Summary
Matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) may soon replace routine electrophoretic methods for monitoring monoclonal proteins in patients with multiple myeloma. To further evaluate the clinical utility of this assay, we compared the performance of MALDI-TOF-MS head-to-head with an established bone marrow-based measurable residual disease assay by flow cytometry (Flow-BM-MRD) using Memorial Sloan Kettering Cancer Center’s 10-color, single-tube method. Our results suggest that MALDI-TOF-MS adds value to bone-marrow based MRD testing and may be most useful for early detection of relapse in peripheral blood compared to current electrophoretic methods.
In multiple myeloma (MM), the absence of measurable residual disease (MRD) after completed therapy is associated with longer progression-free survival (Kumar et al, 2016; Perrot et al, 2018). Different techniques are available to detect low levels of plasma cells in bone marrow (BM) either by flow cytometry analysis (Roshal et al, 2017; Flores-Montero et al, 2017) or by next-generation sequencing (NGS) as a gold standard of molecular methods (Korde et al, 2015; Perrot et al, 2018). These techniques are limited because they require a representative BM sample obtained by an invasive procedure and may have sampling biases due to the patchy nature of disease infiltration. A sensitive serum-based test would be ideal because serial sampling is much easier, and it would allow for the detection of extramedullary disease. Protein electrophoresis (SPEP), immunofixation electrophoresis (IFE) and free light chain (FLC) immunoassays have long been used to detect the monoclonal protein (M-protein) in serum but are currently not sensitive enough to be used in the setting of MRD (Thoren, 2018). Considering the tremendous progress in the treatment and outcome of MM patients in the past few years (Moreau, 2017), a more responsive technique is needed. Mass spectrometry techniques that detect M-proteins in serum have recently been developed and shown to be more sensitive compared to current electrophoretic methods (Mills et al, 2016). MS techniques are based on the principle that each immunoglobulin has a unique amino acid sequence and, consequently, a unique mass (Barnidge et al, 2014; Mills et al, 2016; Thoren, 2018). This patient-specific mass is stable over time and can be used as a disease biomarker (Barnidge et al, 2014; Mills et al, 2016; Mills et al, 2017; Thoren, 2018). In particular, Matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) may soon replace current electrophoretic methods for routinely monitoring MM patients due to its relatively low cost, high throughput and improved analytical sensitivity and specificity. In order to further evaluate the clinical utility of MALDI-TOF-MS, we compared the performance of this technique with an established Flow-BM-MRD assay using the Memorial Sloan Kettering Cancer Center’s (MSKCC) 10-color, single-tube method (Roshal et al, 2017).
This study included 71 patients treated at MSKCC since 2010, who had serum samples available at 2 timepoints including during active disease and within + 60 days of Flow-BM-MRD measurement. Active disease was defined as detectable M-protein in serum or urine by immunofixation. The cohort was composed of 26 females and 45 males with a median age of 61 years (range 37–78). Of them, 48 had been newly diagnosed at MSKCC. Active disease samples were taken at baseline for 42 patients and during follow-up or relapse for 29. Cytogenetic analyses at baseline identified 27 high risk patients and 43 standard risk. According to the International Scoring System (ISS), patients were classified in 3 groups: ISS1 (n=38), ISS2 (n=18), ISS3 (n=6). ISS data were missing for 9 patients (Table SI). SPEP, IFE and FLC were performed at both considered time points (active disease and MRD). At baseline, both SPEP and IFE were positive for 63/69 patients. Five patients were positive by IFE only and one with FLC MM was positive by urine electrophoresis only. The median time between diagnosis and MRD timepoint was 13.4 months (range: 3.4–91) (table I).
Table I.
Immunologic results at baseline and at MRD time points, including MALDI-TOFF result and Flow cytometry analysis
| N=71 | |||
|---|---|---|---|
| Baseline | |||
| SPEP | + | 63 | |
| − | 6 | ||
| missing | 2 | ||
| IFE | + | 68 | |
| − | 1 | ||
| missing | 2 | ||
| Free κ/λ ratio | abnormal | 60 | |
| normal | 4 | ||
| missing | 7 | ||
| MRD time points | |||
| Median time (month) between MRD and Diagnosis (range) | 13.4 (3.4–91) | ||
| SPEP | + | 7 | |
| − | 64 | ||
| IFE | + | 17 | |
| − | 54 | ||
| Free K/Λ ratio | abnormal | 24 | |
| normal | 47 | ||
| MALDI-TOF-MS | + | 25 | |
| − | 46 | ||
| Flow cytometry | + | 18 | |
| − | 53 |
SPEP: serum protein electrophoresis, IFE: immunofixation electrophoresis, MRD: measurable residual disease
MALDI-TOF-MS was performed according to the method published by Mills et al (2016). Briefly, immunoglobulins were purified from serum samples using CaptureSelect beads specific for IgG and IgA heavy chains or total kappa and lambda light chains (Thermo Fisher Scientific). Immunoglobulins were eluted and light and heavy chains separated by the addition of a reducing agent. Samples were analyzed using a Microflex LT MALDI-TOF mass spectrometer (Bruker). Samples obtained during active disease were used to identify the mass to charge ratio of the M-protein which served as surrogate marker in subsequent analyses. MALDI-TOF-MS results were compared to the Flow-BM-MRD assay.
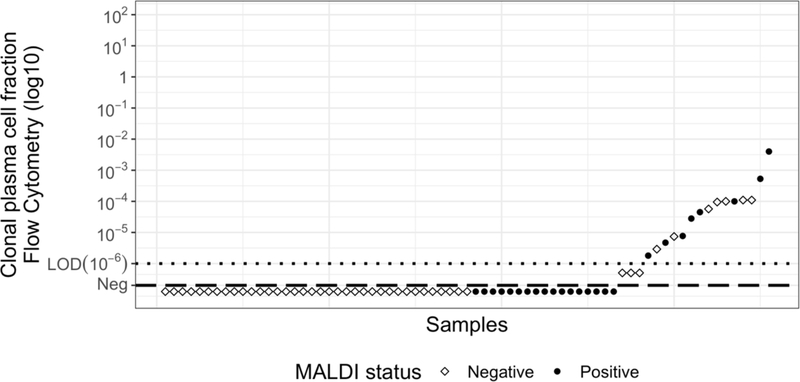
MALDI-TOF-MS detected an M-protein in all 71 active disease samples and in 25 MRD samples (Table I, Fig S1). MALDI-TOF-MS results at the MRD timepoint were concordant with Flow-BM-MRD results for 44/71 (62%) patients (p=0.342, χ2 test). Eight patients were positive by both techniques and 36 patients negative by both techniques (Fig 1). Twenty-seven patients were discordant, including 17 detectable only by MALDI-TOF-MS and 10 detectable only by Flow-BM-MRD (Fig 1). Regarding the 17 patients positive only by MALDI-TOF-MS, the BM sample for flow analysis was not suitable for 3 (evidence of marked hemodilution or not enough cells acquired) including one with only 9 abnormal plasma cells (below the level of 10 cells in cluster) while the others reached the target of sensitivity with a limit of detection of 0.0001%. Eleven of these 17 patients also had positive IFE, of whom 6 also had positive SPEP. Of those 6, only 1 was in CR at last news, suggesting that true MRD was detected (table SII). All the other 6 patients (only positive in MALDI) were in steady stringent CR at MRD time point and at last follow-up (median 10 months; range 6–12). Although BM-based MRD analysis (either flow-cytometry or NGS-based assays) is considered the gold standard measurement for disease detection in MM, these tests can have false negatives for a few reasons: i)sample quality, ii)MM patchy characteristic, iii)BM sampling unable to detect extramedullary disease. Alternately, MALDI-TOF results could be falsely positive in terms of disease detection. The mass spectrometer is likely not falsely detecting M-proteins; indeed, immunofixation was also positive in 11/17 of these samples. However, low levels of M-protein may not indicate the presence of active disease. A confounding factor is that immunoglobulins, especially IgG and IgA, have a long half-life in serum (Mills et al, 2017). This would cause a time lag between tumor lysis and M-protein elimination, especially at low immunoglobulin concentrations (Kendrick et al, 2017). The lag may be especially pronounced with the use of new, very potent therapies (Moreau, 2017). Among the 10 patients detectable by flow cytometry but not by MALDI, the median detection level was 0.00092% (+<0.0001% - 0.011%). All were in stringent complete response (sCR) at the MRD time point and at last news, only one relapsed 6 months after MRD analysis. The M-protein may thus remain present but below the polyclonal background. Of note, 3/10 of these patients had free kappa light chain MM, which can be challenging to detect by this MALDI assay (Sepiashvili et al, 2019).
Fig 1:
Comparison of Flow-BM-MRD and MALDI-TOF-MS results obtained at the MRD timepoint.
To determine the clinical utility of a more sensitive M-protein detection, we focused on the clinical outcome of a subgroup of our cohort which included only newly diagnosed MM patients in CR at the MRD timepoint (n=40). In this subgroup, the median follow-up was 11 months and the residual detection rate by MALDI-TOF-MS was 22.5% (9/40) compared to 27.5% (11/40) by flow-cytometry. Of note, 2 of the 3 patients that were positive by both techniques relapsed during follow-up. Yet, only one out of 23 patients that were negative by both techniques relapsed. However, none of the 6 patients who were positive only by MALDI-TOF relapsed, which is probably linked to the clearance of the M-protein, while only 1/8 patients who were positive only by Flow-BM-MRD relapsed. Although this subgroup has a limited number of patients and follow-up after the MRD time points, our results suggest that PB MALDI-TOF-MS adds value to BM results patients positive by both techniques being more likely to relapse.
This study is an important step in understanding how a more sensitive routine blood test can be used in the follow-up of MM patients. MALDI-TOF analysis may provide complementary results to Flow-BM-MRD especially for the follow-up of patients in CR and during maintenance therapy to detect poor responders that would be positive by both techniques. A previous study has compared the performance of a mass spectrometry assay to detect the M-protein in serum to a flow-cytometry BM based assay (Mills et al, 2010). This study compared liquid chromatography quadrupole (LC-QTOF-MS) and a 6-color flow cytometry assay with a sensitivity of 10−4 to 10-5. Although LC-QTOF-MS has superior sensitivity compared to MALDI-TOF-MS, the instrumentation is more complex and expensive, and it remains to be seen how the QTOF will be incorporated into clinical laboratories for M-protein monitoring (Thoren et al, 2018). Timing of these tests is clearly important. Sensitive MS assays that track the M-protein biomarker in serum can be useful after the disease is cleared and patients are being monitored for relapse. Following serum M-protein levels with a highly sensitive technique may be a good approach since circulating tumor DNA may not serve as a sufficient analyte for monitoring (Mazotti et al, 2018). MALDI-TOF-MS is also able to detect other monoclonal proteins such as therapeutic antibodies (Moore et al, 2019), which sometimes prevents the accurate monitoring of disease status when patients are receiving targeted therapy. This may be especially important in clinical trials and in accurately defining CR and sCR (Kumar et al, 2016; Dejoie et al, 2019) which should incorporate the use of more sensitive serum-based techniques like MALDI-TOF-MS as they are integrated into clinical practice.
Supplementary Material
Table SI. Patient characteristics at baseline
Fig S1. Example of MALDI-TOF results for a patient with IgG lambda multiple myeloma during active disease (green) and at the MRD time point (blue).
Acknowledgments
The authors would like to thank Sun Cho for her technical expertise. This work was supported by a grant from the Society of Memorial Sloan Kettering Cancer Center (to K.L.T.) and by the Memorial Sloan Kettering Core Grant (P30 CA008748) funded by the National Cancer Institute.
Conflicts of Interest:
Ola C Landgren:
Grant support: NIH, FDA, MMRF, IMF, LLS, Perelman Family Foundation, Rising Tides Foundation, Amgen, Celgene, Janssen, Takeda, Glenmark, Seattle Genetics, Karyopharm
Honoraria/ad boards: Adaptive, Amgen, Binding Site, BMS, Celgene, Cellectis, Glenmark, Janssen, Juno, Pfizer
Independent Data Monitoring Committee (IDMC): Takeda, Merck, Janssen, Theradex
Hani Hassoun:
Takeda: funding support for clinical trials
Janssen: funding support for clinical trials
Novartis: consultant in advisory boards
Neha Korde:
Amgen: research funding
Eric Smith
Honoraria/ad boards: Celgene, Fate therapeutics
Research founding: Celgene
Katie Thoren:
research support: Binding Site
Marion Eveillard, Even Rustad, Yanming Zhang, Amanda Ciardiello Malin Hultcrantz, Alexander Lesokhin, Sham Mailankody and Mikhail Roshal have nothing to disclose.
Footnotes
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- Barnidge DR, Dasari S, Botz CM, Murray DH, Snyder MR, Katzmann JA, Dispenzieri A & Murray DL (2014) Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. Journal of Proteome Research, 13, 1419–1427. [DOI] [PubMed] [Google Scholar]
- Dejoie T, Corre J, Caillon H, Moreau P, Attal M & Loiseau HA (2019) Responses in multiple myeloma should be assigned according to serum, not urine, free light chain measurements. Leukemia, 33, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Montero J, Sanoja-Flores L, Paiva B, Puig N, García-Sánchez O, Böttcher S, van der Velden VHJ, Pérez-Morán JJ, Vidriales MB, García-Sanz R, Jimenez C, González M, Martínez-López J, Corral-Mateos A, Grigore GE, Fluxá R, Pontes R, Caetano J, Sedek L, Del Cañizo MC, Bladé J, Lahuerta JJ, Aguilar C, Bárez A, García-Mateo A, Labrador J, Leoz P, Aguilera-Sanz C, San-Miguel J, Mateos MV, Durie B, van Dongen JJM & Orfao A (2017) Next Generation Flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia, 31, 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick F, Evans ND, Arnulf B, Avet-Loiseau H, Decaux O, Dejoie T, Fouquet G, Guidez S, Harel S, Hebraud B, Javaugue V, Richez V, Schraen S, Touzeau C, Moreau P, Leleu X, Harding S & Chappell MJ (2017) Analysis of a Compartmental Model of Endogenous Immunoglobulin G Metabolism with Application to Multiple Myeloma. Frontiers in Physiology, 8, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde N, Roschewski M, Zingone A, Kwok M, Manasanch EE, Bhutani M, Tageja N, Kazandjian D, Mailankody S, Wu P, Morrison C, Costello R, Zhang Y, Burton D, Mulquin M, Zuchlinski D, Lamping L, Carpenter A, Wall Y, Carter G, Cunningham SC, Gounden V, Sissung TM, Peer C, Maric I, Calvo KR, Braylan R, Yuan C, Stetler-Stevenson M, Arthur DC, Kong KA, Weng L, Faham M, Lindenberg L, Kurdziel K, Choyke P, Steinberg SM, Figg W & Landgren O (2015) Treatment With Carfilzomib-Lenalidomide-Dexamethasone With Lenalidomide Extension in Patients With Smoldering or Newly Diagnosed Multiple Myeloma. JAMA Oncology, 1, 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, Munshi N, Lonial S, Bladé J, Mateos MV, Dimopoulos M, Kastritis E, Boccadoro M, Orlowski R, Goldschmidt H, Spencer A, Hou J, Chang WJ, Usmani SZ, Zamagni E, Shimizu K, Jagannath S, Johnsen HE, Terpos E, Reiman A, Kyle RA, Sonneveld P, Richardson PG, McCarthy P, Ludwig H, Chen W, Cavo M, Harousseau JL, Lentzsch S, Hillengass J, Palumbo A, Orfao A, Rajkumar SV, Miguel JS & Avet-Loiseau H (2016) International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncology, 17, e328–e346. [DOI] [PubMed] [Google Scholar]
- Mazzotti C, Buisson L, Maheo S, Perrot A, Chretien ML, Leleu X, Hulin C, Manier S, Hébraud B, Roussel M, Do Souto L, Attal M, Avet-Loiseau H & Corre J (2018) Myeloma MRD by deep sequencing from circulating tumor DNA does not correlate with results obtained in the bone marrow. Blood Advances, 2, 2811–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Barnidge DR & Murray DL (2015) Detecting monoclonal immunoglobulins in human serum using mass spectrometry. Methods, 15, 56–65. [DOI] [PubMed] [Google Scholar]
- Mills JR, Barnidge DR, Dispenzieri A & Murray DL (2017) High sensitivity blood-based M-protein detection in sCR patients with multiple myeloma. Blood Cancer Journal, 7, e590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LM, Cho S & Thoren, K.L. MALDI-TOF mass spectrometry distinguishes daratumumab from M-proteins. (2019) Clinica Chimica Acta, 18, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P (2017) How I treat myeloma with new agents. Blood, 130, 1507–1513. [DOI] [PubMed] [Google Scholar]
- Perrot A, Lauwers-Cances V, Corre J, Robillard N, Hulin C, Chretien ML, Dejoie T, Maheo S, Stoppa AM, Pegourie B, Karlin L, Garderet L, Arnulf B, Doyen C, Meuleman N, Royer B, Eveillard JR, Benboubker L, Dib M, Decaux O, Jaccard A, Belhadj K, Brechignac S, Kolb B, Fohrer C, Mohty M, Macro M, Richardson PG, Carlton V, Moorhead M, Willis T, Faham M, Anderson KC, Harousseau JL, Leleu X, Facon T, Moreau P, Attal M, Avet-Loiseau H & Munshi N (2018) Minimal residual disease negativity using deep sequencing is a major prognostic factor in multiple myeloma. Blood, 132, 2456–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshal M, Flores-Montero JA, Gao Q, Koeber M, Wardrope J, Durie BGM, Dogan A, Orfao A & Landgren O (2017) MRD detection in multiple myeloma: comparison between MSKCC 10-color single-tube and EuroFlow 8-color 2-tube methods. Blood Advances, 1, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepiashvili L, Kohlhagen MC, Snyder MR, Willrich MAV, Mills JR, Dispenzieri A & Murray DL (2019) Direct Detection of Monoclonal Free Light Chains in Serum by Use of Immunoenrichment-Coupled MALDI-TOF Mass Spectrometry. Clinical Chemistry, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Thoren KL (2018) Mass spectrometry methods for detecting monoclonal immunoglobulins in multiple myeloma minimal residual disease. Seminar Hematology, 55, 41–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SI. Patient characteristics at baseline
Fig S1. Example of MALDI-TOF results for a patient with IgG lambda multiple myeloma during active disease (green) and at the MRD time point (blue).