Abstract
HIV-1 cure remains elusive with only one reported case a decade ago1,2. Termed the “Berlin Patient”, the individual underwent 2 allogeneic Hematopoietic Stem Cell Transplantation (allo-HSCT) procedures using a donor with a homozygous mutation in the HIV coreceptor CCR5 (CCR5Δ32/Δ32) to treat his acute myeloid leukemia. Total body irradiation was given with each HSCT. Critically, it is unclear which treatment or patient parameters contributed to this only documented case of long term HIV remission. Here we show that HIV-1 remission may be possible with a less aggressive and toxic approach. An HIV-1-infected adult underwent allo-HSCT for Hodgkin’s Lymphoma using cells from a CCR5Δ32/Δ32 donor. He experienced mild gut graft versus host disease. Antiretroviral therapy was interrupted 16 months after transplantation. HIV-1 remission has been maintained through a further 18 months. Plasma HIV-1 RNA has been undetectable at <1 copy/ml along with undetectable HIV-1 DNA in peripheral CD4 T lymphocytes. Quantitative viral outgrowth assay from peripheral CD4 T lymphocytes shows no reactivatable virus using a total of 24 million resting CD4 T cells. CCR5-tropic, but not CXCR4-tropic viruses were identified in HIV-1 DNA from CD4 T cells of the patient prior to transplant. CD4 T cells isolated from peripheral blood post-transplant did not express CCR5 and were only susceptible to CXCR4-tropic virus ex vivo. HIV-1 Gag-specific CD4 and CD8 T cell responses were lost after transplantation whilst Cytomegalovirus (CMV)-specific responses were detectable. Likewise, HIV-1-specific antibodies and avidities fell to levels comparable to those in the Berlin patient following transplantation. Although at 18 months post-treatment interruption it is premature to conclude that this patient has been cured, these data suggest that single allo-HSCT with homozygous CCR5Δ32 donor cells may be sufficient to achieve HIV-1 remission with reduced intensity conditioning and no irradiation, and the findings further support the development of HIV remission strategies based on preventing CCR5 expression.
The HIV-1 epidemic continues with nearly 37 million infected.3 Although over 21 million are accessing lifelong antiretroviral therapy (ART)3, drug resistant HIV in both untreated4 and treated5,6 individuals is significant in low and middle-income countries and sustainability of ART programs is uncertain.7 Drug-free durable HIV-1 suppression is therefore an urgent global priority.
Thus far, the only documented case of sustained HIV remission is the ‘Berlin Patient’, who received two allogeneic hematopoietic stem cell transplantations (allo-HSCT) using cells from a homozygous CCR5Δ32 (CCR5Δ32/Δ32) donor1. This 32-base pair deletion prevents CCR5 expression rendering these cells resistant to infection with HIV variants utilising the CCR5 co-receptor8.
The only other case of an HIV-infected patient transplanted with CCR5Δ32/Δ32 cells who interrupted ART was the ‘Essen Patient’9. In this case in which ART was interrupted one week before allo-HSCT, a rapid viral rebound of a pre-existing minority HIV-1 variant able to infect cells via the alternative CXCR4 co-receptor was observed three weeks later9,10. Such pre-existing CXCR4 variants were not observed in the ‘Berlin patient’.11 Three other cases transplanted with wildtype CCR5 cells experienced a viral rebound 12, 32 or 41 weeks after ART interruption despite profound reduction of the HIV reservoir.12,13
We report an individual diagnosed with HIV infection in 2003, with a CD4 nadir of 290 cells/mm3 and baseline HIV-1 plasma viral load (pVL) of 180,000 copies/ml. ART was initiated with tenofovir disoproxil fumarate (TDF), emtricitabine (FTC) and efavirenz (EFV) in 2012.
In December 2012 Stage IVB (nodular sclerosing) Hodgkin’s Lymphoma (HL) was diagnosed. HL was refractory to first-line chemotherapy (ABVD) and a number of salvage regimens including ESHAP, anti-CD30 monoclonal antibody (Brentuximab) and mini-LEAM were used. ART was switched to TDF/FTC/raltegravir (RAL) during periods of chemotherapy for HL; there was a 5-day episode of ART interruption in late 2015 with HIV-1 pVL of 1,500 copies/ml that did not reach viral set-point. Based on resistance mutations K65R and M184V in reverse transcriptase as well as E157Q in integrase, the regimen was switched to rilpivirine (RPV), lamivudine (3TC) and dolutegravir (DTG), with viral suppression subsequently achieved.
Mobilisation of autologous peripheral blood stem cells failed despite the use of CXCR4 antagonists, thereby precluding standard autologous HSCT. A complete metabolic remission by CT/PET criteria was achieved with IGEV chemotherapy in March 2016.
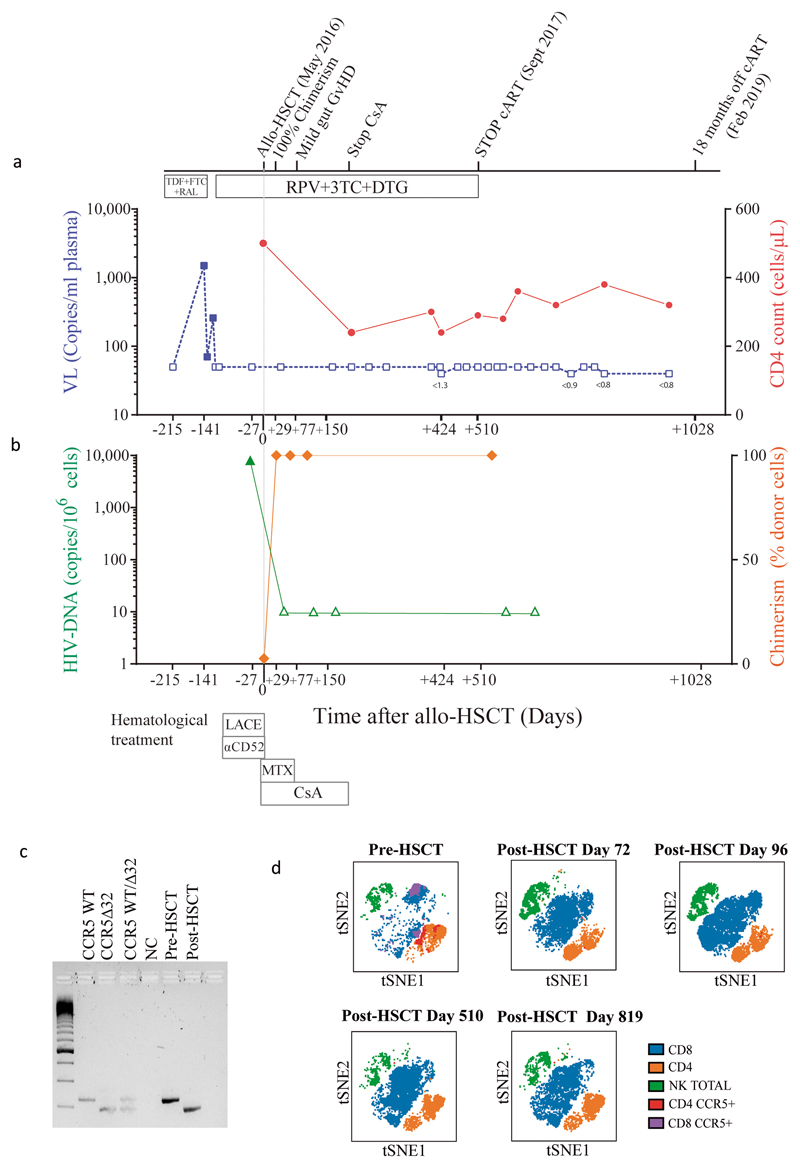
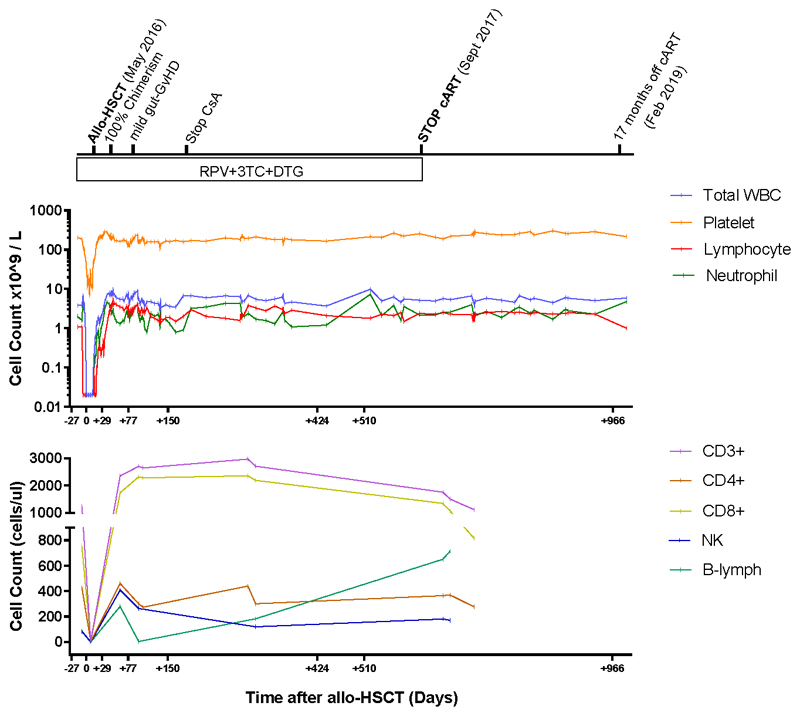
An unrelated (9/10) donor was identified from an international registry with one allelic mismatch at HLA-B by high-resolution HLA typing that also was CCR5Δ32/Δ32 (Extended data table 1 and methods). No fully matched donors were identified in the registry. The patient underwent conditioning with Lomustine, Cyclophosphamide, Ara-C and Etoposide (LACE) followed by infusion of 3.6 × 106 CD34+ cells/kg. In vivo T-cell depletion employed anti–CD52 (Alemtuzumab), 10 mg daily for 5 days (days -7 to -3) and GvHD prophylaxis used Cyclosporine-A (CsA) with a short-course of methotrexate (MTX). ART was continued throughout with RPV/3TC/DTG (Figure 1a). Allo-HSCT was relatively uncomplicated and the patient was discharged on Day+31. Both Epstein-Barr Virus (EBV) and cytomegalovirus (CMV) reactivation occurred at day +85 requiring treatment with anti-CD20 monoclonal antibody (Rituximab) and ganciclovir respectively. At day +77 the patient presented with fever and gastrointestinal symptoms. Gastric, duodenal and colonic biopsies were consistent with grade 1 GvHD, which resolved without intervention. Full-donor chimerism was achieved in the whole leukocyte and in CD3+ T cell fractions from day +30 and maintained in both cell fractions throughout (Figure 1b). Host genotype was CCR5wt/wt before allo-HSCT, and became CCR5Δ32/Δ32 after transplant (Figure 1c), with loss of CCR5 surface expression from circulating CD4 and CD8 T cells (Figure 1d). At +180 days post-transplant CsA was discontinued. CT/PET scan at +120 days and +365 days post-transplant confirmed complete metabolic remission with no subsequent relapse. Post-transplant white cell counts and lymphocyte subsets returned to pre-transplant levels (Extended data figure 1), except for CD4 counts which have been slower to recover (Figure 1a).
Figure 1. Clinical course before and after allogeneic Hematological Stem Cell Transplantation.
a. Antiretroviral treatment and chemotherapy/immunosuppression associated with allogeneic HSCT along with plasma viral load (HIV-1 RNA) and CD4 count over time. Small numbers below blue data points indicate results of ultra sensitive viral load assay. b. HIV-1 DNA in PBMC and donor chimerism in T cell fraction c. Genotyping of CCR5 alleles with agarose gel electrophoresis of PCR amplified DNA fragments using a 100 base pair DNA ladder; NC negative control. d. tSNE plots of PBMC pre and post HSCT showing CCR5 expression changes and cell population changes over time. Abbreviations: HSCT: haematopoietic stem cell transplantation LACE: lomustine Ara-C cyclophosphamide etoposide; MTX methotrexate; CsA ciclosporin A; ART antiretroviral therapy; RPV rilpivirine; DTG dolutegravir; 3TC lamivudine; RAL raltegravir; TDF tenofovir disoproxil fumarate; FTC emtricitabine. These experiments were carried out once only (a-d) and sample size is n=1 for all panels.
ART was maintained post-HSCT and analytical treatment interruption (ATI) was initiated at day +510 (September 2017). Weekly plasma viral load was performed for the first 3 months and then monthly thereafter. HIV-1 pVL remained undetectable thereafter with limit of detection (LOD) <1 copy RNA/ml (Figure 1a). Plasma concentrations of TDF, 3TC and DTG were negative by HPLC at day +648 and a panel of all currently available antiretroviral drugs tested negative by LC-MS at +973 days. Total PBMC associated HIV-1 DNA fell to below the limit of detection after transplant (Figure 1b). Total DNA in CD4+ T cells at day +876 was undetectable in all replicates by ultra-sensitive qPCR (<0.65 HIV LTR copies/million cells and <0.69 HIV-1 Gag copies /million cells) and in 7/8 replicates of the ultra-sensitive HIV-1 LTR ddPCR14 ; in one replicate a low-level signal was observed. Such occasional positive signals were also observed in the Berlin patient15 and may reflect a false ddPCR signal, potential contamination, or evidence of very low levels of persistence of HIV infected cells that either did not harbor fully replication competent virus or were unable to lead to recrudescence given that the vast majority of target cells are incapable of being infected with this patient’s HIV CCR5 tropic variants (Figure 2). HIV-1 DNA and RNA were also repeatedly undetectable in whole blood when tested with SAMBA II, a CE marked point-of-care isothermal amplification method (LOD: 284 copies/ml; 95% CI: 214-378 copies/ml)16.
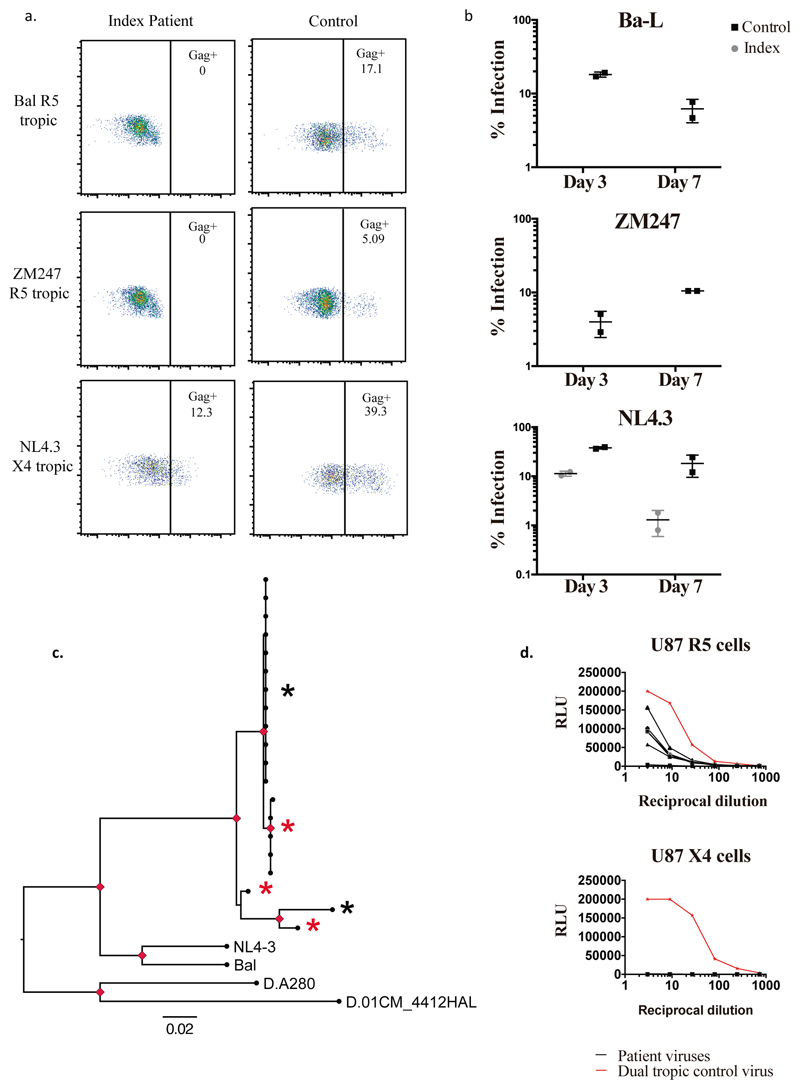
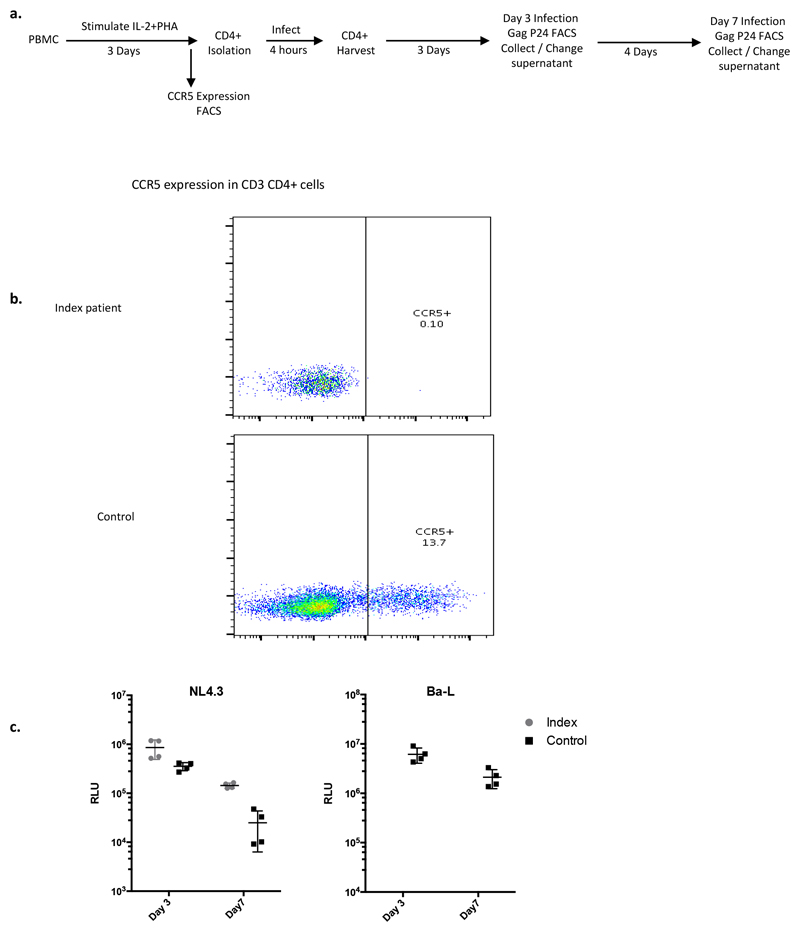
Figure 2. Susceptibility of index patient CD4 T cells to CCR5 tropic and CXCR4 tropic HIV-1 and coreceptor usage by index patient viruses prior to HSCT.
a. Representative plots of intracellular p24 gag staining within CD4+ T cell populations three days post infection of isolated CD4+ cells by CCR5 (R5) tropic viruses Bal and ZM247 and CXCR4 (X4) tropic virus NL4.3 b. Percentage infection in CD3+ CD4+ T cells determined by p24 staining at day 3 and day 7 post infection using R5 and X4 tropic viruses. Error bars represent standard error of the mean. Data are representative of 3 independent experiments each conducted in duplicate. c. Maximum likelihood phylogenetic tree showing single genome env C2-V5 sequences (HXB2 env 367-1533 nt) from PBMC prior to HSCT. NL4.3, Ba-L and two subtype D sequences from Genbank were also used. Red nodes indicate >70% bootstrap support. Gp120 amplicons from sequences marked with an asterix were cloned into a clade B gp160 Env plasmid. Virus env sequences marked by a red asterix generated infectious virus particles when co-transfected into HEK293T cells with the envelope deficient full-length HIV plasmid encoding luciferase (NL4-3 delta Env Luc). Black asterix indicates a sequence that did not generate infectious virus. d. Pseudoviruses from c. were used to infect U87 cells expressing either CXCR4 (X4) or CCR5 (R5). A dual tropic pseudovirus (WEAU-d15.410.787) was produced in parallel as a positive contro l for infection (red line). Experiments in a, b, c and d. were performed independently three times with similar results (sample size n=1 healthy control donor and n=1 index patient for a and b; sample size n=9 clones tested in d.).
Blood was obtained for a modified Quantitative Viral Outgrowth Assay (QVOA)17,18 at three time points post-HSCT at day +217 (on ART), and days +678 and +876 (off ART). QVOA was undetectable on all occasions, giving a reservoir estimation in infectious units per million (IUPM) resting CD4 T cells of <0.286, <0.309 and <0.063 respectively. Pooling the results from the three qVOA tests on a total of 24 million resting CD4 T cells gives an estimate of <0.029 IUPM.
We next sought to confirm that the post-transplant CD4 cells lacked expression of CCR5 and were resistant to HIV-1 infection (Figure 2 and Extended data Figure 2). These CD4 T cells from the study patient were challenged in vitro with the CCR5-tropic viruses Ba-L and ZM247 and productive HIV-1 infection over 7 days was measured by (i) intracellular staining for HIV-1 p24 protein (Figure 2a-b) and (ii) infectivity of culture supernatants on indicator cell lines that respond to expression of HIV Tat protein (Extended data Figure 2). In contrast to a HIV-negative donor, post-transplant cells from the study patient could not be infected with either CCR5 tropic virus (Figure 2a-b). The study patient cells were then challenged with the canonical CXCR4-tropic HIV-1NL4-3. As expected, infection was observed in cells and supernatants from both the patient and an HIV-negative donor (control) (Figure 2a-b and extended data Figure 2).
In order to determine whether the study patient was infected with CCR5- or CXCR4-using virus (or both), we deep-sequenced the V3 loop in HIV-1 envelope (the key determinant of coreceptor usage) and computational algorithms predicted CCR5-tropism19,20For phenotypic verification we first performed single genome sequencing (SGS) of gp120 from pre-transplant PBMC by limiting dilution PCR21,22. (Figure 2c). These single genomes were cloned into expression vectors and used to generate virions pseudotyped with patient derived gp120 envelope protein (Figure 2d)21, before infection of indicator cells expressing either CCR5 or CXCR4. As predicted from the genotype, robust infection was observed in CCR5 but not CXCR4 expressing cells (Figure 2d).
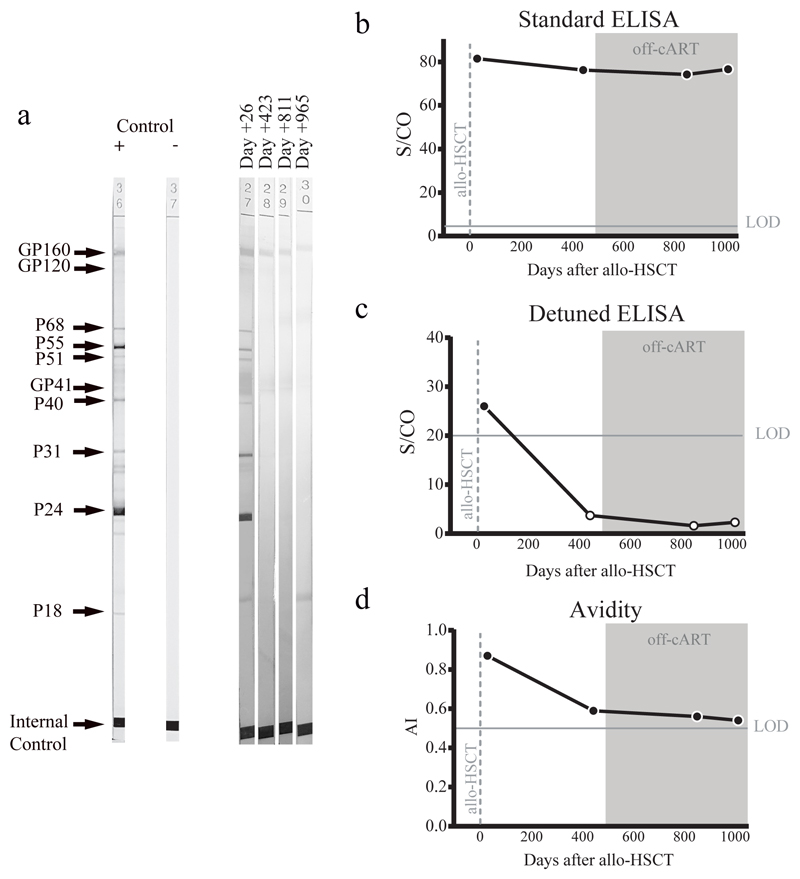
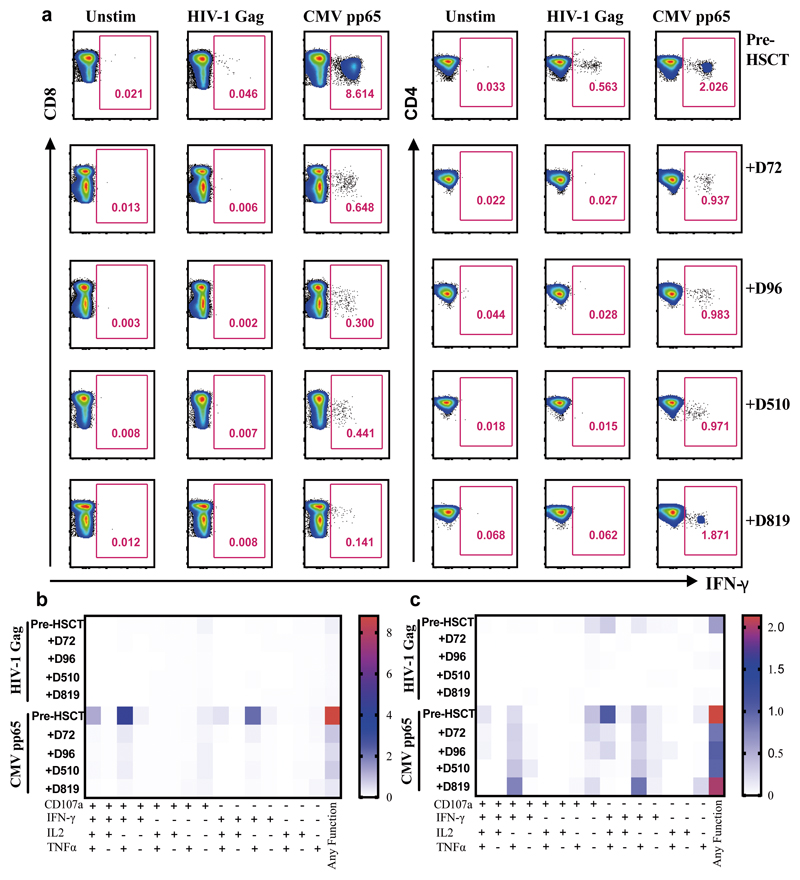
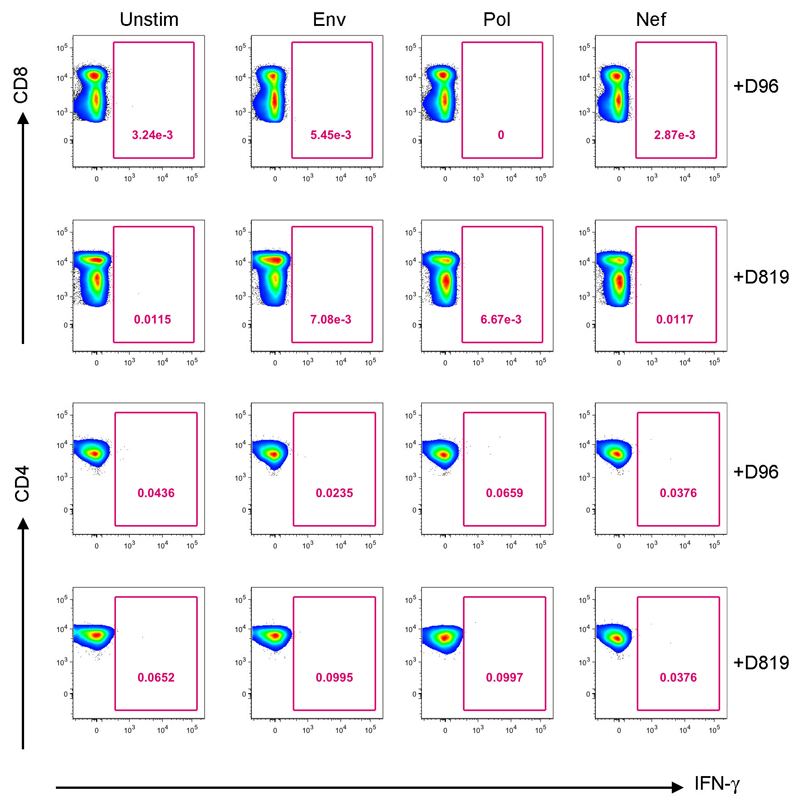
Analyses of both antibody and T cell responses were undertaken in order to further investigate absence of persistent HIV-1 infection and antigenic stimulation. Western blot analysis of antibodies demonstrated loss of p24, p31 and multiple other bands between pre- and post-transplant time points in a similar pattern seen previously in other transplanted patients who have remained on ART23 (Figure 3a and extended data table 2), whilst antibodies to envelope protein gp160 and standard ELISA for Env antibodies persisted (Figure 3a-c). Low-sensitivity VITROS analysis and antibody avidity assays were also consistent with loss of HIV antigen following allo-HSCT (Figure 3d-e)15. CD8 and CD4 T cell virus-specific responses were determined following stimulation with HIV-1 Gag and CMV pp65 overlapping peptide pools (Figure 4a). Small Gag-virus specific T-cell responses were identified by intracellular staining and multifunctional responses (IFN-γ, CD107a, TNF-α and IL-2) were detected prior to HSCT (day -35) but not at days +72, 96, 510 and 819 after the procedure (Figure 4a-b). No HIV-1 specific T cell responses were detected to Nef, Pol and Env peptide pools at days +96 and +819 post-transplant (Extended data Figure 3). By contrast CMV- specific T-cell responses were detected both before and after HSCT, albeit at reduced frequencies (Figure 4a-c).
Figure 3. HIV Specific Antibodies.
Humoral response dynamics were tested at days +26, +423, +811 and +965 after HSCT (last two time points in absence of ART). Antibody levels were measured using Western blot (a), the standard HIV-1 VITROS assay (b), a detuned low sensitive (LS) version of the HIV-1 VITROS assay (c), and the Limiting Antigen avidity assay (d). Open symbols represent values under the limit of detection. AI: Avidity Index; S/CO: Ratio signal/cut off; Allo-HSCT: allogeneic hematopoietic stem cell transplantation; ART: antiretroviral therapy. a, c, d were repeated twice independently with similar results.
Figure 4. HIV-Gag specific and CMV specific T cell responses.
a. Representative FACS plots showing percentage of virus specific CD8+ T cells (left panel) and CD4+ T cells (right panel) identified via intracellular staining for IFN-g, following stimulation with HIV-1 Gag or CMV pp65 peptide pools pre and post-HSCT at indicated days (+D72, +D96, +D510, +D819). A negative control containing PBMC from the same subject but without peptide mix was included (unstim) for each assay. b. Heat maps of levels of expression of CD107a, IFN-g, IL-2 and TNF-a in CD8+ T cells and (C) CD4+ T cells in response to HIV-1 Gag and CMV pp65 peptide stimulation pre and post-HSCT at indicated days subsequent to Boolean gating. Functions are listed beneath the heat maps with each of their respective combinations or any function detected. a. Experiment performed once due to limited cell numbers.
Both antibody and T cell responses observed here are highly reminiscent of observations in the Berlin patient.1,15 Important similarities between the two cases were CCR5-tropic HIV-1 infection and receipt of a CCR5Δ32/Δ32 transplant. The GvHD prophylaxis was very similar and utilized standard regimens (CsA + MMF versus CsA + MTX) that in randomized studies have shown similar outcomes.24 In both cases there was probable mild GvHD, which may have contributed to the loss of HIV-infected cells. Finally, both achieved and maintained full-donor chimerism in peripheral blood that might have contributed to reduced reservoir size23,25.
Notable differences were that before allo-HSCT our patient was homozygous CCR5 wildtype as compared to the heterozygous CCR5WT/Δ32 genotype observed in the “Berlin Patient”. Our patient received a reduced intensity conditioning regimen consisting exclusively of chemotherapy agents with known activity against lymphoma26 while the Berlin patient received total body irradiation in conjunction with cyclophosphamide as the conditioning regimen. For lymphodepletion our patient received anti-CD52 Campath while the Berlin patient was treated with antithymocyte globulin. Our patient was treated with a short course of anti CD20 for EBV reactivation. There has been recent interest in the potential for anti-B cell therapy to disrupt B cell follicles that are known to harbour persistent HIV in lymphoid tissues27, though we think this mechanism is unlikely to be related to remission observed here. Finally, this patient achieved full remission after a single allo-HSCT while the Berlin patient experienced a relapse of the AML and received further chemotherapy with an anti-CD33 monoclonal conjugate before a second allo-HSCT.
In terms of a road map forward we speculate that CCR5 gene therapy strategies using stem cells could conceivably be a scalable approach to remission. As significant graft-versus-host effect is likely important, manifesting as early and sustained full donor chimerism in T cells, an autologous approach needs to achieve high levels of re placement by CCR5 depleted cells. Depletion of the CD4+ CCR5+ T cells other potential HIV reservoirs is a major challenge given the toxicities associated with anti-thymocyte globulin and anti-CD52 and the unknown role of anti-proliferative agents such as cyclosporin or methotrexate in clearing infected cells.
This report demonstrates (i) that the ‘Berlin’ patient was not an anomaly; (ii) that remission of HIV infection can be achieved with reduced intensity drug regimens (iii) that a single CCR5Δ32/Δ32 allo-HSCT is sufficient and (iv) that total body irradiation is not required. Our observation supports the development of HIV cure strategies based on preventing the expression of the CCR5 coreceptor.
Methods
Patient consent pathway
Following the clinical decision in 2015 to undertake allogeneic HSCT for treatment of aggressive lymphoma, the possibility of a CCR5 delta 32/delta 32 match was discussed with the patient. International registry searches identified no fully matched (10/10) donors. The only available donors on the international registries were the selected 9/10, and two 8/10. The 9/10 donor selected was therefore the best available match, and subsequent testing confirmed that the potential donor was also homozygous for CCR5 d32. Ultimately because the best available donor happened to be CCR5 negative, there were no deviations from the standard hospital consent protocol for HSCT, and therefore standard written informed consent for ‘LACE conditioned allogenic stem cell transplant' was obtained from the patient.
In Dec 2015, six months before HSCT, co-receptor usage was inferred from sequencing of genomic DNA that reported CCR5 tropic virus. The potential for remission was then discussed with the patient and that a supervised treatment interruption would be needed to demonstrate successful remission. Ethical approval for treatment interruption, frequent HIV viral load monitoring and tissue sampling was sought using the standard NHS framework in mid-late 2016. The protocol specified that at 12 months post-transplant, if viral load was consistently < 50 copies/ml on treatment with ‘target not detected’ for at least 6 months on the 2 most recent consecutive visits ART would be withdrawn and thereafter viral load would be monitored weekly for the first 3 months, monthly for a further 9 months if undetectable at all time points and three monthly between years 1 and 4. The protocol specified that if assays for the HIV reservoir were deemed negative by a group of UK experts from CHERUB we would thereafter monitor viral load 6 monthly. The protocol stated that if viral rebound occurs then moderate or severe symptoms would not be expected due to the likely low viral load where the donor chimerism is close to 100%. For the assessment of and management of risk the protocol stated that there is a risk of rebound viremia and symptoms such as fever and other flu like symptoms and that this would be mitigated by performing regular viral load testing in order to detect viral rebound early, re-starting ART where we observe at least 2 viral loads above 200 copies/ml one week apart in the absence of any other cause of viral blips, and monitoring CD4 cells (%) at least monthly for the first year. UK NHS Health Research Authority Research Ethics Committee approval was obtained in April 2017 UK with the reference number 17/SW/0021, protocol number 16/0594. and the patient provided full written, informed consent in July 2017. The interruption did not occur until Sept 2017, 16 months after transplantation. The patient was registered to the IciStem consortium as IciStem#36.
Chimerism testing
We measured whole leukocyte and also T-cell specific (CD3 selected) chimerism by short tandem repeats (STR) analysis with the PowerPlex16 system.
Primary cell culture and CD4 isolation
Peripheral blood mononuclear cells were isolated from whole blood by centrifugation on Lymphoprep (Axis-Shield UK, UK) gradient. The cells collected at the interface were washed with PBS and stimulated for 3 days in the presence of PHA and IL-2 (at 10pg/ml) in RPMI 1640 supplemented with 10% FCS. Stimulated PBMC were used for CD4+ negative selection with antibody-coated magnetic beads (Biolegend, Mojosort UK).
Extracellular receptor staining
Stimulated PBMC were washed with FACS buffer (PBS + 1% FCS) then stained with viability dye (Ghost Dye Violet 450, Tonbo Bioscience), followed by anti CD3-APC, anti CD4-FITC and anti CD195-PE (CCR5) (BioLegend, UK). Cells were washed and fixed in 2% PFA (paraformaldehyde). At least 105 lymphocytes were acquired on a BD LSRFortessa flow cytometer and analyzed using FlowJo (Tree Star Inc).
Spreading infection
Infection of primary cells using full-length CCR5- and CXCR4-tropic HIV-1 viruses
CXCR-4 tropic hybrid clone NL4-3 and CCR5 tropic viruses Bal and ZM247 were used to infect CD4+ T-cells for 4h (623, 1150 and 430pg per well respectively), before cells were washed twice and incubated in culture medium RPMI 1640 supplemented with 10% FCS and IL-2 (at 10pg/ml) for a period of 7 days. Culture medium was changed on day 3 and the collected supernatants were stored for analysis. Cells were stained with viability dye (Ghost Dye Violet 450, Tonbo Bioscience) then fixed in 4% PFA, permeabilized (BD Perm/Wash™, BD Biosciences) and stained for intracellular p24 using anti-p24 KC57-RD1 conjugated antibody (Beckman Coulter, USA). The percentage of infected cells was monitored by flow cytometry using BD LSRFortessa flow cytometer (BD Biosciences, UK) and analyzed by FlowJo software (Tree Star, OR, USA). ZM2471 was a gift from Beatrice Hahn.
Supernatant infectivity assay on HeLa TZM-bl reporter cells
In this assay, infectivity is measured as Tat-induced firefly luciferase reporter gene expression after a single round of virus infection. Briefly, 104 TZM-bl cells/well are seeded in a 96 well white plate in culture medium DMEM supplemented with 10% FCS for 24 h. After this, medium was replaced with supernatant collected from the infection assay at a final dilution of 1/3 and 1/6. After 48 h incubation cells were lysed by addition of Steady-Glo luciferase reagent (Promega, UK) and luminescence read using a GloMax 96 Luminometer (Promega, UK). For analysis, the relative luminescence units (RLU) are determined following the subtraction of background luciferase activity from cell control wells (average of eight replicates) and subtraction of RLU from uninfected supernatant wells (average of 6 replicates).
Next generation sequencing of HIV-1 Env V3 from genomic DNA
Nested polymerase chain reaction (PCR) was performed on the isolated DNA to amplify the HIV-1 env region{Symons et al, Clinical Microbiolo and Infection, 2011). Subsequently, PCR products were cleaned using the QiaQuick PCR purification kit (Qiagen) and amplicons were sequenced using the MiSeq v2 reagent kit (500 cycles) to yield paired-end reads of 250 bases each. Co-receptor tropism could be predicted by aligning reads to the consensus B sequence and isolating and trimming reads that overlap the entire V3 region. Unique V3 sequences that are supported by 1.75% were used for HIV-1 co-receptor tropism predicted using geno2pheno and PSSM algorithms.
Single genome sequencing from PBMC pre HSCT
Single genomes were generated as follows. Genomic DNA was extracted from total peripheral PBMC using the Qiagen DNAeasy kit. Two rounds of nested PCR were then performed using previously validated primers and Invitrogen High Fidelity platinum taq polymerase. 2kb Env amplicons were visualised by agarose gel electrophoresis and sanger sequenced using a primer internal to Env.
Sequence alignment and phylogenetic tree construction
Env amplicons generated by SGS were Sanger-sequenced. Prior to alignment sequences were trimmed at the 5’ and 3’ ends until base calls consistently reached a quality score ≥30, resulting in 99.9% accuracy of base call. Patient env nucleotide sequences were aligned at the protein sequence level using MUSCLE v3.8.31 (1) and mapped back to nucleotide, with minor manual adjustments. Sequences from laboratory isolates NL4.3 and Bal were included for reference, together with two HIV-1 subtype D reference isolates from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/) to serve as an outgroup. A maximum likelihood (ML) phylogeny was estimated using RAxML 8 (https://raxml-ng.vital-it.ch/#/), with the general time-reversible nucleotide substitution model and gamma-distributed rate heterogeneity. Stationary frequencies of nucleotides were estimated from their counts in the sequences. Clade support was estimated from 1000 non-parametric bootstrap replicate datasets. The ML phylogeny was rooted on the outgroup branch and visualised using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Phenotypic determination of CCR5 or CXCR4 usage by patient-derived Env clones
Selected amplicons were cloned into an expression vector deleted for env and co-transfected into HEK293T cells with the envelope deficient full length HIV plasmid encoding lucifese (NL4-3 delta Env Luc). Pseudoviruses were harvested, fitered and titrated 3-fold in U87 cells expressing either CXCR4 (X4) or CCR5 (R5). A dual tropic pseudovirus (WEAU-d15.410.787) was produced in parallel as a positive control for infection. After 48 h incubation cells were lysed by addition of Steady-Glo luciferase reagent (Promega, UK) and luminescence read using a GloMax 96 Luminometer (Promega, UK).
Modified Quantitative Viral Outgrowth Assay (QVOA)
Virus outgrowth assay was performed as previously described with modifications. Briefly, total CD4+ T cells were isolated from PBMCs by immune-magnetic negative selection (StemCell Technologies). Cells displaying activation markers (CD25/CD69 / HLA-DR) were labelled with FITC-conjugated antibodies (Biolegend) and depleted by FITC selection kit (StemCell Technologies). T cells with activation markers adhering to the beads were recovered, additionally activated with phytohemagglutinin-L (PHA-L, Sigma), seeded in a limiting dilution and co-cultured with SupT1-CCR5 for at least 21 days.
Resting CD4+ T cells were recovered from the eluant and cultured in the presence of 100nM raltegravir and 20nM efavirenz for 1-3 days to allow unintegrated viral DNA to degrade. After which resting CD4+ T cells were counted, activated with PHA-L, 10 fold excess of irradiated allogeneic PBMC and 10 units/ml interleukin-2 (IL-2, NIBSC) and seeded in a limiting dilution. The activation mix was washed away 24h later and SupT1-CCR5 cells were added to each co-culture well.
The dilution series include replicates at 2.5million, 0.5million and 0.1 million CD4+ T cells per well as appropriate. Co-cultures were maintained for at least 21 days. Cells were monitored for syncytia formation, and the supernatant was sampled regularly for p24. The infectious unit per million cells were calculated by limiting dilution statistics.
Reagents for QVOA
Interleukin-2 (NIBSC repository reference ARP901) was obtained from the Centre for AIDS reagents, National Institute of Biological Standards and Control (NIBSC), United Kingdom. Raltegravir was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Raltegravir (Cat # 11680) from Merck & Company, Inc. Efavirenz was also obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Efavirenz.
Residual viremia by single copy assay / ultra sensitive VL (usVL)
Residual viremia (HIV-RNA) was measured by ultracentrifugation of up to 6.5 ml of plasma (4-6.5ml) at 43100 rpm at 4°C for 30 minutes, followed by viral RNA extraction using the m2000sp Abbot RealTime HIV-1 Assay device and laboratory-defined applications software from the instrument. HIV-1 RNA copies in the low range were determined by an in-house calibration curve set (range, 10-103 absolute copies), which had previously been validated using a standard HIV-1 DNA control from the WHO in the range of 128–0.5 copies/mL. Limit of detection was calculated relative to the plasma volume used in each sample.
Quantification of HIV Antibodies
Specific HIV-1 antibodies in longitudinal sera samples were tested in a qualitative western blot assay (New Lav Blot I, Bio-Rad). Standard and low sensitive (LS) versions of the Vitros anti-HIV-1 assay (Ortho-Clinical Diagnostics) as well as the limiting avidity antigen assay were also measured in same samples as previously described2. Briefly, four recombinant antigens (HIV-1 Env 13, HIV-1 Env 10, HIV-1 p24, and HIV-2 Env AL) derived from HIV-1 core, HIV-1 envelope, and HIV-2 envelope proteins were quantified. The optimized version of the LS-Vitros assay (detuned or LS version) uses a 1:400 dilution of HIV-positive sample. The cut off was set up at 20 S/CO. The avidity assay measures the capacity of the guanidine to elute low-avidity and low-affinity antibodies after antigen-antibody bonds have formed. The results are reported as an avidity index (AI), which was calculated as a ratio of the S/CO of the sample incubated in guanidine to the S/CO of the sample incubated in PBS. Cut off was established at 0.51.
Flow Cytometry
The following fluorochrome-conjugated antibodies were used in this study: CD14 BV510, CD19 BV510, CD3 APC Fire 750 or CD3 BV605, CD4 PE/Dazzle 594, or CD4 APC Fire 750, CD8 BV421, CCR5 PE/Dazzle 594, CD56 PeCy7 (Biolegend) for surface antigens; and IFN-γ PeCy7, TNF-α FITC (Biolegend) and IL-2 PercP eFluor710 (eBioscience), for intracellular staining. PBMC were washed in PBS, and surface stained at 4°C for 20 min with saturating concentrations of different combinations of antibodies in the presence of fixable live/dead stain (Invitrogen). Cells were then fixed and permeabilized for detection of intracellular antigens. Cells were acquired on a BD Fortessa X20 using BD FACSDiva8.0 (BD Bioscience) and analysed using FlowJo 10 (Tree Star). Stochastic neighbor embedding (SNE) analysis was performed using the mrc.cytobank platform.
Intracellular cytokine staining
PBMCs were thawed and resuspended in RPMI complete media. Following, overnight rest at 370C and 5%CO2, PBMCs were stimulated for 6 hours with 2μg/ml HIV-1 Gag pools or CMV pp65 (JPT Peptide Technologies) in the presence of 1ug/mL anti-CD28 and anti-CD49d CoStimtm antibodies (BD Biosciences), 2 uM Monensin (BD biosciences), 10 ug/mL Brefeldin A (Sigma) and anti-CD107a BV605 (Biolegend). Where indicated and cell numbers permitted PMBCs were stimulated with 2μg/ml HIV-1 Env, Pol and Nef peptide pools. Following stimulation virus specific T cells were identified via intracellular cytokine staining (ICS) as previously described. In brief, cells were surface, fixed and permeabilised (CytoFix/CytoPerm™ BD Biosciences) followed by ICS for IFN-γ PeCy7, TNF-α FITC, and IL-2 PercP eFluor710 (eBioscience). Stimulation with 0.005% DMSO in the presence of costimulatory antibodies, protein transport inhibitors and CD107a was performed as a negative control.
Extended Data
Extended data figure 1. Blood Cell populations over time.
Abbreviations: HSCT: haematopoietic stem cell transplantation; CsA ciclosporin A; ART antiretroviral therapy; RPV rilpivirine; DTG dolutegravir; 3TC lamivudine
Extended data figure 2. Susceptibility of index patient CD4 T cells to R5 tropic and X4 tropic HIV-1.
a. Experimental flow for measurement of infection by intracellular p24 gag staining. Control cells were from a healthy HIV- CCR5+ donor. b. Flow cytometry analysis of PBMC following 3 days of stimulation exhibiting the expression pattern of CCR5 receptor within CD3+ CD4+ T cells in both healthy donor (control) and index patient. c. Culture supernatants from CD4 T cells infected with R5 and X4 tropic viruses were collected on days 3 and 7 to measure infectivity on HeLa TZM-bl reporter cells. Infectivity is measured as a reduction in Tat-induced firefly luciferase reporter gene expression in TZM-bl. Error bars represent standard error of the mean. N=2: one donor and one index patient Experiments were repeated 3 times with similar results.
Extended data figure 3. CD8+T cell responses and CD4 T cell responses to HIV.
Representative FACS plots showing percentage of virus specific CD8+ T cells (top panel) and CD4+ T cells (bottom panel) identified via intracellular staining for IFN-g, following stimulation with HIV Pol, Env and Nef peptide pools post-HSCT at days +D96 and +D819.
Extended data table 1. Comparison of blood group and tissue typing between stem cell donor and index case.
| Donor | Recipient | |
|---|---|---|
| Blood group (Rhesus) | A(+) | A(+) |
| CMV Status | + | + |
| HLA-A | 11:01,24:02 | 11:01,24:02 |
| HLA-B | 14:02,51:01 | 14:02,51:07 |
| HLA-Cw | 08:02,14:02 | 08:02,14:02 |
| HLA-DR | 01:02,13:01 | 01:02,13:01 |
| HLA-DQ | 05:01,06:03 | 05:01,06:03 |
Extended data table 2. Detection of bands on western blot pre- and post-transplantation.
| Days post allo-HSCT | Observed Bands | Bands left |
|---|---|---|
| 26 | GP160, GP110/120, GP41, p68, p55, p40, p31, p24, p18 | None |
| 423 | GP160, GP110/120 faint, GP41, p31 faint, p24 very faint | p68, p55, p52, p40, p18 |
| 811 | GP160, GP110/120 faint, GP41, p31 very faint | p68, p55, p52, p40, p24, p18 |
| 965 | GP160, GP110/120 very faint, GP41, p18 | p68, p55, p52, p40, p24, p31 |
Acknowledgements
This study was funded by a Wellcome Trust Senior Fellowship in Clinical Science to RKG, research capability funding (RCF) from UCLH BRC to RKG, as well as funding from Oxford and Cambridge Biomedical Research Centres (BRC), amfAR (The Foundation for AIDS Research), through the amfAR Research Consortium on HIV Eradication (ARCHE) program (AmfAR 109858-64-RSRL), the MRC (MR/R008698/1 to LEM and MRM008614/2 to DP). AJI is supported by an NIHR Clinical Lectureship, and acknowledges support from the NIHR and Imperial Biomedical Research Centre (BRC). We would like to thank the CHERUB (http://www.cherub.uk.net) and IciStem Consortia (www.icistem.org) for support and continuous discussion of results.
We would also like to thank Nina Parmahand, Mikaila Bandara, Isobel Jarvis, Axel Fun, Kirit Ardeshna, Alison Hill, Neha Goel, Richard Szydlo, David Slade, Sarah Griffith and Cristina Gálvez. Águeda Hernández Rodríguez, Victoria González Soler and Belén Rivaya Sánchez from the Microbiology Department of the Hospital Germans Trias i Pujol. Erik van Maarseveen from the department of Pharmacology, and Laura Huyveneers, Pauline Schipper and Dorien de Jong from the Translational Virology Group of the department of Medical Microbiology of the UMC Utrecht. Finally, we would also like to thank Professor Jane Apperley, Zoe Allwood and Sandra Loaiza from Imperial College Healthcare NHS Trust and all the nurses in the BMT Unit that looked after the patient.
Footnotes
Author contributions
Conception and design: RKG, SAJ, SGE, EO, HPM and IHG
Designed and/or performed experiments; SAJ, JMP, AMW, MS, MN, FS, SA, LEM, IHG, LMc, DP, HPM, PG, EN, AL, LW, SGE, MP, EN, PG, CM
Analysis and interpreted data: RKG, JMP, AMW, SAJ, MS, MN, FS, SA, LEM, LMc, EO, HL, JF, MP, IHG, HPM, SGE, EN, PG, JL, AJI,CM
Drafting of the article: RKG, MN, AMW, JMP, AL, JL, HPM, JL, AJI
Critical revision of the article for important intellectual content: JMP, AMW, MS, MN, FS, SA, LEM, LMc, DP, HPM, PG, EN, AL, SGE, LW, MP, IHG, HPM, AJI
Competing interests
The authors have no conflicts of interest
Data availability
SGA Sequences are available via GenBank under accession numbers MK493056-MK493075.
The following figures have associated raw data: Figure 1, 2, 3, 4.
References
- 1.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 2.Allers K, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 3.UNAIDS. UNAIDS DATA. 2017 < http://www.unaids.org/en/resources/documents/2017/20170720_Data_book_2017>.
- 4.Gupta RK, et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis. 2018;18:346–355. doi: 10.1016/S1473-3099(17)30702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TenoRes Study G. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis. 2016;16:565–575. doi: 10.1016/S1473-3099(15)00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barth RE, van der Loeff MF, Schuurman R, Hoepelman AI, Wensing AM. Virological follow-up of adult patients in antiretroviral treatment programmes in sub-Saharan Africa: a systematic review. Lancet Infect Dis. 2010;10:155–166. doi: 10.1016/S1473-3099(09)70328-7. [DOI] [PubMed] [Google Scholar]
- 7.Avert. FUNDING FOR HIV AND AIDS. 2017 < https://www.avert.org/professionals/hiv-around-world/global-response/funding>.
- 8.Simmons G, et al. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. Journal of virology. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kordelas L, et al. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 10.Verheyen J, et al. Rapid rebound of a preexisting CXCR4-tropic HIV variant after allogeneic transplantation with CCR5 delta32 homozygous stem cells. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2018 doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
- 11.Symons J, et al. Dependence on the CCR5 coreceptor for viral replication explains the lack of rebound of CXCR4-predicted HIV variants in the Berlin patient. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2014;59:596–600. doi: 10.1093/cid/ciu284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrich TJ, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummins NW, et al. Extensive virologic and immunologic characterization in an HIV-infected individual following allogeneic stem cell transplant and analytic cessation of antiretroviral therapy: A case study. PLoS medicine. 2017;14:e1002461. doi: 10.1371/journal.pmed.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosman KJ, et al. Development of sensitive ddPCR assays to reliably quantify the proviral DNA reservoir in all common circulating HIV subtypes and recombinant forms. J Int AIDS Soc. 2018;21:e25185. doi: 10.1002/jia2.25185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yukl SA, et al. Challenges in detecting HIV persistence during potentially curative interventions: a study of the Berlin patient. PLoS pathogens. 2013;9:e1003347. doi: 10.1371/journal.ppat.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie AV, et al. Performance evaluation of the point-of-care SAMBA I and II HIV-1 Qual whole blood tests. J Virol Methods. 2016;237:143–149. doi: 10.1016/j.jviromet.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS pathogens. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fun A, Mok HP, Wills MR, Lever AM. A highly reproducible quantitative viral outgrowth assay for the measurement of the replication-competent latent HIV-1 reservoir. Sci Rep. 2017;7 doi: 10.1038/srep43231. 43231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen MA, Coetzer M, van 't Wout AB, Morris L, Mullins JI. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. Journal of virology. 2006;80:4698–4704. doi: 10.1128/JVI.80.10.4698-4704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nature biotechnology. 2007;25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 21.Smith NM, et al. Proof-of-Principle for Immune Control of Global HIV-1 Reactivation In Vivo. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2015 doi: 10.1093/cid/civ219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watters SA, et al. Sequential CCR5-Tropic HIV-1 Reactivation from Distinct Cellular Reservoirs following Perturbation of Elite Control. PloS one. 2016;11:e0158854. doi: 10.1371/journal.pone.0158854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salgado M, et al. Mechanisms That Contribute to a Profound Reduction of the HIV-1 Reservoir After Allogeneic Stem Cell Transplant. Ann Intern Med. 2018;169:674–683. doi: 10.7326/M18-0759. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton BK, et al. Long-term follow-up of a prospective randomized trial comparing CYA and MTX with CYA and mycophenolate mofetil for GVHD prophylaxis in myeloablative sibling donor hematopoietic cell transplantation. Bone Marrow Transplant. 2013;48:1578–1580. doi: 10.1038/bmt.2013.89. [DOI] [PubMed] [Google Scholar]
- 25.Duarte RF, et al. CCR5 Delta32 homozygous cord blood allogeneic transplantation in a patient with HIV: a case report. Lancet HIV. 2015;2:e236–242. doi: 10.1016/S2352-3018(15)00083-1. [DOI] [PubMed] [Google Scholar]
- 26.Pavlu J, et al. LACE-conditioned autologous stem cell transplantation for relapsed or refractory diffuse large B-cell lymphoma: treatment outcome and risk factor analysis from a single centre. Hematol Oncol. 2011;29:75–80. doi: 10.1002/hon.956. [DOI] [PubMed] [Google Scholar]
- 27.Bronnimann MP, Skinner PJ, Connick E. The B-Cell Follicle in HIV Infection: Barrier to a Cure. Front Immunol. 2018;9:20. doi: 10.3389/fimmu.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]