Abstract
The aims of this work were to screen isolated bacteria with a dual capacity: to inhibit Fusarium solani and to promote plant growth. Also, volatile compounds that would be responsible for that effect were identified. Seventy bacterial strains from the air, agricultural soils, hydrocarbons-contaminated soils, and extremophile soils were tested. The former were identified by Matrix-Assisted Laser Desorption/Ionization-time of flight mass spectrometry and 16S rDNA sequencing. The plant growth-promoting bacteria (PGPB) and their capability for phosphate solubilization, siderophores production, and indole production were determined. Twenty isolates from Bacillus and Pseudomonas genera inhibited the mycelial growth up to 40% in direct assays. Eleven isolates significantly inhibited mycelial growth in 18–24% via volatile emissions. Volatile compounds related to antifungal activity or stress response include ketones, sesquiterpenes, monoterpenoids, alkanes, and fatty acids. Our results support the potential of these PGPB to act as biocontrol agents against fungal pathogens via volatile emissions.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02290-6) contains supplementary material, which is available to authorized users.
Keywords: Biocontrol, Fusarium solani, Plant-growth promoting bacteria, Bacillus spp., Volatile compounds
Introduction
The ability of plants to adapt to different environments is conditioned by their associated rhizosphere microbiota (Peiffer et al. 2013; Philippot et al. 2013). Plant growth-promoting bacteria (PGPB) are of particular importance in the search for successful biological agents since they may benefit crop performance through direct or indirect mechanisms. Direct effects include stimulating plant development through production of plant regulators, improving nutrient uptake, seed germination, degrading phytotoxic compounds, and inducing systemic resistance (Compant et al. 2010; Peiffer et al. 2013; Philippot et al. 2013). For instance, indol-acetic phytohormone may improve root formation and fruit development; some organic acids may act as quorum quenchers for other microorganism and, at the same time, improve metal uptake by plants. Some bacteria using these mechanisms belong to Pseudomonas, Bacillus, Azospirillum, Rhizobium, and Serratia genera (Siddiqui, 2006). Indirect effects include suppression of pathogens through nutrient competition, production of diffusible antifungal compounds, and the emission of bacterial volatile compounds (BVCs) (Cazorla et al. 2007; Guevara-Avendaño et al. 2018; Vinodkumar et al. 2017). The suppression of phytopathogens is especially relevant for agricultural crops growing in intensive monocultures and, therefore, highly susceptible to fast-spreading fungal diseases. Depending on the strategy of the fungal pathogen, competing microorganisms can impact negatively their survival, spore germination, hypha growth, or penetration and colonization in plant tissues (Weller et al. 2004; Whipps 2001). The genera Pseudomonas and Bacillus play an active role in the suppression of soil-borne pathogens (Siddiqui 2006; Cazorla et al. 2007; Dunlap et al. 2017; Guevara-Avendaño et al. 2018). Although BVCs have been described for Pseudomonas (Hunziker et al. 2015), more research on BVCs from Bacillus capable of inhibiting the mycelial growth of Fusarium euwallaceae, F. oxysporum and Rosellinia necatrix is needed. The ability of bacteria to secrete a wide range of antifungal BVCs makes them useful in the search of potential biological control agents (Bais et al. 2006; Umeda et al. 2016). It is necessary to consider biocontrol as an option to prevent the impact of fungal diseases since the use of chemical pesticides is restricted for certain crops (Stout et al. 2003). Therefore, PGPB constitute a source of potential biological control agents useful as sustainable alternatives for agriculture. The objectives of this study were to: (1) assess the antagonistic activity of isolated bacteria from different environments against Fusarium solani and evaluate the potential of the isolates with highest IP (IP ≥ 40%) as PGPB, (2) conduct a preliminary screening of the presence of BVCs produced by those bacteria with IP ≥ 40%, and (3) evaluate the volatile profile by SPME–GC–MS of that isolate with the best IP against F. solani.
Materials and methods
Bacterial isolation
Bacterial isolates used for the present study are derived from the Environmental Biotechnology Laboratory of CIATEJ, Guadalajara, state of Jalisco, Mexico. They were isolated in previous works from agricultural soil (S) (20° 46′ 7″ N and 103° 41′ 18″ W), soils contaminated with hydrocarbons (H) (20° 27′ 22.6″ N and 97° 20′ 8.5″ W), air samples (A) (20º 37′ 53.76″ N and 103º 20′ 42.72″ W), and extremophile soils (E) (20° 3′ 36.03″ N and 102° 36′ 58.49″ W).
Bacterial isolates identification by mass spectrometry (MALDI-TOF)
Bacterial isolates identification was carried out by Matrix-Assisted Laser Desorption/Ionization-time of flight mass spectrometry (MALDI-TOF) performed on a MICROFLEX LT mass spectrometer (Bruker Daltonics, Bremen, Germany) by applying two different extraction protocols: 1) using biomass from a single colony (DT Method, Bruker Daltonics), 2) an ethanol/formic acid protein extraction procedure (EX Method, Bruker Daltonics). The taxonomic identification was achieved by comparing the mass spectra with reference spectra from the BDAL database employing the MALDI-Biotyper® 3.4 software (Bruker Daltonics, Billerica, MA, USA). The spectrometer was calibrated using the Bacterial Test Standard purchased from Bruker Daltonics. The following identification reliability criteria were used: 2.300–3.000 score, reliable at the species level; 2.000–2.299, at genus (presumable at species level); 1.700–1.999, presumable identification at genus level; < 1.700, not reliable identification (Rocca et al. 2018).
In vitro antagonism screening assays against Fusarium solani
Fusarium solani was inoculated onto Potato Dextrose Agar medium (PDA, BD Bioxon) and incubated at 30 °C for 5 days before dual plating. An agar plug of 1 cm in diameter was taken from the border of the mycelial growth with a sterile cork borer and placed on the center of a PDA (pH 7.2) Petri dish. Bacterial isolates were taken from a single colony with a sterile loop and inoculated at a 1.5-cm distance from the mycelial plug. The antagonism assays were carried out in triplicate and incubated at 30 °C. After 5 days, the mycelium radial growth was measured for the bacterial and control treatments. The percentage of inhibition (IP) of mycelial growth was calculated using the following formula: % inhibition = [(D − d)/D] * 100, where D is the diameter of fungal growth for the control treatment, and d is the diameter of fungal growth for the bacterial treatment (Idris et al. 2007). The selection criterion was arbitrarily set at IP ≥ 40%. After that, the 20 best isolates were selected for molecular identification by 16S rDNA sequencing, PGPB evaluation, and antifungal activity of their emitted BVCs, as described below.
Molecular identification of bacterial isolates with antagonistic effect against F. solani
Bacterial isolates showing antagonistic activity were identified through 16S rDNA gene sequencing. DNA was extracted from each bacterial isolate using the phenol:chloroform:isoamyl alcohol technique described by Bollet et al. (1991). The 16S rDNA region was amplified by PCR using universal primers 63F (5′- CAG GCC TAA CAC ATG CAA GTC-3′) and 1387R (5′- GGG CGG WGT GTA CAA GGC-3′) (Hugenholtz and Goebel 2001) in 100 μL reactions containing 300 ng of template DNA, 10X of Taq buffer, 10 mM of each dNTP, 2 mM of MgCl2, 10 μM of each primer, 1 μL BSA (3%), 0.5 μL DMSO, and 1U of Taq DNA polymerase. Reactions were performed in a ProFlex PCR System thermal cycler under the following conditions: initial denaturation at 95 °C for 5 min; 40 cycles of denaturation at 95 °C for 1 min, annealing at 54 °C for 1 min, extension at 72 °C for 1 min, and a final extension step at 72 °C for 5 min. PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System Purification kit (Promega), following the manufacturer’s instructions. Purified PCR products were sent to Macrogen Inc. (South Korea) for sequencing. Sequences were manually checked and trimmed in Sequencher 5.4.6 (Gene Codes Corporation). The edited sequences were BLAST-analyzed (Altschul et al. 1990) and deposited in the GenBank nucleotide database. Sequences were uploaded to NCBI and received the accession numbers MN262681-MN262699, and MN298762.
Evaluation of the properties of plant growth-promoting bacteria
Bacterial isolates (n = 20) showing antagonistic effect against F. solani were evaluated for their capacity as PGPB. Phosphate solubilization was evaluated using Pikovskaya’s culture medium (Sundara Rao and Sinha 1963a, b) supplemented with 5 g L−1 phosphorite rock. One single colony was taken with a sterile toothpick and inoculated into Pikovskaya’s culture broths. The high-range reagent set kit for phosphate determination (HANNA Instruments, Mexico city, Mexico) was used and phosphate concentration was evaluated in a HI83200 Bench multiparameter spectrophotometer (HANNA Instruments) at 420 nm (APHA/AWWA/WEF 2012). For indolacetic acid (IAA) production, two trypticase soy (TSA) broths with or without 0.1 g L−1 tryptophan were prepared. IAA was evaluated based on Salkowsky’s reagent (Salkowsky’s, 1889, modified by Rodríguez, 2013: 1 mL of 0.5 m FeCl3, 50 mL of water and 30 mL of concentrate H2SO4). Concentration was determined on a Microplate xMark spectrophotometer (Bio-Rad Laboratories® Hercules, CA, USA) at 530 nm, using a standard curve of indoleacetic acid (mg IAA L−1). Both assays were carried out in triplicate and incubated for 2 days at 30 °C. Sterile medium was inoculated with a sterile toothpick and used as a negative control. Siderophore production was determined using Chromeazurol S (CAS) medium (Alexander and Zuberer 1991). Solutions were prepared as described in the methodology described by the former authors without solution 4. One single colony was inoculated with a sterile toothpick by picking the agar. The assay was carried out with four replicates. The appearance of an orange coloration, after 4 days of incubation at 30 °C, was considered as a positive result (+). Additionally, hemolysis test was performed to identify bacteria that could be pathogenic to humans. Bacterial isolates were inoculated into TSA and incubated at 20 °C for 24 h. Blood agar base was prepared with 5% of human blood. One single isolated colony was taken and transferred with a sterile toothpick. The assay was carried out in triplicate with negative control (sterile toothpick) and incubated at 30 °C for 48 h. Complete β-hemolysis was observed as a clear zone around the colony, and partial α-hemolysis as a dark-green coloration. Bacteria with γ-hemolysis do not exhibit any alteration of color or opacity in the medium, indicating the absence of hemolysis (Forbes et al. 2002).
Bacterial volatile compounds emission
The bacterial isolates that could inhibit F. solani mycelial growth by more than 40% in the first antagonism assay (n = 20) were selected for additional evaluation; the antifungal activity of their emitted BVCs was further tested. The indirect antagonism assays were carried out using the two-sealed-base-plates method described in Méndez-Bravo et al. (2018). A 5-day-old fungal culture was placed facing, but not touching. A 24-h-old bacterial culture on TSA. Briefly, each bacterial isolate was streaked to prepare a bacterial lawn onto a base plate containing PDA medium. Lids were removed and replaced by another base plate containing in its center a disc of 1-cm diameter of fungal mycelium. Both base plates were assembled and sealed with two layers of Parafilm® and incubated for 5 days at 30 °C. Each bacterial isolate was tested in triplicate with a control of the fungus. After 5 days, the IP was measured. The isolate with the highest IP was evaluated for BVCs identification by solid-phase microextraction (SPME) coupled to gas chromatography and mass spectrometry (GC–MS) as described below.
Preliminary screening of bacteria volatile compounds (BVCs) by SPME-GC–MS
Bacillus S18 was grown 24 h prior to the experimental set up on LB medium at 30 °C with 200 rpm agitation at an optical density of 0.300 at 600 nm. Then 50 μL of the culture was re-inoculated onto Petri plates containing PDA medium and a 5-day-old fungal culture plug of 1-cm diameter was placed facing the sample (bacteria and fungi: BF). Both base plates were sealed face-to-face with two layers of Parafilm® and an additional layer of plastic wrap (Kleenpack®). Controls of only bacteria (B), fungus alone (F), and PDA medium were also evaluated in triplicate. SPME fibers [50–30 μm Carboxen®/DVB/PDMS (2 cm), Agilent Technologies, Inc., Palo Alto, CA, USA] were inserted into the middle space of the two-Petri dish assembly through a hole and allowed to rest for 20 min to adsorb volatile compounds. This BVCs trapping was performed at 3, 4, 5 days of growth for the BF assay, and at 0, 3, 4, and 5 days of growth for B and F at 30 °C of incubation. SPME fibers were injected into the GC port and volatile compounds were thermally desorbed at 250 °C in a gas chromatograph (Agilent Technologies 6890 N) coupled to a mass analyzer (Agilent Technologies 5975). Helium gas was used as carrier gas (1.0 mL min−1) and a DB-5MS column (30 m length × 0.25 mm inner diameter × 0.25 μm film thickness; J&W) was used. A splitless injector was used to introduce the sample and the GC was operated following the conditions described in Méndez-Bravo et al. (2018). The MS was operated in electron impact (70 eV) mode with a source temperature of 230 °C and a continuous scan from 40 to 550 m/z. The mass spectra of volatile compounds were compared with those in the NIST/EPA/NIH Mass Spectrometry Library v2.0 d 2004 using a range of 70–99% similarity values. The relative abundance of the putatively annotated BVCs was expressed as adjusted peak area after subtracting peaks from control average chromatogram.
Statistical analysis
Data were processed with Kruskal–Wallis’s test for non-parametric data (p ≤ 0.01) for the mycelial growth inhibition (cm) of F. solani using the Wizard Software for Solutions 1.9.31 (250). For BVCs presence evaluation, one-way analyses of variance (ANOVA) were performed. Mycelial growth inhibition (cm) data were previously normalized with log (x + 1) function and analysis was performed with the Statgraphic Centurion XVI. Principal component analysis (PCA) and heat map designs were performed on XLSTAT 2018.7 statistical and data analysis solution software (Boston, USA), using the average values of the relative abundance (RA) of the peak area obtained for each compund.
Results and discussion
Isolation, identification, and antifungal activity against Fusarium solani
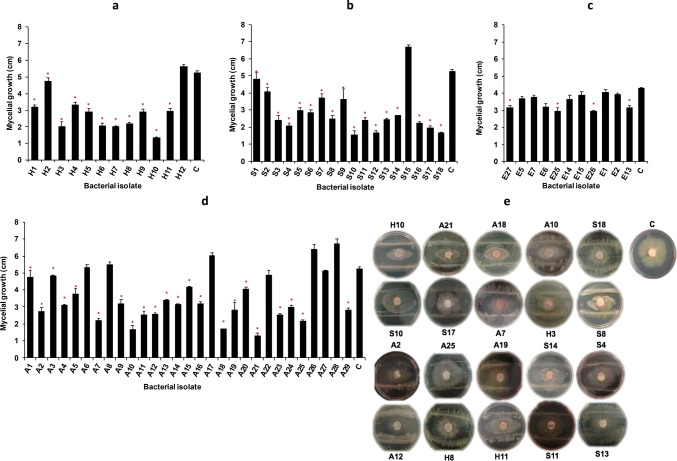
Seventy isolates were obtained from different environments: 18 from agricultural soil (S), 12 from hydrocarbon-contaminated soil (H), 29 from air (A), and 11 from soils of extremophile environments (E) (Table 1). For this first approach, Matrix-Assisted Laser Desorption/Ionization-time of flight mass spectrometry (MALDI-TOF MS) was used to identify the genus of the 70 isolates. Only 37 isolates, belonging to the Bacillus, Pseudomonas, Arthrobacter, Stenotrophomonas, Micrococcus, and Staphylococcus genera could be identified by MALDI-TOF MS, with scores ranging from 1.489 to 2.261. It should be mentioned that none of the samples from the extremophile environment (E) was identified by this technique, despite obtaining good quality mass spectra. The mycelial radial growth of F. solani was significantly reduced by 54 bacterial isolates (p ≤ 0.01) (Fig. 1). Some of the isolates (36%) from E showed antagonistic capacity (Fig. 1a), as well as 94% of the isolates from S (Fig. 1b), 97% of the isolates from H (Fig. 1c), and 76% of the isolates from A (Fig. 1d). Ten isolates showed > 60% fungal inhibition (IP), four of Bacillus (scores > 1.7), one Staphylococcus (score > 1.8), and the other five could not be identified. Isolates H10, A21, and A18 inhibited F. solani mycelial growth by more than 70%, two belong to Bacillus (score > 1.7), and A18 could not be identified (Supplementary Table S1). On the other hand, isolates A8, A17, A26, A28, S15, and H12 stimulated mycelial growth of F. solani and 35 isolates were not identified. Some Bacillus strains inhibit F. graminearum in 50–62.7% (Pan et al. 2015). In another study, 62 bacterial strains, belonging to Pseudomonas sp., Bacillus sp, and Microbacterium sp., inhibited F. culmorum growth by 15–40% (Mnasri et al. 2017). These percentages of inhibition are similar to some obtained in our work, with the exception of some isolates from this work, which presented more than 70% inhibition. This phenomenon could be due to volatile emission and/or difussible compounds (Cazorla et al. 2007; Compant et al. 2010; Kai et al. 2009).
Table 1.
Molecular identification by 16S rDNA sequencing, plant growth promotion, and hemolysis traits of bacterial isolates with antagonistic activity. Sequence closest matches were based on the NCBI database 16S ribosomal DNA sequences
| ID Bacteria | NCBI best match | Accesion number | Identity (%) | mg PO−34 L−1 | S | mg IAA L−1 | Hemolysis | |
|---|---|---|---|---|---|---|---|---|
| With l-tryptophan | Without l-tryptophan | |||||||
| H3 | Bacillus subtilis | MN298762 | 100 | 36.3 ± 2.0 | + | ND | 2.4 ± 3.4 | γ |
| H8 | Bacillus pacificus | MN262697 | 100 | 28.7 ± 2.3 | + | ND | ND | – |
| H10 | Bacillus subtilis | MN262696 | 100 | 22.3 ± 4.6 | + | ND | ND | – |
| H11 | Bacillus velezensis | MN262684 | 99 | 10.5 ± 1.5 | – | ND | 11.7 ± 0.8 | γ |
| A2 | Bacillus wiedmannii | MN262689 | 100 | 16.7 ± 3.7 | – | ND | ND | β |
| A7 | Bacillus subtilis | MN262690 | 100 | 8.3 ± 0.9 | – | ND | ND | β |
| A10 | Bacillus wiedmannii | MN262686 | 100 | 22.7 ± 6.8 | – | 7.8 ± 10.2 | 6.6 ± 1.7 | β |
| A12 | Bacillus wiedmannii | MN262688 | 100 | 21.0 ± 4.0 | – | 26.3 ± 6.8 | 67.7 ± 4.0 | β |
| A18 | Bacillus wiedmannii | MN262681 | 97 | 19.0 ± 2.6 | – | ND | ND | β |
| A19 | Bacillus circulans | MN262682 | 99 | 7.3 ± 0.3 | – | 7.6 ± 4.2 | 7.5 ± 0.6 | γ |
| A21 | Bacillus amyloliquefaciens | MN262693 | 100 | 10.7 ± 0.7 | – | ND | ND | γ |
| A25 | Bacillus pacificus | MN262699 | 100 | 12.7 ± 0.7 | + | ND | 3.4 ± 4.8 | γ |
| S4 | Bacillus subtilis | MN262694 | 100 | 15.3 ± 2.3 | + | ND | 2.4 ± 1.8 | β |
| S8 | Pseudomonas lactis | MN262691 | 95 | 17.7 ± 2.3 | + | ND | ND | β |
| S10 | Bacillus tequilensis | MN262685 | 100 | 9.0 ± 2.5 | – | ND | ND | γ |
| S11 | Bacillus amyloliquefaciens | MN262698 | 99 | 24.0 ± 2.6 | – | ND | ND | γ |
| S13 | Bacillus wiedmannii | MN262695 | 99 | 31.7 ± 9.9 | – | ND | ND | β |
| S14 | Bacillus wiedmannii | MN262687 | 100 | 33.0 ± 1.0 | + | 44.9 ± 4.3 | 3.9 ± 3.3 | γ |
| S17 | Bacillus tequilensis | MN262683 | 100 | 26.3 ± 3.2 | – | ND | ND | β |
| S18 | Bacillus wiedmannii | MN262692 | 100 | 17.0 ± 2.3 | + | ND | 6.5 ± 0.3 | β |
Average in mg of PO−34 L−1 or IAA L−1 with its standard error are shown. For siderophores (S) a qualitative result was taken (+)
ND no detected. Isolate H3 was sequenced two times MN298762-MN298763
Fig. 1.
Inhibition of F. solani mycelial growth (diameter in cm) by bacterial isolates from: a extremophile soils (strains’ names with E), b agricultural soil (S), c soils contaminated with hydrocarbons (EH), and d air samples (A). Average values are shown with standard error, * indicates significant difference relative to control (c at the extreme right of each plot) (Kruskal–Wallis’s test p ≤ 0.01). e Five-day culture assays of fungus (circle at the center) with bacterial isolates (inoculated in the lines 1.5 cm above and below the fungus circle). These isolates were based on a ≥ 40% inhibition
Molecular identification of bacterial isolates
After antagonism assays, 20 isolates presenting an IP ≥ 40% were selected for the second stage in which they were subjected to molecular identification (16S) and evaluated for their PGPB traits (Fig. 1e). None of the samples from E presented an IP higher than 40%. Hence, they were discarded for the following trials. Closest sequence matches, based on BLASTn similarity analysis, are presented in Table 1. Most of the isolates belonged to the Bacillus genus. Isolates H10, A21, and A18 with an IP > 70% were identified as Bacillus sp., with H10 and A21 corresponding to the genus identified by MALDI-TOF analysis. We found 19 strains of the Bacillus genus and one Pseudomonas. In addition to this, nine of the isolates that were not identified previously by MALDI-TOF were identified as Pseudomonas sp. and Bacillus sp. Isolates H8 and A12 with MALDI-TOF scores > 2, belong to the Bacillus genus as identified in both 16S and MALDI-TOF database, but the two techniques did not agree at the species level. Isolate H11 (score > 2) did not correspond with either the genus or the species determined previously by MALDI-TOF. Examples of bacterial antagonists of fungal pathogens isolated from ecological niches other than plant habitats include P. fluorescens, P. luteola, and B. brevis, isolated from the rhizosphere of Physalis peruviana, which effectively prevent F. oxysporum growth. Strain P. fluorescens significantly delayed the appearance of disease symptoms on P. peruviana plants (Urrea et al. 2011). In another study, 14 morphologically different strains (typical and not typical of Pseudomonadaceae) inhibited the growth of F. culmorum mycelia by more than 50% (Przemieniecki et al. 2019). Guevara et al. (2018) performed dual culture assays, using bacteria isolated from the rhizosphere of avocado, and identified species of Bacillus sp. with a high antagonistic capacity against F. euwallaceae, Graphium euwallaceae, Graphium sp, and Phytophthora cinnamomi (Guevara et al. 2018).
Evaluation of plant growth-promoting capacity and hemolysis analysis
Isolates H3, H8, S13, S14, and S17 solubilized 25 mg PO−34 L−1 or more, and the other 15 isolates solubilized 7.3–24.0 mg PO−34 L−1. Isolates A25, S8, S4, S18, S14, H3, H8, and H10 produced siderophores. Isolates A10, A12, A19, and S14 produced 7.6–44.9 mg IAA L−1 in a medium supplemented with l-tryptophan, and isolates A25, A10, A12, A19, H3, H11, S14, S18, and S4 produced 2.4–67.7 mg IAA L−1 in a medium without l-tryptophan (Table 1). These results indicate that Trp-dependent and Trp-independent pathways are used (Woodward and Bartel 2005). Species of Bacillus and Pseudomonas have been widely reported for their ability to improve plant growth, through the production of siderophores (Singh et al. 2014), phytohormones or signaling molecules (López-Bucio et al. 2007), phosphate solubilization, nitrogen fixation, and for their potential antimicrobial activity (Sarbadhikary and Mandal 2017). Bacillus spp., Pseudomonas spp. and Paenibacillus spp. strains have been shown to inhibit F. verticillioides growth by 45–85%, as well as being able to produce siderophores and auxins, with the strain Paenibacillus polymyxa an auxin-like compounds producer with 40 µmol L−1 (Figueroa-López et al. 2016). An analysis of bacterial properties among the growth-promoting properties, metabolic activity, and fungistatic properties against F. culmorum revealed that Pseudomonas putida PSDM3, Enterobacter sp. PSDM16, Advenella sp. PSDM17, and Proteus sp. PSDM21 were able to improve plant growth due to one or more of these mechanisms. Two of the tested bacterial strains, Proteus penneri PSDM6 and Proteus sp. PSDM7, produced IAA in a medium enriched with l-tryptophan at concentrations > 20 μl mL−1 (Przemieniecki et al. 2019). On the other hand, the hemolytic test in our study revealed 10 isolates with non-hemolytic (−) or γ-hemolytic effects (H3, H8, H10, H11, A19, A21, A25, S10, S11, and S14). None of the isolates exhibited α-hemolysis. The isolates exhibiting β-hemolysis were A2, A7, A10, A12, A18, S4, S8, S13, S17, and S18 (Table 1).
Bacterial volatile compounds activity evaluation
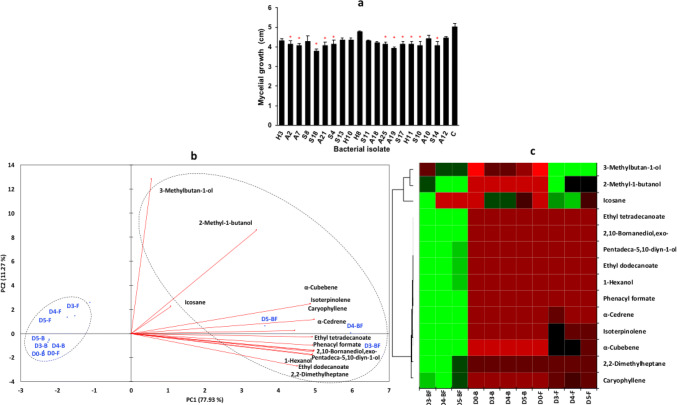
Of the 20 best bacterial isolates and with capacity as PGPB, only 11 strains significantly inhibited the mycelial growth (cm) of F. solani (p ≤ 0.05) (Fig. 2a), with an IP between 18–24%. Bacillus sp. S18 showed the highest IP (24%). These results suggest that its antagonistic capacity can be originated from its capacity to emit BVCs. Mnasri et al. (2017) reported bacteria of Pseudomonas, Bacillus, and Microbacterium genera capable of inhibiting (62–75%) the growth of F. culmorum via BVCs. On the other side, Pseudomonas sp. and Arthrobacter sp. have also been reported to present an antagonistic activity against P. cinnamomi, simultaneously promoting in vitro growth of Arabidopsis thaliana via BVCs (Méndez et al. 2018).
Fig. 2.
a Inhibition of F. solani mycelial growth (diameter in cm) by bacterial isolates. Mean values are shown with standard error. * indicates significant difference relative to control (c at the extreme right of each plot) (Kruskal–Wallis’s test p ≤ 0.01). b Principal component analysis (PCA) of the volatile compounds of the Bacillus S18 strain on day zero, 3, 4, and 5 (D0, D3, D4, D5). Some compounds were present in cultures of bacteria alone (B), fungus alone (F), or both together (BF). c Heatmap of the compounds abundance found in each treatment of the same strain at times D0, D3, D4, and D5. Green corresponds to the highest abundance, black medium and red, to the lowest
Analysis of BVCs emitted by isolate S18
From the analysis of the S18 strain volatile profile facing F. solani (BF) and S18 strain alone (B), 14 compounds appeared, i.e., they arose from the bacteria-fungi interaction (BF). This group comprises alcohols, ketones, sesquiterpenes, monoterpenoids, alkanes, and fatty acids with a similar percentage > 60%. Additionally, F. solani alone (F) produced its own profile of volatile compounds but at a lower abundance as compared with BF. These compounds are sesquiterpenes tentatively identified as α-cedrene, α-cubebene, and caryophyllene (Fig. 2b, c). Some of them have been reported in Fusarium sp. when induced by stress factors such as the presence of predators, parasites, and competitors (Kramer and Abraham 2012). In total, seven compounds were identified only in BF and were tentatively identified as ethyl tetradecanoate, ethyl dodecanoate, 2,10-bornanediol,exo-, pentadeca-5,10-diyn-1-ol, 2,2-dimethylheptane, 1-hexanol, and phenacyl formate. However, 3-methylbutan-1-ol, isoterpinolene, and icosane were found in volatile profiles from all treatments (B, F, and BF) within the same days but with different abundance. None of the compounds was found in samples from B, or at the beginning of the experiment (day 0). The PCA showed similar tendency among the compounds found in BF, although in some cases, they appeared in B or F, i.e., those from BF were grouped and appeared close to the total amount of detected compounds on the positive side of Component 1 (PC1 (+)) and B and F samples were placed on PC1 (−). Days 3, 4, and 5 of BVCs evaluation and day 0 for samples of F and B were grouped on PC1 (−), whereas samples from BF were grouped on PC1 (+) (Fig. 2b). The heatmap shows that the sesquiterpenes increased their abundance in the bacteria-fungi interaction (BF) (Fig. 2c) as a response to the antagonistic activity.
In another study, the main volatile compounds produced by a strain closely related to the genus Bacillus were ketones, aldehydes, alkyls, sulfoxides, pyrazines, and alcohols, which were able to inhibit P. cinnamomi mycelial growth by 76% (Méndez-Bravo et al. 2018). On the other hand, volatile compounds that are effective as antifungals, such as benzothiazole, cyclohexanol, n-decanal, dimethyl trisulfide, 2-ethyl-1-hexanol, and nonanal, inhibit mycelial growth, ascospore germination, and survival of Sclerotinia sclerotiorum, which causes white rot (Zhang and Fernando 2004). Some Bacillus species have a high capacity to emit BVCs against Ralstonia solanacearum and F. oxysporum, such as benzene, pentadecane, tetradecane, and some ketones (Raza et al. 2016; Yuan et al. 2012). Pseudomonas spp., due to their ability to emit antifungal and anti-oomycete BVCs such as 1-undecene and hydrogen cyanide, have been reported to inhibit the mycelial growth of Phytophthora infestans, Rhizoctonia solani, and Sclerotinia sclerotiorum (Elkahoui et al. 2015; Hunziker et al. 2015; Nandi et al. 2015).
Conclusion
Twenty bacterial isolates, belonging to the Bacillus and Pseudomonas genera, from different environments showed antagonistic activity versus F. solani. Bacillus amyloliquefaciens A21, Bacillus subtilis H10, and Bacillus wiedmannii A18 inhibited > 70% F. solani growth. Bacillus spp. S18, A19, and A21 produced volatile compounds that reduced F. solani mycelial growth in ≥ 20%. The volatile profile of Bacillus sp. S18 facing F. solani comprised ketones, sesquiterpenes, monoterpenoids, alkanes, and fatty acids. It is worth mentioning that the obtained volatile profiles were different among B, F, and BF treatments, suggesting a possible response to the interaction between bacteria and fungi via volatile compounds. These results emphasize the relevance of PGPB and their volatile compounds as biological control agents of phytopathogenic fungi and the potential use of Bacillus and Pseudomonas species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
A. Gutierrez-Santa Ana acknowledges the support through fellowship (720742) of Consejo Nacional de Ciencia y Tecnología (CONACYT), Mexico.
Author contributions
Conceptualization and experimental design were made by SMC-R and JBV-F. Experiments were mainly performed by AG-SA with the guidance, experiment design and help of JR-C (for chromatographical analysis and compound identification), MK (for MALDI-TOF identificaction) and JBV-F (MALDI-TOF identification, 16S identification and statistical analysis). HAC-C helped with microbial culture and confrontation of bacteria against F. solani. First draft was written by AG-SA and all authors read, revised, commented on previous versions. All authors whose names appear on the submission. (1) made substantial contributions to the design of the work, the acquisition, analysis, or interpretation of data; (2) approved the version to be published; and (3) agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
The present study was funded by the Fondo Institucional de Fomento Regional para el Desarrollo Científico, Tecnológico y de Innovación (FORDECYT) under the project 292399.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Alexander DB, Zuberer DA. Use of chromeazurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol Fertil Soils. 1991;12(1):39–45. doi: 10.1007/BF00369386. [DOI] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- APHA/AWWA/WEF (2012) Standard Methods for the Examination of Water and Wastewater. Standard Methods, 51. ISBN 9780875532356
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57(1):233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Bollet C, Gevaudan MJ, de Lamballerie X, Zandotti C, de Micco P. A simple method for the isolation of chromosomal DNA from gram positive or acid-fast bacteria. Nucleic Acids Res. 1991;19(8):8893. doi: 10.1093/nar/19.8.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla FM, Romero D, Pérez-García A, Lugtenberg BJJ, Vicente A De, Bloemberg G. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J Appl Microbiol. 2007;103(5):1950–1959. doi: 10.1111/j.1365-2672.2007.03433.x. [DOI] [PubMed] [Google Scholar]
- Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42(5):669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- Dunlap CA, Lueschow S, Carrillo D, Rooney AP. Screening of bacteria for antagonistic activity against phytopathogens of avocados. Plant Gene. 2017;11:17–22. doi: 10.1016/j.plgene.2016.11.004. [DOI] [Google Scholar]
- Elkahoui S, Djébali N, Yaich N, Azaiez S, Hammami M, Essid R, Limam F. Antifungal activity of volatile compounds-producing Pseudomonas P2 strain against Rhizoctonia solani. World J Microbiol Biotechnol. 2015;31(1):175–185. doi: 10.1007/s11274-014-1772-3. [DOI] [PubMed] [Google Scholar]
- Figueroa-López AM, Cordero-Ramírez JD, Martínez-Álvarez JC, López-Meyer M, Lizárraga-Sánchez GJ, Félix-Gastélum R, Maldonado-Mendoza IE. Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioides. SpringerPlus. 2016;5(1):330. doi: 10.1186/s40064-016-1780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott’s diagnostic microbiology. 11. St. Louis: Mosby Inc.; 2002. [Google Scholar]
- Guevara-Avendaño E, Carrillo JD, Ndinga-Muniania C, Moreno K, Méndez-Bravo A, Guerrero-Analco JA, Reverchon F. Antifungal activity of avocado rhizobacteria against Fusarium euwallaceae and Graphium spp., associated with Euwallacea spp. nr. fornicatus, and Phytophthora cinnamomi. Antonie van Leeuwenhoek Int J Gen Mol Microbiol. 2018;111(4):563–572. doi: 10.1007/s10482-017-0977-5. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM (2001) The polymerase chain reaction as a tool to investigate microbial diversity. In Rochelle PA (ed) Environmental molecular microbiology: protocols and applications.
- Hunziker L, Bönisch D, Groenhagen U, Bailly A, Schulz S, Weisskopf L. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl Environ Microbiol. 2015;81(3):821–830. doi: 10.1128/AEM.02999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris HA, Labuschagne N, Korsten L. Screening rhizobacteria for biological control of Fusarium root and crown rot of sorghum in Ethiopia. Biol Control. 2007;40(1):97–106. doi: 10.1016/j.biocontrol.2006.07.017. [DOI] [Google Scholar]
- Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81(6):1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- Kramer R, Abraham W. Volatile sesquiterpenes from fungi: what are they good for? Phytochem Rev. 2012;11(March):15–37. doi: 10.1007/s11101-011-9216-2. [DOI] [Google Scholar]
- López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farias-Rodríguez R, Macías-Rodríguez L. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant-Microbe Interact. 2007;20(2):207–217. doi: 10.1094/MPMI-20-2-0207. [DOI] [PubMed] [Google Scholar]
- Méndez-Bravo A, Cortazar-Murillo EM, Guevara-Avendaño E, Ceballos-Luna O, Rodríguez-Haas B, Kiel-Martínez AL, Reverchon F. Plant growth-promoting rhizobacteria associated with avocado display antagonistic activity against Phytophthora cinnamomi through volatile emissions. PLoS One. 2018;13(3):1–18. doi: 10.1371/journal.pone.0194665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnasri N, Chennaoui C, Gargouri S, Mhamdi R, Hessini K, Elkahoui S, Djébali N. Efficacy of some rhizospheric and endophytic bacteria in vitro and as seed coating for the control of Fusarium culmorum infecting durum wheat in Tunisia. Eur J Plant Pathol. 2017;147(3):501–515. doi: 10.1007/s10658-016-1018-3. [DOI] [Google Scholar]
- Nandi M, Selin C, Brassinga AKC, Belmonte MF, Fernando WGD, Loewen PC, De Kievit TR. Pyrrolnitrin and hydrogen cyanide production by Pseudomonas chlororaphis strain PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans. PLoS One. 2015;10(4):1–19. doi: 10.1371/journal.pone.0123184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Mionetto A, Tiscornia S, Bettucci L. Endophytic bacteria from wheat grain as biocontrol agents of Fusarium graminearum and deoxynivalenol production in wheat. Mycotoxin Res. 2015;31(3):137–143. doi: 10.1007/s12550-015-0224-8. [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110(16):6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- Przemieniecki SW, Pawe T, Kotlarz K, Krawczyk K, Damszel M, Pszczó A, Mastalerz J. Bacteria isolated from treated wastewater for biofertilization and crop protection against Fusarium spp. pathogens. J Soil Sci Plant Nutr. 2019;19:1–11. doi: 10.1007/s42729-018-0001-9. [DOI] [Google Scholar]
- Raza W, Ling N, Yang L, Huang Q, Shen Q. Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci Rep. 2016;6:24856. doi: 10.1038/srep24856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca M, Prieto M, Almuzara M, Barberis C, Vay C. Manual for interpretation of MALDI-TOF results (Bruker Daltonics) Argentina: Buenos Aires; 2018. [Google Scholar]
- Sarbadhikary SB, Mandal NC. Field application of two plant growth promoting rhizobacteria with potent antifungal properties. Rhizosphere. 2017;3(1):170–175. doi: 10.1016/j.rhisph.2017.04.014. [DOI] [Google Scholar]
- Siddiqui ZA. PGPR: biocontrol and biofertilization. Aligarh: Department of Botany, Aligarh Muslim University; 2006. [Google Scholar]
- Singh S, Gupta G, Khare E, Behal KK, Arora NK. Effect of enrichment material on the shelf life and field efficiency of bioformulation of Rhizobium sp. and P-solubilizing Pseudomonas fluorescens. Sci Res Rep. 2014;4(1):44–50. [Google Scholar]
- Stout J, Huang SW, Calvin L, Lucier G, Perez A, Pollack S (2014) NAFTA trade in fruits and vegetables. In: Wu HS (eds) Global trade patterns in fruits and vegetables. Agriculture and Trade Report Number: WRS-04-06; United States Department of Agriculture: Washington, DC, USA, pp 39–51
- Sundara Rao WVB, Sinha MK. Phosphate dissolving micro-organisms in the soil and rhizosphere. Indian J Agric Sci. 1963;33(1963):272–278. [Google Scholar]
- Sundara Rao WVB, Sinha MK. Phosphate dissolving organisms in the soil and the rhizosphere. Indian J Agric Sci. 1963;33:272–278. [Google Scholar]
- Umeda C, Eskalen A, Paine T (2016) Chapter 26. Polyphagous shot hole borer and Fusarium dieback in California. In: Paine TD, Lieutier F (eds) Insects and diseases of mediterranean forest systems. Springer International Publishing, Switzerland. 10.1007/978-3-319-24744-1_26
- Urrea R, Cabezas L, Sierra R, Cárdenas M, Restrepo S, Jiménez P. Selection of antagonistic bacteria isolated from the Physalis peruviana rhizosphere against Fusarium oxysporum. J Appl Microbiol. 2011;111(3):707–716. doi: 10.1111/j.1365-2672.2011.05092.x. [DOI] [PubMed] [Google Scholar]
- Vinodkumar S, Nakkeeran S, Renukadevi P, Malathi VG. Biocontrol potentials of antimicrobial peptide producing Bacillus species: multifaceted antagonists for the management of stem rot of carnation caused by Sclerotinia sclerotiorum. Front Microbiol. 2017;8(MAR):1–13. doi: 10.3389/fmicb.2017.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM, Van Pelt JA, Mavrodi DV, Pieterse CMJ, Bakker PAHM, Van Loon LC. Induced systemic resistance (ISR) in Arabidopsis against Pseudomonas syringae pv. tomato by 2, 4-diacetylphloroglucinol (DAPG)-producing Pseudomonas. Phytopathology. 2004;94:S108. doi: 10.1094/PHYTO-08-11-0222. [DOI] [PubMed] [Google Scholar]
- Whipps JM. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005 doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Raza W, Shen Q, Huang Q. Antifungal Activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl Environ Microbiol. 2012;78(16):5942–5944. doi: 10.1128/aem.01357-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fernando WGD. Presence of biosynthetic genes for phenazine-1-carboxylic acid and 2,4-diacetylphloroglucinol and pyrrolnitrin in Pseudomonas chlororaphis strain PA-23. Can J Plant Pathol. 2004;26:430–431. doi: 10.1080/07060660409507173. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.