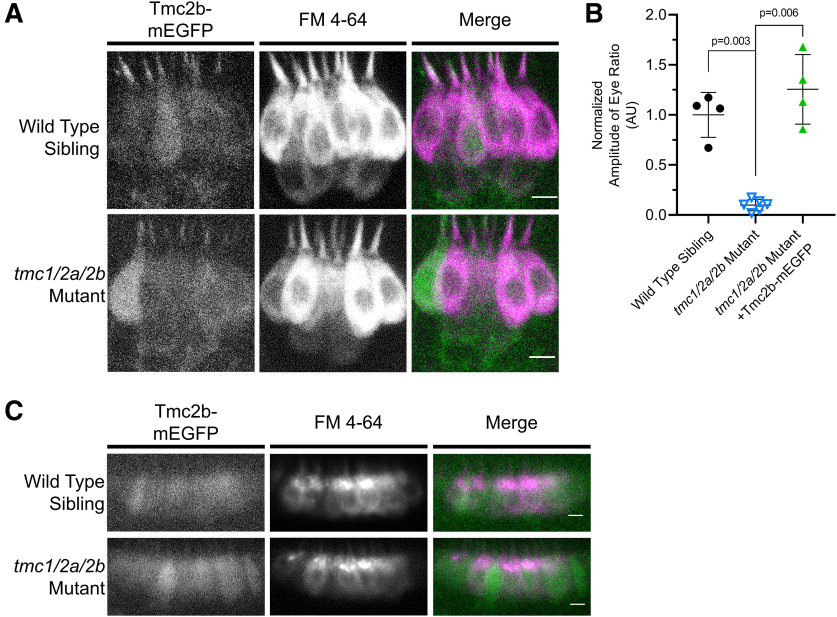
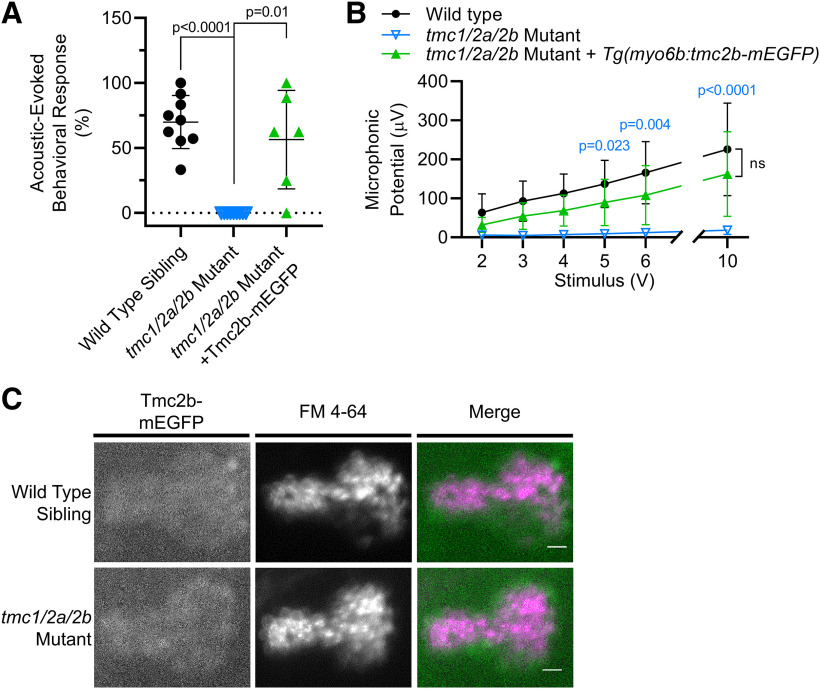
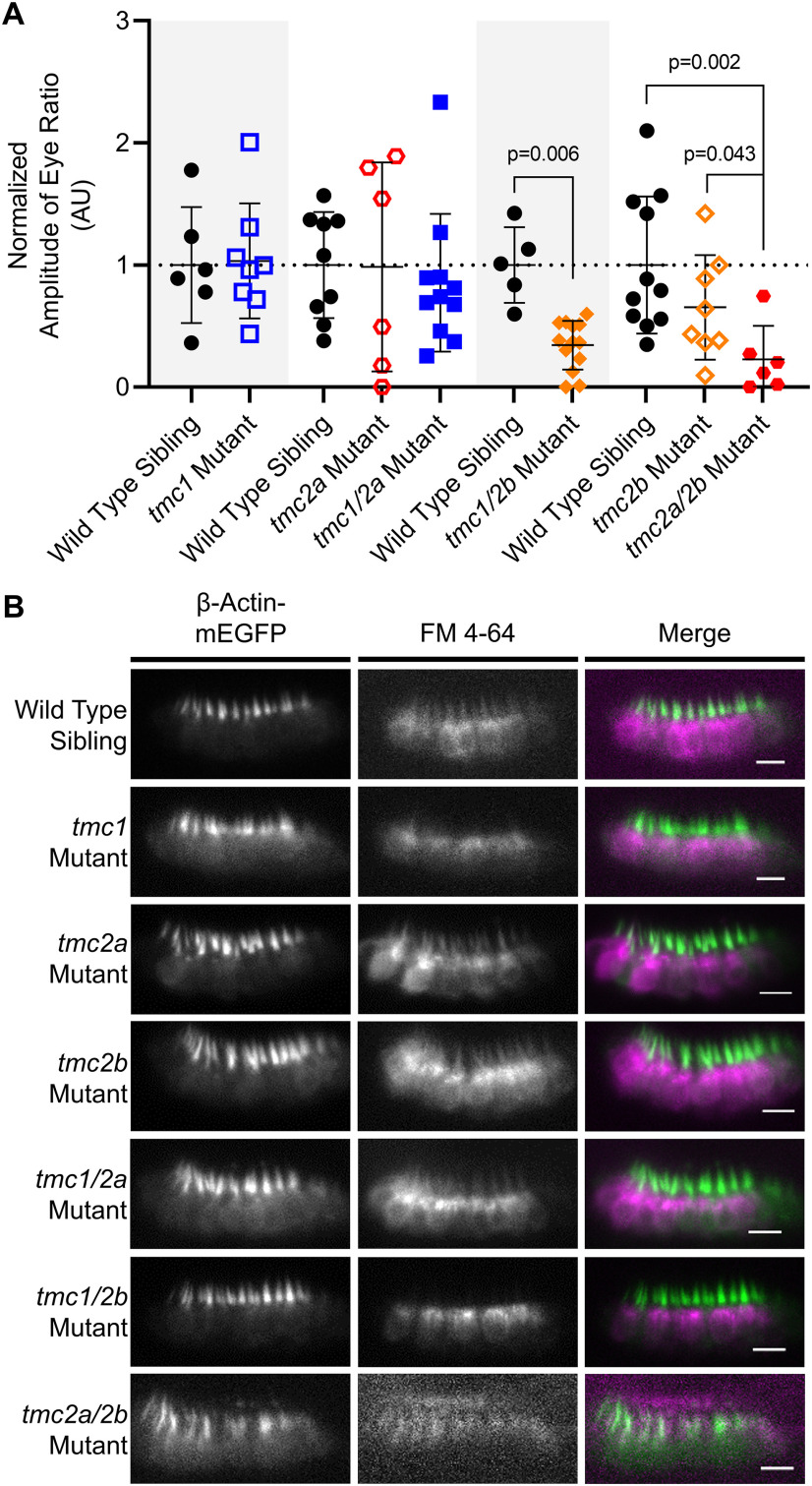
Abstract
Gap junctions are ubiquitous throughout the nervous system, mediating critical signal transmission and integration, as well as emergent network properties. In mammalian retina, gap junctions within the Aii amacrine cell-ON cone bipolar cell (CBC) network are essential for night vision, modulation of day vision, and contribute to visual impairment in retinal degenerations, yet neither the extended network topology nor its conservation is well established. Here, we map the network contribution of gap junctions using a high-resolution connectomics dataset of an adult female rabbit retina. Gap junctions are prominent synaptic components of ON CBC classes, constituting 5%–25% of all axonal synaptic contacts. Many of these mediate canonical transfer of rod signals from Aii cells to ON CBCs for night vision, and we find that the uneven distribution of Aii signals to ON CBCs is conserved in rabbit, including one class entirely lacking direct Aii coupling. However, the majority of gap junctions formed by ON CBCs unexpectedly occur between ON CBCs, rather than with Aii cells. Such coupling is extensive, creating an interconnected network with numerous lateral paths both within, and particularly across, these parallel processing streams. Coupling patterns are precise with ON CBCs accepting and rejecting unique combinations of partnerships according to robust rulesets. Coupling specificity extends to both size and spatial topologies, thereby rivaling the synaptic specificity of chemical synapses. These ON CBC coupling motifs dramatically extend the coupled Aii-ON CBC network, with implications for signal flow in both scotopic and photopic retinal networks during visual processing and disease.
SIGNIFICANCE STATEMENT Electrical synapses mediated by gap junctions are fundamental components of neural networks. In retina, coupling within the Aii-ON CBC network shapes visual processing in both the scotopic and photopic networks. In retinal degenerations, these same gap junctions mediate oscillatory activity that contributes to visual impairment. Here, we use high-resolution connectomics strategies to identify gap junctions and cellular partnerships. We describe novel, pervasive motifs both within and across classes of ON CBCs that dramatically extend the Aii-ON CBC network. These motifs are highly specific with implications for both signal processing within the retina and therapeutic interventions for blinding conditions. These findings highlight the underappreciated contribution of coupling motifs in retinal circuitry and the necessity of their detection in connectomics studies.
Keywords: bipolar cell, circuit, connectomics, coupling, gap junction, retina
Introduction
Gap junctions are intercellular channels electrically and metabolically coupling cells through direct exchange of ions and small molecules. Ubiquitous throughout invertebrate and vertebrate nervous systems, gap junctions provide the anatomic substrate for electrical synapses. Like their chemical synapse counterparts, gap junctions exhibit developmentally regulated tissue- and cell-specific expression (Lin et al., 2005; Kihara et al., 2006), divergent conductance and selectivity (Veenstra et al., 1995), and extensive plasticity through gating modulation (O'Brien, 2019).
Historically believed to simply facilitate rapid bidirectional signal propagation, electrical synapses are increasingly acknowledged as key network components with diverse roles in the transmission and integration of signals (Nagy et al., 2018). Gap junctions are well known for mediating synchronization of oscillatory activity among neuronal ensembles, the fundamental mechanism underlying emergence of the central pattern generator in the embryonic zebrafish spinal cord (Saint-Amant and Drapeau, 2001), and implicated in cognition, including attention, learning, and memory (Hormuzdi et al., 2001; Buhl et al., 2003; Long et al., 2004; Frisch et al., 2005; Coulon and Landisman, 2017). Additional emergent network properties afforded by synchrony (Marder, 1998) include the following: coincidence detection (Das et al., 2017), enhanced signal saliency (DeVries et al., 2002), pathway sensitization (Yang et al., 1990; Rash et al., 2013), low-pass filtering triggering desynchronization (Vervaeke et al., 2010), and shunting of presynaptic signals for regulatory feedback (Kawano et al., 2011). Despite repeated findings of critical roles for gap junctions in the formation and function of neural networks, the expressing cells, participating circuits, and function remain largely unknown, and oft ignored.
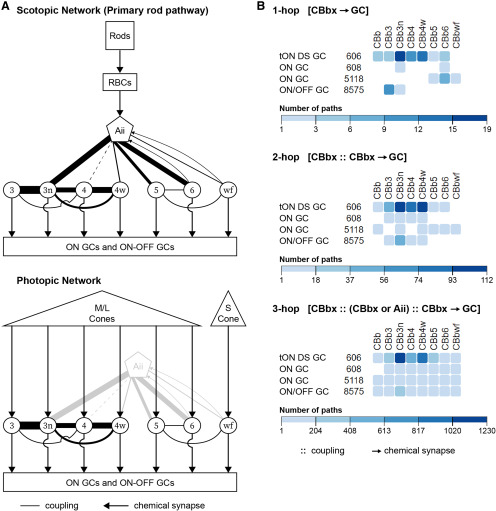
Gap junctions in mammalian retina are best known for their role in scotopic night vision as obligate components of the primary pathway. In dim light, signals produced by light-sensing rod photoreceptors are collected by rod bipolar cells and transferred through chemical synapses to Aii amacrine cells (ACs). Aii cells distribute these scotopic signals into the ON/OFF pathways of photopic day vision established by the opposing functional responses of ON and OFF cone bipolar cell (CBC) superclasses to light. Using sign-inverting chemical synapses onto OFF CBCs and sign-conserving electrical synapses with ON CBCs, Aii cells thereby maintain opposing light responses (Kolb and Famiglietti, 1974; Marc et al., 2014). CBCs transmit these scotopic signals (or photopic signals from cone photoreceptors) to ganglion cells (GCs) for projection to the brain. Absence of the connexin proteins supporting Aii-ON CBC gap junctions strongly impairs scotopic signaling (Guldenagel et al., 2001; Deans et al., 2002; Maxeiner et al., 2005). The 5-7 classes of mammalian ON CBCs are believed to serve parallel processing channels and appear to receive differential input from Aii cells (McGuire et al., 1984; Cohen and Sterling, 1990; Veruki and Hartveit, 2002a; Tsukamoto and Omi, 2017). Gap junctions or coupling have also been reported between CBCs (Kolb, 1979; Marc et al., 1988; Cohen and Sterling, 1990; Umino et al., 1994; Luo et al., 1999; Mills, 1999; Dacey et al., 2000; Jacoby and Marshak, 2000; Arai et al., 2010; Kántor et al., 2017; Tsukamoto and Omi, 2017). Unfortunately, partner identification largely remains unresolved, and species-specific divergence is suggested. Understanding ON CBC coupling motifs is critical as they contribute to shaping visual processing in both the scotopic and photopic networks (Guldenagel et al., 2001; Deans et al., 2002; Maxeiner et al., 2005; Demb and Singer, 2012; Farrow et al., 2013; Kuo et al., 2016; Seilheimer et al., 2020). Moreover, these gap junctions subserve aberrant hyperactivity contributing to visual impairment in retinal degenerative disease (Trenholm et al., 2012; Ivanova et al., 2016).
Here, we used serial section transmission electron microscopy (TEM)-based ultrastructural connectomics to map the ground truth for gap junction contribution to retinal circuits. We report class-specific coupling patterns for all 7 rabbit ON CBC classes that are richer and more extensive than previously appreciated. These motifs reveal extensive coupling both within and across parallel processing streams of the rabbit retina, with implications for photopic and scotopic visual networks and their disruption in disease.
Materials and Methods
Connectomics volume RC1
Volume construction
Retinal Connectome 1 (RC1) is an ultrastructural dataset acquired at 2.18 nm/pixel resolution from the retina of a light-adapted 13-month-old female Dutch Belted rabbit (Oregon Rabbitry). Dataset is freely available at https://connectomes.utah.edu. RC1 spans the inner nuclear layer (INL) through GC layer of a 0.25-mm-diameter field of mid-peripheral retina. Methods concerning tissue acquisition and processing, volume assembly, visualization, and annotation have been extensively detailed (Anderson et al., 2009, 2011a,b; Lauritzen et al., 2013, 2019; Marc et al., 2013, 2014). In short, the RC1 dataset was constructed from 371 serial TEM sections (70-90 nm thick), captured via Automated Transmission Electron Microscopy at 5000×, combined with 11 optical sections intercalated through the inner plexiform layer (IPL), 6 capstone optical sections in the INL, and another 12 capstone optical sections in the GC layer. These optical sections were probed for small-molecule signals for computational molecular phenotyping (Marc et al., 1995). Sections were aligned into a single volume using the NCR ToolSet, which has since been replaced by Nornir (RRID:SCR_016458). All protocols were in accord with Institutional Animal Care and Use protocols of the University of Utah, the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research, and the Policies on the Use of Animals and Humans in Neuroscience Research of the Society for Neuroscience.
Dataset viewing and annotation
Dataset visualization and annotation were performed using the Viking Viewer for Connectomics (RRID:SCR_005986; referred to hereafter as “Viking”), available via free license for educational use through the University of Utah. Viking allows concurrent volume annotation by multiple users across html-compliant protocols. Our manual annotation process involves users converting the raw 3D image data into a set of 2D shape geometries stored in a central Microsoft SQL Server Spatial Database. Each cell is tracked through the cross-sectional images of the 3D volume while recording the boundaries of the cell membrane and cellular features (e.g., ribbon presynapse, postsynaptic density, etc.) into the database. An “annotation” is a single disconnected 2D outline of a cell or cellular feature on a single section image and any user-defined data. Each cellular feature, or “structure,” is described by a set of annotations interconnected across sections that collectively describe its 3D morphology. For legacy reasons, RC1 cell boundaries were encoded using a 2D disk sized to the largest diameter that can be completely contained within the continuous shape of the cell's membrane on a given section. Membrane-associated structures, such as gap junctions and postsynaptic densities, were encoded as poly-lines in the database and visualized as curves in the Viking user interface. Viking organizes structures hierarchically and relationally. All cellular features are internal features of a cell and organized in a parent-child hierarchy. Relational connections are encoded as links between structures. For example, a ribbon presynapse in a bipolar cell would be a “child” of that bipolar cell “parent.” In turn, the ribbon presynapse could be linked to a postsynaptic density of the opposed cell to record the relationship between the structures. Importantly, a gap junction instance requires 2 gap junction annotations, each a child of their respective parent cells, with a bidirectional link. As some candidate gap junctions present with oblique orientations, confidence values were assigned to each annotation to reflect their confirmed versus candidate status and results of reimaging. Reimaging to obtain optimized section tilt for structure validation was performed at 40,000× magnification (0.27 nm/pixel resolution) with goniometric tilt.
Dataset analysis
All cells and structures are automatically assigned unique identifiers in the Microsoft SQL Server Spatial Database during annotation and numerically indexed to their location, shapes, and connectivity within RC1. This enables direct queries of the SQL database (SQL queries) to extract descriptions of cellular composition and network interactions, as well as evaluation in a host of custom, open-source software tools for visualization or analysis (for references, links, and RRIDs, see Software accessibility). Cell morphology was visualized on a per-cell basis in ball-and-stick plots through morphology export functions in Viking to the network graph visualization application Tulip. The morphology of individual cells and spatial organization of large groups of cells were visualized in the 3D rendering environments VikingPlot and VikingView or through import into Blender. Morphometric features were retrieved or computed using Microsoft SQL queries. Use of a spatial database allows easy access to the established computational geometry analysis methods referred to throughout the paper. Synaptology and connectivity were explored or analyzed using Microsoft SQL queries, Cell Sketches, Graffinity, and/or through network export for Tulip using the custom Python-based TulipPaths plug-in developed to query the connectivity graphs.
Cell identification
The small-molecule signals from immunolabeled capstone and intercalated sections through the cell somas in the INL of the RC1 dataset for glutamate, glycine, and 4-aminobutyrate (GABA) enabled classification of every soma in the INL as belonging to an OFF CBC, ON CBC, rod bipolar cell, narrow-field glycinergic AC (GAC), wide-field GABAergic AC, or Müller cells (Anderson et al., 2009, 2011b). The characteristic signature of high glutamate, medium glycine, and no GABA initially identified the subset of ON CBC candidates. ON CBC identity was confirmed by the presence of ribbon-type and absence of conventional-type presynapses. Classification of these cells proceeded as described in Results based on axonal arbor features, including field area and the depth and breadth of stratification within the IPL, tiling, and synaptology. This process was performed iteratively until every candidate ON CBC with arbors fully contained within the volume was classified.
Synaptology and coupling profiles
Tabulations of synapses and other child structures, as well as details regarding the partner cell and child structure, were obtained from Microsoft SQL queries or Cell Sketches. Cell Sketches software was developed to support the visual analysis of neuron statistics and spatial properties. It pulls data from RC1's public interface (http://connectomes.utah.edu/export/odata.html) and is implemented in JavaScript using AngularJS (http://www.angularjs.org) and D3.js (https://d3js.org). Cell Sketches counts synapse statistics through iterative database queries.
Cell morphometrics and distribution
Aii and ON CBC stratification
To control for the inherent volume curvature and local variability in the position of the IPL, the section corresponding to the top and bottom of the IPL was determined based on the emergence or disappearance of neuronal processes at the XY somal position for 22 RC1 Aii cells directly from volume visualization in Viking. For Aii cells impacted by the block refacing event in strata 4/5 of the IPL, the bottom of the IPL was estimated using the average IPL thickness calculated from nonimpacted Aii cells (n = 8 of 22 Aii cells, 225 ± 4 sections). Using vertical views of 3D reconstructions in VikingView of Aii cell morphology and synaptology, the ON/OFF boundary position was assessed at the XY somal position as the bottom of the lobular dendrites and top of gap junction compartmentalization (Marc et al., 2014). The consistency of the calculated ON/OFF boundary position within the IPL (45% ± 3% IPL; n = 22 Aii cells) suggests that our normalization method accounted well for volume curvature and local variation is limited. Stratification of ON CBC arbors was evaluated from histograms of annotation frequency by RC1 section, using a 5-section bin size. To control for volume curvature, the top and bottom of a cell's axonal arbor were determined as the start or end section, respectively, of the bin situated at the inflection points of the histogram curve. As the block refacing event also impacted determination of the bottom of the IPL for ON CBC axonal stratification measures, the position of these sections within the IPL was calculated as the average section difference from the top or bottom of the IPL determined for the nearest Aii cell(s).
Branch path complexity
Using the 3D reconstructions rendered in VikingView or VikingPlot, binary trees were manually drawn to capture the branching topology, ignoring branch lengths. Tree elements (Uylings and van Pelt, 2002) were marked and the number of branch terminals derived. Branch path complexity was determined using the parameterization and calculation reported by Elliott et al. (2015).
Mosaic analysis
Class mosaics were evaluated for regularity using the conformity ratio (mean/SD of the nearest neighbor distances), as this measure is least impacted by boundary effects (Cook, 1996). Nearest neighbor distances were obtained using a Microsoft SQL query that calculates the distance between the geometric centroids of annotations on a given section in the RC1 database. Because of slight differences in somal depths across the volume, sections were chosen to optimize soma position and remove annotation ambiguity (section 0001-CBb5, CBb6; section 0020-CBb4, CBb4w; section 0030-CBb3, CBb3n).
Intersection analysis
The area of spatial overlap between cell arbors was calculated using a Microsoft SQL query (Anderson et al., 2019). First, the 2D convex hulls of all cells in 2 specified classes were determined. The intersection of the convex hulls for all pairwise combinations between the 2 classes was identified and the overlap fraction determined. The area of spatial overlap was then computed by multiplying the fraction by the area of the cell's convex hull. Using the R software environment, the data were fitted to a linear model with a y intercept of zero and adjusted R2 values obtained. An ANCOVA was then run using package “car” and the post hoc Scheffé test run using package “agricolae.”
Size distribution of gap junctions
The areas of the individual gap junction plaques formed by the 37 ON CBCs examined in this study were estimated by summing the products of the annotation lengths on each section multiplied by section thickness. Polyline annotations for gap junctions were centered on the inner leaflet of the parent cell's plasma membrane and extended the length of the zippered gap junction. In oblique and en face views, annotations were centered in the middle of the density and extended the length of the darkest continuous portion. Area values were obtained through a Microsoft SQL query of the spatial database, which assumed a 90 nm section thickness. Freeze-fracture studies have revealed that gap junctions of the rodent IPL exist in a variety of configurations (Kamasawa et al., 2006). Our analyses are restricted to the ON sublamina, where string and ribbon configurations are virtually absent. Thus, the gap junctions described in this report likely exist in plaque (conventional crystalline or noncrystalline) or reticular (containing small voids) configurations, supporting our methodology for size estimation. Plasticity evoked by light adaptation and neurotransmission primarily alters the open conductance of existing channels by controlling their phosphorylation state, rather than promoting changes in the abundance or type of channels. Furthermore, while light-dependent changes in gap junctional area (arising from changes in connexon density) have been reported for gap junctions coupling horizontal cells in teleosts (Kurz-Isler and Wolburg, 1986, 1988; Baldridge et al., 1989; Kohler et al., 1990; Washioka et al., 1991; Kurz-Isler et al., 1992), no such changes in connexon density or changes in the number or distribution of gap junction configurations were reported in the IPL of rat or mice under different light adaptation conditions and points in the circadian cycle (Kamasawa et al., 2006). As each gap junction is represented by 2 linked gap junction annotations, 1 belonging to each of the coupled parent cells, size estimates for only 1 annotation was used to prevent overrepresentation of homocellular gap junctions during analyses, as appropriate. The size distributions for gap junctions mediating various coupling modes both in sum and within classes were compared using pairwise Wilcoxon rank-sum tests with continuity correction using the R software environment. All reported p values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure (α = 0.5).
Spatial distribution of gap junctions
Positions of the 1339 gap junctions formed by the 37 ON CBCs detailed in this study were assessed in the horizontal plane as a function of distance from the geometric center of the parent cell's convex hull using the spatial properties derived by Cell Sketches. Using the 2D (XY) position of location annotations in the RC1 database, Cell Sketches computes the convex hull of structures using an implementation of Andrew's Monotone Chain Algorithm (Bostock et al., 2011). Geometric centroids of these convex hulls are then used to compute the 2D distance between a child structure and the parent cell's axonal territory. As bipolar cell axonal arbors are best described as star domains, to evaluate the distribution of gap junctions relative to arbor boundaries, the vertices of the convex hull for each cell were located, annotated as child structures, and distances computed as for gap junctions. As each gap junction is represented by 2 linked gap junction annotations, 1 belonging to each of the coupled parent cells, distance measures for only 1 annotation were used to prevent overrepresentation of homocellular gap junctions in the analyses, as appropriate. The spatial distributions for gap junctions mediating various coupling modes and convex hull vertices were compared using pairwise Wilcoxon rank-sum tests with continuity correction using the R software environment. All reported p values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure (α = 0.5).
Glycine signatures
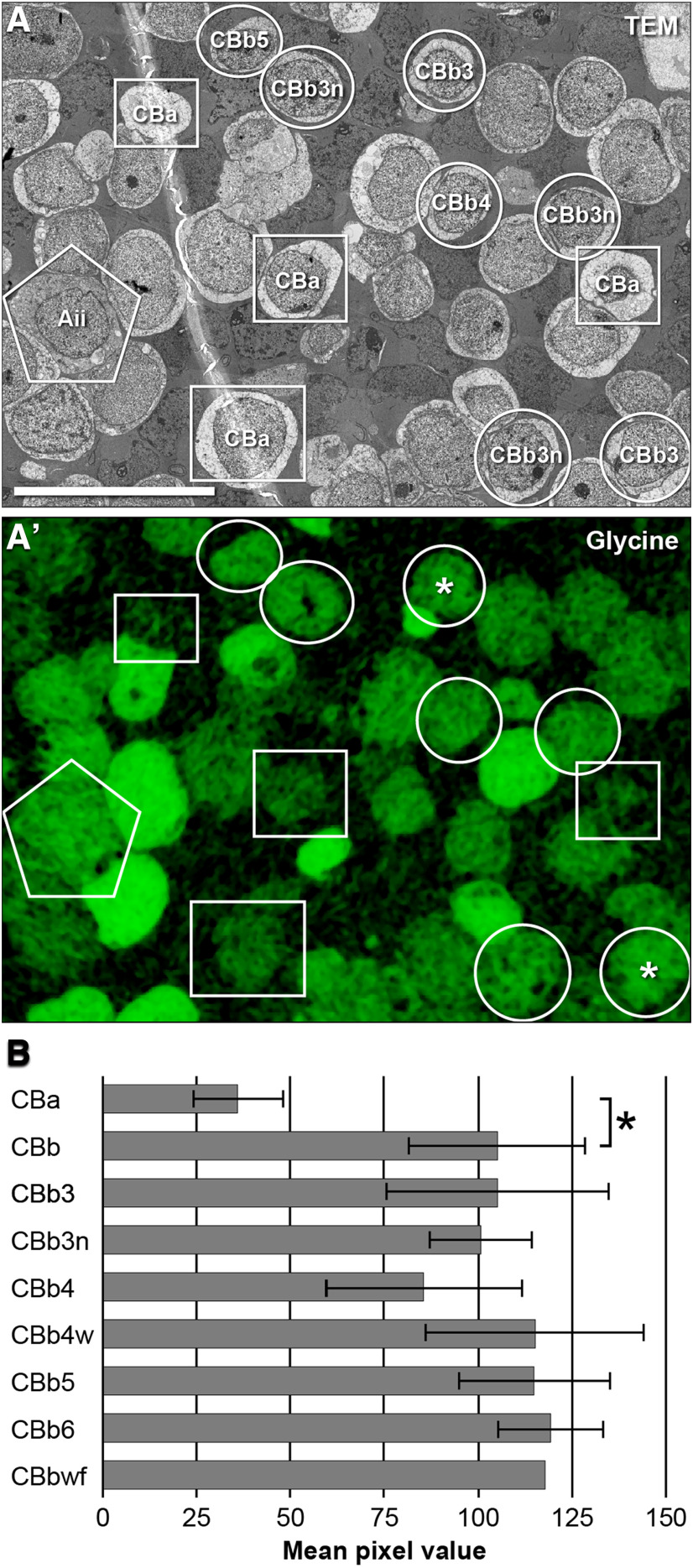
The use of quantitative small-molecule signatures, including glycine to define ON CBC cells coupled to GACs and Aii cells, has been described previously (Marc, 1999; Anderson et al., 2011b; Marc et al., 2014). Every ON CBC in RC1 has its soma or axon aligned with sections 1, 30, and/or 152 containing quantitative grayscale imagery of glycine immunoreactivity (Anderson et al., 2011b), spanning intracellular concentrations ranging from ≈ 0.4 to 10 mm (Jones et al., 2003; Marc and Jones, 2003). All retinal cells contain measurable levels of glycine. Most cells (e.g., rod bipolar cells, OFF CBCs, Müller cells) maintain very low levels at ≤0.1 mm, whereas true GACs reach 0.6–10 mM (Kalloniatis et al.,1996; Marc et al., 2014). ON CBCs lie in between, with pixel values corresponding to 0.2–0.5 mm. Such glycine signals have been interpreted as arising from gap junctional coupling (Marc and Liu, 1984; Cohen and Sterling, 1986; Pourcho and Goebel, 1987; Vaney et al., 1998; Pow and Hendrickson, 2000; Petrides and Trexler, 2008; Marc et al., 2014). Histograms of quantitative glycine signal were obtained for each bipolar cell in RC1, and the mean pixel value was calculated using ImageJ (RRID:SCR_003070). Glycine values reported here are higher than those reported previously (Anderson et al., 2011b) due to sampling using a larger ROI that better matches full cell histograms, but changes no cell assignments. Glycine signal in ON versus OFF CBCs was compared using conventional parametric statistics (Welch two-sample t test, two-tailed, heteroscedastic) using the R software environment.
Image preparation
Viking (Anderson et al., 2009, 2011a; Lauritzen et al., 2013, 2019; Marc et al., 2014) was used for all TEM image viewing, small-molecule overlays, and image annotation. Small-molecule optical overlays combined TEM grayscale data with hue and saturation of the small-molecule optical signal as described previously (Anderson et al., 2011b). Pseudocoloring of cells in TEM images for figures was accomplished in Adobe Photoshop CC 2018 (Adobe; RRID:SCR_014199) by creating separate color layers in either color or overlay blend mode with an opacity of 40%-70% that were then merged with the original TEM image. High-resolution TEM recaptures (40,000× magnification, 0.27 nm/pixel resolution) and enlarged native views of structures directly from Viking were min-max contrast-stretched and γ remapped to 1.3 in Adobe Photoshop CC 2018 to improve visualization, unless noted otherwise. Density plots were acquired with Fiji (ImageJ 1.52p; RRID:SCR_002285) for the ROI denoted in the figures. Graphs were generated in Microsoft Excel 2016. 3D cell reconstructions were rendered in MATLAB (RRID:SCR_001622) from Viking annotations (Anderson et al., 2011a,b) and displayed using VikingPlot. Cell features (synapses, adherens junctions, etc.) are shown unscaled, unless noted otherwise, and rendered in 3D visualizations as volumetric shapes from circles with a diameter equal to that of the summed polyline lengths on each section. Cells in Figures 1A and 7A illustrating stratification were chosen for their close proximity and rendered together but with only 1 cell visible at a time to maintain spatial relationships. Renderings were layered and cells moved laterally to facilitate visibility and ordering. Inherent volume curvature was corrected for using 2 CBb3 cells on either side for the y axis and the CBbwf arbor along the x axis based on reference to Aii cells. IPL and ON/OFF boundaries were defined using neighboring Aii cell morphology and connectivity compartmentalization. Connectivity graphs were generated using Graffinity. All graphs and images were imported into Adobe Illustrator CC 2018 (Adobe; RRID:SCR_010279) for final figure organization.
Figure 1.
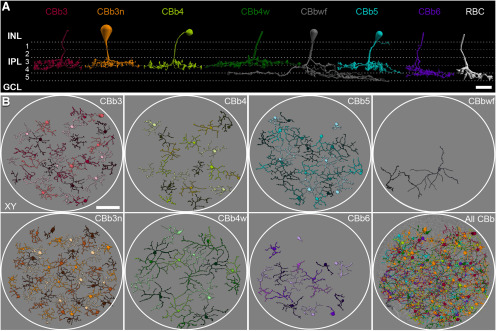
Classification of rabbit ON CBCs from connectome RC1. A, 3D reconstructions of representative cells for each class. All cells stratify in the ON sublamina (strata 3-5) of the IPL. A rod bipolar cell is shown for reference. Dendrites and some somas are not shown. B, Horizontal views of identified ON CBCs by class. Cells within class sheets are differentially colored for clarity. Cells in “All CBb” are colored by class label according to the scheme in A, and overlaid onto TEM ultrastructure at the level of the ON sublamina of the IPL. White circles approximate the volume boundaries of RC1. Scale bars: A, 10 µm; B, 50 µm. GCL, Ganglion cell layer; RBC, rod bipolar cell.
Figure 7.
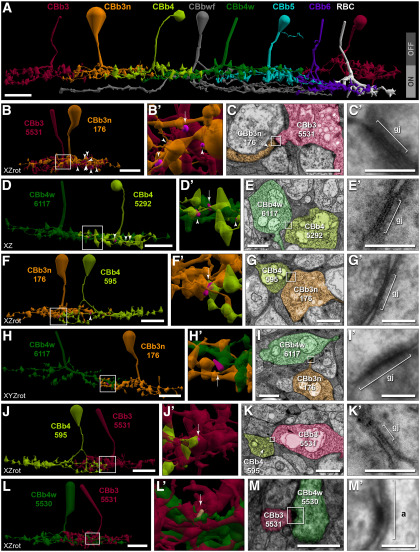
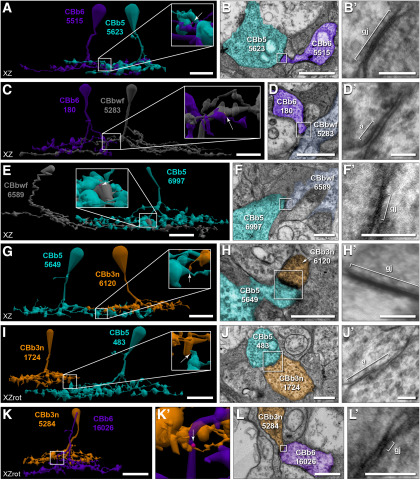
Cross-class homocellular coupling topologies of CBb3/3n/4/4w. A, Extensive costratification of ON CBC axonal arborizations. B, C, Extensive CBb3n coupling with CBb3. B, Twelve of the 22 cross-class gap junctions (magenta; arrow and arrowheads in B,B′) between CBb3n 176 (orange) and CBb3 5531 (maroon) are visible. B′, Enlarged view of boxed region in B. C, TEM image of gap junction 54567 (arrow in B′). C′, Reimaging of boxed region in C at 0.27 nm/pixel resolution with goniometric tilt. D, E, Frequent CBb4w coupling with CBb4. D, Six of the 10 cross-class gap junctions (magenta; arrow, arrowheads in D,D′) between CBb4w 6117 (green) and CBb4 5292 (lime) are visible. D′, Enlarged view of boxed region in D. E, TEM image showing gap junction 51346 (arrow in D′). E′, Reimaging of boxed region in E at 0.27 nm/pixel resolution with goniometric tilt. F, G, Frequent CBb3n coupling with CBb4. F, Two of 3 cross-class gap junctions (magenta; arrow, arrowheads in F,F′) between CBb4 595 (lime) and CBb3n 176 (orange) are visible. F′, Enlarged view of boxed region in F. G, TEM image showing gap junction 46663 (arrow in F′). G′, Reimaging of boxed region in G at 0.27 nm/pixel resolution with goniometric tilt. H, I, CBb4w coupling with CBb3n. H, One of 2 cross-class gap junctions (magenta, arrow in H′) between CBb4w 6117 (green) and CBb3n 176 (orange). H′, Enlarged view of boxed region in H. I, TEM image showing gap junction 58716 (arrow in H′). I′, Reimaging of boxed region in I at 0.27 nm/pixel resolution with goniometric tilt. J, K, Sparse CBb4 coupling with CBb3. J, Single cross-class gap junction (magenta, arrow in J′) between CBb4 595 (lime) and CBb3 5531 (maroon). J′, Enlarged view of boxed region in J. K, TEM image showing gap junction 91490 (arrow in J′). K′, Reimaging of boxed region in K at 0.27 nm/pixel resolution with goniometric tilt. L, M, Rejection of coupling between CBb3 and CBb4w. L, Costratification of CBb4w 5530 (green) and CBb3 5531 (maroon) axonal arbors. L′, Enlarged view of boxed region showing position of adherens junction 81621 (tan; arrow). M, TEM image showing a strong density at the location of contact indicated by the arrow in L′. M′, Reimaging of boxed region in M at 0.27 nm/pixel resolution with goniometric tilt reveals an adherens junction with strong asymmetric densities, but no gap junction. Gap junctions scaled by a factor of 4 (B,B′,D,D′,F,F′,H,H′,J,J′) for visualization. Most cells form additional gap junctions within these specific motifs, but with additional cells (Fig. 3-1). Scale bars: A, B, D, F, H, J, L, 10 µm; C, E, G, I, K, M, 1 µm; C′, E′, G′, I′, K′, M′, 100 nm. a, Adherens junction; gj, gap junction; RBC, rod bipolar cell.
Experimental design and statistical analysis
All results were derived from the RC1 dataset, a single volume of retinal tissue containing at least 16 cells for every ON CBC class (except CBbwf). At least 5 fully reconstructed and mapped cells from each class were used to compile synaptology and coupling profiles. Data from 2 incomplete cells were reported, where possible, for the wide-field ON cone bipolar (CBbwf) class. Analyses used the 1339 gap junctions formed by the 37 ON CBCs or 455 gap junctions formed by the 4 Aii cells detailed in this paper. The numbers of cells, synapses, or gap junctions for each statistic are provided in Results. All values are reported as the mean ± SD, coefficient of variation (CV), unless otherwise noted. Statistical analyses of network features are detailed in the Materials and Methods sections describing the analyses. Statistical analyses were performed in Microsoft Excel 2016 (RRID:SCR_016137) or R version 3.6.1 (RRID:SCR_001905), as indicated. The R software environment reports p values < 2.2E−16 as zero. We report these in figures (***) and in the text (p < 2.2E−16). The following sections detail the analyses performed in this publication.
Software accessibility
The custom, open-source software tools are freely available: Viking (RRID:SCR_005986) (Anderson et al., 2009), VikingPlot (https://zenodo.org/record/3234870#.XO7Y7IhKguU) (Anderson et al., 2011a,b), VikingView (https://zenodo.org/record/3267451#.XSUW1OhKguU; available for download here: https://connectomes.utah.edu/export/vikingview.html), Tulip (http://tulip.labri.fr/TulipDrupal/), TulipPaths (https://zenodo.org/record/1346342) (Lauritzen et al., 2019), Blender (http://www.blender.org/), Cell Sketches (https://zenodo.org/record/1346344), and Graffinity (https://zenodo.org/record/1422851) (Kerzner et al., 2017). Export of Viking data for these and other programs (e.g., Microsoft Excel) is available here: http://connectomes.utah.edu/export/toctree.html.
Code accessibility
All Microsoft SQL queries used in this paper are freely available (https://github.com/connectomes/Archive/tree/2018_OnBipolarCoupling) (Anderson et al., 2019). Specific cell numbers, labels, and synapse types contained within these queries can be changed according to the provided database schema. For specific cell and structure identification numbers, see the figure legends.
Results
During reconstruction of the ON CBCs in RC1 in our previous work (Marc et al., 2014; Lauritzen et al., 2019), we discovered that gap junctions between ON CBCs were more common than those formed with Aii cells. As this contradicts the general belief that Aii cells are indeed the dominate coupling partner of ON CBCs and have implications on visual processing in the retina, we sought to investigate these specific motifs in more detail. Doing so first required a comprehensive classification of the ON CBCs in RC1. This classification required additional parameters beyond conventional morphologic measures, namely, connectivity, in which several coupling motifs proved valuable in achieving complete classification. Therefore, we first describe the classification of ON CBCs in rabbit retina into 7 classes. We then detail their coupling profiles, including the prevalence and distribution of gap junctions and the specificity of coupling partnerships, size, and spatial topologies. Lastly, we explore the implications of these coupling motifs on retinal networks, particularly on the distribution of scotopic signals by Aii cells, and the potential mixing of parallel processing streams for input to GCs.
Classification of ON CBCs
Here, we use terminology based on computational classification theory where a class is a collection of members sharing certain attributes and separable from other classes. If further separation is not possible, these may reflect natural classes (Marc and Jones, 2002; Marc et al., 2018). Accordingly, we have distinguished 7 classes of ON CBCs in rabbit retina: CBb3, CBb3n, CBb4, CBb4w, CBb5, CBb6, and CBbwf (Fig. 1A). We previously adopted a nomenclature (Marc et al., 2018; Lauritzen et al., 2019) that merged our early classifications (Lauritzen et al., 2013; Marc et al., 2014) with those of MacNeil et al. (2004) and McGillem and Dacheux (2001), and retain it here for purposes of consistency. One exception is the renaming of the CBb7 class from our previous publication (Marc et al., 2014) to CBb6. We stress, however, that our numbers do not reflect positioning within the historical, 5 equal strata divisions of the retinal IPL as was common in many early classification schemes.
Manual classification in RC1 began with a small set of ∼40 fully reconstructed and centrally located bipolar cells. Historically, classification of ON CBCs relied heavily on axon arbor area and the stratification depth and breadth of the axonal arbors within the IPL (Famiglietti, 1981; McGuire et al., 1984; Cohen and Sterling, 1990; Strettoi et al., 1994; Jeon and Masland, 1995; Massey and Mills, 1996; Brown and Masland, 1999; McGillem and Dacheux, 2001; Casini et al., 2002; MacNeil et al., 2004; Pignatelli and Strettoi, 2004; MacNeil and Gaul, 2008; Wässle et al., 2009; Helmstaedter et al., 2013; Greene et al., 2016; Shekhar et al., 2016). In RC1 (mid-peripheral rabbit retina), only 3 ON CBC classes could be readily distinguished using these variables (Fig. 1A; Table 1). CBbwf was easily identified due to its notably large arbor size. Its axonal arbor begins to spread laterally at the top of stratum 4 with processes terminating deep within stratum 5. The CBb5 and CBb6 classes could be distinguished by their smaller arbors and distinct, albeit overlapping, depth of axonal stratification. CBb5 cells stratify largely within stratum 4, whereas CBb6 cells span strata 4 and 5. The remaining 4 classes (CBb3/3n/4/4w) fully costratify within the proximal IPL, occupying the lower three-fourths of stratum 3 and extending into the top half of stratum 4. Detailed analysis of Aii cell morphology and connectivity compartmentalization are consistent with the slightly deeper ON/OFF boundary implied by the top of these arborizations (Aii cell ON/OFF boundary = 46 ± 3%, n = 8 Aii cells).
Table 1.
Morphometric features of rabbit ON CBC classesa
| Class | No. of identified cells | Density (cells/mm2) | Stratification depth (% IPL) | Axonal field area (μm2) | Maximum axonal diameter (μm) | No. of branch terminals/cell | Branch path complexityb | CFc | CR |
|---|---|---|---|---|---|---|---|---|---|
| CBb3 | 29 (5 incomplete) | 784 | 45–75 | 1059 ± 192, 0.18 | 44 ± 5, 0.11 | 51.4 ± 12.6, 0.26 | 8.6 ± 1.0, 0.11 | 0.83 | 3.52 |
| CBb3n | 47 (8 incomplete) | 1101 | 44–69 | 895 ± 200, 0.22 | 40 ± 5, 0.12 | 28 ± 6.5, 0.23 | 7.1 ± 0.8, 0.11 | 0.98 | 3.92 |
| CBb4 | 18 (4 incomplete) | 505 | 44–73 | 1215 ± 328, 0.27 | 49 ± 9, 0.18 | 18.5 ± 3.6, 0.20 | 6.0 ± 0.8, 0.14 | 0.61 | 5.72 |
| CBb4w | 19 (9 incomplete) | 394 | 45–72 | 2186 ± 297, 0.14 | 67 ± 7, 0.10 | 21.7 ± 3.6, 0.17 | 6.5 ± 0.4, 0.06 | 0.92 | 3.32 |
| CBb5 | 24 (5 incomplete) | 725 | 59–86 | 1676 ± 318, 0.19 | 59 ± 5, 0.09 | 29.5 ± 5.6, 0.19 | 7.4 ± 1.3, 0.18 | 1.25 | 2.45 |
| CBb6 | 17 (7 incomplete) | 719 | 65–95 | 1223 ± 112, 0.09 | 51 ± 6, 0.11 | 24.2 ± 8.6, 0.35 | 6.8 ± 1.5, 0.22 | 0.88 | 3.44 |
| CBbwf | 2 (2 incomplete) | ND | 62–105 | >3800 | >100 | ND | ND | ND | ND |
aData are mean ± SD, CV. CR, Conformity ratio; ND, not determined.
bBranch path complexity per Elliott et al. (2015).
cCoverage factor (CF) for axonal arbors within the IPL was computed as the product of the average axonal field area (mm2/cell) and the class density (cells/mm2). Idealized patterns include the following: (1) packings: CF < 1, arbors do not overlap, but possible gaps; (2) coverings: CF > 1, arbors do not leave gaps, but may overlap; (3) tilings: CF = 1, arbors neither overlap nor leave gaps (Marc, 2009).
Much like the 4 costratifying classes of mouse Type 5 ON CBCs (Helmstaedter et al., 2013; Greene et al., 2016; Shekhar et al., 2016; Tsukamoto and Omi, 2017), the costratified rabbit CBb3/3n/4/4w classes lack obvious discriminating morphologic features. Therefore, additional variables were needed for further classification. Recent CBC classification schemes in mouse found the frequency or distribution of synaptic ribbons provided discriminatory power (Tsukamoto and Omi, 2014, 2017), but we found no obvious trends. Target-specific connectivity has also aided CBC classification, including GC target specificity and coupling with Aii cells (Cohen and Sterling, 1990; Helmstaedter et al., 2013; Greene et al., 2016). In RC1, GC dendritic processes are sufficiently spaced such that some ON CBCs may not have an opportunity for connectivity, despite class specificity. Aii cells are narrow field, and their arbors highly overlapping, providing ample contact opportunity. Thus, much like the separability achieved for the b3 and b4 classes using Aii connectivity in cat retina (Cohen and Sterling, 1990), connectivity with Aii cells divided the shallowly stratifying ON CBCs of RC1 into 2 groups based on a virtually binary presence/absence of gap junctions with Aii cells. Cells within the resulting groups still exhibited significant arbor overlap (lack of tiling), suggesting that at least 2 classes remained intermingled in both the Aii-coupled and -noncoupled groups. The Aii-coupled group showed some differences in arbor size among the cells, but cells of the Aii-noncoupled group were very similar. Examining synaptology and non-Aii-coupling partners revealed that cells of both groups formed frequent gap junctions with some cells of the other group, but rare or no coupling with other cells of the group, despite opportunity afforded by costratified and overlapping arbors. Using these coupling patterns, together with 3D plots to assess tiling and compare arbor sizes, the cells within each resultant group were finally resolved: the Aii-coupled group resolved into the narrow field CBb3n and wide-field CBb4w classes, whereas the Aii-noncoupled group resolved into the CBb3 and CBb4 classes. The discriminating coupling patterns were therefore the high frequency of coupling of CBb3 cells with CBb3n cells and of CBb4 cells with CBb4w cells, and the virtual lack of coupling of CBb3 cells with CBb4w and of CBb4 cells with CBb3n (detailed in the following sections).
The virtually binary nature of discriminating coupling relationships (with Aii, CBb4w and CBb3n) enabled us to rapidly extend classification across the entire RC1 volume after an initial segregation based on stratification. Notably, this scheme often allowed classification of even partial cells. Thus, of the 178 whole or partial ON CBCs identified in RC1, 156 (88%) were classified as 1 of the 7 classes, 40 of which had at least some portion of their arbors extending beyond volume boundaries. Of the remaining 22 unclassified cells, 14 somas had descending axons that extended outside of the volume boundaries and another 8 had significant portions of their arbors lost due to a block refacing event in strata 4/5 of the IPL that affected tracing of some ON CBCs in the northeast corner of the volume.
Notably, every cell with complete arbor reconstruction was readily classified as 1 of the 7 classes, and no intermediate forms were identified (Cohen and Sterling, 1990). The conformity ratios (Table 1) of the resulting class mosaics are consistent with those reported for identified neuronal mosaics in the mammalian retina (Wässle and Riemann, 1978; Cook, 1996; Reese, 2008; Marc, 2009). Tiling of neighboring arbors without (or with only minimal) overlap is a fundamental rule that holds for many neuronal classes and has been shown for all known CBC classes where evaluated (Wässle et al., 2009; Helmstaedter et al., 2013; Euler et al., 2014; Tsukamoto and Omi, 2014, 2016, 2017; Greene et al., 2016; Kántor et al., 2017). With the possible exception of the CBbwf class, which exhibited some arbor overlap, our classification resulted in sheets of cells that tiled the volume (Fig. 1B), further supporting each as a pure class.
Is this class list complete? Such a “parts list” is essential in the study of networks. Our classifications reveal that RC1 contains >15 cells of all but one ON CBC class and, importantly, at least 2 of the rarest class (CBbwf). Targeted searches using spatial queries and brute force surveys for unannotated or unclassified bipolar processes within the ON sublamina failed to identify additional candidates. Therefore, any additional classes must (1) be extremely sparse, (2) lack coupling with Aii cells, (3) lack coupling with all other ON CBC classes, and/or (4) stratify outside the ON sublamina of the IPL. There is no evidence suggesting a sparser class than CBbwf. Of the synaptic ribbon-forming cells fitting criteria 2–4, all exhibit Aii connectivity characteristic of either OFF CBCs or rod bipolar cells.
Ultrastructural identification of intercellular contacts
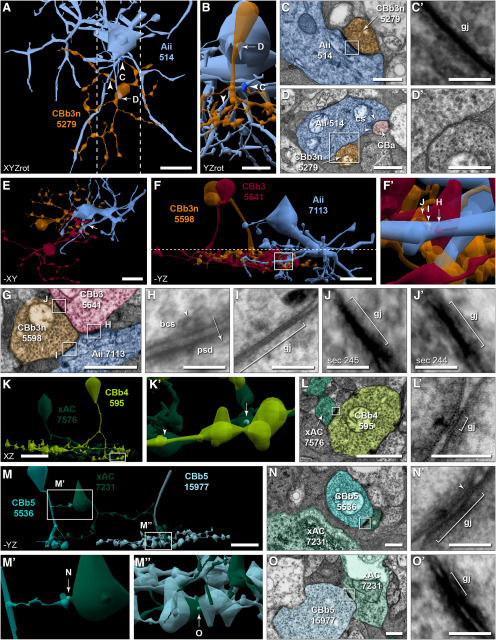
ON CBC axon terminals participate in a number of synapse types within the retinal IPL. In addition to electrical synapses mediated by gap junctions, these terminals provide excitatory glutamatergic drive via ribbon- and bipolar conventional-type synapses to GC and AC processes, and receive inhibitory conventional-type input from ACs. The connectomics strategy used in the construction and annotation of RC1 provides unparalleled visualization and quantification of connectivity. Figure 2A,A′,B,B′ are 3D reconstructions of the soma and axonal arbor of an ON CBC from RC1, illustrating the relative size and distribution of all identified synaptic contacts. At 2.18 nm/pixel (native RC1 resolution), all types of synaptic and structural contacts are directly discernable (Fig. 2C–G). Importantly, even intercellular synaptic and structural contacts at nonoptimal orientations are discernable from nonsynaptic membrane-to-membrane appositions, enabling us to take advantage of the unique capabilities of serial section TEM to perform high magnification reimaging with goniometric tilt. This is particularly relevant for the validation of gap junctions as they are indistinguishable from adherens junctions at oblique angles. At native resolution, a gap junction in RC1 oriented perpendicularly to the section plane appears as a single, thin, electron-dense band between 2 cells (Fig. 2F,F′) (Anderson et al., 2011b; Marc et al., 2014, 2018). The membranes of the 2 cells appear to “pinch” together at this location, obliterating any visible extracellular space between the 2 cells. This presentation has been confirmed through extensive reimaging at high resolution and goniometric tilt (n = 40; 40,000× magnification = 0.27 nm/pixel resolution) to observe the pentalaminar profile (alternating dark-light-dark-light-dark bands; Fig. 2H,H′) characteristic of gap junctions under these staining conditions (Peters, 1962; Nagy et al., 2018). An electrical synapse density is often observed within the cytoplasm beneath the membrane at sites of gap junctions and can be symmetric or asymmetric, usually correlating with symmetry/asymmetry of participating cell classes and believed to arise as a consequence of differences in molecular composition of the connexons formed by each cell class (Strettoi et al., 1992; Anderson et al., 2011b; Pereda et al., 2013; Marc et al., 2014; Pereda, 2014; Marsh et al., 2017; Miller et al., 2017; Tsukamoto and Omi, 2017; Nagy and Lynn, 2018). In contrast, adherens junctions present as feathery membrane densities of variable thickness and symmetry, but with a distinct extracellular space between the 2 cells that is wider, more uniform, and more electron-dense than surrounding nonjunctional extracellular space (Fig. 2G,H′′). Often, gap junctions are flanked by adherens junctions (Fig. 2H) (Nagy and Lynn, 2018). For the purposes of this paper, all densities with oblique native orientations between CBCs and Aii cells or other CBCs were annotated as gap junction candidates until reimaging proved otherwise. Of the 1339 gap junction instances described in this report, 484 are confirmed, 429 (32%) without the need for reimaging. Between this and our previous studies, 41 of 160 candidate RC1 gap junctions have been refuted (12 remain inconclusive due to insufficient tilt capacity).
Figure 2.
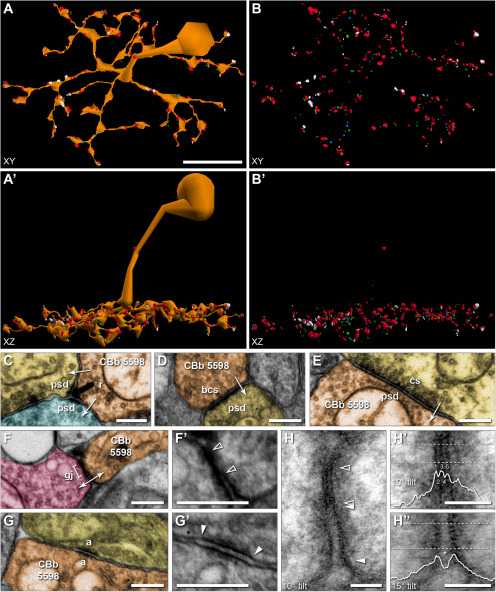
Ultrastructural identification and visualization of connectivity. A, Horizontal view of a 3D rendering of the soma and axonal arbor reconstruction of cell 5598 and its 413 synaptic structures. The synaptology includes ribbons (n = 90, green), bipolar conventional presynapses (n = 16, light blue), postsynaptic densities to presynaptic conventional synapses from ACs (n = 226, red), and gap junctions (n = 81, white). Synapses are rendered as volumetric shapes using circles having a diameter equal to the summed segment length of the open curve annotation for the visible structure on each section. A′, Vertical view of A. B, B′, Cell morphology rendered transparent to visualize synaptic structures from A and A′, respectively. C-G, TEM images of representative synaptic structures from Viking at native 2.18 nm resolution. Arrows indicate direction of signaling. C, Ribbon 38955 mediates signaling from cell 5598 (orange) to 2 postsynaptic cells (yellow, blue) in a classic dyad arrangement. D, Bipolar cell conventional presynapse 91577 exhibits all the features of a traditional ribbon synapse, including vesicles present at the plasma membrane, but lacks the ribbon structure. E, Postsynaptic density 114606 to an AC (yellow) conventional presynapse. F, Gap junction 56921 mediates coupling with an ON CBC (pink). F′, Enlarged view of F. Gap junction located between open arrowheads. G, Adherens junction 124245 formed with a cell of unknown identity (yellow). G′, Enlarged view of G. Arrowheads indicate adherens junction boundaries. H, Gap junction 56921 from F reimaged at 40,000× magnification (resolution = 0.27 nm/pixel) with goniometric tilt (10°). Open arrowheads indicate gap junction boundaries. Arrowheads indicate adjacent adherens junction boundaries. H′, Enlarged view of gap junction from H, with an overlay of the corresponding density plot obtained from the region between the dashed lines. Peaks (1, 3, 5) and valleys (2, 4) in the density plot that generate the characteristic pentalaminar profile are labeled. H′′, Enlarged view of adjacent adherens junction from H at 15° tilt. Corresponding density plot obtained from the region between the dashed lines and overlaid. Note the prominent extracellular space (asterisk) between the membrane peaks. Scale bars: A, 10 µm; C-G, F′, G′, 250 nm; H-H′′, 50 nm. a, Adherens junction; bcs, bipolar conventional synapse; cs, conventional presynapse; gj, gap junction; psd, postsynaptic density; r, ribbon. E, F′, G′, Image max-min contrast stretched.
Prominence of gap junctions in ON CBC circuitry within the IPL
To assess the contribution of gap junctions to ON CBC axonal circuitry, we focused on a total of 37 cells, 5-7 per class, reconstructed to statistical completion. The CBbwf class remains an exception, as we have only identified 2 class members within RC1 due to their low frequency, and neither is completely contained within the volume due to their large size. We find that all rabbit ON CBCs form gap junctions, at least 20 per cell, with some cells forming upwards of 80 (Table 2). Thus, electrical synapses can constitute upward of 25% of an ON CBC's total synaptic contacts within the IPL, similar to that observed for Aii cells (total gap junctions = 114 ± 11, 0.09 CV; % total synaptic contact = 24.2% ± 2.8%, 0.11 CV; n = 4 cells). For 4 of the 7 ON CBC classes, electrical synapses constitute at least 10% of their total synaptic contacts. Assuming these coupling sites are indeed functionally bidirectional (Xin and Bloomfield, 1999; Trexler et al., 2001; Veruki and Hartveit, 2002a,b; compare Vaney, 1997), electrical synapses may constitute upwards of 50% of a given cell's total potential synaptic input or 40% of its total potential synaptic output. Similarly, electrical synapses constitute nearly two-thirds of an Aii cell's total potential synaptic output (64.0% ± 4.4%, 0.07 CV, n = 4), but only one-fourth of its total potential input (28.0% ± 2.9%, 0.10 CV, n = 4). In terms of a cell's potential electrical conductivity, gap junctions account for 4%-25% of the total channel-occupied membrane area in ON CBCs and 40%-42% in Aii cells. While class-specific differences exist in both the frequency of gap junctions and their overall contribution to a cell's synaptic contacts, it is clear that electrical synapses are a prevalent form of synaptic communication and can represent a significant component of ON CBC circuitry within the IPL.
Table 2.
Synaptology of core set of ON CBC axonal arborsa
| Cell ID | Class | No. of PSDs | No. of ribbons | No. of BCSs | No. of GJs (confirmed)b | % Synaptic contact | % Total input | % Total output | % Total channel areac (μm2) |
|---|---|---|---|---|---|---|---|---|---|
| 909 | CBb3 | 131 | 177 | 47 | 48 (19) | 11.9 | 26.8 | 17.6 | 7.0 |
| 3756 | CBb3 | 161 | 215 | 39 | 64 (16) | 13.4 | 28.4 | 20.1 | 8.1 |
| 5278 | CBb3 | 111 | 136 | 28 | 31 (10) | 10.1 | 21.8 | 15.9 | 4.6 |
| 5297 | CBb3 | 111 | 166 | 21 | 45 (10) | 13.1 | 28.8 | 19.4 | 7.6 |
| 5531 | CBb3 | 131 | 215 | 34 | 51 (13) | 11.8 | 28.0 | 17.0 | 9.0 |
| CBb3 Class | 129 ± 20, 0.16 | 182 ± 34, 0.19 | 34 ± 10, 0.30 | 48 ± 12, 0.25 | 12.3 ± 1.4, 0.11 | 27.3 ± 2.9, 0.11 | 18.4 ± 1.9, 0.10 | 7.2 ± 1.7, 0.23 | |
| 142 | CBb3n | 223 | 106 | 15 | 59 (21) | 14.6 | 20.9 | 32.8 | 9.8 |
| 176 | CBb3n | 273 | 112 | 32 | 64 (19) | 13.3 | 19.0 | 30.8 | 10.3 |
| 5279 | CBb3n | 163 | 86 | 8 | 45 (15) | 14.9 | 21.6 | 32.4 | 13.8 |
| 5284 | CBb3n | 210 | 110 | 13 | 69 (27) | 17.2 | 24.7 | 35.9 | 12.9 |
| 5561 | CBb3n | 147 | 76 | 15 | 59 (18) | 19.9 | 28.6 | 39.3 | 12.0 |
| 5598 | CBb3n | 226 | 90 | 16 | 81 (26) | 19.6 | 26.4 | 43.3 | 17.3 |
| 6050 | CBb3n | 236 | 95 | 14 | 60 (26) | 14.8 | 20.3 | 35.5 | 11.6 |
| CBb3n Class | 211 ± 43, 0.20 | 96 ± 13, 0.14 | 16 ± 7, 0.46 | 62 ± 11, 0.18 | 16.3 ± 2.6, 0.16 | 23.1 ± 3.6, 0.15 | 35.7 ± 4.4, 0.12 | 12.5 ± 2.5, 0.20 | |
| 595 | CBb4 | 82 | 175 | 14 | 75 (16) | 21.7 | 47.8 | 28.4 | 21.5 |
| 1021 | CBb4 | 110 | 198 | 14 | 62 (16) | 16.1 | 36.0 | 22.6 | 16.2 |
| 4877 | CBb4 | 98 | 179 | 17 | 62 (20) | 17.4 | 38.8 | 24.0 | 15.7 |
| 5292 | CBb4 | 63 | 125 | 6 | 67 (25) | 25.7 | 51.5 | 33.8 | 21.0 |
| 5499 | CBb4 | 99 | 183 | 18 | 63 (20) | 17.4 | 38.9 | 23.9 | 17.9 |
| 20136 | CBb4 | 90 | 133 | 19 | 35 (12) | 12.6 | 28.0 | 18.7 | 8.7 |
| CBb4 Class | 90 ± 16, 0.18 | 166 ± 29, 0.18 | 15 ± 5, 0.32 | 61 ± 14, 0.22 | 18.5 ± 4.6, 0.25 | 40.2 ± 8.4, 0.21 | 25.2 ± 5.2. 0.21 | 16.8 ± 4.7, 0.28 | |
| 170 | CBb4w | 152 | 98 | 4 | 48 (24) | 15.9 | 24.0 | 32.0 | 21.5 |
| 5530 | CBb4w | 182 | 109 | 2 | 72 (31) | 19.7 | 28.3 | 39.3 | 23.7 |
| 5601 | CBb4w | 179 | 122 | 3 | 57 (29) | 15.8 | 24.2 | 31.3 | 24.2 |
| 5650 | CBb4w | 178 | 129 | 9 | 55 (27) | 14.8 | 23.6 | 28.5 | 17.5 |
| 6117 | CBb4w | 179 | 125 | 5 | 50 (24) | 13.9 | 21.8 | 27.8 | 17.6 |
| 7024 | CBb4w | 160 | 103 | 2 | 57 (15) | 17.7 | 26.3 | 35.2 | 17.6 |
| CBb4w Class | 172 ± 12, 0.07 | 114 ± 13, 0.11 | 4 ± 3, 0.63 | 56 ± 8, 0.15 | 16.3 ± 2.1, 0.13 | 24.7 ± 2.3, 0.09 | 32.4 ± 4.3, 0.13 | 20.3 ± 3.2, 0.16 | |
| 485 | CBb5 | 264 | 230 | 65 | 35 (12) | 5.9 | 11.7 | 10.6 | 4.9 |
| 593 | CBb5 | 305 | 211 | 109 | 23 (13) | 3.5 | 7.0 | 6.7 | 4.4 |
| 5562 | CBb5 | 288 | 206 | 73 | 39 (19) | 6.4 | 11.9 | 12.3 | 5.8 |
| 5649 | CBb5 | 200 | 184 | 79 | 30 (15) | 6.1 | 13.0 | 10.2 | 5.3 |
| 6115 | CBb5 | 256 | 203 | 59 | 35 (12) | 6.3 | 12.0 | 11.8 | 4.6 |
| 6997 | CBb5 | 283 | 244 | 54 | 34 (15) | 5.5 | 10.7 | 10.2 | 4.3 |
| CBb5 Class | 266 ± 37, 0.14 | 213 ± 21, 0.10 | 73 ± 20, 0.27 | 33 ± 6, 0.17 | 5.6 ± 1.1, 0.19 | 11.1 ± 2.1, 0.19 | 10.3 ± 2.0, 0.19 | 4.9 ± 0.6, 0.12 | |
| 180 | CBb6 | 185 | 163 | 27 | 22 (8) | 5.5 | 10.6 | 10.4 | 7.2 |
| 419 | CBb6 | 239 | 251 | 34 | 34 (13) | 6.1 | 12.5 | 10.7 | 7.9 |
| 6156 | CBb6 | 220 | 211 | 32 | 26 (7) | 5.3 | 10.6 | 9.7 | 6.1 |
| 9693 | CBb6 | 149 | 182 | 33 | 21 (11) | 5.5 | 12.4 | 8.9 | 5.2 |
| 16026 | CBb6 | 228 | 203 | 45 | 28 (12) | 5.6 | 10.9 | 10.1 | 7.4 |
| CBb6 Class | 204 ± 37, 0.18 | 202 ± 33, 0.16 | 34 ± 7, 0.19 | 26 ± 5, 0.20 | 5.6 ± 0.3, 0.05 | 11.4 ± 0.9, 0.08 | 9.9 ± 0.7, 0.07 | 6.8 ± 1.1, 0.12 | |
| 5283d | CBbwf | 138 | 149 | 19 | 26 (2) | 7.8 | 15.9 | 13.4 | 5.0 |
| 6589d | CBbwf | 97 | 126 | 19 | 20 (7) | 7.6 | 17.1 | 12.1 | 6.0 |
aDetailed synaptology and class designations for each of the 37 completely reconstructed (core) ON CBCs as well as the percentage contribution of gap junctions to the total synaptic contact, total input, and total output of each ON CBC axonal arbor. Bolded values indicate class summary statistics (mean ± SD, CV). BCSs, Bipolar cell conventional presynapses; GJs, gap junctions; PSDs, postsynaptic densities.
bNumber of confirmed gap junctions, determined either from native capture resolution or through recapture at 0.27 nm/pixel resolution with goniometric tilt.
cChannel area is defined as the total membrane area occupied by both gap junctions and postsynaptic densities.
dIncomplete arbors.
Coupling partnerships of ON CBCs
Before examining the specific coupling partnerships of ON CBC classes, we first wish to clarify terminology regarding modes of coupling. Here, we define heterocellular coupling as that between cells of different superclasses. In the retina, these are the categories of photoreceptors, horizontal cells, bipolar cells, ACs, and GCs. Homocellular coupling occurs between cells of the same superclass. Historically, heterologous/homologous has been used in place of heterocellular/homocellular, but these terms evoke evolutionary considerations. This is also distinct from the uses of heteromeric/homomeric to describe the connexin protein composition of individual hexameric connexons and of heterotypic/homotypic to describe the composition of the 2 connexons comprising a gap junction channel. Homocellular coupling requires further distinctions. Cross-class homocellular coupling refers to coupling between 2 cells of same superclass, but different class (e.g., a CBb3n cell coupling with CBb3 cell). In-class involves coupling between pairs of cells from the same class (e.g., CBb3n cell coupling with a neighbor CBb3n cell). Lastly, self gap junctions, formed between processes originating from the same neuron, often called “sister” branches, are a specific type of in-class coupling (e.g., gap junction between 2 branches of CBb5 593). For ON CBCs in RC1, the sister branch often originates locally or is an extension of an adjacent varicosity. Such self gap junctions are worth differentiating as these may have limited contribution to the lateral spread of signals between cells, and many neurons have been shown to distinguish processes arising from sister branches versus another cell of the same class and to initiate divergent growth behaviors accordingly, which underlie tiling and self-avoidance (Cameron and Rao, 2010; Grueber and Sagasti, 2010; Jan and Jan, 2010; Lefebvre, 2017; Mountoufaris et al., 2018).
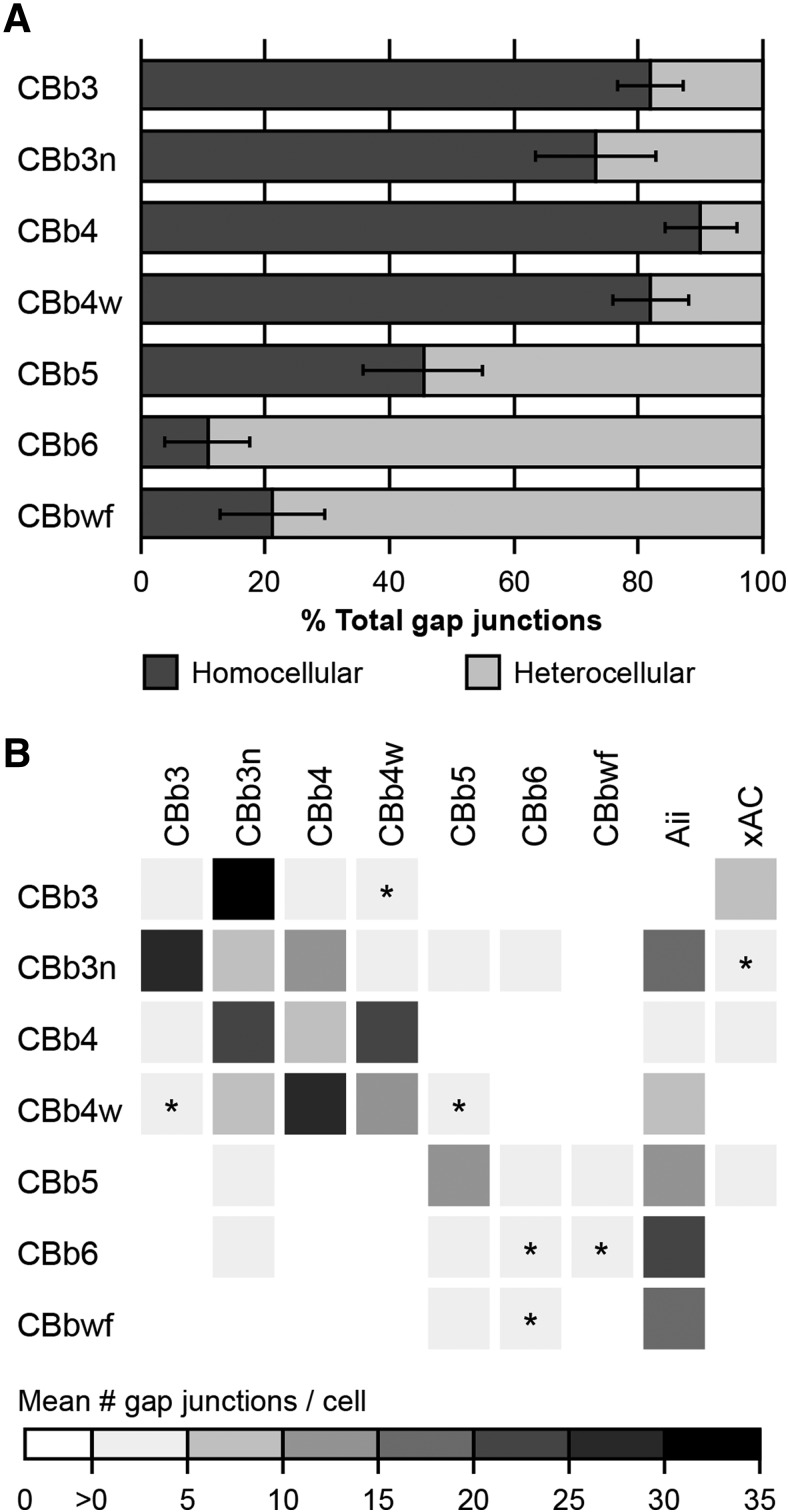
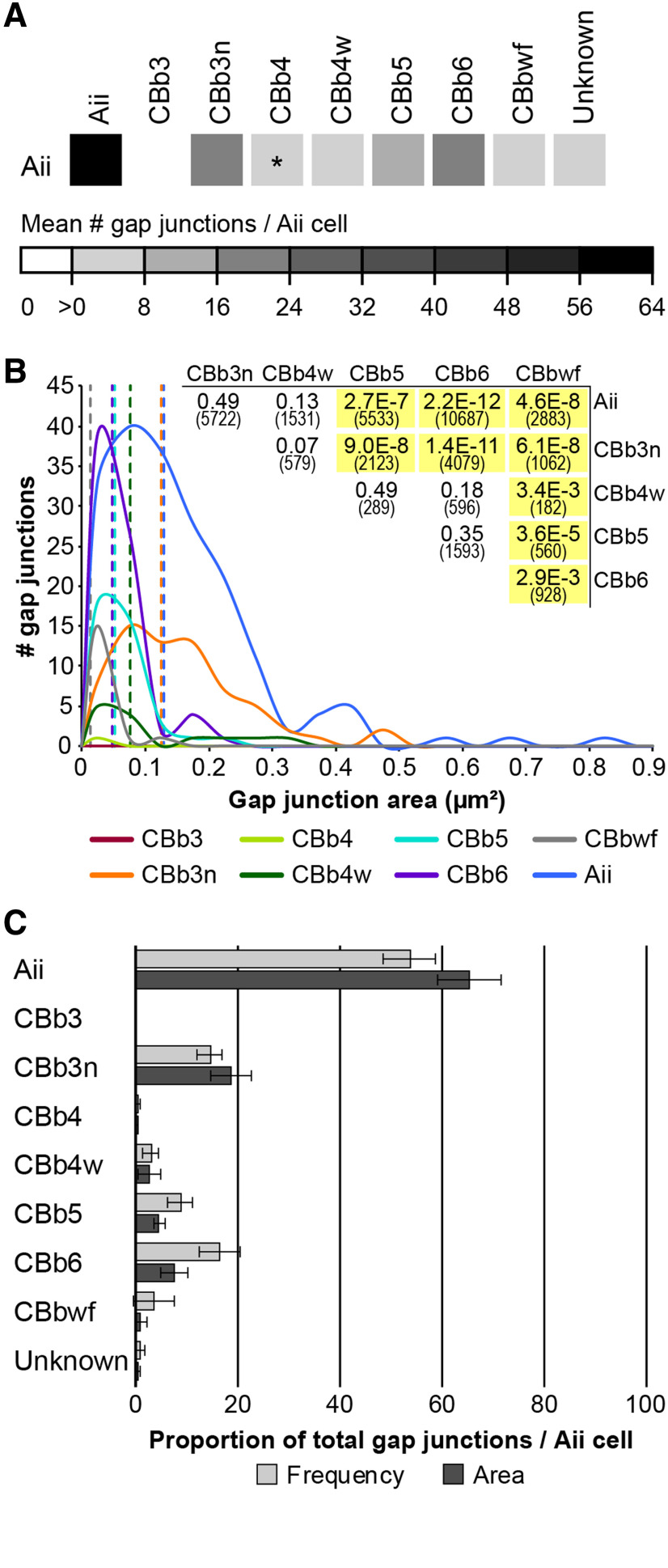
Evaluation of ON CBC coupling partnerships in RC1 revealed 2 critical points. First, noncanonical partnerships dominate ON CBC coupling in the IPL. Of the 1339 gap junctions involving at least 1 of the 37 ON CBCs previously described, only 30% are the canonical heterocellular gap junctions with Aii cells responsible for transmission of scotopic signals through the mammalian rod pathway (Kolb and Famiglietti, 1974; McGuire et al., 1984; Strettoi et al., 1992; Anderson et al., 2011b; Marc et al., 2014). The other 70% mediate homocellular coupling between ON CBCs (62%) and heterocellular coupling with other noncanonical AC partners (8%). At the individual class level, homocellular coupling dominates for 4 of the 7 ON CBC classes, with a fifth class exhibiting a roughly equal distribution (Fig. 3A). Detailed examination reveals that cells of each ON CBC class form coupling partnerships according to specific rulesets: a unique combination of partners and/or frequencies (Fig. 3B; Fig. 3-1). This underscores the second point: that ON CBC coupling partnerships are class-specific. In the following sections, we expand on these partnerships in turn.
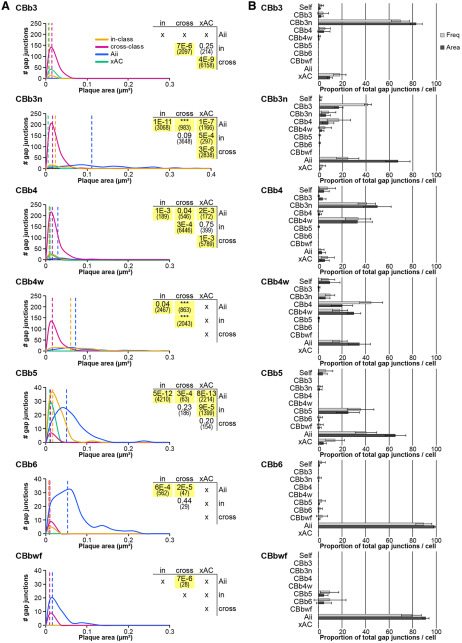
Figure 3.
Graphical summaries of ON CBC coupling. A, Proportion of homocellular versus heterocellular gap junctions by class based on frequency counts. B, Coupling profiles of ON CBC classes. The matrix reports the mean number of gap junctions formed by a given ON CBC class (row) with each partner class (column), as assessed from the core set of 37 ON CBCs in RC1 detailed in this paper. The matrix is therefore asymmetrical, as classes differ in arbor diameter, coverage factors, etc. Values are encoded by the linear heat map scale below. *Class pairings where coupling has not been confirmed. The profile for the CBbwf class is preliminary due to the incompleteness of available arbors. Detailed coupling profiles for individual cells and class statistics are provided in Figure 3-1.
Detailed individual cell coupling profiles and class statistics for the core set of 37 ON CBC and four Aii cells. 1The CBb label contains all partial cells or fragments that could not be classified beyond the ON superclass. The number of coupled cells and number of gap junctions per cell were not quantified. 2Most partner xACs exist as fragments, only sufficiently traced and annotated to confirm they were not Aii cells. Thus, the number of coupled cells and number of gap junctions per cell were not quantified. 3Two Aii cell partners could not be traced far enough to determine whether they were of amacrine or bipolar cell identity. Download Figure 3-1, XLSX file (22.3KB, xlsx) .
Canonical heterocellular coupling with Aii cells
Coupling with Aii cells has been presented as a hallmark of mammalian ON CBC identity, despite reports to the contrary in various species (cat: Cohen and Sterling, 1990; rabbit: Petrides and Trexler, 2008; mouse: Tsukamoto and Omi, 2017). Indeed, most rabbit ON CBCs in RC1 form gap junctions with Aii cells (Fig. 4A–C). However, we also confirm that some rabbit ON CBCs lack direct coupling with Aii cells (Fig. 3B; detailed in Fig. 3-1). Cells of the CBb3 class appear to never form gap junctions with Aii cells. This is not due to a lack of opportunity. We have identified 6 validated contacts between CBb3 axonal processes and the arboreal dendrites of Aii cells. Recaptures of these sites confirm the absence of gap junctions at all 6 (Fig. 4E–H). Indeed, 2 of these contacts lacked any membrane density, whereas adherens junctions (n = 2) or bipolar conventional synapses (n = 2) were identified at the other 4. Both the CBb6 (previously CBb7 class) (Marc et al., 2014) and CBbwf cells are also sparsely presynaptic to Aii cells via ribbon synapses. CBb3 cells also lack the canonical inhibition from Aii lobules characteristic of OFF CBCs, clearly stratify below these Aii lobules within the ON sublamina (Fig. 4F), and fully costratify (Fig. 4F) and couple with (Fig. 4J) other Aii-coupled ON CBC classes (Fig. 4I), supporting their classification within the ON CBC superclass. Three of the 6 fully reconstructed CBb4 cells are also devoid of gap junctions with Aii cells and the 3 that do form gap junctions with Aii cells do so infrequently (1-3 gap junctions per cell). These CBb4 cells fully stratify with CBb3 arbors below Aii lobules in the ON sublamina, couple with other Aii-coupled ON CBCs, and are also never postsynaptic to inhibitory conventional synapses from Aii lobules, arguing that they too belong within the ON CBC superclass.
Figure 4.
Heterocellular coupling topologies. A-D, Canonical heterocellular coupling between ON CBCs and Aii cells. A, Contact sites between CBb3n 5279 (orange) and Aii 514 (light blue), including 2 canonical heterocellular gap junctions (dark blue; arrowheads). B, Vertical view of the region between the dotted lines in A. C, Gap junction 6171 (arrowhead 'C' in B) between CBb3n 5279 axonal terminal (orange) and Aii 514 arboreal dendrite (light blue). C′, Enlarged view of boxed region in C. D, TEM showing membrane contact (arrow 'D' in B) between the descending axon of CBb3n 5279 and lobule of Aii 514. The Aii lobule forms a conventional presynapse (between arrowheads) onto a passing OFF CBC (CBa) process. D′, Enlarged view of boxed region in D. E-J, CBb3 rejects canonical heterocellular coupling with Aii cells, but couples with the Aii-coupled CBb3n class. E, F, CBb3 5641 (maroon) exhibits territory overlap (E) and costratification (F) below the lobular dendrites of Aii 7113 (light blue) and fully costratifies with CBb3n 5598 (orange). F, Horizontal dotted line indicates the ON/OFF boundary of the IPL. F′, Enlarged view of the boxed region in F (arrow in E). G, Single TEM image showing bipolar conventional synapse 124055 from CBb3 5641 onto Aii 7113, cross-class homocellular gap junction 114999 between CBb3 5641 and CBb3n 5598, and canonical heterocellular gap junction 115004 between CBb3n 5598 and Aii 7113. H, I, Reimaging of boxed regions in G at 0.27 nm/px resolution with goniometric tilt. Synaptic vesicle (arrowhead, H), direction of signaling (arrow, H), and gap junction boundaries (bracket, I) indicated. J, J′, Enlarged view of boxed region in G on section 245 (J) and adjacent section 244 (J′). K–O, Noncanonical coupling between ON CBCs and xACs. K, Two of 5 noncanonical gap junctions (mint; arrow and arrowhead in K′) between CBb4 595 (lime) and xAC 7576 (teal) are visible. K′, Enlarged view of boxed region in K. L, TEM image of gap junction 54868 (arrow in K′). L′, Reimaging of boxed region in E at 0.27 nm/pixel resolution with goniometric tilt. M, Noncanonical heterocellular gap junctions (mint, arrows in M′,M′′) between xAC 7231 (teal) and 2 CBb5 cells, 5536 (cyan), and 15977 (powder blue). M′, M′′, Enlarged views of the boxed regions in M. N, TEM image of gap junction 116945 (arrow in M′) between a branch of CBb5 5536 in the OFF sublamina and small extension off the soma of xAC 7231. N′, Reimaging of boxed region in N at 0.27 nm resolution with goniometric tilt. Small region of pentalaminar structure visible (arrowhead). Tilt reveals full extent (bracket). O, TEM image of gap junction 116987 (arrow in M′′) between CBb5 15977 and xAC 7231 in the ON sublamina. O′, Enlarged view of boxed region in O. Gap junctions scaled by a factor of 2 (A,B) or 4 (K,K′,M,M′,M′′) for visualization. Scale bars: A, E, F, K, M, 10 µm; B, 5 µm; D, G, L, 1 µm; C, D′, N, O, 500 nm; C′, H, I, J, J′, L′, N′, O′, 100 nm. bcs, Bipolar conventional presynapse; cs, conventional presynapse; gj, gap junction; psd, postsynaptic density.
Noncanonical heterocellular coupling with unclassified ACs
Approximately 8% of the 1339 gap junctions formed by the 37 ON CBCs detailed in this study mediate coupling with AC targets lacking the classical morphology and synaptology of Aii cells (Fig. 4K–Q), referred to hereafter as xACs. To date, we have confirmed gap junctions between xACs and only 3 ON CBC classes (CBb3, CBb4, and CBb5) (Fig. 3B; detailed in Fig. 3-1). CBb3 cells are the primary partner, forming gap junctions with xACs nearly twice as often as any other ON CBC class (8.4 ± 2.2 gap junctions with xACs/CBb3 cell; CV = 0.26; n = 5 CBb3 cells). Interestingly, xACs provide the sole source of heterocellular coupling in which the CBb3 class participates. Similarly, CBb4 cells, many of which lack coupling with Aii cells, also form gap junctions with xACs (4.7 ± 3.4 gap junctions with xACs/CBb4 cell; CV = 0.73; n = 6 CBb4 cells; Fig. 4K–M). Thus, as a consequence of this xAC coupling, all ON CBC classes participate in heterocellular coupling motifs. CBb5 cells were also observed to form gap junctions with xACs (Fig. 4N–Q), making them the only class of ON CBCs that participates in both canonical and noncanonical heterocellular coupling.
The processes of xACs present with medium-to-dark electron-dense cytoplasm, irregular lobular varicosities, and/or very thin, tortuous extensions. Reconstruction of 3 of these (and partial reconstruction of several more) suggest at least 2 separate classes of small-diameter (50–100 μm), diffusely stratified arbors with somas containing high levels of glycine, consistent with narrow-field glycinergic ACs (Fig. 4K,N). However, none of these resembles the characteristic morphology of the narrow-field glycinergic A8 AC shown to couple with ON CBCs in mouse (Kolb and Nelson, 1996; Lee et al., 2015; Yadav et al., 2019), despite identification of a homologous A8 cell in rabbit (DAPI-3: Vaney, 1990; Bloomfield, 1992; Wright et al., 1997; or flat bistratified: MacNeil and Masland, 1998; MacNeil et al., 1999). Moreover, these reconstructed xACs are also inconsistent with the non-GABAergic non-glycinergic Gbx2+ ACs recently reported to couple with presumed ON CBCs in stratum 3 and/or 5 of mouse retina (Kerstein et al., 2019). The coupling profiles of these xACs also support at least 2 classes: one that couples with at least CBb3 and CBb4 cells (CBb3n are also candidate partners, but no gap junctions have been confirmed) and a second that may couple exclusively with CBb5 cells. Further classification of these xACs and detailing of their connectomes are beyond the scope of this manuscript but will be the subject of a future publication.
In-class homocellular coupling among ON CBCs
Coupling among neighboring cells of the same class is a common motif within the retina (Völgyi et al., 2013), although a rarer occurrence among mammalian bipolar cells (Raviola and Gilula, 1975; Kolb, 1979; Cohen and Sterling, 1990; Luo et al., 1999; Mills, 1999; Dacey et al., 2000; Jacoby and Marshak, 2000; Arai et al., 2010; Kántor et al., 2016, 2017; Tsukamoto and Omi, 2017) than those of lower vertebrates (Marc et al., 1988; Umino et al., 1994; Arai et al., 2010). Here, we find that 5 of the 7 rabbit ON CBC classes form in-class gap junctions (Fig. 3B; detailed in Fig. 3-1). Overall, in-class gap junctions tend to be less frequent than other coupling modes, but this is not surprising given the limited contact opportunities afforded by tiling among neighboring cells. Both CBb5 and CBb4w classes exhibit the highest frequency of gap junctions mediating in-class partnerships, and both have high coverage factors (CF = 1.25 and CF = 0.92, respectively; Table 1) and nearly perfectly tile the retinal plane (Fig. 1D). Recapture of all candidate in-class contacts for a CBb5 cell reveals a single gap junction at virtually every contact with each of its 5 immediate CBb5 neighbors (n = 8 of 9 contacts; 1 of 9 inconclusive; Fig. 5A–I). Likewise, for 2 of the 6 completed CBb4w cells, all in-class contacts with neighbors contain confirmed gap junctions, whereas the remaining 4 cells have over 80% of their in-class gap junctions with neighbors confirmed (the remaining 20% present obliquely). Based on the very high coverage factor (0.98; Table 1) and observed near-perfect tiling (Fig. 1D), the CBb3n class was also predicted to have a high frequency of in-class gap junctions. Although branches of neighboring CBb3n cells often contact and form gap junctions (Fig. 5J,K), CBb3n cells form half as many gap junctions with in-class neighbors than either the CBb5 or CBb4w classes (Fig. 3B). Closer investigation revealed that, in many cases, Aii coupling appeared to bridge CBb3n in-class coupling, in which the branches of 2 neighboring CBb3n cells terminate with gap junctions onto an intervening arboreal dendrite of an Aii cell, often immediately opposite each other, or a short distance apart (Fig. 5L–O). In contrast, the relatively high frequency of CBb4 in-class coupling was unexpected in light of the low coverage factor (0.61) leaving large gaps and few opportunities for contact with neighboring CBb4 cells (Fig. 1B; Table 1). In this case, we discovered that many of the in-class gap junctions are indeed self-gap junctions formed between sister branches of the same cell (Fig. 3-1). Self-gap junctions within other ON CBC classes are rare, except the CBb4w class, which exhibit similar self-gap junction frequencies as the CBb4 class. Thus, it appears that these ON CBC classes will participate in in-class motifs whenever the opportunity presents. It was therefore surprising to find that, while cells of the CBb3 class participate in in-class coupling motifs, they may decline a large portion of coupling opportunities, both with in-class neighbors and between sister branches. Recapture of representative contact sites confirmed the presence of gap junctions at only 2 of 5 validated encounters between CBb3 processes originating from neighboring CBb3 cells (Fig. 5P,Q) and 1 of 3 validated encounters between sister branches.
Figure 5.
In-class homocellular coupling topologies. A-I, CBb5 in-class coupling. A, Horizontal view showing the locations of 8 in-class gap junctions (yellow) between cell 593 (cyan) and its immediate CBb5 neighbors, cells 166 (forest green), 6115 (powder blue), 5649 (teal), 5562 (powder blue), and 6997 (forest green). One gap junction (72657, white circle) remains inconclusive, even at maximum tilt (± 65°). B-I, TEM images for regions indicated in A (white boxes). CBb5 593 forms gap junctions 60046 (E) and 60010 (F) with cell 166, 55890 (G) with cell 6115, 53818 (H) with cell 5649, 49434 (I) and 79658 (J) with cell 5562, and 52441 (K) and 59747 (L) with cell 6997. B′-I′, B-I, Reimaging of boxed regions at 0.27 nm/pixel resolution with goniometric tilt. Brackets notate gap junction boundaries based on tilt series. J, K, In-class coupling between neighboring CBb3n cells. J, Five in-class gap junctions (yellow; arrow and arrowheads) between CBb3n cells 5284 (brown) and 6050 (tan). J′, Enlarged and rotated view of circled region in J. Arrowheads in J′ indicate different gap junctions than in J. K, TEM image of gap junction 57071 (arrow in J′). K′, Reimaging of boxed region in K at 0.27 nm/pixel resolution with goniometric tilt confirms gap junction identity. L-O, Aii coupling bridging CBb3n in-class coupling. L, Some branches of neighboring CBb3n cells 6118 (tan) and 6120 (orange) terminate in tip-to-tip topology with in-class gap junctions (yellow, arrowheads), whereas others terminate without contact, forming gap junctions with Aii cells (dark blue) at their tips instead (asterisks). Cross-class (magenta) and heterocellular xACs (mint) gap junctions also shown. Rare case of tiling violation (open arrowhead). L′, Rotated view of the circled area in L rendered with coupling partner Aii 7904 (light blue). M, TEM image showing gap junction 111592 (N) between CBb3n 6120 and Aii 7904 directly opposite gap junction 119474 (O) between Aii 7904 and CBb3n 6118. N, Reimaging of boxed region in M at 0.27 nm resolution with goniometric tilt. O, Enlarged view of boxed region in M. P, Q, CBb3 in-class coupling. P, Two in-class gap junctions (yellow) formed at a single contact site between neighboring CBb3 cells 909 (maroon) and 5297 (peach). P′, Enlarged and rotated view of encircled region in P. Q, TEM image of gap junction 121062 (arrow in P′). Q′, Reimaging of boxed region in Q at 0.27 nm/pixel resolution with goniometric tilt. Gap junctions scaled by a factor of 6 (A), 4 (J,P,P′), or 2 (J′,L,L′) for visibility. Scale bars: A, J, L, P, 10 µm; L′, 5 µm; Q, 1 µm; B-I, K, M, 500 nm; B′-I′, K′, N, O, Q′, 100 nm. gj, Gap junction.
This propensity for in-class coupling is not shared among all classes of ON CBCs, and indeed, is in stark contrast to the CBb6 class, which expressly decline in-class coupling opportunities. CBb6 cells also tile the retinal plane affording in-class opportunities; but when processes of neighboring CBb6 cells contact each other, gap junctions are never present (n = 5 validated instances; Fig. 6A–E). Such contact sites often contain adherens junctions (Fig. 6C), indicative of cell-cell recognition and the ability to recruit and assemble protein complexes at these sites. The absence of gap junctions at these sites is not due to a lack of connexin trafficking, as gap junctions with Aii cells are often found nearby or even immediately adjacent within the same varicosity (Fig. 6A′,B,D,E). Importantly, these Aii processes do not intervene, possibly preventing or obviating the contact of CBb6 processes, as seen for many potential CBb3n in-class coupling opportunities. Moreover, unlike the CBb4 class, we have yet to confirm the formation of self-gap junctions by CBb6 cells, and only a few candidate sites have been identified (n = 8 contacts). Like the CBb6 class, CBbwf cells may also decline in-class coupling. With only 2 identified CBbwf cells, both of which having arbors that extend beyond the boundaries of the RC1 volume, only one in-class contact site has been identified. Despite a discernable membrane density at this site, recaptures of both serial sections containing membrane density clearly revealed the presence of an adherens junction lacking any gap junctional zippering (Fig. 6F–H). This lack of in-class coupling is a feature the CBb6 and CBbwf classes share with rod bipolar cells, which were also never observed to form gap junctions within the IPL of RC1 (n = 104 cells), despite having a number of contact opportunities within their incomplete packing of the retinal plane (Lauritzen et al., 2019).
Figure 6.
ON CBC classes lacking in-class homocellular coupling. A-E, Rejection of in-class coupling by the CBb6 class. A, Presence of an adherens junction (tan) at site of contact between neighboring CBb6 cells 400 (magenta) and 9693 (purple). A′, Enlarged view of the boxed area in A rendered with the processes of Aii cells 2610 (slate) and 304 (blue) and relevant gap junctions (arrowheads; canonical heterocellular in blue and homocellular Aii in white). B-E, TEM images of select serial sections. B, Section 240: heterocellular gap junction 18732 between CBb6 9693 (purple) and Aii 2610 (slate). C, Section 242: adherens junction 59565 between CBb6 9693 (purple) and CBb6 400 (pink) (arrow in A′). D, Section 244: heterocellular gap junction 59564 between CBb6 9693 (purple) and Aii 304 (blue). E, Section 254: heterocellular gap junction 130578 between CBb6 400 (pink) and Aii 304 (blue). Homocellular gap junction also present between Aii 2610 (slate) and Aii 304 (blue) (bracket in E). B′, D′, E′, Reimaging of boxed regions in B, D, and E at 0.27 nm/pixel resolution with goniometric tilt. Brackets notate structure boundaries. C′, Enlarged view of boxed region in C. F-H, Absence of in-class coupling between CBbwf cells. F, Adherens junction (tan, arrow) present at the only contact site between CBbwf cells 5283 (slate) and 6589 (gold). Inset, Enlarged and rotated view of the site of interaction. G, H, TEM images from serial sections 267 and 268 showing membrane contact between the branches of CBbwf 6589 (gold) and CBbwf 5283 (slate) at the site indicated by the arrow in F. A density is present in both sections. G′, H′, Reimaging of boxed regions in G, H at 0.27 nm/pixel resolution with goniometric tilt reveals adherens junction 123847 and no evidence of an adjacent gap junction. Scale bars: F, 20 µm; A, 10 µm; B-E, 1 µm; G, H, 500 nm; B′-E′, G′, H′, 100 nm. a, Adherens junction; gj, gap junction; sec, section.
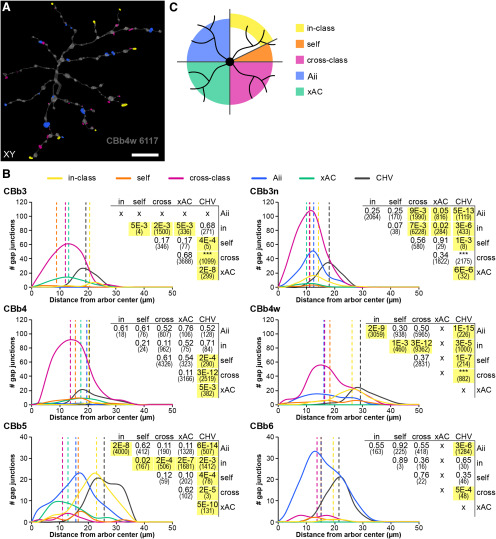
Cross-class homocellular coupling among ON CBCs
Cross-class coupling among ON CBCs is extensive, accounting for 78% of the homocellular motifs. Every class of ON CBCs participates in cross-class homocellular coupling with at least 1 and up to 5 other classes, although typically dominated by a single cross-class partner (Fig. 3B; detailed in Fig. 3-1). Consistent with all reported cross-class coupling motifs in other mammalian species (Raviola and Gilula, 1975; Kolb, 1979; Cohen and Sterling, 1990; Luo et al., 1999; Mills, 1999; Dacey et al., 2000; Jacoby and Marshak, 2000; Arai et al., 2010; Kántor et al., 2016, 2017; Tsukamoto and Omi, 2017), cross-class coupling motifs of ON CBCs in RC1 respect ON/OFF functional segregation. Coupling between ON CBCs and OFF CBCs was never observed. Importantly, both the CBb3 and CBb4 classes, which exhibit few to zero canonical heterocellular gap junctions with Aii cells, routinely form a large number of cross-class gap junctions with other classes that do participate in this classic ON CBC coupling motif, further supporting the classification of the CBb3 and CBb4 classes within the ON superclass.
Cross-class coupling opportunities are afforded by the costratification of ON CBC axonal arborizations within the rabbit IPL (Fig. 7A). The extensive costratification of the CBb3/3n/4/4w classes within strata 3 and 4 is reflected in their coupling partnerships. All 4 classes couple with each other (Fig. 7B–K), with the possible exception of the CBb3 and CBb4w classes with each other (Fig. 7L,M). Coupling frequencies among the other class pairings are variable, ranging from averaging a few gap junctions per cell to over 30 per cell (Fig. 3B), and suggests that these classes preferentially select some classes over others as coupling partners. In contrast, the more deeply stratifying CBb5/6/wf classes in strata 4 and 5 infrequently form gap junctions with each other, despite also exhibiting significant costratification. For example, the CBb5 and CBb6 arbors theoretically overlap by ∼75%, yet CBb5 cells average 0.3 gap junctions with the CBb6 class (± 0.5 gap junctions; 1.55 CV; n = 6 CBb5 cells) and CBb6 cells average 0.8 gap junctions with the CBb5 class (± 0.8 gap junctions; 1.05 CV; n = 5 CBb6 cells). Recapture of 3 instances revealed gap junctions at only 2 of these sites (Fig. 8A,B), suggesting that not all sites of membrane contact contain gap junctions. CBb6 cross-class coupling appears restricted to the CBb5 class, despite costratification with the majority of the CBbwf arbor and 5 validated contacts (Fig. 8C,D). CBbwf arbors also overlap with CBb5 arbors, albeit to a lesser extent than with CBb6, yet exhibit rare instances of coupling (Fig. 8E,F). Although the CBb5 class shares ∼50% arbor overlap with the CBb3, CBb3n, CBb4, and CBb4w classes, formation of gap junctions has only been confirmed with CBb3n cells, and at only 1 of 4 validated contacts (Fig. 8G–J). A single instance of CBb3n coupling with CBb6 was also found (Fig. 8K,L), suggesting that the CBb3n class may be fairly indiscriminate toward coupling partners. Together, these data demonstrate that all ON CBC classes participate in cross-class coupling according to clear class-specific rulesets. However, not all class pairings couple, even when opportunity is afforded by the laminar organization of the retinal IPL, further demonstrating the specificity of these connections.
Figure 8.
Cross-class homocellular coupling topologies of CBb5/6/wf. A, B, Rare CBb5 coupling with CBb6. A, Single cross-class gap junction (magenta; arrow in inset) between CBb6 5515 (purple) and CBb5 5623 (cyan). Inset, Enlarged view of boxed region. B, TEM image of gap junction 119263 (arrow in A). B′, Reimaging of boxed region in B at 0.27 nm/pixel resolution with goniometric tilt. C, D, Rejection of coupling between CBb6 and CBbwf. C, Costratification and overlapping arbors affords 2 sites of membrane contact, but only a single adherens junction (tan; arrow in inset) between the axonal arbors of CBb6 180 (purple) and CBbwf 5283 (gray) and no gap junctions. Inset, Enlarged view of boxed region. D, TEM image of adherens junction 66309 (arrow in C). D′, Reimaging of boxed region in D at 0.27 nm/pixel resolution with goniometric tilt. E, F, Rare CBb5 coupling with CBbwf. E, Single cross-class gap junction (magenta; arrow in inset) between CBb5 6997 (cyan) and CBbwf 6589 (gray). Inset, Enlarged view of boxed region. F, TEM image of gap junction 130564 (arrow in E). F′, Reimaging of boxed region in F at 0.27 nm/pixel resolution with goniometric tilt. G, H, Rare CBb5 coupling with CBb3n. G, Single cross-class gap junction (magenta; arrow in inset) between CBb5 5649 (cyan) and CBb3n 6120 (orange). Inset, Enlarged view of boxed region. H, TEM image of gap junction 111023 (arrow in G). H′, Reimaging of boxed region in H at 0.27 nm/pixel resolution with goniometric tilt. I, J, Rejection of coupling between CBb5 and CBb3n. I, Presence of an adherens junction (tan; arrow in inset) between CBb5 483 (cyan) and CBb3n 1724 (orange), but no gap junctions. Inset, Enlarged view of boxed region. J, TEM image of adherens junction 101900 (arrow in I). J′, Reimaging of boxed region in J at 0.27 nm/pixel resolution with goniometric tilt. K, L, Rare CBb6 coupling with CBb3n. K, Single cross-class gap junction (magenta; arrow in K′) between CBb3n 5284 (orange) and CBb6 16 026 (purple). K′, Enlarged view of boxed region in K. Gap junction scaled by a factor of 4 for visualization. L, TEM image of gap junction 113383 (arrow in K′). L′, Reimaging of boxed region in L at 0.27 nm/pixel resolution with goniometric tilt. Scale bars: A, C, E, G, I, 10 µm; K, 20 µm; B, D, H, J, 1 µm; F, L, 500 nm; B′, D′, F′, H′, J′, L′, 100 nm. a, Adherens junction; gj, gap junction.
Geometric and anatomic opportunities only partially define ON CBC specificity
In the previous section, we detailed the rulesets of ON CBC coupling in the rabbit retina, revealing class-specific differences both in partner choice and frequency. We next sought to examine what confers this specificity. A common strategy for synaptic precision in chemical synapses is the sequential constraint of potential targets, turning connection availability on and off over time (Yogev and Shen, 2014). Initially, developmental processes regulating cell fate determination, cell migration, and the growth and branching of neuronal arbors restrict the pool of potential targets based on geometric and anatomic opportunity, which are later refined through the addition or removal of synapses at contact sites in response to molecular discrimination of partners and/or neural activity. Indeed, the restriction of geometric opportunity by cell spacing and tiling appears to account for much of the specificity observed for in-class coupling patterns. However, the specific rejection of in-class coupling by CBb6 cells, despite opportunity, demonstrates that additional molecular or activity-dependent mechanisms are involved.
Costratification of source and target arbors at specific depths of the IPL is often cited as a driving factor for synaptic precision within the retina. However, the observed stratification patterns in RC1 likely only limits connection availability between CBb6 and CBbwf classes with the shallowly stratified CBb3/3n/4/4w classes and cannot account for the divergence in coupling frequencies among the fully costratifying CBb3/3n/4/4w classes or CBb5/6/wf classes. To further assess the coupling specificity of cross-class homocellular and heterocellular modes, we examined the frequency of coupling as a function of geometric opportunity afforded by the degree of planar overlap in axonal territory between pairs of cells (the intersection area illustrated in Fig. 9A). Importantly, all class pairings exhibited a broad range of intersection areas, even when cell spacing produced large gaps in the tile. Many class pairings exhibited a trend of increased coupling with increased intersection area (Fig. 9B), demonstrating that these coupling motifs are often generated according to specific patterns. By extension, this also suggests that the variability in coupling frequencies observed among individual cells of the same class (Fig. 3-1) may in part be due to variability in arbor size or distribution within the mosaic or tile. In support of the former, CBb4 595 forms 34 gap junctions with the CBb4w class, whereas CBb4 5292 only forms 19. This 56% reduction in coupling frequency can largely be explained by the fact that CBb4 5292 has ∼50% of the field area as CBb4 595. More importantly, the rate at which coupling frequency increases with intersection area often differs among the different partners of a given ON CBC class, demonstrating that ON CBC classes can have unique patterns with each of their partners, even within coupling modes. Arbor branching patterns differ among the various partner classes and can influence anatomic encounter rates that would in turn contribute to the specific patterns of coupling. While this is difficult to quantify, qualitative evaluation suggests that, for at least some class pairings, anatomic encounter rates cannot fully account for the observed specificity. For example, the CBb3n coupling partners, CBb4 and CBb4w, exhibit nearly identical stratification features, but CBb4 cells have fewer branch terminals and an overall lower branch path complexity, suggesting a lower potential anatomic encounter rate than CBb4w cells with CBb3n. Yet, CBb3n cells are 6 times as likely to couple with the CBb4 class compared with the CBb4w class. Thus, while the contributions of molecular discrimination and activity-dependent refinement cannot be evaluated here, neither geometric opportunity nor anatomic encounter rates are sufficient to fully account for the observed coupling specificity of ON CBC classes, arguing that these other mechanisms are likely involved.
Figure 9.
Geometry accounts for the variation in, but not specificity of, ON CBC coupling. A, Calculating the 2D (XY) intersection of the convex hulls for each CBb4 cell with that of each member of the CBb3 class. Left, Convex hulls of CBb4 cells. Right, Convex hulls of CBb3 cells. Middle, The portion of each CBb4 cell's convex hull that intersects in XY with the convex hull of a CBb3 cell. B, Comparisons of coupling frequency as a function of the degree of axonal field intersection between cell pairings (computed in A) for each class. Each point represents a single calculated area of intersection between 2 cells, regardless of the presence/absence of coupling. Thus, many points have an intersection area > 0, but no gap junctions. Since each point represents a single cell-cell pairing (e.g., CBb4 4877 with CBb4w 5601), every cell contributes multiple points due to multiple pairings. Note the different x axis scaling used for the CBb3 and CBb3n graphs. Data were fitted to a linear model with y intercept of zero and the adjusted R2 values reported adjacent to each model. Significance (p) values from the post hoc Scheffé test are presented in the corresponding matrix for each class' graph with the difference (D) statistic in parentheses below. Significance (p < 0.05) is highlighted in yellow. ***Instances where the R software environment computed p < 2.2E − 16.
Specificity beyond numerical fractions
Synaptic specificity extends beyond partner choice and frequency (Yogev and Shen, 2014). Class-specific differences in postsynaptic density sizing among chemical synapses based on input source identity or output target identity exist (Lauritzen et al., 2019) and can influence synaptic strength (Minerbi et al., 2009). The positioning of inhibitory synapses can function as powerful veto synapses (Sivyer and Williams, 2013) or provide differential regulation of sodium-dependent versus calcium-dependent action potentials in the same neuron (Miles et al., 1996). Likewise, gap junction size has been shown to correlate with coupling strength (Flores et al., 2012; Szoboszlay et al., 2016), while positioning can facilitate interactions with chemical synapses and regulate current flow (Yang et al., 1990; Pereda, 2014).
Size specificity
ON CBCs form gap junctions varying in plaque area over 200-fold (minimum = 0.0022 µm2; maximum = 0.46 µm2; mean ± SD = 0.041 ± 0.053 µm2, 1.3 CV; n = 1339). The smallest validated gap junction formed by an ON CBC measures 43 nm in diameter and was present on a single section. Freeze-fracture analyses of connexon spacing within gap junction plaques in the IPL of rodent retinas (Raviola and Gilula, 1973, 1975; Reale et al., 1978; Kamasawa et al., 2006) suggest that this small gap junction corresponds to a plaque comprised of only 3 or 4 connexons across and 1-9 connexons deep (3-36 connexons in total). In contrast, the largest identified gap junction exhibits a maximum diameter of 680 nm and spans 11 sections (minimally 900 nm), corresponding to a plaque containing 2050-4600 connexons.
Parsing the size distribution by coupling mode reveals mode-dependent sizing rules. The median size of gap junction plaques formed by ON CBCs with Aii cells is 3 times greater than those formed with other ON CBCs (Aii partners: median = 0.061 µm2, n = 404; ON CBC partners: median = 0.017 µm2, n = 825; W = 277973, p < 2.2E−16, Wilcoxon rank-sum with continuity correction) and 7 times greater than those formed with xACs (median = 0.0084 µm2, n = 110; W = 42 532, p < 2.2E−16, Wilcoxon rank-sum with continuity correction). Among homocellular gap junctions, those mediating in-class coupling are larger than their cross-class counterparts (in-class: median = 0.022 µm2, n = 211; cross-class: median = 0.016 µm2, n = 605; W = 72 205, p = 0.0045, Wilcoxon rank-sum with continuity correction). Individual ON CBC classes also exhibit differences in the size distributions for gap junctions mediating different coupling modes (Fig. 10A), suggesting that these sizing rules are common across all CBC classes. For several classes (CBb3, CBb4, CBb6, and possibly CBbwf), these mode-dependent sizing rules had little effect on a class' relative distribution of gap junctions among its partners (Fig. 10B), possibly because these classes are dominated by either a single coupling partner or those of the same mode (e.g., cross-class homocellular partners only). The most striking change was for the CBb3n class, where homocellular partners accounted for 73% of the gap junctions formed by this class, but only 33% of the total gap junctional area. Likewise, Aii coupling accounts for a greater proportion of CBb5 and CBb4w gap junctions when measured as a function of gap junctional area rather than frequency, resulting in canonical heterocellular coupling dominating the CBb5 profile. Additionally, the relatively smaller plaque size of cross-class versus in-class gap junctions formed by the CBb4w class, combined with the larger representation by Aii gap junctions, completely reorders the representation of coupling modes within the CBb4w profile.
Figure 10.
Size distribution of gap junction plaques. A, Histograms comparing the frequency distributions of gap junction plaque size (area) by coupling mode for each ON CBC class. Vertical dashed lines indicate median values. Note the change in y axis scaling for the CBb5, CBb6, and CBbwf class graphs. Adjusted significance (p) values (corrected for multiple comparisons) from the Wilcoxon rank-sum tests with continuity correction are presented in the corresponding matrix for each class' graph with the test statistic (W) in parentheses below; n (in-class, cross-class, xAC, Aii) = CBb3 (13, 183, 42, 0), CBb3n (32, 279, 11, 106), CBb4 (30, 297, 28, 7), CBb4w (68, 188, 0, 60), CBb5 (63, 8, 29, 79), CBb6 (5, 9, 0, 116), CBbwf (0, 9, 0, 36). Yellow shading represents significance (p < 0.05). ***Instances where the R software environment computed p < 2.2E − 16. B, Proportion of gap junctions with different partners based on numerical frequency or cumulative area. Data are mean ± SD. cross, Cross-class; in, in-class.
Subcellular targeting
Subcellular synaptic specificity is abundant in the laminar architecture of the mammalian retina. Synapses and partnerships of the Aii cell are highly compartmentalized. Gap junctions with ON CBCs are restricted to the waist and arboreal dendrites of Aii cells that are positioned within the ON sublamina (Marc et al., 2014). Consistent with this, gap junctions with Aii cells are never observed on the descending axon proximal to the bifurcation that initiates ON CBC terminal arborization (n = 404 gap junctions). Indeed, such coupling opportunities are specifically rejected between known coupling partners when contact occurs with inappropriate topology (n = 2 contacts; Fig. 4A–D). In addition, several ON CBCs (n = 8 cells: 1 CBb4w, 6 CBb5, and 1 CBb6) extend a single, often short process within the OFF sublamina, well above their primary axonal arborization within the ON sublamina (e.g., CBb5 cell in Figs. 1A, 4M; several CBb5 and CBb6 cells in Fig. 8), which has also been observed in the cat retina (Cohen and Sterling, 1990). These branches participate in both input and output synapses but appear to not form gap junctions with Aii cells. However, since we have yet to identify a validated contact, it remains unclear whether these branches would specifically reject these as coupling sites.
The positioning of in-class gap junctions at tip-to-tip or tip-to-shaft topologies is striking (Fig. 5). At least one branch terminates in virtually every instance, creating near-perfect tiling for several ON CBC classes. Violation of tiling and tip-to-tip or tip-to-shaft topologies is rare, with only 3 instances of significant in-class branch overlap identified in RC1 (CBb5: n = 1; CBb3n: n = 2; Fig. 5L, open arrowhead). In 2 of the 3 cases, the offending branches share membrane contact and gap junction at the site of crossing; but in all 3 cases, these branches then gap junction and terminate on contact with another process from the same or another class. This observation prompted us to evaluate the potential for spatial rules to govern coupling specificity across ON CBC arbors. Gap junction formation occurs along the full central to peripheral extent of ON CBC arbors (Fig. 11). As expected, the distribution of in-class gap junctions tends to be shifted farther from the geometric center than other coupling modes. This is likely a consequence of the tip-to-tip or tip-to-shaft topologies of in-class gap junctions obviating the opportunity of an individual branch to form another gap junction after an in-class. The distribution of in-class gap junctions was occasionally more central than the vertices of the convex hull itself, but this likely reflects the poor approximation of ON CBC axonal territories by convex hulls (Marc et al., 2014; Lauritzen et al., 2019). Star domains provide better approximations but are difficult to assess mathematically. Interestingly, the distributions of gap junctions mediating canonical heterocellular coupling with Aii cells were shifted more centrally than those mediating in-class coupling for the CBb4w and CBb5 classes, but not for the CBb3n class, consistent with the previously noted tendency for Aii processes to be positioned between branches of in-class neighbors. Additionally, in the absence of in-class coupling, the distribution of gap junctions mediating Aii coupling appeared to occur with a higher frequency toward the hull vertices. Together, these findings support at least a spatial rule for the positioning of in-class gap junctions, although whether their positioning at CBC axonal boundaries is a consequence of, or provides direction for, axonal tiling remains unclear.
Figure 11.
Spatial distribution of gap junctions across ON CBC axonal arbors. A, Spatial distribution of gap junctions across a representative ON CBC arbor, color-coded by coupling mode: heterocellular with Aii (dark blue) or xACs (mint, none), homocellular in-class (yellow), self (orange), or cross-class (magenta). All gap junctions scaled by a factor of 4 for visualization. B, Histograms comparing the frequency distributions of gap junction distance by coupling mode for each ON CBC class. Distance is calculated in the XY plane from the geometric center of the parent cell's convex hull. The distribution of distances for the vertices forming the convex hull (CHV) for the parent cells is included for comparison as an approximation of the arbor boundaries. Vertical dashed lines indicate median values. Note the change in y axis scaling for the CBb5 and CBb6 class graphs. The CBbwf class is not shown due to the incompleteness of the representative cells' arbors. Adjusted significance (p) values from the Wilcoxon rank-sum tests with continuity correction are presented in the corresponding matrix for each class' graph with the test statistic (W) in parentheses below; n (in-class, self, cross-class, xAC, CHV, Aii) = CBb3 (10, 6, 183, 42, 50, 0), CBb3n (34, 5, 279, 11, 66, 106), CBb4 (4, 27, 297, 28, 49, 7), CBb4w (62, 27, 188, 0, 59, 60), CBb5 (68, 10, 8, 29, 62, 75), CBb6 (2, 4, 9, 0, 46, 116). Yellow shading represents significance (p < 0.05). ***Instances where the R software environment computed p < 2.2E − 16. C, Model illustrating the spatial distribution of gap junctions according to coupling mode across a generalized ON CBC arbor. cross, Cross-class; in, in-class.
Glycine as a marker of ON CBCs
An important feature of gap junctional coupling is the diffusion of small molecules. Indeed, accumulation of the neurotransmitter glycine in ON CBCs is dependent on heterocellular coupling with glycinergic ACs (Vaney et al., 1998), and glycine is commonly used as a marker to distinguish ON CBCs from OFF CBCs and rod bipolar cells (Cohen and Sterling, 1986; Yang and Yazulla, 1988; Marc et al., 1995, 2007; Vaney et al., 1998; Haverkamp and Wässle, 2000; Deans et al., 2002; compare Petrides and Trexler, 2008; Jones et al., 2011, 2016; Lauritzen et al., 2013, 2019). The Aii cell has long been considered the source of this glycine signal. Evidence for glycine-negative and/or Aii-noncoupled populations of rabbit ON CBCs prompted the suggestion that only directly Aii-coupled ON CBCs accumulate glycine (Petrides and Trexler, 2008). Here, we demonstrate that the rabbit CBb3 and CBb4 classes virtually lack gap junctions with Aii cells, but we also reveal that both classes have 2 alternative routes for glycine diffusion: (1) through direct coupling with glycinergic ACs other than the Aii cell (e.g., glycinergic xACs; Fig. 4K–O), or (2) through indirect coupling with Aii cells via gap junctions formed with other ON CBC classes (Fig. 3B; Fig. 3-1). We therefore wondered whether CBb3 and CBb4 cells would exhibit glycine accumulation. The RC1 dataset uniquely allows quantification of glycine and other small-molecule signals in the same cells for which connectivity is mapped (Fig. 12A,A′). We observed robust glycine signals in the somas of cells from all 7 ON CBC classes, including both the CBb3 and CBb4 classes (Fig. 12B). Indeed, of all 99 evaluated ON CBC somas, which included 20 CBb3 and 11 CBb4 cells, only 1 cell did not contain glycine at levels above at least 1 SD from the mean observed for OFF CBCs. Coupling of this cell (CBb4 364) with an Aii or other glycinergic AC has not been detected, but annotation remains incomplete. The stratification of CBb4 364 and frequent coupling with cells of the CBb4w and CBb3n classes, however, argues that it is not a misclassified OFF CBC or rod bipolar cell and that its classification as a CBb4 is robust. Although the connexin composition and modulation of these gap junctions remain ambiguous, it is clear that at least one of these routes is functional and sufficient to allow accumulation of glycine, consistent with previous discoveries in mouse of functional coupling between ON CBCs and other non-Aii ACs (Farrow et al., 2013; Lee et al., 2015; Kerstein et al., 2019).
Figure 12.
Glycine accumulation in ON CBCs. A, TEM image from Viking of section 30 of the RC1 volume displaying the somas of 7 ON CBCs (circles) and one Aii (pentagon). A′, Section 30 glycine channel showing intermediate glycine signals in the somas of all ON CBCs (circles) and the Aii (pentagon), including members of the CBb3 and CBb4 classes (*). B, Plot quantifying the glycine signals recorded for the somas of ON CBCs. Glycine signals are reported as the average mean pixel value (0-255) ± SD; n = 17 (CBa, CBb5), 20 (CBb3), 34 (CBb3n), 11 (CBb4), 10 CBb4w), 6 (CBb6), 1 (CBbwf), 105 (all CBb). *Significance (t(38) = 19.08, p < 2E − 16, Welsh two-sample t test, two-tailed). Scale bars: A, 20 µm.
ON CBC coupling in retinal networks
In the previous sections, we have detailed the class-specific coupling patterns of rabbit ON CBCs. We next sought to place these findings in the context of the larger retinal networks, particularly on the distribution of scotopic signals and the potential mixing of parallel processing streams before input to GCs.
Distribution of rod signals through the Aii cell
Previously, we reported a large-scale connectivity and partnership mapping of Aii cells, including their collection of rod-generated scotopic signals from rod bipolar cell terminals (Marc et al., 2014). However, we did not detail Aii cell output to CBCs with class specificity. Our discovery of class-specific coupling patterns between ON CBCs and Aii cells, together with anatomic and physiological reports of differentially weighted Aii output in other species (cat: McGuire et al., 1984; Cohen and Sterling, 1990; rat: Veruki and Hartveit, 2002a; mouse: Tsukamoto and Omi, 2017), prompted a reexamination of the potential signal flow through the scotopic network in the rabbit retina.
The distribution of Aii output to the various ON CBC classes in RC1 was assessed by reanalyzing the coupling partnerships of Aii cells 476, 514, 2610, and 3679 from our previous study (Marc et al., 2014). Each Aii cell forms an average of 114 ± 11 gap junctions (0.12 CV; n = 4 Aii cells). Over 53% of these gap junctions mediate homocellular coupling among Aii cells and therefore dominate the coupling profile (Fig. 13A). This homocellular coupling was found to occur both between neighboring Aii cells and between sister branches of the same Aii cell (Fig. 3-1). Although Aii lobular dendrites tile the OFF sublamina, their arboreal dendrites within the ON sublamina extensively overlap, such that any point may be sampled by up to 4 separate Aii cells (Marc et al., 2014). This affords numerous in-class contact opportunities for homocellular coupling among the arboreal dendrites; and accordingly, these gap junctions are typically not found among the lobular dendrites. Heterocellular coupling with ON CBCs was dominated by 3 classes, together representing over 86% of the heterocellular gap junctions (36% by CBb6, 32% by CBb3n, and 19% by CBb5). Although the distributions of plaque sizes diverge across the coupled partners, including between different classes of ON CBCs (Fig. 13B), these same 3 ON CBC classes accounted for almost 90% of the total heterocellular gap junction area (Fig. 13C). However, the relative representation by these 3 classes shifted, such that the CBb3n class accounts for over half of the heterocellular area, rather than more equal representations by the CBb3n and CBb6 classes when based on gap junction frequency alone (Fig. 13C). No gap junctions were found between any of these Aii cells and the CBb3 class, and only one candidate has been identified with the CBb4 class (Fig. 13A; detailed in Fig. 3-1). Importantly, the identity of all but 3 coupled partners was determined. Of these, only one could be identified as a bipolar cell, but all were found deep within the IPL, making it unlikely that any of these partners belong to the CBb3 or CBb4 class.
Figure 13.
Differential distribution of Aii output to ON CBCs. A, Coupling profile of the Aii cell class. Each column reports the mean number of gap junctions formed by an Aii cell with ON CBC classes and other Aii cells, as assessed from 4 Aii cells in RC1. Values are encoded by the linear heat map scale below. *Pairing where coupling has not been confirmed. This is not the reciprocal of the Aii column presented in Figure 3, as the frequencies here are calculated per Aii cell, whereas Figure 3 calculates Aii coupling frequencies per ON CBC. Detailed coupling profiles for individual Aii cells and extended class statistics are provided in Figure 3-1. B, Partner class-dependent sizing rules for Aii cell gap junction plaques. Histogram comparing the frequency distributions of gap junction plaque size (area) by partner ON CBC class. Vertical dashed lines indicate median values. Adjusted significance (p) values from the Wilcoxon rank-sum tests with continuity correction are presented in the corresponding matrix with the test statistic (W) in parentheses below; n = 184 Aii, 66 CBb3n, 13 CBb4w, 39 CBb5, 73 CBb6, 17 CBbwf. Yellow shading represents significance (p < 0.05). C, The proportion of gap junctions with different partners based on numerical frequency or cumulative area. Data are mean ± SD.
The Aii cell is also a component of all but one known motif predicted to mediate rod-driven cross-suppression of photopic signals (Lauritzen et al., 2019). In motifs R1, R2, and R25, the activation of Aii-coupled ON CBCs drives narrow and wide-field ACs to directly suppress ON CBCs. While the preference of rod inhibitory motifs for the suppression of particular ON CBC classes will require further evaluation, the preferential coupling of Aii cells with specific ON CBC classes, by either numerical counts or cumulative area metrics, suggests that the ON CBC targets of CBb3n, CBb6, and possibly CBb5-driven ACs should provide important insight.
Potential mixing of ON CBC input to GCs
Coupling between ON CBC classes provides numerous potential lateral paths for signal flow (Fig. 14A), yet individual CBC classes have long been thought to transmit separate parallel channels of visual information to GCs (Masland, 2001; Euler et al., 2014). While connexin composition and modulation of gating will undoubtably regulate the strength and effect of these lateral paths, we sought to explore their potential impact on GC input. Exploration of this question required analysis beyond the direct coupling of the 37 ON CBCs described in this study and, rather, use of the entire annotated connectivity in the RC1 dataset. Graffinity was developed for rapid visualization and exploration of complex connectivity in large datasets (Kerzner et al., 2017). In an initial exploration of the potential mixing of ON CBC signals cross-class coupling may afford, we chose 3 distinct and well-annotated GCs in RC1. Each receives unique combinations of direct ON CBC drive (→) via ribbon- or bipolar conventional-type synapses (1-hop path: [CBbx → GC]; Fig. 14B). Expanding the path length to include direct coupling (::) between ON CBCs (2-hop path: [CBb :: CBb → GC]) potentially alters the input profile of these GCs in 2 important ways: (1) the number of paths for each direct ON CBC input increased due to the recruitment of neighboring cells via in-class homocellular coupling, indicative of greater synaptic input; and, more importantly, (2) cross-class homocellular coupling provides potential paths for indirect signal flow from an additional 2 or 3 ON CBC classes, which likely contribute to the shaping of the output response of the GC's direct ON CBC input class (Demb and Singer, 2016; Kuo et al., 2016). In the case of the transient ON directionally selective GC 606, which already directly samples broadly from the ON CBC classes, cross- and in-class coupling has the potential to alter the relative path contribution among the ON CBC classes. Further extension of the path length by additional hops eventually reveals paths from all ON CBC classes, as early as 3 hops for the selected GCs, especially as Aii and CBb3n hubs are incorporated through heterocellular and cross-class coupling, respectively.
Figure 14.
ON CBC coupling motifs within retinal networks. A, Models of the scotopic and photopic networks of the light-adapted rabbit retina involving ON CBCs. Arrows indicate chemical synapses (ribbon). Plain lines indicate gap junctions. In the primary rod pathway, rod photoreceptor signals are unevenly distributed through canonical heterocellular gap junctions to the 7 classes of ON CBCs. Multipoint coupling mediated by in-class homocellular gap junctions among neighboring cells of the same class (not shown) produces sheets of coupled cells (albeit to differing degrees). Cross-class homocellular coupling potentially allows lateral spread of signals across parallel ON CBC channels. CBb6 and CBbwf classes contact Aii cells with both electrical (plain lines) and chemical (arrows) synapses. These ON CBCs then transfer these processed rod signals to ON and ON-OFF GCs. In the photopic network, M/L cone signals are collected by 6 classes of ON CBCs, whereas S cone signals are collected by the CBbwf class. Mixing of these parallel ON CBC channels may occur directly through cross-class homocellular coupling or indirectly through the Aii cell, despite rectification of these canonical heterocellular gap junctions. ON CBCs then transfer the processed signals on to ON and ON-OFF GCs. B, Potential mixing of parallel ON CBC channel input to GCs via gap junctional coupling (::). Connectivity matrices generated by Graffinity for paths of different lengths (hops) between ON CBCs and GCs. The data include all cells and synapses currently annotated in connectome RC1, including incompletely annotated cells. The CBb label contains all partial cells or fragments that could not be classified beyond the ON superclass. Empty tiles indicate instances of zero identified paths. The path count is encoded using the linear color scale below. :: Coupling; RBC, rod bipolar cell; tON DS GC, transient ON directionally selective GC.
Discussion
We report the participation of rabbit ON CBCs in extensive coupling, each forming 20-81 candidate gap junctions (5-25% total axonal synaptic contacts; ≤40% total IPL input/output). Reports in other species suggest ≤20 per cell (≤15% synaptic contact) (cat: McGuire et al., 1984; Cohen and Sterling, 1990; mouse: Tsukamoto and Omi, 2017). Is rabbit the exception or do species truly differ in their extent of ON CBC coupling? Our approach has the highest base resolution, recapture ability with goniometric tilt, a large contiguous volume for high copy numbers, and a coherent toolset for network exploration, leaving the possibility that extensive coupling is the rule. However, preliminary findings in our comparable mouse volume support species-specific differences in retinal connectivity. More importantly, how do these models compare to human? Ultrastructure-based analysis of coupling motifs in humans or nonhuman primates is lacking, and we suggest that 63% of ON CBC gap junctions (77% homocellular; 98% xAC; 24% Aii) are unresolvable by traditional light or confocal microscopy. Nonetheless, CX36 colocalization suggests that homocellular gap junctions are prominent in human OFF CBC circuitry and possibly ON CBC circuitry as well (Kántor et al., 2017).
Within coupling modes, some features appear conserved whereas others are species-specific. Like cat (Cohen and Sterling, 1990) and mouse (Tsukamoto and Omi, 2017), rabbit ON CBCs exhibit class-specific Aii coupling frequencies and areas, suggesting differential Aii output to the various ON CBC classes. Similarities with mouse are striking. 3 classes (rabbit: CBb3n, CBb6, CBb5; mouse: T6, T7, T5a) collect the vast majority of Aii output, one completely lacks Aii coupling (rabbit: CBb3; mouse: T5b), and a second rarely couples (rabbit: CBb4; mouse: T5c). Moreover, these classes exhibit similar stratification (shallow = strata 3/4 vs deep = strata 4/5). In cat, only one shallow class (b3) lacks Aii coupling, but uniquely, so does a deep (b5). Additionally, frequency counts and area measures suggest a differential output preference to deep versus shallow classes in cat, but the converse in rabbit and mouse. Assessments of noncanonical heterocellular AC partners are incomplete, but we identified novel xAC class(es) inconsistent with known mouse AC partners (Kolb and Nelson, 1996; Lee et al., 2015; Kerstein et al., 2019; Yadav et al., 2019). Homocellular coupling varies more. In rabbit, 5 of 7 classes in-class couple according to axonal tiling patterns, in contrast to a single class in cat and none in mouse, owing to lack of contact. Tiling in macaque (Tsukamoto and Omi, 2016) suggests abundant opportunities for 3 of 6 classes, and in-class coupling was reported for at least one human ON CBC class, despite limited opportunity (Kántor et al., 2017), suggesting that in-class coupling may be the rule. Cross-class coupling appears considerably more extensive in rabbit, with all 7 classes participating to some extent. Cross-class coupling is generally more pervasive among shallow classes, although robust coupling between the deep T6 and T7 classes in mouse is a clear exception. Even in RC1, such gap junctions among the deep classes are sparse (average: <2/cell) and small, possibly causing them to have been missed in previous datasets. Colocalization of CX36 interior to DB6 branch tips and not with Aii processes supports cross-class dominancy in human ON CBC coupling as well (Kántor et al., 2017).
Despite possible divergence in overall coupling extent, the underlying motifs appear fairly conserved, facilitating class correlation across species (Table 3). We define 7 classes in rabbit, consistent with previous classification schemes, but contrasting with 8 in mouse and 5 in cat. Notably, comparisons with mouse using morphology or Aii coupling alone yielded incongruent schemes. We prioritized connectivity, as this more likely correlates with function (Vlasits et al., 2019), and underlies the discrepancy between our alignment of cat b2 with mouse T7 rather than mouse T8, as proposed by Tsukamoto and Omi (2017). We also predict the existence of 2 additional classes in cat retina (b3b and b4b), proposing their resolution by Cohen and Sterling (1990) was limited by small sample sizes and similar Aii connectivity. True functional correlation across species requires additional features. It will be interesting to see how these hold up as additional connectivity and physiology features become available.
Table 3.
Class alignment with literature and across speciesa
| Class | Historicalb | Reference | Mousetypec | Mouse type(morphology)d | Mouse type(Aii output)e | Cattypef |
|---|---|---|---|---|---|---|
| CBb3 | nab CBnb3-4 CBb3/3-4 |
Famiglietti, 1981 McGillem and Dacheux, 2001 MacNeil et al., 2004 |
5b | 5c | 5b | b3(a) |
| CBb3n | nab CBnb3 CBb3n |
Famiglietti, 1981 McGillem and Dacheux, 2001 MacNeil et al., 2004 |
5a | 5b | 6 | b4(a) |
| CBb4 | CBnb4 CBb4 |
McGillem and Dacheux, 2001 MacNeil et al., 2004 |
5c | 5a | 5c?/9? | b3(b) |
| CBb4w | CBmb4 NK1+ CBb4 |
McGillem and Dacheux, 2001 Casini et al., 2002 MacNeil et al., 2004 |
5d | 5d | 8 | b4(b) |
| CBb5 | nb1, nb2 S4 CD15+ CBmb4-5 |
Famiglietti, 1981 Strettoi et al., 1994 Brown and Masland, 1999 McGillem and Dacheux, 2001 |
7 | 7 | 5a | b2 |
| CBb6 | nb1, nb2 CaBP+ CBmb5 CBb5 |
Famiglietti, 1981 Massey and Mills, 1996 McGillem and Dacheux, 2001 MacNeil et al., 2004 |
6 | 8 | 7 | b1 |
| CBbwf | Wb Biocytin+ |
Famiglietti, 1981 Jeon and Masland, 1995; MacNeil and Gaul, 2008 |
9 | 9 | 5d | b5g |
| NCC | 8 | 6 |
aNCC, No comparable class.
bAlignment to nomenclature in previous rabbit ON CBC classification schemes or descriptions of classes identified using cell-specific markers.
cAlignment with mouse ON CBC types using the unified nomenclature proposed by Tsukamoto and Omi (2017). The alignment used class descriptions from additional relevant references (Haverkamp et al., 2005; Wässle et al., 2009; Helmstaedter et al., 2013; Greene et al., 2016; Shekhar et al., 2016; Tsukamoto and Omi, 2017).
dAlignment with mouse ON CBC types based on morphologic features alone (e.g., axonal stratification, arbor breadth, and diameter).
eAlignment with mouse ON CBC types based on the differential output of Aii cells to the various ON CBC classes (Tsukamoto and Omi, 2017).
fAlignment with cat ON CBC types as described by Cohen and Sterling (1990). Note our proposal for the splitting of the original b3 class into b3a and b3b and original b4 class into b4a and b4b.
gCohen and Sterling (1990) described b5 as a wide-field ON CBC lacking input from overlying pedicles, but may align with the blue CBC described by Famiglietti (1981).
Irrespective of species, coupling profiles are class-specific: cells accept and reject unique combinations of Aii and ON CBC partnerships according to robust rulesets. The differences reported here in the frequency, size, and spatial distributions of gap junctions across motifs argue that coupling specificity is as rigorous as chemical synaptic specificity. Costratification is often touted as the critical mechanism conferring specificity within retina, but, as shown here, is a poor indicator of coupling specificity and synaptic specificity in general (e.g., GC 8575; Fig. 14). Geometric and anatomic opportunity also fails to confer specificity. Thus, as demonstrated for chemical synaptic precision, including in ON CBCs (Morgan et al., 2011; Yogev and Shen, 2014), coupling specificity requires molecular partner discrimination or activity-dependent refinement. Invertebrate innexin-based gap junctions play dual roles, conferring target specificity by acting as “lock and key” recognition factors (Baker and Macagno, 2014). A general lack of connexin diversity and extensive Cx36/45-mediated motifs (Lin et al., 2005; Li et al., 2008; Kántor et al., 2017) opposes a similar role in mammalian ON CBCs.
How might ON CBC coupling influence retinal networks? Uneven distribution of Aii signals to ON CBC classes has clear implications on scotopic signaling (Fig. 14A). Some GCs exhibiting high scotopic thresholds appear to lack primary pathway input, relying instead on secondary (direct rod-cone photoreceptor coupling) and/or tertiary (direct rod-to-CBC synapses) pathways (Deans et al., 2002; Völgyi et al., 2004; Seilheimer et al., 2020). Non-Aii-coupled CBb3 and CBb4 classes may provide the predominant input for such GCs. Alternatively, these classes may not participate in scotopic retinal circuitry at all, as some GCs exhibit only photopic responses (Deans et al., 2002). However, rod-cone coupling is extensive (Owen, 1985; Wu and Yang, 1988; Tsukamoto et al., 2001; Massey, 2008; Asteriti et al., 2017), making it difficult to conceive of CBCs that would not participate in the secondary pathway. Moreover, both CBb3 and CBb4 indirectly couple with Aii cells through other ON CBC classes, although differential regulation, as described for homocellular Aii-Aii and heterocellular Aii-ON CBC coupling (Mills and Massey, 1995; Xia and Mills, 2004; Petrides and Trexler, 2008), may reconfigure these circuits.
ON CBC coupling also has implications for photopic networks (Fig. 14A). Gap junction-mediated photopic signal spread among ON CBCs generates nonlinear amplification of CBC output at chemical synapses to some GCs, increasing GC sensitivity to spatiotemporally correlated stimuli (Kuo et al., 2016). Although we find some ON CBC gap junctions exist in “mixed” (chemical plus electrical) synapses, none occur with GCs, consistent with the proposition that this modulation occurs through lateral motifs within the coupled Aii-ON CBC network (Demb and Singer, 2016; Kuo et al., 2016). Indirect coupling mediated by Aii cells may contribute, as Aii-ON CBC gap junctions are bidirectional (Xin and Bloomfield, 1999; Trexler et al., 2001; Demb and Singer, 2012), despite ∼2-fold rectification (Veruki and Hartveit, 2002a). However, we reveal potential direct paths through homocellular motifs. Contribution by in-class coupling is intuitive, or these motifs may facilitate enhanced signal saliency as reported for cone photoreceptors (DeVries et al., 2002). The mixing of parallel channels afforded by cross-class coupling could also contribute. Alternatively, mixing may shape more complex temporal output (Euler et al., 2014) and/or desynchronization (Vervaeke et al., 2010), although sustained and transient classes tend to segregate with stratification depth (Baden et al., 2013), similar to cross-class coupling. In support of the latter, oscillations emerge from the Aii-ON CBC network when photoreceptor input is blocked (Trenholm et al., 2012).
Our method provides no indication of open probability. Moreover, connexin composition [which remains ambigous, but likely differs between coupling modes (Lin et al., 2005; compare Li et al., 2008)] and modulatory mechanisms [which can differ for the same connexin in different cell classes (O'Brien, 2019)], will clearly regulate when and to what degree signals flow along these paths (Veenstra, 1996; Nielsen et al., 2012). However, effects on network activity are often poorly predicted by coupling coefficients (Haas, 2015), and even a small fraction of open channels can confer network effects (Curti et al., 2012; Szoboszlay et al., 2016), including in the coupled CBC network of the retina (Farrow et al., 2013). Furthermore, coupling effects on networks depend on the spatial and temporal spread of signals and are therefore dynamic and context-dependent (Alcami and Pereda, 2019). In degenerated retinal networks, Cx36-expressing gap junctions within the extended Aii-ON CBC network mediate aberrant hyperactivity contributing to visual impairment (Trenholm et al., 2012; Ivanova et al., 2016), and an elevated fraction of phosphorylated CX36 (Ivanova et al., 2015) suggests aberrant opening. Whether this reflects deregulation of normative or novel motifs remains unknown.
Our findings significantly expand the repertoire of motifs within the Aii-ON CBC network, providing additional sites for disruption in disease and possibly therapeutic targeting. Deciphering their contributions is vital to our understanding of visual processing in the retina. Here, we detail the available partnerships, frequencies, sizing, and topology for future physiology and modeling studies assessing function.
Footnotes
R.E.M. is a principal of Signature Immunologics, Inc., manufacturer of some antibodies used in this manuscript. The remaining authors declare no competing financial interests.
This work was supported by the National Institutes of Health Grants R01 EY02576 to R.E.M., R01 EY015128 to B.W.J., R01 EY028927 to B.W.J., P30 EY014800 to Core, and T32 EY024234 to C.L.S.; the Calvin and JeNeal Hatch Presidential Endowed Chair to R.E.M.; and Research to Prevent Blindness (New York) Unrestricted Grant to the Department of Ophthalmology & Visual Sciences, University of Utah. Funding for the JEOL JEM-1400 was generously provided by the late Martha Ann Healy, a friend of the Moran Eye Center. We thank Hope Morrison, John Hoang, Alex Sessions, Nicholas McCarthy, Jeebika Dahal, Jessica Garcia, and Andrea Bordt for cell tracing.
References
- Alcami P, Pereda AE (2019) Beyond plasticity: the dynamic impact of electrical synapses on neural circuits. Nat Rev Neurosci 20:253–271. 10.1038/s41583-019-0133-5 [DOI] [PubMed] [Google Scholar]
- Anderson JR, Jones BW, Yang JH, Shaw MV, Watt CB, Koshevoy P, Spaltenstein J, Jurrus E, Whitaker RT, Mastronarde D, Tasdizen T, Marc RE (2009) A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol 7:e1000074. 10.1371/journal.pbio.1000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Mohammed S, Grimm B, Jones BW, Koshevoy P, Tasdizen T, Whitaker R, Marc RE (2011a) The Viking viewer for connectomics: scalable multi-user annotation and summarization of large volume data sets. J Microsc 241:13–28. 10.1111/j.1365-2818.2010.03402.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Jones BW, Watt CB, Shaw MV, Yang JH, Demill D, Lauritzen JS, Lin Y, Rapp KD, Mastronarde D, Koshevoy P, Grimm B, Tasdizen T, Whitaker R, Marc RE (2011b) Exploring the retinal connectome. Mol Vis 17:355–379. [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Sigulinsky CL, Nelson NT, Kerzner E (2019) On bipolar coupling code [Query code]. Available at https://github.com/connectomes/Archive/tree/2018_OnBipolarCoupling.
- Arai I, Tanaka M, Tachibana M (2010) Active roles of electrically coupled bipolar cell network in the adult retina. J Neurosci 30:9260–9270. 10.1523/JNEUROSCI.1590-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asteriti S, Gargini C, Cangiano L (2017) Connexin 36 expression is required for electrical coupling between mouse rods and cones. Vis Neurosci 34:E006. 10.1017/S0952523817000037 [DOI] [PubMed] [Google Scholar]
- Baden T, Berens P, Bethge M, Euler T (2013) Spikes in mammalian bipolar cells support temporal layering of the inner retina. Curr Biol 23:48–52. 10.1016/j.cub.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Baker MW, Macagno ER (2014) Control of neuronal morphology and connectivity: emerging developmental roles for gap junctional proteins. FEBS Lett 588:1470–1479. 10.1016/j.febslet.2014.02.010 [DOI] [PubMed] [Google Scholar]
- Baldridge WH, Ball AK, Miller RG (1989) Gap junction particle density of horizontal cells in goldfish retinas lesioned with 6-OHDA. J Comp Neurol 287:238–246. 10.1002/cne.902870207 [DOI] [PubMed] [Google Scholar]
- Bloomfield SA. (1992) Relationship between receptive and dendritic field size of amacrine cells in the rabbit retina. J Neurophysiol 68:711–725. 10.1152/jn.1992.68.3.711 [DOI] [PubMed] [Google Scholar]
- Bostock M, Ogievetsky V, Heer J (2011) D(3): data-driven documents. IEEE Trans Vis Comput Graph 17:2301–2309. 10.1109/TVCG.2011.185 [DOI] [PubMed] [Google Scholar]
- Brown SP, Masland RH (1999) Costratification of a population of bipolar cells with the direction-selective circuitry of the rabbit retina. J Comp Neurol 408:97–106. [DOI] [PubMed] [Google Scholar]
- Buhl DL, Harris KD, Hormuzdi SG, Monyer H, Buzsaki G (2003) Selective impairment of hippocampal gamma oscillations in connexin-36 knock-out mouse in vivo. J Neurosci 23:1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, Rao Y (2010) Molecular mechanisms of tiling and self-avoidance in neural development. Mol Brain 3:28. 10.1186/1756-6606-3-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini G, Sabatini A, Catalani E, Willems D, Bosco L, Brecha NC (2002) Expression of the neurokinin 1 receptor in the rabbit retina. Neuroscience 115:1309–1321. 10.1016/s0306-4522(02)00408-6 [DOI] [PubMed] [Google Scholar]
- Cohen E, Sterling P (1986) Accumulation of (3H)glycine by cone bipolar neurons in the cat retina. J Comp Neurol 250:1–7. 10.1002/cne.902500102 [DOI] [PubMed] [Google Scholar]
- Cohen E, Sterling P (1990) Demonstration of cell types among cone bipolar neurons of cat retina. Philos Trans R Soc Lond B Biol Sci 330:305–321. 10.1098/rstb.1990.0201 [DOI] [PubMed] [Google Scholar]
- Cook JE. (1996) Spatial properties of retinal mosaics: an empirical evaluation of some existing measures. Vis Neurosci 13:15–30. 10.1017/s0952523800007094 [DOI] [PubMed] [Google Scholar]
- Coulon P, Landisman CE (2017) The potential role of gap junctional plasticity in the regulation of state. Neuron 93:1275–1295. 10.1016/j.neuron.2017.02.041 [DOI] [PubMed] [Google Scholar]
- Curti S, Hoge G, Nagy JI, Pereda AE (2012) Synergy between electrical coupling and membrane properties promotes strong synchronization of neurons of the mesencephalic trigeminal nucleus. J Neurosci 32:4341–4359. 10.1523/JNEUROSCI.6216-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey D, Packer OS, Diller L, Brainard D, Peterson B, Lee B (2000) Center surround receptive field structure of cone bipolar cells in primate retina. Vision Res 40:1801–1811. 10.1016/s0042-6989(00)00039-0 [DOI] [PubMed] [Google Scholar]
- Das S, Trona F, Khallaf MA, Schuh E, Knaden M, Hansson BS, Sachse S (2017) Electrical synapses mediate synergism between pheromone and food odors in Drosophila melanogaster. Proc Natl Acad Sci USA 114:E9962–E9971. 10.1073/pnas.1712706114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR, Völgyi B, Goodenough DA, Bloomfield SA, Paul DL (2002) Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron 36:703–712. 10.1016/s0896-6273(02)01046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, Singer JH (2012) Intrinsic properties and functional circuitry of the AII amacrine cell. Vis Neurosci 29:51–60. 10.1017/S0952523811000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, Singer JH (2016) Mind the gap junctions: the importance of electrical synapses to visual processing. Neuron 90:207–209. 10.1016/j.neuron.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries SH, Qi X, Smith R, Makous W, Sterling P (2002) Electrical coupling between mammalian cones. Curr Biol 12:1900–1907. 10.1016/s0960-9822(02)01261-7 [DOI] [PubMed] [Google Scholar]
- Elliott H, Fischer RS, Myers KA, Desai RA, Gao L, Chen CS, Adelstein RS, Waterman CM, Danuser G (2015) Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat Cell Biol 17:137–147. 10.1038/ncb3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Haverkamp S, Schubert T, Baden T (2014) Retinal bipolar cells: elementary building blocks of vision. Nat Rev Neurosci 15:507–519. 10.1038/nrn3783 [DOI] [PubMed] [Google Scholar]
- Famiglietti EV., Jr (1981) Functional architecture of cone bipolar cells in mammalian retina. Vision Res 21:1559–1563. 10.1016/0042-6989(81)90032-8 [DOI] [PubMed] [Google Scholar]
- Farrow K, Teixeira M, Szikra T, Viney TJ, Balint K, Yonehara K, Roska B (2013) Ambient illumination toggles a neuronal circuit switch in the retina and visual perception at cone threshold. Neuron 78:325–338. 10.1016/j.neuron.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Flores CE, Nannapaneni S, Davidson KG, Yasumura T, Bennett MV, Rash JE, Pereda AE (2012) Trafficking of gap junction channels at a vertebrate electrical synapse in vivo. Proc Natl Acad Sci USA 109:E573–E582. 10.1073/pnas.1121557109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch C, De Souza-Silva MA, Sohl G, Guldenagel M, Willecke K, Huston JP, Dere E (2005) Stimulus complexity dependent memory impairment and changes in motor performance after deletion of the neuronal gap junction protein connexin36 in mice. Behav Brain Res 157:177–185. 10.1016/j.bbr.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Greene MJ, Kim JS, Seung HS, EyeWirers (2016) Analogous convergence of sustained and transient inputs in parallel On and Off pathways for retinal motion computation. Cell Rep 14:1892–1900. 10.1016/j.celrep.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Sagasti A (2010) Self-avoidance and tiling: mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol 2:a001750. 10.1101/cshperspect.a001750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldenagel M, Ammermuller J, Feigenspan A, Teubner B, Degen J, Sohl G, Willecke K, Weiler R (2001) Visual transmission deficits in mice with targeted disruption of the gap junction gene connexin36. J Neurosci 21:6036–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas JS. (2015) A new measure for the strength of electrical synapses. Front Cell Neurosci 9:378. 10.3389/fncel.2015.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wässle H (2000) Immunocytochemical analysis of the mouse retina. J Comp Neurol 424:1–23. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wässle H, Duebel J, Kuner T, Augustine GJ, Feng G, Euler T (2005) The primordial, blue-cone color system of the mouse retina. J Neurosci 25:5438–5445. 10.1523/JNEUROSCI.1117-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W (2013) Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500:168–174. 10.1038/nature12346 [DOI] [PubMed] [Google Scholar]
- Hormuzdi SG, Pais I, LeBeau FE, Towers SK, Rozov A, Buhl EH, Whittington MA, Monyer H (2001) Impaired electrical signaling disrupts gamma frequency oscillations in connexin 36-deficient mice. Neuron 31:487–495. 10.1016/s0896-6273(01)00387-7 [DOI] [PubMed] [Google Scholar]
- Ivanova E, Yee CW, Sagdullaev BT (2015) Increased phosphorylation of Cx36 gap junctions in the AII amacrine cells of RD retina. Front Cell Neurosci 9:390. 10.3389/fncel.2015.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Yee CW, Baldoni R Jr, Sagdullaev BT (2016) Aberrant activity in retinal degeneration impairs central visual processing and relies on Cx36-containing gap junctions. Exp Eye Res 150:81–89. 10.1016/j.exer.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby RA, Marshak DW (2000) Synaptic connections of DB3 diffuse bipolar cell axons in macaque retina. J Comp Neurol 416:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY (2010) Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 11:316–328. 10.1038/nrn2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Masland RH (1995) A population of wide-field bipolar cells in the rabbit's retina. J Comp Neurol 360:403–412. 10.1002/cne.903600304 [DOI] [PubMed] [Google Scholar]
- Jones BW, Watt CB, Frederick JM, Baehr W, Chen CK, Levine EM, Milam AH, Lavail MM, Marc RE (2003) Retinal remodeling triggered by photoreceptor degenerations. J Comp Neurol 464:1–16. 10.1002/cne.10703 [DOI] [PubMed] [Google Scholar]
- Jones BW, Kondo M, Terasaki H, Watt CB, Rapp K, Anderson J, Lin Y, Shaw MV, Yang JH, Marc RE (2011) Retinal remodeling in the Tg P347L rabbit, a large-eye model of retinal degeneration. J Comp Neurol 519:2713–2733. 10.1002/cne.22703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Pfeiffer RL, Ferrell WD, Watt CB, Marmor M, Marc RE (2016) Retinal remodeling in human retinitis pigmentosa. Exp Eye Res 150:149–165. 10.1016/j.exer.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalloniatis M, Marc RE, Murry RF (1996) Amino acid signatures in the primate retina. J Neurosci 16:6807–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasawa N, Furman CS, Davidson KG, Sampson JA, Magnie AR, Gebhardt BR, Kamasawa M, Yasumura T, Zumbrunnen JR, Pickard GE, Nagy JI, Rash JE (2006) Abundance and ultrastructural diversity of neuronal gap junctions in the OFF and ON sublaminae of the inner plexiform layer of rat and mouse retina. Neuroscience 142:1093–1117. 10.1016/j.neuroscience.2006.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kántor O, Benkő Z, Énzsöly A, Dávid C, Naumann A, Nitschke R, Szabó A, Pálfi E, Orbán J, Nyitrai M, Németh J, Szél Á, Lukáts Á, Völgyi B (2016) Characterization of connexin36 gap junctions in the human outer retina. Brain Struct Funct 221:2963–2984. 10.1007/s00429-015-1082-z [DOI] [PubMed] [Google Scholar]
- Kántor O, Varga A, Nitschke R, Naumann A, Enzsoly A, Lukats A, Szabo A, Nemeth J, Völgyi B (2017) Bipolar cell gap junctions serve major signaling pathways in the human retina. Brain Struct Funct 222:2603–2624. 10.1007/s00429-016-1360-4 [DOI] [PubMed] [Google Scholar]
- Kawano T, Po MD, Gao S, Leung G, Ryu WS, Zhen M (2011) An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron 72:572–586. 10.1016/j.neuron.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Kerstein PC, Leffler J, Sivyer B, Taylor WR, Wright KM (2019) Gbx2 identifies and regulates the development of an atypical amacrine cell in the mouse retina. Invest Ophthalmol Vis Sci 60:541. [Google Scholar]
- Kerzner E, Lex A, Sigulinsky CL, Umess T, Jones BW, Marc RE, Meyer M (2017) Graffinity: visualizing connectivity in large graphs. Comput Graph Forum 36:251–260. 10.1111/cgf.13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara AH, Mantovani de Castro L, Belmonte MA, Yan CY, Moriscot AS, Hamassaki DE (2006) Expression of connexins 36, 43, and 45 during postnatal development of the mouse retina. J Neurobiol 66:1397–1410. 10.1002/neu.20299 [DOI] [PubMed] [Google Scholar]
- Kohler K, Kolbinger W, Kurz-Isler G, Weiler R (1990) Endogenous dopamine and cyclic events in the fish retina: II. Correlation of retinomotor movement, spinule formation, and connexon density of gap junctions with dopamine activity during light/dark cycles. Vis Neurosci 5:417–428. 10.1017/s0952523800000547 [DOI] [PubMed] [Google Scholar]
- Kolb H. (1979) The inner plexiform layer in the retina of the cat: electron microscopic observations. J Neurocytol 8:295–329. 10.1007/bf01236124 [DOI] [PubMed] [Google Scholar]
- Kolb H, Famiglietti EV (1974) Rod and cone pathways in the inner plexiform layer of cat retina. Science 186:47–49. 10.1126/science.186.4158.47 [DOI] [PubMed] [Google Scholar]
- Kolb H, Nelson R (1996) Hyperpolarizing, small-field, amacrine cells in cone pathways of cat retina. J Comp Neurol 371:415–436. [DOI] [PubMed] [Google Scholar]
- Kuo SP, Schwartz GW, Rieke F (2016) Nonlinear spatiotemporal integration by electrical and chemical synapses in the retina. Neuron 90:320–332. 10.1016/j.neuron.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz-Isler G, Wolburg H (1986) Gap junctions between horizontal cells in the cyprinid fish alter rapidly their structure during light and dark adaptation. Neurosci Lett 67:7–12. 10.1016/0304-3940(86)90199-0 [DOI] [PubMed] [Google Scholar]
- Kurz-Isler G, Wolburg H (1988) Light-dependent dynamics of gap junctions between horizontal cells in the retina of the crucian carp. Cell Tissue Res 251:641–649. 10.1007/bf00214013 [DOI] [PubMed] [Google Scholar]
- Kurz-Isler G, Voigt T, Wolburg H (1992) Modulation of connexon densities in gap junctions of horizontal cell perikarya and axon terminals in fish retina: effects of light/dark cycles, interruption of the optic nerve and application of dopamine. Cell Tissue Res 268:267–275. 10.1007/bf00318795 [DOI] [PubMed] [Google Scholar]
- Lauritzen JS, Anderson JR, Jones BW, Watt CB, Mohammed S, Hoang JV, Marc RE (2013) ON cone bipolar cell axonal synapses in the OFF inner plexiform layer of the rabbit retina. J Comp Neurol 521:977–1000. 10.1002/cne.23244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen JS, Sigulinsky CL, Anderson JR, Kalloniatis M, Nelson NT, Emrich DP, Rapp C, McCarthy N, Kerzner E, Meyer M, Jones BW, Marc RE (2019) Rod-cone crossover connectome of mammalian bipolar cells. J Comp Neurol 527:87–116. 10.1002/cne.24084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Meyer A, Schubert T, Huser L, Dedek K, Haverkamp S (2015) Morphology and connectivity of the small bistratified A8 amacrine cell in the mouse retina. J Comp Neurol 523:1529–1547. 10.1002/cne.23752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL. (2017) Neuronal territory formation by the atypical cadherins and clustered protocadherins. Semin Cell Dev Biol 69:111–121. 10.1016/j.semcdb.2017.07.040 [DOI] [PubMed] [Google Scholar]
- Li X, Kamasawa N, Ciolofan C, Olson CO, Lu S, Davidson KG, Yasumura T, Shigemoto R, Rash JE, Nagy JI (2008) Connexin45-containing neuronal gap junctions in rodent retina also contain connexin36 in both apposing hemiplaques, forming bihomotypic gap junctions, with scaffolding contributed by zonula occludens-1. J Neurosci 28:9769–9789. 10.1523/JNEUROSCI.2137-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Jakobs TC, Masland RH (2005) Different functional types of bipolar cells use different gap-junctional proteins. J Neurosci 25:6696–6701. 10.1523/JNEUROSCI.1894-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MA, Landisman CE, Connors BW (2004) Small clusters of electrically coupled neurons generate synchronous rhythms in the thalamic reticular nucleus. J Neurosci 24:341–349. 10.1523/JNEUROSCI.3358-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ghosh KK, Martin PR, Grunert U (1999) Analysis of two types of cone bipolar cells in the retina of a New World monkey, the marmoset, Callithrix jacchus. Vis Neurosci 16:707–719. 10.1017/s0952523899164101 [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Masland RH (1998) Extreme diversity among amacrine cells: implications for function. Neuron 20:971–982. 10.1016/s0896-6273(00)80478-x [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Gaul PA (2008) Biocytin wide-field bipolar cells in rabbit retina selectively contact blue cones. J Comp Neurol 506:6–15. 10.1002/cne.21491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH (1999) The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J Comp Neurol 413:305–326. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH (2004) The population of bipolar cells in the rabbit retina. J Comp Neurol 472:73–86. 10.1002/cne.20063 [DOI] [PubMed] [Google Scholar]
- Marc RE. (1999) Kainate activation of horizontal, bipolar, amacrine, and ganglion cells in the rabbit retina. J Comp Neurol 407:65–76. [DOI] [PubMed] [Google Scholar]
- Marc RE. (2009) Functional neuroanatomy of the retina. In: Duane's ophthalmology (Tasman W, Jaeger EA, eds). Philadelphia: Lippincott Williams and Wilkins. [Google Scholar]
- Marc RE, Liu WL (1984) Horizontal cell synapses onto glycine-accumulating interplexiform cells. Nature 312:266–269. 10.1038/312266a0 [DOI] [PubMed] [Google Scholar]
- Marc RE, Jones BW (2002) Molecular phenotyping of retinal ganglion cells. J Neurosci 22:413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW (2003) Retinal remodeling in inherited photoreceptor degenerations. Mol Neurobiol 28:139–147. 10.1385/MN:28:2:139 [DOI] [PubMed] [Google Scholar]
- Marc RE, Liu WL, Muller JF (1988) Gap junctions in the inner plexiform layer of the goldfish retina. Vision Res 28:9–24. [PubMed] [Google Scholar]
- Marc RE, Murry RF, Basinger SF (1995) Pattern recognition of amino acid signatures in retinal neurons. J Neurosci 15:5106–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Anderson JR, Kinard K, Marshak DW, Wilson JH, Wensel T, Lucas RJ (2007) Neural reprogramming in retinal degeneration. Invest Ophthalmol Vis Sci 48:3364–3371. 10.1167/iovs.07-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Jones BW, Watt CB, Anderson JR, Sigulinsky C, Lauritzen S (2013) Retinal connectomics: towards complete, accurate networks. Prog Retin Eye Res 37:141–162. 10.1016/j.preteyeres.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Anderson JR, Jones BW, Sigulinsky CL, Lauritzen JS (2014) The AII amacrine cell connectome: a dense network hub. Front Neural Circuits 8:104. 10.3389/fncir.2014.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Sigulinsky CL, Pfeiffer RL, Emrich D, Anderson JR, Jones BW (2018) Heterocellular coupling between amacrine cells and ganglion cells. Front Neural Circuits 12:90. 10.3389/fncir.2018.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. (1998) Electrical synapses: beyond speed and synchrony to computation. Curr Biol 8:R795–R797. 10.1016/s0960-9822(07)00502-7 [DOI] [PubMed] [Google Scholar]
- Marsh AJ, Michel JC, Adke AP, Heckman EL, Miller AC (2017) Asymmetry of an intracellular scaffold at vertebrate electrical synapses. Curr Biol 27:3561–3567.e3564. 10.1016/j.cub.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. (2001) The fundamental plan of the retina. Nat Neurosci 4:877–886. 10.1038/nn0901-877 [DOI] [PubMed] [Google Scholar]
- Massey SC. (2008) Circuit functions of gap junctions in the mammalian retina. In: The senses: vision (Masland RH, Albright T, eds), pp 457–472. Oxford: Elsevier. [Google Scholar]
- Massey SC, Mills SL (1996) A calbindin-immunoreactive cone bipolar cell type in the rabbit retina. J Comp Neurol 366:15–33. [DOI] [PubMed] [Google Scholar]
- Maxeiner S, Dedek K, Janssen-Bienhold U, Ammermuller J, Brune H, Kirsch T, Pieper M, Degen J, Kruger O, Willecke K, Weiler R (2005) Deletion of connexin45 in mouse retinal neurons disrupts the rod/cone signaling pathway between AII amacrine and ON cone bipolar cells and leads to impaired visual transmission. J Neurosci 25:566–576. 10.1523/JNEUROSCI.3232-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillem GS, Dacheux RF (2001) Rabbit cone bipolar cells: correlation of their morphologies with whole-cell recordings. Vis Neurosci 18:675–685. 10.1017/s0952523801185019 [DOI] [PubMed] [Google Scholar]
- McGuire BA, Stevens JK, Sterling P (1984) Microcircuitry of bipolar cells in cat retina. J Neurosci 4:2920–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles R, Toth K, Gulyas AI, Hajos N, Freund TF (1996) Differences between somatic and dendritic inhibition in the hippocampus. Neuron 16:815–823. 10.1016/s0896-6273(00)80101-4 [DOI] [PubMed] [Google Scholar]
- Miller AC, Whitebirch AC, Shah AN, Marsden KC, Granato M, O'Brien J, Moens CB (2017) A genetic basis for molecular asymmetry at vertebrate electrical synapses. Elife 6:e25364 10.7554/eLife.25364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SL. (1999) Unusual coupling patterns of a cone bipolar cell in the rabbit retina. Vis Neurosci 16:1029–1035. 10.1017/s0952523899166057 [DOI] [PubMed] [Google Scholar]
- Mills SL, Massey SC (1995) Differential properties of two gap junctional pathways made by AII amacrine cells. Nature 377:734–737. 10.1038/377734a0 [DOI] [PubMed] [Google Scholar]
- Minerbi A, Kahana R, Goldfeld L, Kaufman M, Marom S, Ziv NE (2009) Long-term relationships between synaptic tenacity, synaptic remodeling, and network activity. PLoS Biol 7:e1000136. 10.1371/journal.pbio.1000136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Soto F, Wong RO, Kerschensteiner D (2011) Development of cell type-specific connectivity patterns of converging excitatory axons in the retina. Neuron 71:1014–1021. 10.1016/j.neuron.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, Maniatis T (2018) Writing, reading, and translating the clustered protocadherin cell surface recognition code for neural circuit assembly. Annu Rev Cell Dev Biol 34:471–493. 10.1146/annurev-cellbio-100616-060701 [DOI] [PubMed] [Google Scholar]
- Nagy JI, Lynn BD (2018) Structural and intermolecular associations between Connexin36 and protein components of the adherens junction-neuronal gap junction complex. Neuroscience 384:241–261. 10.1016/j.neuroscience.2018.05.026 [DOI] [PubMed] [Google Scholar]
- Nagy JI, Pereda AE, Rash JE (2018) Electrical synapses in mammalian CNS: past eras, present focus and future directions. Biochim Biophys Acta Biomembr 1860:102–123. 10.1016/j.bbamem.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Axelsen LN, Sorgen PL, Verma V, Delmar M, Holstein-Rathlou NH (2012) Gap junctions. Compr Physiol 2:1981–2035. 10.1002/cphy.c110051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J. (2019) Design principles of electrical synaptic plasticity. Neurosci Lett 695:4–11. 10.1016/j.neulet.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen WG. (1985) Chemical and electrical synapses between photoreceptors in the retina of the turtle, Chelydra serpentina. J Comp Neurol 240:423–433. 10.1002/cne.902400410 [DOI] [PubMed] [Google Scholar]
- Pereda AE. (2014) Electrical synapses and their functional interactions with chemical synapses. Nat Rev Neurosci 15:250–263. 10.1038/nrn3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereda AE, Curti S, Hoge G, Cachope R, Flores CE, Rash JE (2013) Gap junction-mediated electrical transmission: regulatory mechanisms and plasticity. Biochim Biophys Acta 1828:134–146. 10.1016/j.bbamem.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. (1962) Plasma membrane contacts in the central nervous system. J Anat 96:237–248. [PMC free article] [PubMed] [Google Scholar]
- Petrides A, Trexler EB (2008) Differential output of the high-sensitivity rod photoreceptor: AII amacrine pathway. J Comp Neurol 507:1653–1662. 10.1002/cne.21617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli V, Strettoi E (2004) Bipolar cells of the mouse retina: a gene gun, morphological study. J Comp Neurol 476:254–266. 10.1002/cne.20207 [DOI] [PubMed] [Google Scholar]
- Pourcho RG, Goebel DJ (1987) A combined Golgi and autoradiographic study of 3H-glycine-accumulating cone bipolar cells in the cat retina. J Neurosci 7:1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pow DV, Hendrickson AE (2000) Expression of glycine and the glycine transporter Glyt-1 in the developing rat retina. Vis Neurosci 17:1R–9R. [PubMed] [Google Scholar]
- Rash JE, Curti S, Vanderpool KG, Kamasawa N, Nannapaneni S, Palacios-Prado N, Flores CE, Yasumura T, O'Brien J, Lynn BD, Bukauskas FF, Nagy JI, Pereda AE (2013) Molecular and functional asymmetry at a vertebrate electrical synapse. Neuron 79:957–969. 10.1016/j.neuron.2013.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviola E, Gilula NB (1973) Gap junctions between photoreceptor cells in the vertebrate retina. Proc Natl Acad Sci USA 70:1677–1681. 10.1073/pnas.70.6.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviola E, Gilula NB (1975) Intramembrane organization of specialized contacts in the outer plexiform layer of the retina: a freeze-fracture study in monkeys and rabbits. J Cell Biol 65:192–222. 10.1083/jcb.65.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale E, Luciano L, Spitznas M (1978) Communicating junctions of the human sensory retina: a freeze-fracture study. Albrecht Von Graefes Arch Klin Exp Ophthalmol 208:77–92. 10.1007/bf00406984 [DOI] [PubMed] [Google Scholar]
- Reese BE. (2008) Mosaics, tiling and coverage by retinal neurons. In: The senses: vision (Masland RH, Albright T, eds), pp 439–456. Oxford: Elsevier. [Google Scholar]
- Saint-Amant L, Drapeau P (2001) Synchronization of an embryonic network of identified spinal interneurons solely by electrical coupling. Neuron 31:1035–1046. 10.1016/s0896-6273(01)00416-0 [DOI] [PubMed] [Google Scholar]
- Seilheimer RL, Sabharwal J, Wu SM (2020) Genetic dissection of rod and cone pathways mediating light responses and receptive fields of ganglion cells in the mouse retina. Vision Res 167:15–23. 10.1016/j.visres.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR (2016) Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166:1308–1323.e1330. 10.1016/j.cell.2016.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivyer B, Williams SR (2013) Direction selectivity is computed by active dendritic integration in retinal ganglion cells. Nat Neurosci 16:1848–1856. 10.1038/nn.3565 [DOI] [PubMed] [Google Scholar]
- Strettoi E, Raviola E, Dacheux RF (1992) Synaptic connections of the narrow-field, bistratified rod amacrine cell (AII) in the rabbit retina. J Comp Neurol 325:152–168. 10.1002/cne.903250203 [DOI] [PubMed] [Google Scholar]
- Strettoi E, Dacheux RF, Raviola E (1994) Cone bipolar cells as interneurons in the rod pathway of the rabbit retina. J Comp Neurol 347:139–149. 10.1002/cne.903470111 [DOI] [PubMed] [Google Scholar]
- Szoboszlay M, Lőrincz A, Lanore F, Vervaeke K, Silver RA, Nusser Z (2016) Functional properties of dendritic gap junctions in cerebellar Golgi cells. Neuron 90:1043–1056. 10.1016/j.neuron.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Borowska J, Zhang J, Hoggarth A, Johnson K, Barnes S, Lewis TJ, Awatramani GB (2012) Intrinsic oscillatory activity arising within the electrically coupled AII amacrine-ON cone bipolar cell network is driven by voltage-gated Na+ channels. J Physiol 590:2501–2517. 10.1113/jphysiol.2011.225060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trexler EB, Li W, Mills SL, Massey SC (2001) Coupling from AII amacrine cells to ON cone bipolar cells is bidirectional. J Comp Neurol 437:408–422. 10.1002/cne.1292 [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Omi N (2014) Some OFF bipolar cell types make contact with both rods and cones in macaque and mouse retinas. Front Neuroanat 8:105. 10.3389/fnana.2014.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Omi N (2016) ON bipolar cells in macaque retina: type-specific synaptic connectivity with special reference to OFF counterparts. Front Neuroanat 10:104. 10.3389/fnana.2016.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Omi N (2017) Classification of mouse retinal bipolar cells: type-specific connectivity with special reference to rod-driven AII amacrine pathways. Front Neuroanat 11:92. 10.3389/fnana.2017.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Morigiwa K, Ueda M, Sterling P (2001) Microcircuits for night vision in mouse retina. J Neurosci 21:8616–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umino O, Maehara M, Hidaka S, Kita S, Hashimoto Y (1994) The network properties of bipolar-bipolar cell coupling in the retina of teleost fishes. Vis Neurosci 11:533–548. 10.1017/s0952523800002443 [DOI] [PubMed] [Google Scholar]
- Uylings HB, van Pelt J (2002) Measures for quantifying dendritic arborizations. Network 13:397–414. [PubMed] [Google Scholar]
- Vaney DI. (1990) The mosaic of amacrine cells in the mammalian retina. In: Progress in retinal research (Osborne NN, Chader G, eds), pp 49–100. Oxford: Pergamon; 10.1016/0278-4327(90)90004-2 [DOI] [Google Scholar]
- Vaney DI. (1997) Neuronal coupling in rod-signal pathways of the retina. Invest Ophthalmol Vis Sci 38:267–273. [PubMed] [Google Scholar]
- Vaney DI, Nelson JC, Pow DV (1998) Neurotransmitter coupling through gap junctions in the retina. J Neurosci 18:10594–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra RD. (1996) Size and selectivity of gap junction channels formed from different connexins. J Bioenerg Biomembr 28:327–337. 10.1007/bf02110109 [DOI] [PubMed] [Google Scholar]
- Veenstra RD, Wang HZ, Beblo DA, Chilton MG, Harris AL, Beyer EC, Brink PR (1995) Selectivity of connexin-specific gap junctions does not correlate with channel conductance. Circ Res 77:1156–1165. 10.1161/01.res.77.6.1156 [DOI] [PubMed] [Google Scholar]
- Veruki ML, Hartveit E (2002a) Electrical synapses mediate signal transmission in the rod pathway of the mammalian retina. J Neurosci 22:10558–10566. 10.1523/JNEUROSCI.22-24-10558.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veruki ML, Hartveit E (2002b) AII (rod) amacrine cells form a network of electrically coupled interneurons in the mammalian retina. Neuron 33:935–946. 10.1016/S0896-6273(02)00609-8 [DOI] [PubMed] [Google Scholar]
- Vervaeke K, Lorincz A, Gleeson P, Farinella M, Nusser Z, Silver RA (2010) Rapid desynchronization of an electrically coupled interneuron network with sparse excitatory synaptic input. Neuron 67:435–451. 10.1016/j.neuron.2010.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasits AL, Euler T, Franke K (2019) Function first: classifying cell types and circuits of the retina. Curr Opin Neurobiol 56:8–15. 10.1016/j.conb.2018.10.011 [DOI] [PubMed] [Google Scholar]
- Völgyi B, Deans MR, Paul DL, Bloomfield SA (2004) Convergence and segregation of the multiple rod pathways in mammalian retina. J Neurosci 24:11182–11192. 10.1523/JNEUROSCI.3096-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völgyi B, Kovács-Oller T, Atlasz T, Wilhelm M, Gábriel R (2013) Gap junctional coupling in the vertebrate retina: variations on one theme? Prog Retin Eye Res 34:1–18. 10.1016/j.preteyeres.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Washioka H, Watanabe H, Negishi K, Tonosaki A (1991) Horizontal-cell gap junction in the goldfish retina: area and density of particles as revealed by complementary freeze replicas. Arch Histol Cytol 54:181–188. 10.1679/aohc.54.181 [DOI] [PubMed] [Google Scholar]
- Wässle H, Riemann HJ (1978) The mosaic of nerve cells in the mammalian retina. Proc R Soc Lond B Biol Sci 200:441–461. 10.1098/rspb.1978.0026 [DOI] [PubMed] [Google Scholar]
- Wässle H, Puller C, Muller F, Haverkamp S (2009) Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J Neurosci 29:106–117. 10.1523/JNEUROSCI.4442-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LL, Macqueen CL, Elston GN, Young HM, Pow DV, Vaney DI (1997) The DAPI-3 amacrine cells of the rabbit retina. Vis Neurosci 14:473–492. 10.1017/s0952523800012141 [DOI] [PubMed] [Google Scholar]
- Wu SM, Yang XL (1988) Electrical coupling between rods and cones in the tiger salamander retina. Proc Natl Acad Sci USA 85:275–278. 10.1073/pnas.85.1.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XB, Mills SL (2004) Gap junctional regulatory mechanisms in the AII amacrine cell of the rabbit retina. Vis Neurosci 21:791–805. 10.1017/S0952523804215127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin D, Bloomfield SA (1999) Comparison of the responses of AII amacrine cells in the dark- and light-adapted rabbit retina. Vis Neurosci 16:653–665. 10.1017/s0952523899164058 [DOI] [PubMed] [Google Scholar]
- Yadav SC, Tetenborg S, Dedek K (2019) Gap junctions in A8 amacrine cells are made of Connexin36 but are differently regulated than gap junctions in AII amacrine cells. Front Mol Neurosci 12:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Yazulla S (1988) Light microscopic localization of putative glycinergic neurons in the larval tiger salamander retina by immunocytochemical and autoradiographical methods. J Comp Neurol 272:343–357. 10.1002/cne.902720305 [DOI] [PubMed] [Google Scholar]
- Yang XD, Korn H, Faber DS (1990) Long-term potentiation of electrotonic coupling at mixed synapses. Nature 348:542–545. 10.1038/348542a0 [DOI] [PubMed] [Google Scholar]
- Yogev S, Shen K (2014) Cellular and molecular mechanisms of synaptic specificity. Annu Rev Cell Dev Biol 30:417–437. 10.1146/annurev-cellbio-100913-012953 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed individual cell coupling profiles and class statistics for the core set of 37 ON CBC and four Aii cells. 1The CBb label contains all partial cells or fragments that could not be classified beyond the ON superclass. The number of coupled cells and number of gap junctions per cell were not quantified. 2Most partner xACs exist as fragments, only sufficiently traced and annotated to confirm they were not Aii cells. Thus, the number of coupled cells and number of gap junctions per cell were not quantified. 3Two Aii cell partners could not be traced far enough to determine whether they were of amacrine or bipolar cell identity. Download Figure 3-1, XLSX file (22.3KB, xlsx) .