approach enables taxonomically and spatially targeted bioprospecting and supports the battle against the global antimicrobial crisis.
Keywords: natural products, biodiversity, chemical diversity, phylogenetics, chemoinformatics
Abstract
Antibiotic resistance and viral diseases are rising around the world and are becoming major threats to global health, food security, and development. One measure that has been suggested to mitigate this crisis is the development of new antibiotics. Here, we provide a comprehensive evaluation of the phylogenetic and biogeographic patterns of antiinfective compounds from seed plants in one of the most species-rich regions on Earth and identify clades with naturally occurring substances potentially suitable for the development of new pharmaceutical compounds. Specifically, we combine taxonomic and phylogenetic data for >7,500 seed plant species from the flora of Java with >16,500 secondary metabolites and 6,255 georeferenced occurrence records to 1) identify clades in the phylogeny that are characterized by either an overrepresentation (“hot clades”) or an underrepresentation (“cold clades”) of antiinfective compounds and 2) assess the spatial patterns of plants with antiinfective compounds relative to total plant diversity across the region. Across the flora of Java, we identify 26 “hot clades” with plant species providing a high probability of finding antibiotic constituents. In addition, 24 “cold clades” constitute lineages with low numbers of reported activities but which have the potential to yield novel compounds. Spatial patterns of plant species and metabolite diversity are strongly correlated across Java, indicating that regions of highest species diversity afford the highest potential to discover novel natural products. Our results indicate that the combination of phylogenetic, spatial, and phytochemical information is a useful tool to guide the selection of taxa for efforts aimed at lead compound discovery.
The continued high rates of antibiotic use in healthcare and livestock farming have led to a dramatic increase in antimicrobial resistance, with multidrug-resistant bacteria emerging as a major public health threat worldwide (1). Scientists have warned that we very soon might face a “postantibiotic era” (2), just 90 y after the discovery of penicillin by Sir Alexander Fleming. The major threats of antimicrobial resistance include a longer duration of illnesses as well as an increase of infection-related mortality rates, higher costs of medical treatments, and the inability to perform certain procedures due to the high risk of postoperative infections (2). A number of strategies have been suggested to alleviate these risks, including a more responsible use of antibiotics, improvements of infection control measures, heightened awareness of the risks of increasing resistance, and the development of novel antimicrobial agents (1). However, despite the urgent need for new antibiotics, many pharmaceutical companies have largely suspended their antibiotic drug discovery efforts, mainly due to very high investment and low returns, as well as legislative and other restrictions (1, 3).
Nine antibiotic classes, which can be differentiated based on their scaffolds (the core molecular structure common to each class), contribute to most of the clinically approved antibiotics today. The incremental modification of available natural scaffolds has become the prevailing approach of antibiotic drug discovery (4, 5). Most of these were developed between the mid-1930s and early 1960s, and only three new classes were introduced after an innovation gap of around 40 y (4), despite having already been known for at least two decades. While combinatorial chemistry has allowed the pharmaceutical industry to produce and screen vast numbers of molecules, this approach was only moderately successful in finding compounds with new modes of action (3, 5). Compound libraries produced through combinatorial chemistry might have larger numbers of compounds, but unless focused around an already known lead compound (new chemical entity with the potential to develop a new drug; ref. 6) they contain a lower rate of biologically relevant ones (7). Only 22 to 33% of the antiinfective drugs approved between 1981 and 2014 have active ingredients of synthetic origin, while the vast majority are natural products, or their derivatives or mimetics (5).
Plants have been used medicinally since ancient times (e.g., the use of poppy by Sumerians around 4000 BCE; ref. 8), and—considering the number of people depending on them—still constitute the most important part of healthcare today, as a major part of the developing world depends on traditional medicine as primary health care (9). Of the ∼370,000 seed plant species that are scientifically described and named (10) only a fraction have been studied phytochemically or evaluated for their biological activity (11). Even if most clinically used antibiotics are derived from microbial origin, plants should not be disregarded as a source for antiinfective agents since they have exceptional abilities to produce cytotoxic agents and to resist pathogenic microorganisms (12, 13). More than 200,000 natural products have been discovered from plants to date (14), with estimates of the total number exceeding 500,000 (15). While there should be an abundance of undiscovered bioactive plant natural products (16), selecting target species for screening and phytochemical evaluation is not trivial (but see ref. 17). In the past, random sampling and ethnobotanical approaches were most common (16, 18), whereas approaches incorporating evolutionary thinking or ecological observations have been rare (but see refs. 19 and 20–23).
Throughout their evolutionary history, plants have developed a wide range of chemical compounds associated with their growth and survival as well as with interactions with their abiotic and biotic (intra- and interspecific) environment, which determine their survival and ultimately their evolutionary success (24). Novel metabolites can originate through several processes, such as gene or whole-genome duplication events (25, 26) followed by divergence (18, 27, 28). These events may uncouple gene copies from selective pressures and eventually lead to the emergence of novel metabolites (29). Furthermore, already existing pathways may be modified to produce novel metabolites under natural selection (30). Thus, given that chemical compounds represent an adaptation to a species’ ecological niche, and assuming that ecological niches are at least to some extent conserved (i.e., ancestral ecological tolerances are retained; ref. 31), specific metabolites will not be randomly distributed across species but generally shared among closely related ones (14, 30, 32). This correlation between phylogeny and biosynthetic pathways (phylogenetic signal; e.g., refs. 33 and 34) allows for a more efficient selection of plants for lead compound discovery (20, 22, 35–38) by identifying clusters of species with desired metabolite profiles (39). Zhu et al. (19) showed that families producing “drugs” across plants, bacteria, fungi, and metazoa tend to be concentrated in “drug-producing” clusters in the respective phylogenies: Eighty percent of approved drugs are concentrated in 17 “drug-prolific” families. Furthermore, they suggested that the paucity of drugs outside “drug-productive” families is not necessarily the result of underexploration but indicates a lack of appropriate compounds. However, in contrast to these findings, almost 30% of novel secondary metabolites published in 2001 to 2011 were found in families outside these “drug-productive” clusters, showing that previously unrecognized groups can significantly contribute to novel drug leads (19). At the same time, the shared evolutionary history suggests that biological and chemical diversity might not be correlated linearly as closely related species would tend to produce similar metabolites. In addition, if the number of molecular scaffolds available through common metabolic pathways is limited, so should be the number of possible metabolites (18), suggesting that the rates of evolution of chemical diversity might be lower than that of species diversity (40). Also, similarity in metabolites should not be mistaken as similarity in function or bioactivity, as a single simple structural change can substantially change the biological profile.
In order to streamline and target the search for new antiinfective compounds, we present an approach that integrates phylogenetic and spatial data with information on bioactivity. As only a small portion of known plant species have been thoroughly screened, high biodiversity regions offer a great potential for the discovery of new lead compounds (41). With more than 40,000 species of vascular plants, the Malesian region is one of the global centers of plant species richness (42–44). Within this region, Indonesia stands out particularly, being one of 17 megadiverse countries (45) and containing the world’s third-largest area of rainforest, spread over more than 16,000 islands. At the same time, however, the biodiversity of this region is under severe threat from habitat loss, highlighted by two biodiversity hotspots (Wallacea and Sundaland; refs. 46 and 47). In addition, many medicinal plants are under threat from overexploitation due to unsustainable or destructive harvest (48–50). Thus, we stand not only to lose known medicinal plants but also plants with until-now-unknown properties and potentially new compounds of medicinal value. Focusing on the island of Java, we combine taxonomic, phylogenetic, spatial, and phytochemical information to 1) identify over-/underrepresentation of antiinfective activities across the flora and 2) evaluate the relationship between biodiversity and secondary metabolite diversity.
Results and Discussion
Natural Product Classes.
The most frequent natural product classes that were identified for the 16,072 metabolites of Javanese seed plants were terpenes, phenylpropanoids, phenols, sugars, and lactones, coinciding with the general distribution in plants (Table 1). Remarkably, we found antiinfective metabolites in each of the natural product classes incorporated here. Among these, metabolites with a fluorene motif were observed to have the highest proportion of antiinfectives (75%; Table 1). However, this is also the natural product class with the fewest number of metabolites (eight, with six identified as antiinfective). Other classes with a high ratio of antiinfective metabolites were quinones (29%), xanthones (24%), anthracenes (23%), and coumarins (23%). Indeed, these natural product classes frequently display antiinfective activities (12, 51), and several have distinct redox properties, a trait that usually is less favored for commercial antibiotics. The natural product classes with the lowest proportion of antiinfective metabolites are sugars and glycosylated compounds (6.7%). This is not surprising, since those metabolites will have a growth-promoting effect on pathogens due to their easily accessible carbon source. In addition, their hydrophilicity does not make them very bioavailable as such, although glycosides often act as storage compounds and prodrugs of their aglycones in plants, and thus their potential may be underestimated (52).
Table 1.
Natural product classes found for the 16,072 metabolites present in Javanese seed plants
| No. of metabolites | ||||||
| Natural product class | Antiinfective metabolites | Remaining metabolites | Sum | Antiinfective, % | ||
| Acyclic alkaloid | 130 | (2.6%) | 722 | (2.6%) | 852 | 15.3 |
| Cyclic alkaloid | 463 | (9.3%) | 1,884 | (6.8%) | 2,347 | 19.7 |
| Anthracene | 33 | (0.7%) | 109 | (0.4%) | 142 | 23.2 |
| Benzofuran | 127 | (2.5%) | 599 | (2.2%) | 726 | 17.5 |
| Coumarine | 82 | (1.6%) | 281 | (1.0%) | 363 | 22.6 |
| Fatty acid (ester) | 228 | (4.6%) | 1,828 | (6.6%) | 2,056 | 11.1 |
| Flavonoid | 296 | (6.0%) | 1,909 | (7.0%) | 2,205 | 13.4 |
| Fluorene | 6 | (0.1%) | 2 | (0%) | 8 | 75.0 |
| Isoflavonoid | 100 | (2.0%) | 414 | (1.5%) | 514 | 19.5 |
| Lactam | 44 | (0.9%) | 305 | (1.1%) | 349 | 12.6 |
| Lacton | 420 | (8.4%) | 2,101 | (7.6%) | 2,521 | 16.7 |
| Macrocycle | 72 | (1.4%) | 333 | (1.2%) | 405 | 17.8 |
| Peptide | 19 | (0.4%) | 185 | (0.7%) | 204 | 9.3 |
| Peptoid | 38 | (0.8%) | 253 | (0.9%) | 291 | 13.1 |
| Phenanthrene | 59 | (1.2%) | 288 | (1.0%) | 347 | 17.0 |
| Phenolic | 681 | (13.6%) | 3,499 | (12.7%) | 4,180 | 16.3 |
| Phenylpropanoid | 834 | (16.7%) | 3,669 | (13.3%) | 4,503 | 18.5 |
| Polyacetylene | 145 | (2.9%) | 712 | (2.6%) | 857 | 16.9 |
| Polyketide | 21 | (0.4%) | 112 | (0.4%) | 133 | 15.8 |
| Quinone | 85 | (1.7%) | 204 | (0.7%) | 289 | 29.4 |
| Steroid | 122 | (2.4%) | 871 | (3.2%) | 993 | 12.3 |
| Sugar | 193 | (3.9%) | 2,684 | (9.8%) | 2,877 | 6.7 |
| Terpene | 739 | (14.8%) | 4,328 | (15.7%) | 5,067 | 14.6 |
| Xanthone | 51 | (1.0%) | 165 | (0.6%) | 216 | 23.6 |
| No class assigned | 15 | (0.3%) | 55 | (0.2%) | 70 | 21.4 |
For each natural product class, we provide the total number and proportion of all metabolites (in parentheses) for the 2,859 compounds labeled as antiinfective and the remaining 13,213 metabolites, respectively. In addition, we calculated the sum and proportion of antiinfective metabolites for each natural product class. It should be noted that metabolites can be assigned to multiple natural product classes.
Phylogenetic Analyses.
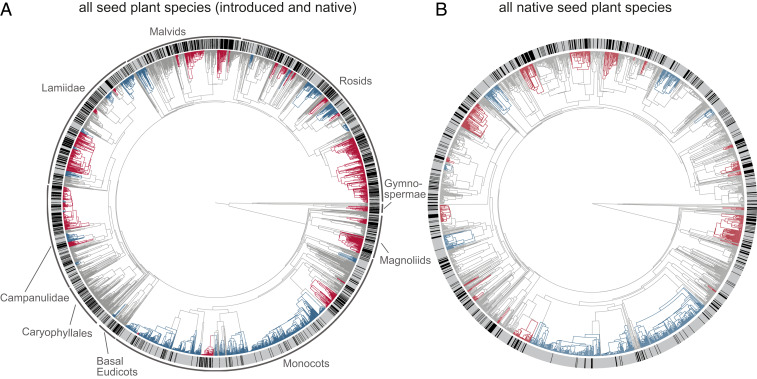
We found significant levels of phylogenetic signal of antiinfective activities for all species found on Java (D = 0.655) as well as for native species only (D = 0.691; SI Appendix, Table S1). The observed values of D differed significantly both from a random distribution as well as from a conserved pattern (Brownian motion). This demonstrates an underlying phylogenetic pattern in the distribution of drug-productive sources in seed plants, which is an important prerequisite for utilizing phylogenetic data for lead compound discovery. Our phylogenetic analyses of antiinfective activities identified 26 and 21 clades across the two phylogenies with a significant degree of clustering (i.e., overrepresentation of antiinfective activities, hereafter referred to as “hot clades”) when considering all species or native taxa only, respectively (Fig. 1). These clades contained 40 seed plant families (25 in whole and 15 in part) in the combined analysis (Fig. 1A) and 21 families (12 in whole and nine in part) in the native-only analysis (Fig. 1B), respectively. Furthermore, we found 24 clades (nine in the native-only data) where antiinfective activities were significantly underrepresented (hereafter “cold clades”; Fig. 1), which included a total of 27 families (13 in the native-only data), of which 16 (15 in the native-only data) were represented as a whole and 11 families (two families in the native-only data) in part (SI Appendix, Tables S2–S5). It is striking that antiinfective activities appear to be underrepresented in the monocotyledons (seven “cold clades” including 10 families across all species, four “cold clades” with five families across the native plant species as a whole or in part), with grasses (Poaceae) and orchids (Orchidaceae) as the most species-rich families within these “cold clades.” Although representatives of both families have been reported to contain bioactive secondary metabolites, they are relatively poorly studied considering their exceptional species richness (53–55). In the analysis of all seed plants on Java, a few “hot” and “cold” clades were found to be nested within each other, meaning that a “cold clade” may be found to be nested within a clade where, as a whole, antiinfective activities were overrepresented (“hot clade”). Four “cold clades” (clades 12, 13, 15, and 16; SI Appendix, Fig. S2 and Table S3) were nested inside “hot clades” (clades 11 and 12; SI Appendix, Fig. S2 and Table S2), while one “hot clade” (clade 9; SI Appendix, Fig. S2 and Table S2) was found to be nested within a “cold clade” (clade 8; SI Appendix, Fig. S2 and Table S3).
Fig. 1.
Phylogenetic patterns of antiinfective activities across the flora of Java. Overrepresentation (“hot clades,” red) and underrepresentation (“cold clades,” blue) were identified using the presence of known antiinfective activities (black bars at tips) for all seed plant species on Java (A) and the native species only (B). Details of each “hot” and “cold” clade can be found in SI Appendix, Figs. S2 and S3 and Tables S2–S5.
The overlap between the two analyses (all vs. native taxa only) is considerable, with seven “hot clades” and six “cold clades” being consistently recovered in both analyses, for example Orchidaceae as a “cold clade” in both the combined and native-only analysis (clades 2 and 1; see SI Appendix, Figs. S1 and S2 and Tables S2 and S3, respectively). Furthermore, eight “hot clades” (e.g., clades 11 [Asteraceae] and 13 [Lamiales]; SI Appendix, Fig. S2 and Table S5) and one “cold clade” (clade 3; SI Appendix, Fig. S2 and Table S4) were largely congruent in both analyses. Despite the high degree of overlap between both analyses, there are also some striking differences. Ten “hot clades” found in the analysis of the entire flora are not recovered when considering native taxa only (Fig. 1B), which is attributable to two possible phenomenona: Either a clade comprises only introduced taxa or the ratio between bioactive and inactive species is substantially reduced due to the exclusion of the nonnative species with antiinfective effects, thus causing the clade to lose the significant overrepresentation. Indeed, many nonnative plant species will have likely been introduced specifically because of their use in traditional medicine (56). The “hot clade” comprising Amaryllidaceae, Asparagaceae, and Liliaceae (clade 5 in SI Appendix, Fig. S1 and Table S2) is a case in point of the second scenario. With ca. 90% introduced species in total, most antiinfective species are excluded in the native-only analysis. Other “hot clades” like the Asteraceae (clade 11 in SI Appendix, Figs. S2 and S3 and Tables S2 and S4) remain but are substantially reduced in size. Here, only 33 of the 131 species in the “hot clade” remain in the native-only analysis. Over a third of the species that comprise the new “hot clade” (clade 11 in SI Appendix, Fig. S3 and Table S4) belong to the following genera: Blumea, Carpesium, Pluchea, Pterocaulon, and Sphaeranthus. All of these are native to Java (without any introduced species) and for over 50% of the species antiinfective effects have been described. The genera are widely distributed and relatively rich in species (ca. 10 to 100 species per genus), which might contribute to their prevalence in phytochemical and ethnobotanical research. Widespread, species-rich, and herbaceous taxa are often found to contain bioactive compounds, as larger geographical and wider ecological ranges potentially allow for more opportunities for adaptation to different conditions (57–59), as well as easier access for researchers. In contrast, four additional “hot clades” are recovered in the native-only data: Menispermaceae (clade 7), Polygonaceae (clade 8), Molluginaceae (clade 9), and Cornaceae (clade 10; SI Appendix, Fig. S3 and Table S4). Similar patterns are observed with regard to the “cold clades”: In total, 15 clades disappear and one clade increases in size from the family Araliaceae (clade 11; SI Appendix, Fig. S2 and Table S3) to the whole order Apiales (clade 5; SI Appendix, Fig. S3 and Table S4), when only native taxa are considered. Again, human preselection may account for the result, as, for example, food plants are often introduced which commonly lack significant bioactivity (other than nutritional and flavor/fragrance/color properties).
The taxonomic analyses (SI Appendix, Fig. S4 and Table S6) showed that the average ratio of antiinfective species per family was 26% and 20% for the analyses with all taxa and natives-only, respectively. When compared with the phylogenetic analyses, we found a large degree of overlap. Just as in the phylogenetic analyses, Amaryllidaceae and Asparagaceae lay above the 95% CIs when all species were considered but fell within the CIs when only the native species were analyzed. The fourth family in this clade (clade 5; SI Appendix, Fig. S1 and Table S2), Liliaceae, however, did not fall above the 95% CI in both analyses. We also found a few notable differences between the analyses, as a number of families that are above the 95% CI in the taxonomic analyses (e.g., Polygonaceae and Rhizophoraceae) do not appear in “hot clades.” In some cases, the phylogenetic analyses revealed a more complex pattern of over- and underrepresentation of antiinfective activities than the taxonomic analyses. Rubiaceae fell below the 95% CI in the taxonomic analyses (despite having a number of species with documented antiinfective activities), while in the phylogenetic analyses two small “hot clades” (clade 14; SI Appendix, Fig. S2 and Table S2 and clade 17; SI Appendix, Fig. S3 and Table S4) are recovered with 11 and 9 species, respectively. These all belong to the subfamily Cinchonoidae (tribe Naucleeae), which is characterized by the occurrence of oxidized indole alkaloids. Thus, the low percentage of antiinfective species in the taxonomic analysis is due to the restriction of antiinfective activities to one subfamily within the very species-rich Rubiaceae.
In addition, the taxonomic analyses indicate that Fabaceae have significantly more taxa with antiinfective activities compared to the rest of the flora, corroborating the phylogenetic analysis of all species, which identified two “hot clades,” covering over 90% of Fabaceae (230 species; clades 25 and 26; SI Appendix, Fig. S2 and Table S2). The analysis of the native taxa on the other hand found three small “hot clades” (clades 16 to 18; SI Appendix, Fig. S2 and Table S5) with a total of 37 species, with two-thirds of the family not diverging significantly from the overall pattern. Given the >19,500 species in this family (60), this should not come as a surprise, but it highlights how our approach avoids any bias that focusing on any specific taxonomic level might introduce, and argues for the importance of a phylogenetic approach instead of following a strict taxonomic one. Within Fabaceae, the antiinfective activities might be correlated with the presence of isoflavonoids. This compound class was not reported for any of the genera forming the “cold clade,” but was overrepresented in genera of the “hot clade.” Finally, the taxonomic analyses for families that were represented by only a few species on Java (usually fewer than four) need to be interpreted with great caution, as the proportion of bioactive species can quickly reach extreme values, because of the small sample size. Thus, these will not be discussed further here.
Spatial Analyses.
We modeled the spatial distribution of 1,895 Javanese seed plant species. The relative contribution of the environmental variables used for the species distribution modeling showed no substantial differences between climatic and soil variables (SI Appendix, Table S7). The evaluation of the model (SI Appendix, Table S8) showed a relatively high species richness error (988.32 ± 306.90), which is mainly due to the data type (few presence data with randomly generated absences). The specificity measure shows that absence data are predicted correctly in almost 50% of the cases (which again reflects the random distribution of absence data), while 97% of the occurrences are predicted correctly (sensitivity).
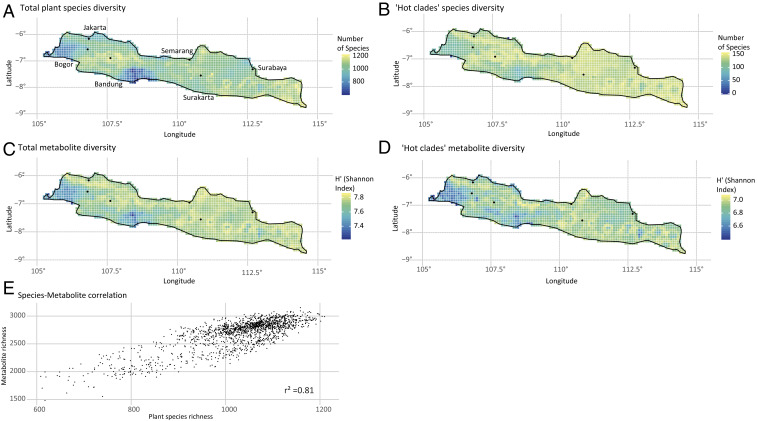
Areas with the highest number of species are found in mountainous regions across Java, and the lowest species richness can be found in the lowlands of central and west Java (Fig. 2A). The Shannon index of metabolite diversity shows a pattern very similar to that of species diversity (Fig. 2C). Indeed, plant diversity and metabolite diversity appear to be strongly correlated across Java (r2 = 0.81; Fig. 2E) and our analyses did not exhibit a saturation of metabolite diversity with increasing plant diversity, which is probably due to the limited geographic (only Java) and taxonomic (only seed plants) extent. If sampling was extended to continental to global scales or with complete taxonomic sampling of individual clades, we would expect to observe saturation of the species richness–metabolite relationship. In addition, metabolite information was available for only 1,001 of the modeled species, and it is unlikely that all metabolites occurring in a species will be known. Classical natural product chemists and pharmaceutical biologists usually pick only selected compounds (easily separable, abundant, or bioactive), while modern metabolic profiling approaches across species (61, 62) are still unable to extract and detect all metabolites—and are far from identifying more than half of them.
Fig. 2.
Spatial patterns of seed plants and secondary metabolites on Java. Spatial patterns of all modeled seed plant species (A) and of seed plant species found in “hot clades” (B) are contrasted with the overall metabolite diversity (C) and the diversity of secondary metabolites of species found in “hot clades” (D). The latter two are calculated using the Shannon index, which takes the relative abundance of each metabolite into account. Plant species richness and metabolite diversity are closely correlated across Java (E).
Several factors have to be kept in mind with regard to the phylogenetic and spatial patterns of antiinfective activities. First, convergent evolution may explain irregular patterns (63) observed by some studies (32, 38), in which the distribution of metabolites or activities does not closely match the phylogeny of the investigated plant taxa. However, for medicinal chemistry such events can be of high interest in that they can provide additional evidence for a relevant lead (convergence into similar structures) or even provide the equivalent to scaffold hopping with functional convergence (convergence onto a target or effect with different structures). A similar pattern can be caused by horizontal gene transfers between plants and pathogens (64). Second, not all metabolites are constitutively expressed but may be produced as a response to herbivory or other damage, and the biosynthesis may vary with the developmental stage, organ, or the abiotic environment (65). Finally, the documentation of taxonomic and geographic data in natural product research is often lacking or inconclusive (66), which can hinder effective bioprospecting (see also Materials and Methods). In a large survey such as this one, the potential of any single species for its antibiotic effects remains unpredictable, but for larger clades statistically supported guidelines can be derived to help researchers in the field and laboratory. Our study provides an approach to identify geographic areas for maximum levels of metabolite diversity (ideally also structural and functional), as well as taxa based on their metabolite prevalence (e.g., antiinfectives). Furthermore, the search for new lead compounds has implications for the conservation of natural resources as well as intellectual property rights and benefit sharing (67). Traditional medicinal plants are still mainly sourced from natural populations, so overexploitation and habitat destruction pose a major threat to the sustained availability of these resources. Some of these issues can be addressed with the help of our approach, in that alternative sources or less threatened habitats may be identified. This might be extended to biotechnological analyses, for example of pathways suggested to produce the valuable compounds (68, 69).
Conclusion
We detected strong phylogenetic and spatial patterns in the distribution of antiinfective activities across the flora of Java. Our results indicate that the combination of phylogenetic, spatial, and phytochemical information is a useful tool to guide the selection of taxa for directing efforts aimed at lead compound discovery. We suggest two major paths for future lead compound discovery from natural products. First, species within “hot clades” without documented antiinfective activities have a high probability of providing bioactive compounds of a given type. Importantly, given the sometimes considerable variation of chemical compounds among closely related species (38), these species might still provide new compounds or activities. The Rubiaceae constitute a particularly interesting group for further analyses, since the family is species-rich, with only a few species having a documented antiinfective activity (but enough to make them a “hot clade”). Second, species within “cold clades” (“high risk–high reward” clades) have an overall low probability for antiinfectives, but at the same time any compound found has a higher probability of being new for the screened activity, and potentially even structurally novel. Both strategies, the exploitation of new sources as well as the discovery of new derivatives or structurally novel compounds acting as new leads on antiinfective targets, are urgently needed, and we suggest that this approach should be extended to other geographical regions and biological activities.
Materials and Methods
An overview of the data and analysis pipeline can be found in SI Appendix, Fig. S1.
Taxonomic and Metabolite Information/Bioactivity Profiles.
We extracted all taxonomic information from the Flora of Java volumes 1 through 3 (70), a systematic account of all extant seed plants on the island of Java. We further incorporated additional seed plant species described for Java since the completion of the Flora by downloading available data from the major collections of plants for the region (i.e., the collections of Naturalis [L, WAG, and U], the herbarium at the Royal Botanic Gardens, Kew [K], the Australian National Herbarium [CANB], and the herbarium at the Smithsonian Institution [US]) and identified all species found on Java (between 1960 and 2014) that were not included in the Flora. Finally, we incorporated all seed plants from available regional checklists (e.g., ref. 71) not otherwise included. We then identified all seed plants that were designated as introduced in the Flora of Java to account for the effect of introduced species on the observed patterns. All taxon names were checked and, where necessary, corrected using the Taxonomic Name Resolution Service (TNRS) v4.0 (72, 73), with the Tropicos database, the Plant List, US Department of Agriculture’s (USDA’s) Plants database, the Global Compositae Checklist, and the International Legume database as sources. Family classifications follow the latest account of the Angiosperm Phylogeny Group (APG IV; ref. 74). The final dataset contained 7,573 seed plant species, 5,392 of which were native to Java, belonging to 2,352 genera (1,652 native).
Information on natural products, isolated from seed plant species present on Java, were collected from the KNApSAcK (75) and NPASS (Natural Product Activity and Species Source) (76) databases, which contain natural sources, their natural products, and biological activities. The databases were queried with the names of seed plant species (including their synonyms) on Java for their metabolites and corresponding activities. For 1,883 and 1,889 species metabolite information was gained from the KNApSAcK and NPASS database, respectively, with an intercept of 1,776 species. The 10,612 (KNApSAcK) and 9,894 (NPASS) metabolites were first normalized using the metabolite names, and afterward their chemical structures were compared to identify duplicate structures stored with different names to gain in total 17,117 unique metabolites. Metabolic pathways accessed via the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (77–79) were used to decide whether a metabolite was primary or secondary, and subsequently all primary metabolites (e.g., pathways from the photosynthesis, carbon metabolism, citrate cycle or fatty acid biosynthesis domains, as well as those taking part in hormone or nucleic acid synthesis; see SI Appendix, Table S9) were excluded. The remaining 16,503 secondary metabolites were used to query the NPASS and KNApSAcK databases again to obtain their biological activities. For the analyses, we specifically focused on antiinfective activities, including antifungal, antibacterial, antiparasitic, and antiviral (3,705 metabolites in total). Those were further reduced to only contain those activities affecting vertebrate pathogens (2,859 metabolites), resulting in 1,640 species (796 native species) with antiinfective activity belonging to 941 genera (501 genera considering only native species). All metabolites were classified into natural product classes using substructure matching.
Metabolites may group into several classes if they contained the corresponding substructures. For example, a glycosylated steroid was classified as steroid and sugar-containing. To be classified as terpene, a metabolite had to match two isoprenoid units as well as possess at least one chain of connected carbons with a multiple of 5 as length. Finally, the proportion of species with antiinfective effects (metabolites with known activity) was determined for each seed plant family on Java (hereafter referred to as “taxonomic analysis”). The overall mean across all families was then used to calculate the 95% and 99% CIs depending on the number of species per family (due to the central limit theorem). Thus, seed plant families above or below the 95% CI contain a higher or lower proportion of species with antiinfective effects, respectively, than expected given the overall prevalence across the flora.
Reports on bioactivities of plant extracts or fractions in publications (and hence databases) need to be treated with great caution, as they may constitute a source of false positive results. Many publications often state a multitude of unrelated activities, for example from antiviral to antidiabetic properties, for the same plant extract. Commonly, this is related to a detection of general toxicity (e.g., through redox system disturbance) or tanning properties (e.g., of unspecific polyphenol–protein interactions), or the effects are only observed at very high concentrations (80, 81). In a chemoinformatic survey, such false positive effects cannot (yet) easily be separated from specific effects useful for drug development. Finally, chemo-systematic data are scattered in the literature and negative results are often not reported or published, contributing to a potentially substantial bias. Nonactivities can be of high importance in chemoinformatics, for example in medicinal chemistry, and if done properly and performed based on good experimental design and analyses, they are more valuable than the “pseudopositive” results described above. Absence or presence of a compound or activity may also depend on the amount of plant material investigated as well as the analytical methods (82, 83). This also points toward a more general problem, as some families are more extensively studied than others, often due to biases of researchers regarding preferred taxa, geographic regions, or previous successes, as well as the specific assays that might be done (which are also biased by general availability or ease of performance). Ideally, comparable metabolic information for the different species and negative activity data (the latter would be especially important to distinguish between families which indeed show less activity and those that are understudied) would significantly enhance future studies. To date, only 5 to 15% of vascular plants have been investigated for their natural products (84, 85), often not in a systematic way, with around 250,000 natural product structures in virtual databases (86).
Phylogenetic Data.
We used a dated phylogenetic tree from Smith and Brown (87), which comprises GenBank sequence data for 79,874 seed plant species with a backbone provided by Magallón et al. (88). This tree was pruned to exclude all species not present on Java and outside the taxonomic focus of this study (i.e., nonseed plants). To assess the impact of introduced taxa, we generated two phylogenetic trees for further analyses: one including all seed plant species native to Java and one tree that also included introduced taxa (discussed above); 3,545 native species (4,305 in total including introduced lineages) were not represented in the tree and were thus excluded from the phylogenetic analyses. The final trees consisted of 1,847 seed plant species (native) and 3,268 species (introduced and native), respectively.
We first evaluated the strength of phylogenetic signal of antiinfective activities across the flora using the D statistic (89) implemented in the R package caper. D measures the phylogenetic signal of a binary trait by calculating the sum of sister-clade differences. The observed value is then compared to two distributions: a clumped pattern and a random distribution of traits. For a given phylogeny and prevalence of the trait (proportion of tips in character state 1), these serve as reference estimates under a conserved model of evolution (approximated by Brownian motion, but see refs. 31 and 89) and a trait that is distributed randomly with respect to the phylogeny. Second, to identify target clades for bioactivity screening, we used a phylogenetic clustering approach (nodesig) as implemented in phylocom (90). Originally developed as a metric of phylogenetic community structure, the approach allows us to identify clades where antiinfective effects are over- or underrepresented. Significance of the observed patterns is evaluated using a randomization of antiinfective effects across the tips of the phylogeny.
Spatial Data.
To evaluate the spatial patterns of plant species and metabolite diversity, we downloaded the available occurrence data for seed plants in Indonesia from the Global Biodiversity Information Facility (91) database (accessed 3 May 2018). Given that a large number of accessions do not have coordinate information, we used the approach developed by Gratton et al. (92) to automatically georeference the collection localities of each record (where available). The approach utilizes the information available in the locality description to find the coordinates of matching administrative units and/or geographic features (93). In case multiple matches are found, the midpoint as well as minimum distance between the matches are calculated, providing an estimate of the potential error associated with the coordinates. Again, all taxon names were checked using the Taxonomic Name Resolution Service (72, 73), with the Tropicos database, the Plant List, USDA’s Plants database, the Global Compositae Checklist, and the International Legume database as sources. Subsequently, we removed all accessions that were recorded from cultivated sites (e.g., botanical gardens), as well as those with large uncertainties (e.g., accessions that only identified Java as the locality and hence returned coordinates for the center of the island).
We built species-specific distribution models for all Javanese seed plant species represented by more than four unique localities across Indonesia (1,895 species) using an ensemble modeling approach as implemented in the SSDM (94) package in R (95). We employed all 10 algorithms incorporated in SSDM that are commonly used for species distribution modeling, with pseudoabsences selected based on the recommendations for each algorithm. Modeling was repeated 10 times with each algorithm using the k-folds cross-validation method where the data for each species are partitioned into three training sets and one evaluation set. The individual habitat suitability scores were converted into presence/absence data by applying a threshold that maximizes the true skill statistic (96) and then stacked to obtain the species richness data. We used both climate and soil data on a 5 arc-min resolution (approximately 10 km) as environmental predictors. Bioclimatic variables were downloaded from the Worldclim project (97) and soil characteristics were obtained from the SoilGrids database (98) and rescaled to a 5 arc-min resolution (a list of all variables can be found in SI Appendix). An initial analysis on 100 randomly selected species with all 44 environmental variables was used to select the most relevant variables (based on their relative importance in the ensemble model), excluding all variables with a correlation coefficient of >0.7 for the full model run of all species. In total, 14 environmental variables (including 6 climatic and 8 soil variables) were selected for the full model run (SI Appendix, Table S10). The evaluation of the stacked species distribution models is also implemented in the SSDM package and five evaluation metrics were calculated as described by Pottier et al. (99): While the species richness error describes the difference between the predicted and observed species richness, the assemblage kappa gives the proportion of specific agreement. The number of true negatives is given by the assemblage specificity, whereas the assemblage sensitivity gives the true positives. The Jaccard index is a metric describing the similarity and diversity of communities. Following the ensemble modeling, we then incorporated the remaining 1,605 species (with fewer than four unique localities) as point localities and cropped all maps to our focal region (Java).
To visualize patterns of metabolite diversity, we calculated the Shannon index (Eq. 1) for each grid cell, where pi is the proportion of metabolite i (relative to the total number of metabolites), with S being the total number of unique metabolites found in this grid cell. This index thus takes into account the relative abundance of each metabolite within a grid cell.
| [1] |
Data Availability.
The metabolite information (information on natural products, metabolic pathways, and biological activities), phylogenetic and distribution data, as well as the environmental data are publicly available (see above for details). Other data (e.g., taxonomic data and R code) are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank C. Haris Saslis-Lagoudakis for advice and discussion. Two anonymous reviewers and the editors provided constructive comments on an earlier version of this manuscript. The analyses were run on the Leibniz Institute of Plant Biochemistry computational cluster and the High-Performance Computing Cluster EVE, a joint effort of the Helmholtz Centre for Environmental Research (UFZ) and the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, and we thank Christian Krause (iDiv) for his support. This work was supported by grants from the German Federal Ministry of Education and Research to A.N.M.-R. and L.A.W. (16GW0120K and 16GW0123). J.S. was supported by the German Centre for Integrative Biodiversity Research (iDiv) Halle‐Jena‐Leipzig funded by the German Research Foundation (grant FZT 118). We thank OntocChem GmbH, Halle (Saale) Germany for support with the phytochemical search and ontologies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915277117/-/DCSupplemental.
References
- 1.Roca I., et al. , The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 6, 22–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laxminarayan R., et al. , Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Firn R. D., Bioprospecting—Why is it so unrewarding? Biodivers. Conserv. 12, 207–216 (2003). [Google Scholar]
- 4.Fischbach M. A., Walsh C. T., Antibiotics for emerging pathogens. Science 325, 1089–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman D. J., Cragg G. M., Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Cronk D., “High-throughput screening” in Drug Discovery and Development, Hill R. G., Rang H. P., Eds. (Churchill Livingstone, London, ed. 2, 2013), pp. 95–117. [Google Scholar]
- 7.Harvey A. L., Edrada-Ebel R., Quinn R. J., The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Theis N., Lerdau M., The evolution of function in plant secondary metabolites. Int. J. Plant Sci. 164, S93–S102 (2003). [Google Scholar]
- 9.World Health Organization , “Traditional medicine-growing needs and potential” WHO Policy Perspectives on Medicine (WHO, Geneva, 2002). [Google Scholar]
- 10.Kew R. B. G., The State of the World’s Plants Report 2017 (Royal Botanic Gardens, Kew, 2017). [Google Scholar]
- 11.Verpoorte R., Pharmacognosy in the new millennium: Leadfinding and biotechnology. J. Pharm. Pharmacol. 52, 253–262 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Gibbons S., Plants as a source of bacterial resistance modulators and anti-infective agents. Phytochem. Rev. 4, 63–78 (2005). [Google Scholar]
- 13.Das K., Tiwari R., Shrivastava D., Techniques for evaluation of medicinal plant products as antimicrobial agent: Current methods and future trends. J. Med. Plants Res. 4, 104–111 (2010). [Google Scholar]
- 14.Hartmann T., From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 68, 2831–2846 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Mendelsohn R., Balick M. J., The value of undiscovered pharmaceuticals in tropical forests. Econ. Bot. 49, 223–228 (1995). [Google Scholar]
- 16.Fabricant D. S., Farnsworth N. R., The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect. 109 (suppl. 1), 69–75 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyu J., et al. , Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramesha B. T., et al. , Biodiversity and chemodiversity: Future perspectives in bioprospecting. Curr. Drug Targets 12, 1515–1530 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Zhu F., et al. , Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc. Natl. Acad. Sci. U.S.A. 108, 12943–12948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saslis-Lagoudakis C. H., et al. , The use of phylogeny to interpret cross-cultural patterns in plant use and guide medicinal plant discovery: An example from Pterocarpus (Leguminosae). PLoS One 6, e22275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saslis-Lagoudakis C. H., Rønsted N., Clarke A. C., Hawkins J. A., “Evolutionary approaches to ethnobiology” in Evolutionary Ethnobiology, Albuquerque U. P., De Medeiros P. M., Casas A., Eds. (Springer, 2016), pp. 59–72. [Google Scholar]
- 22.Rønsted N., Savolainen V., Mølgaard P., Jäger A. K., Phylogenetic selection of Narcissus species for drug discovery. Biochem. Syst. Ecol. 36, 417–422 (2008). [Google Scholar]
- 23.Lübken T., Schmidt J., Porzel A., Arnold N., Wessjohann L., Hygrophorones A-G: Fungicidal cyclopentenones from Hygrophorus species (Basidiomycetes). Phytochemistry 65, 1061–1071 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Delgoda R., Murray J. E., “Evolutionary perspectives on the role of plant secondary metabolites” in Pharmacognosy, Badal S., Delgoda R., Eds. (Academic Press, Boston, 2017), pp. 93–100. [Google Scholar]
- 25.Edger P. P., et al. , The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. U.S.A. 112, 8362–8366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno S., Evolution by Gene Duplication (Springer, New York, 1970). [Google Scholar]
- 27.Pichersky E., Nomad DNA–A model for movement and duplication of DNA sequences in plant genomes. Plant Mol. Biol. 15, 437–448 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Soltis P. S., Soltis D. E., Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Weng J.-K., Philippe R. N., Noel J. P., The rise of chemodiversity in plants. Science 336, 1667–1670 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Wink M., Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64, 3–19 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Losos J. B., Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Wink M., Mohamed G. I. A., Evolution of chemical defense traits in the Leguminosae: Mapping of distribution patterns of secondary metabolites on a molecular phylogeny inferred from nucleotide sequences of the rbcL gene. Biochem. Syst. Ecol. 31, 897–917 (2003). [Google Scholar]
- 33.Courtois E. A., et al. , Evolutionary patterns of volatile terpene emissions across 202 tropical tree species. Ecol. Evol. 6, 2854–2864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst M., et al. , Evolutionary prediction of medicinal properties in the genus Euphorbia L. Sci. Rep. 6, 30531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bay-Smidt M. G., et al. , Phylogenetic selection of target species in Amaryllidaceae tribe Haemantheae for acetylcholinesterase inhibition and affinity to the serotonin reuptake transport protein. S. Afr. J. Bot. 77, 175–183 (2011). [Google Scholar]
- 36.Bohlin L., Göransson U., Alsmark C., Wedén C., Backlund A., Natural products in modern life science. Phytochem. Rev. 9, 279–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt I., Barker F. K., Phylogenetic methods in natural product research. Nat. Prod. Rep. 26, 1585–1602 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Rønsted N., et al. , Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol. Biol. 12, 182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saslis-Lagoudakis C. H., et al. , Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc. Natl. Acad. Sci. U.S.A. 109, 15835–15840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becerra J. X., Noge K., Venable D. L., Macroevolutionary chemical escalation in an ancient plant-herbivore arms race. Proc. Natl. Acad. Sci. U.S.A. 106, 18062–18066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey A. L., Natural products in drug discovery. Drug Discov. Today 13, 894–901 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Roos M. C., Keßler P. J. A., Robbert Gradstein S., Baas P., Species diversity and endemism of five major Malesian islands: Diversity–area relationships. J. Biogeogr. 31, 1893–1908 (2004). [Google Scholar]
- 43.Barthlott W., Mutke J., Rafiqpoor M. D., Kier G., Kreft H., Global centres of vascular plant diversity. Nova Acta Leopold. 92, 61–83 (2005). [Google Scholar]
- 44.Roos M., State of affairs regarding Flora Malesiana: Progress in revision work and publication schedule. Flora Males. Bull. 11, 133–142 (1993). [Google Scholar]
- 45.Mittermeier R. A., Mittermeier C. G., Robles Gil P., Megadiversity: Earth’s Biologically Wealthiest Nations (CEMEX, Mexico, 1997). [Google Scholar]
- 46.Myers N., Biodiversity hotspots revisited. Bioscience 53, 916–917 (2003). [Google Scholar]
- 47.Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A. B., Kent J., Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Budiharta S., et al. , The processes that threaten Indonesian plants. Oryx 45, 172–179 (2011). [Google Scholar]
- 49.van Beukering P. J. H., Cesar H. S. J., Janssen M. A., Economic valuation of the Leuser National Park on Sumatra, Indonesia. Ecol. Econ. 44, 43–62 (2003). [Google Scholar]
- 50.Padoch C., Jessup T., Soedjito H., Kartawinata K., “Complexity and conservation of medicinal plants: Anthropological cases from Peru and Indonesia” in The Conservation of Medicinal Plants, Akerele O., Heywood V., Synge H., Eds. (Cambridge University Press, Cambridge, 1991), pp. 321–328. [Google Scholar]
- 51.Bérdy J., Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Kren V., Martínková L., Glycosides in medicine: “The role of glycosidic residue in biological activity”. Curr. Med. Chem. 8, 1303–1328 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Babu R. H., Savithramma N., Phytochemical screening of underutilized species of Poaceae. BioMedRx 1, 947–951 (2013). [Google Scholar]
- 54.Sut S., Maggi F., Dall’Acqua S., Bioactive secondary metabolites from orchids (Orchidaceae). Chem. Biodivers. 14, e1700172 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Cordell G. A., Quinn-Beattie M. L., Farnsworth N. R., The potential of alkaloids in drug discovery. Phytother. Res. 15, 183–205 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Randall R. P., A Global Compendium of Weeds (RP Randall, Perth, WA, ed. 3, 2017). [Google Scholar]
- 57.Leonti M., et al. , Bioprospecting: Evolutionary implications from a post-olmec pharmacopoeia and the relevance of widespread taxa. J. Ethnopharmacol. 147, 92–107 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Weckerle C. S., Cabras S., Castellanos M. E., Leonti M., Quantitative methods in ethnobotany and ethnopharmacology: Considering the overall flora–hypothesis testing for over- and underused plant families with the Bayesian approach. J. Ethnopharmacol. 137, 837–843 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Schwikkard S. L., Mulholland D. A., Useful methods for targeted plant selection in the discovery of potential new drug candidates. Planta Med. 80, 1154–1160 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Azani N., et al. , A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). Taxon 66, 44–77 (2017). [Google Scholar]
- 61.Farag M. A., Weigend M., Luebert F., Brokamp G., Wessjohann L. A., Phytochemical, phylogenetic, and anti-inflammatory evaluation of 43 Urtica accessions (stinging nettle) based on UPLC-Q-TOF-MS metabolomic profiles. Phytochemistry 96, 170–183 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Porzel A., Farag M. A., Mülbradt J., Wessjohann L. A., Metabolite profiling and fingerprinting of Hypericum species: A comparison of MS and NMR metabolomics. Metabolomics 10, 574–588 (2014). [Google Scholar]
- 63.Pichersky E., Lewinsohn E., Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Mylonakis E., Podsiadlowski L., Muhammed M., Vilcinskas A., Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gatehouse J. A., Plant resistance towards insect herbivores: A dynamic interaction. New Phytol. 156, 145–169 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Leal M. C., Hilário A., Munro M. H., Blunt J. W., Calado R., Natural products discovery needs improved taxonomic and geographic information. Nat. Prod. Rep. 33, 747–750 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Soejarto D. D., et al. , Ethnobotany/ethnopharmacology and mass bioprospecting: Issues on intellectual property and benefit-sharing. J. Ethnopharmacol. 100, 15–22 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Staniek A., et al. , Natural products—Modifying metabolite pathways in plants. Biotechnol. J. 8, 1159–1171 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Staniek A., et al. , Natural products—Learning chemistry from plants. Biotechnol. J. 9, 326–336 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Backer C. A., Bakhuizen van den Brink R. C., Flora of Java (The Rijksherbarium, Leyden, The Netherlands, 1963–1968). [Google Scholar]
- 71.Priyadi H., et al. , Five Hundred Plant Species in Gunung Halimun Salak National Park, West Java: A Checklist Including Sundanese Names, Distribution, and Use (CIFOR, 2010). [Google Scholar]
- 72.Boyle B., et al. , The taxonomic name resolution service: An online tool for automated standardization of plant names. BMC Bioinf. 14, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.TNRS , iPlant Collaborative. Version 4.0. http://tnrs.iplantcollaborative.org/. Accessed 27 October 2016 (2018).
- 74.Angiosperm Phylogeny Group , An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20 (2016). [Google Scholar]
- 75.Afendi F. M., et al. , KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Zeng X., et al. , NPASS: Natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 46, D1217–D1222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M., New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quideau S., Deffieux D., Douat-Casassus C., Pouységu L., Plant polyphenols: Chemical properties, biological activities, and synthesis. Angew. Chem. Int. Ed. Engl. 50, 586–621 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Michels K., et al. , A fluorescence-based bioassay for antibacterials and its application in screening natural product extracts. J. Antibiot. 68, 734–740 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Nyman T., Julkunen-Tiitto R., Chemical variation within and among six northern willow species. Phytochemistry 66, 2836–2843 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Zidorn C., Sesquiterpene lactones and their precursors as chemosystematic markers in the tribe Cichorieae of the Asteraceae. Phytochemistry 69, 2270–2296 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Dias D. A., Urban S., Roessner U., A historical overview of natural products in drug discovery. Metabolites 2, 303–336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cragg G. M., Newman D. J., Biodiversity: A continuing source of novel drug leads. Pure Appl. Chem. 77, 7–24 (2005). [Google Scholar]
- 86.Chen Y., de Bruyn Kops C., Kirchmair J., Data resources for the computer-guided discovery of bioactive natural products. J. Chem. Inf. Model. 57, 2099–2111 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Smith S. A., Brown J. W., Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 105, 302–314 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Magallón S., Gómez-Acevedo S., Sánchez-Reyes L. L., Hernández-Hernández T., A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Fritz S. A., Purvis A., Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv. Biol. 24, 1042–1051 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Webb C. O., Ackerly D. D., Kembel S. W., Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (2008). [DOI] [PubMed] [Google Scholar]
- 91.GBIF.org , GBIF Occurrence Download. 10.15468/dl.jj7fsg. Accessed 3 May 2018. [DOI]
- 92.Gratton P., et al. , A world of sequences: Can we use georeferenced nucleotide databases for a robust automated phylogeography? J. Biogeogr. 44, 475–486 (2017). [Google Scholar]
- 93.GeoNames ,The GeoNames geographical database. http://www.geonames.org/. Accessed 14 June 2018.
- 94.Schmitt S., Pouteau R., Justeau D., Boissieu F., Birnbaum P., ssdm: An R package to predict distribution of species richness and composition based on stacked species distribution models. Methods Ecol. Evol. 8, 1795–1803 (2017). [Google Scholar]
- 95.R Development Core Team , R: A Language and Environment for Statistical Computing v.3.3.1 (R Foundation for Statistical Computing, Vienna, Austria, 2016). [Google Scholar]
- 96.Allouche O., Tsoar A., Kadmon R., Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232 (2006). [Google Scholar]
- 97.Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005). [Google Scholar]
- 98.Hengl T., et al. , SoilGrids250m: Global gridded soil information based on machine learning. PLoS One 12, e0169748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pottier J., et al. , The accuracy of plant assemblage prediction from species distribution models varies along environmental gradients. Glob. Ecol. Biogeogr. 22, 52–63 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolite information (information on natural products, metabolic pathways, and biological activities), phylogenetic and distribution data, as well as the environmental data are publicly available (see above for details). Other data (e.g., taxonomic data and R code) are available from the corresponding authors upon request.