Abstract
Study objective:
To employ systems biology-based machine learning to identify biologic processes over-represented with genetic variants (gene enrichment) implicated in post-surgical pain.
Design:
Informed systems biology based integrative computational analyses
Setting:
Pediatric research and teaching institution
Interventions:
Pubmed search (01/01/2001-10/31/2017) was performed to identify “training” genes associated with postoperative pain in humans. Candidate genes were identified and prioritized using Toppgene suite, based on functional enrichment using several gene ontology annotations, and curated gene sets associated with mouse phenotype-knockout studies.
Measurements:
Computationally top-ranked candidate genes and literature-curated genes were included in pathway enrichment analyses. Hierarchical clustering was used to visualize select functional enrichment results between the two phenotypes.
Main results:
Literature review identified 38 training genes associated with postoperative pain and 31 with CPSP. We identified 2610 prioritized novel candidate genes likely associated with acute and chronic postsurgical pain, the top 10th percentile jointly enriched (p 0.05; Benjamini-Hochberg correction) several pathways, topmost being cAMP response element-binding protein and ion channel pathways. Heat maps demonstrated enrichment of inflammatory/drug metabolism processes in acute postoperative pain and immune mechanisms in CPSP.
Conclusion:
High interindividual variability in pain responses immediately after surgery and risk for CPSP suggests genetic susceptibility. Lack of large homogenous sample sizes have led to underpowered genetic association studies. Systems biology can be leveraged to integrate genetic-level data with biologic processes to generate prioritized candidate gene lists and understand novel biological pathways involved in acute postoperative pain and CPSP. Such data would be key to informing future polygenic studies with targeted genome wide profiling. This study demonstrates the utility of functional annotation - based prioritization and enrichment approaches and identifies novel genes and unique/shared biological processes involved in acute and chronic postoperative pain. Results provide framework for future targeted genetic profiling of CPSP risk, to enable preventive and therapeutic approaches.
Keywords: postoperative pain, CPSP, genetics, enrichment, systems biology, pathways
1. Introduction
Post-surgical pain (PoP) afflicts more than 230 million people undergoing surgery every year worldwide.[1] Despite significant advances in perioperative medicine, postoperative pain remains undertreated in up to 50% of patients.[2] Inadequately treated acute pain is a strong predictor for development of long-standing chronic post-surgical pain (CPSP) [3] - a known risk factor for prescription opioid abuse [4] and a major socioeconomic problem, with annual direct and indirect costs of ≈$43,000 US annually per patient[5]. Reasons for undertreatment of pain include limited translation of basic and clinical scientific findings into clinical practice [6] as well as inadequate understanding of the interindividual variability in pain perception. Postoperative pain involves widespread transcriptional dysregulation and sensitization throughout the pain neuraxis[7] involving multiple signaling and modulatory pathways[8] influenced by genes. There is a plethora of hypothesis driven candidate gene approaches and few genome wide studies evaluating genetic influences on acute and chronic post-surgical pain. While genetic variants explain some individual differences in pain perception and postoperative pain responses,[9–11] finding the genetic “holy grail” has largely remained elusive,[12, 13] partly due to lack of replicability[14], and inconsistent findings[15, 16] from genetic association studies. This is partly due to difficulty in achieving the large sample cohorts needed to overcome heterogeneity in surgical pain types, surgical cohorts, population differences, treatment differences etc. Moreover, the variability seen in post-surgical pain does not seem to be driven by single variants with large effect. Instead, the genetic landscape suggests minor contributions from a large number of genetic variants.[17] Identification and prioritization of gene sets is hence key to informing future replicable studies evaluating polygenic risk scores, thus allowing improved targeted approaches that can decrease the statistical burden of genome-wide approaches.
To achieve the aforesaid objectives, we used systems biology-based machine learning approaches to identify biologic processes over-represented with genetic variants (gene enrichment) implicated in post-surgical pain and its maintenance. Systems biology is a scientific approach that combines the principles of engineering, mathematics, physics, and computer science with extensive experimental data from open-source data to develop a conceptual understanding of biological phenomena, permitting characterization of complex biological phenotypes.[18] Systems biology approaches integrate genetic-level data with biologic processes, pathways, and networks, and allow potential targets/mechanisms generated by these different platforms to be analyzed in their biological context.[19] Here, we used this approach to develop a comprehensive understanding of the biological processes involved in acute postoperative pain and the maintenance of pain after surgery. Systems-biology based pathway analyses have been previously used to study genetics of chronic pain conditions [17, 20, 21] and traumatic brain injury[22], but not postoperative pain or CPSP. Our goals were to identify and prioritize novel candidate genes in postoperative pain and to gain a better understanding of the overlapping and unique genomic architecture and biologic processes underlying acute and chronic pain following surgery. The rationale is that the results will provide a framework for improved and efficient targeted polygenic profiling to enable prediction and development of preventive and therapeutic strategies for better control of postoperative pain and CPSP.
2. Materials and Methods
In order to perform an informed systems biology based integrative computational analyses, we first compiled a “training set” of genes, which were identified by performing an extensive literature review of published associations of variant association with postoperative pain phenotypes. We focused on the time frame after 2001, given the advent of new technology and increase in genetic association studies that began in the 21st century.[23] Next, we used the Transcriptome Ontology Pathway PubMed based prioritization of Genes (ToppGene) Suite[24] for candidate gene prioritization. This was followed by functional enrichment of top-ranked candidate and training set of genes using ToppFun application of the ToppGene Suite, as detailed below.
2.1. Literature review: “Training set” gene list search strategy
To generate the “training set” of genes for each phenotype, we conducted a search in Pubmed and Medline (accessed in 2018) using the following search string (genetic association OR polymorphism OR variant OR “genotype” OR SNP) AND (“postoperative pain” OR “postsurgical pain” OR “post-operative pain” OR “post-surgical pain” OR “postoperative analgesia” OR “postoperative opioid”) for postoperative pain/CPSP genetic associations. Searches were limited to English language articles and human studies (including clinical study, clinical trial, multicenter study, observational study and twin study) using filters. The search was conducted from 2001/01/01 to 2017/12/31. Inclusion criteria for articles required that each article evaluate the association of single nucleotide polymorphisms (SNPs) with pain outcomes after surgery. Studies with SNPs (and gene names) which were reported as significantly associated with postoperative pain phenotypes were included. Since this step needed to be sensitive, individual study specified critical p-value thresholds were accepted for statistical significance for associations. The authors (VC and MA) reviewed the abstracts and full-texts of the articles to extract information regarding genes studied, the outcomes studied, genetic variants reported to be significantly associated with the outcomes of interest, and surgical cohort in the case of postoperative pain. Post-surgical pain outcomes were classified as acute if the outcome investigated was immediately after or within seven days of surgery, and classified as CPSP in accordance with the definition proposed by Werner and Kongsgaard (presence of postoperative pain at least 3 months after surgery).[25] Cohorts were classified as adult/pediatric according to age groups included in the study. The CPSP genetic association cohorts and studies are further described in a recent meta-analysis published by our group.[26] Articles evaluating the association of genetics with non-clinical outcomes in humans (e.g., pathology post-mortem) or evaluating the association of gene expression or protein/biomarker levels with outcomes were excluded. Data collected was independently checked by another reviewer (SG: see acknowledgements) and discrepancies resolved by discussion.
2.2. Candidate gene discovery and prioritization.
We used the “training set” lists to identify and rank novel genes (hereafter referred to as “candidate set”), based on their similarity (“guilt by association”) to the curated training sets of genes using ToppFun application of the Toppgene suite.[24] ToppGene Suite is a computational platform for gene list enrichment analyses and machine learning based candidate gene ranking. The knowledge base for ToppGene Suite is a comprehensive collection (more than 12 million gene annotations) of diverse gene annotation libraries compiled from a variety of publicly available gene annotation databases and resources. Although there are several enrichment analyses tools available, the strength of ToppGene is its extensive and relatively up to date collection of functional annotations compared to other system biology tools.[27]
The “training set” of genetic loci serves as input to the system. Fourteen gene ontology (GO) annotation categories downloaded from NCBI Entrez Gene ftp site[28] were used by Toppfun to do this ranking, which include 1) molecular function, 2) biological processes, 3) cellular component, 4) human phenotype, 5) mouse phenotype, 6) pathways, by combining data from KEGG,[29], BioCyc,[30] etc.; 7) protein-protein interactions, 8) co-citations (PMIDs) in PubMed, 9) protein functional domains, 10) transcription factor binding sites, 11) microRNAs, 12) gene tissue expressions obtained from the NCBI gene expression Omnibus (GEO), 13) drug-gene and 14) disease-gene associations. Similarity scores were generated for each annotation by comparing the enriched terms in the training set with the annotations of each of the candidate genes. Similarity scores are based on fuzzy measure logic for categorical variables while for continuous variable annotations, similarity scores are based on Pearson correlation coefficients. The similarity scores are combined into an overall score using statistical meta-analysis, as described by Chen et. Al.[24]
2.3. Shared and unique processes/pathways
We used the Toppfun application to perform gene list functional enrichment analysis and determine unique phenotype-specific biological processes and pathways enriched by the “training set and computationally top-ranked (top 10th percentile) “candidate set” of genes. We also used gene enrichment to identify shared pathways, enriched by genes (top 10% candidate set and training) associated with both phenotypes. Biologic processes associated at a p value of 0.05 (FDR-corrected - Benjamini-Hochberg) were considered to be significantly enriched by the gene sets. Hierarchical clustering was then used to visualize select functional enrichment results. Enrichment plot and heat map were generated using Cytoscape (Cytoscape.org) and Morpheus software (https://software.broadinstitute.org/morpheus) respectively.
3. Results
3.1. Literature review - training set
Details of the studies evaluated along with information about outcomes studied and SNPs reported to be significantly associated with the phenotypes of interest, are provided in Supplementary Table 1. We identified 5 genome wide studies evaluating the phenotypes of interest (marked by “GWAS” in bold in Supplementary Table 1).[31–35] The final number of studies that were evaluable was 109 for postoperative pain; of these 8 were conducted in pediatric cohorts and 19 identified the phenotype to be CPSP. The training sets included 38 genes for acute postoperative pain and 31 for CPSP. Literature-curated training gene lists with significant variant associations with acute and chronic postoperative pain phenotypes, and the pathways they enrich, are provided in Figure 1. Twelve genes had significant variant associations with both acute and chronic post-surgical pain - they significantly enriched seven pathways including G-protein signaling, ligand-binding interactions, serotonin receptor and cAMP signaling pathways.
Figure 1:
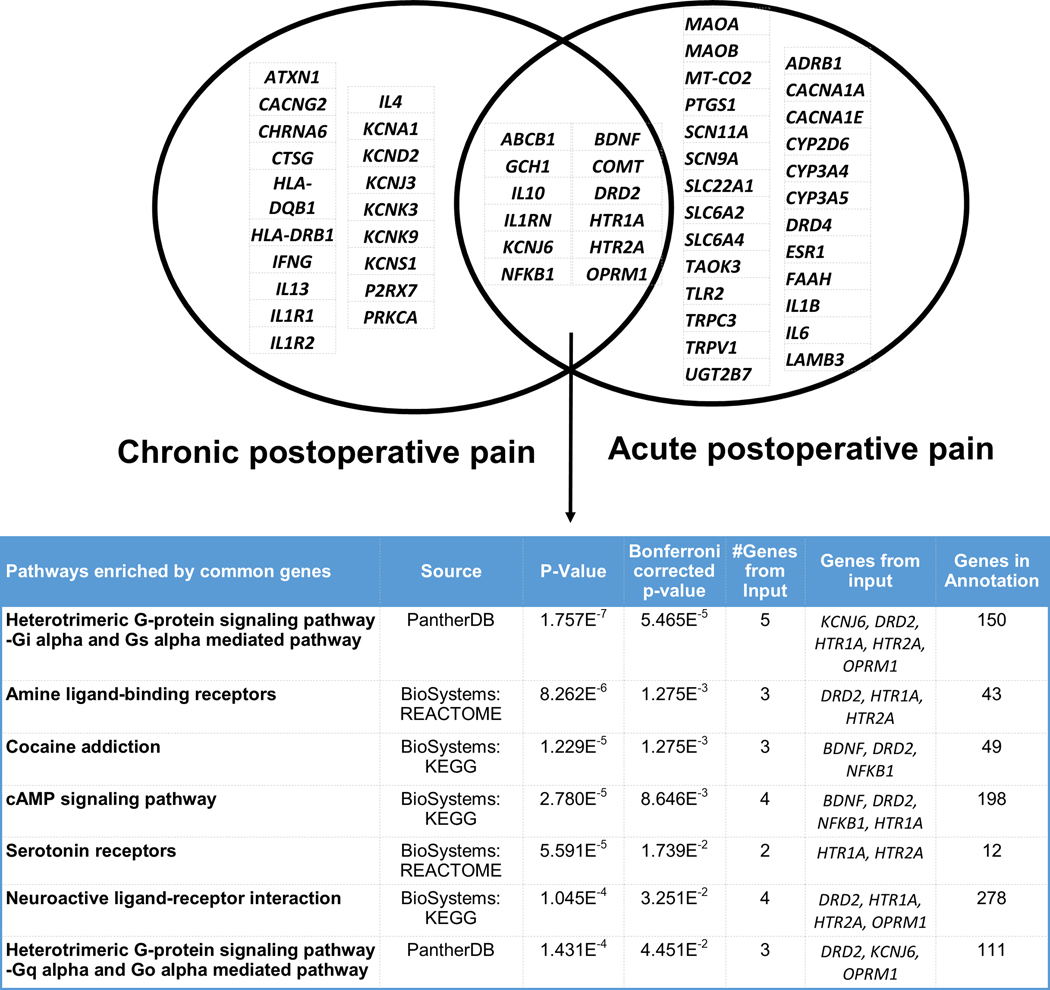
Venn diagram of literature-curated training genes and enriched pathways associated with acute postoperative pain (PoP) and chronic postoperative pain (CPSP) phenotypes. Upper panel shows the manually curated training gene lists, and the lower panel shows the top enriched pathways. Among the genes included solely in PoP, several had functions influencing drug metabolism (UGT2B7, MAOB, CYP3A4, CYP3A5, CYP2D6), inflammation (IL1B, IL6), ion channels (SCN11A, SCN9A, CACNA1A etc.). Other cytokine gene (IL-10/IL-1R) variants were associated with both acute and CPSP, while IL12, IL1 and IL17 variants were involved in CPSP only. There were 12 genes associated with both phenotypes. Common training genes were overrepresented in G-protein signaling, cAMP signaling, neuroactive ligand interaction and serotoninergic pathways (unadjusted p-value and Bonferroni adjusted p values for enrichment are provided). Gene symbol abbreviations: ABCB1 (ATP binding cassette subfamily B member 1), ADRB1 (adrenoceptor beta 1), BDNF (brain derived neurotrophic factor), CACNA1A (calcium voltage-gated channel subunit alpha1 A), CACNA1E (calcium voltage-gated channel subunit alpha1 E), COMT (catechol-O-methyltransferase), CYP2D6 (cytochrome P450 family 2 subfamily D member 6), CYP3A4 (cytochrome P450 family 3 subfamily A member 4), CYP3A5 (cytochrome P450 family 3 subfamily A member 5), DRD2 (dopamine receptor D2), DRD4 (dopamine receptor D4), ESR1 (estrogen receptor 1), FAAH (fatty acid amide hydrolase), GCH1 (GTP cyclohydrolase 1), HTR1A (5-hydroxytryptamine receptor 1A), HTR2A (5-hydroxytryptamine receptor 2A), IL10 (interleukin 10), IL1B (interleukin 1 beta), IL1RN (interleukin 1 receptor antagonist), IL6 (interleukin 6), KCNJ6 (potassium inwardly rectifying channel subfamily J member 6), LAMB3 (laminin subunit beta 3), MAOA (monoamine oxidase A), MAOB (monoamine oxidase B), MT-CO2 (cytochrome c oxidase subunit II), NFKB1 (nuclear factor kappa B subunit 1), OPRM1 (opioid receptor mu 1), PTGS1 (prostaglandin-endoperoxide synthase 1), SCN11A (sodium voltage-gated channel alpha subunit 11), SCN9A (sodium voltage-gated channel alpha subunit 9), SLC22A1 (solute carrier family 22 member 1), SLC6A2 (solute carrier family 6 member 2), SLC6A4 (solute carrier family 6 member 4), TAOK3 (TAO kinase 3), TLR2 (toll like receptor 2), TRPC3 (transient receptor potential cation channel subfamily C member 3), TRPV1 (transient receptor potential cation channel subfamily V member 1), UGT2B7 (UDP glucuronosyl transferase family 2 member B7); ATXN1 (ataxin 1); CACNG2 (calcium voltage-gated channel auxiliary subunit gamma 2); CHRNA6 (cholinergic receptor nicotinic alpha 6 subunit); CTSG (cathepsin G); HLA-DQB1 (major histocompatibility complex, class II, DQ beta 1); HLA-DRB1 (major histocompatibility complex, class II, DR beta 1); IFNG (interferon gamma); IL13 (interleukin 13); IL1R1 (interleukin 1 receptor type 1); IL1R2 (interleukin 1 receptor type 2); IL4 (interleukin 4); KCNA1 (potassium voltage-gated channel subfamily A member 1); KCND2 (potassium voltage-gated channel subfamily D member 2); KCNJ3 (potassium inwardly rectifying channel subfamily J member 3); KCNK3 (potassium two pore domain channel subfamily K member 3); KCNK9 (potassium two pore domain channel subfamily K member 9); KCNS1 (potassium voltage-gated channel modifier subfamily S member 1); P2RX7 (purinergic receptor P2X 7); PRKCA (protein kinase C alpha)
3.2. Candidate sets of genes
Candidate set of genes associated with postoperative pain were identified based on similarity scores for functional annotations using the manually compiled training list of genes. We identified several novel candidate genes not previously reported in human gene-association studies. The top 10% ranked candidate gene sets with highest similarity scores for genes with reported associations with acute and chronic postsurgical pain comprised 130 and 131 genes respectively. The intersection of top ranked candidates for acute and chronic post-surgical pain yielded 82 genes. As expected, the pathways enriched by the 82 genes in the prioritized candidate set common to both phenotypes included pathways that were found to be enriched by previously reported training genes common to the two phenotypes (Table 1). In addition, novel pathways were significantly enriched, notably those involved in immune/inflammatory processes (mitogen-activated protein kinase (MAPK), cytokine, tumor necrosis factor (TNF), T cell, toll like receptor (TLR) and Transforming growth factor beta (TGF-β)-beta signaling) and oxytocin and estrogen signaling.
Table 1:
Pathways significantly enriched by top 10% candidate gene sets common to acute and chronic post-surgical pain. (Benjamini-Hochberg adjusted p<0.05)
| Pathway | Top 10% candidate gene sets enriching the pathway shared by both phenotypes |
|---|---|
| MAPK signaling pathway | AKT1,CACNA1B,CACNA1C,CACNA1S,EGFR,FAS,IL1A,MAP2K1,MAPK1,MAPK3,MAPK8,NTRK2,RELA,TGFB1,TGFB2,TGFB3,TGFBR1,TGFBR2,TNF |
| HIF-1 signaling pathway | AKT1,CYBB,EGFR,MAP2K1,MAPK1,MAPK3,RELA,TLR4 |
| Calcium signaling pathway | ADORA2A,ADRB2,CACNA1B,CACNA1C,CACNA1S,CALM1,CHRM2,EGFR,F2R,GNAQ,GNAS,GRIN1,GRIN2A,GRIN2D,GRM1,GRM5,HTR2C,HTR7,NOS1,NTSR1,P2RX3,P2RX4,RYR1 |
| Th17 cell differentiation | IL2,JAK2,MAPK1,MAPK3,MAPK8,RELA,TGFB1,TGFBR1,TGFBR2 |
| Th1 and Th2 cell differentiation | IL12A,IL12B,IL2,JAK2,MAPK1,MAPK3,MAPK8,RELA |
| Cytokine-cytokine receptor interaction | CCR2,CXCL10,EGFR,FAS,IL12A,IL12B,IL18,IL1A,IL2,NGFR,TGFB1,TGFB2,TGFB3,TGFBR1,TGFBR2,TNF |
| TNF signaling pathway | AKT1,CXCL10,FAS,MAP2K1,MAPK1,MAPK3,MAPK8,PTGS2,RELA,TNF |
| Neuroactive ligand-receptor interaction | ADORA1,ADORA2A,ADRA2C,ADRB2,CHRM2,CHRNA4,CHRNB2,DRD3,F2R,GABBR1,GABRA2,GRIA1,GRIA2,GRIA3,GRIA4,GRIK1,GRIK5,GRIN1,GRIN2A,GRIN2B,GRIN2D,GRM1,GRM5,GRM7,HTR2C,HTR7,NTSR1,OPRD1,OPRK1,P2RX3,P2RX4 |
| Serotonergic synapse | CACNA1B,CACNA1C,CACNA1S,GNAQ,GNAS,HTR2C,HTR3A,HTR3B,HTR7,MAP2K1,MAPK1,MAPK3,PTGS2 |
| Toll-like receptor signaling pathway | AKT1,CXCL10,IL12A,IL12B,IRAK1,MAP2K1,MAPK1,MAPK3,MAPK8,MYD88,RELA,TLR4,TNF |
| cGMP-PKG signaling pathway | ADORA1,ADRA2C,ADRB2,AKT1,CACNA1C,CACNA1S,CALM1,GNAQ,MAP2K1,MAPK1,MAPK3,OPRD1 |
| Oxytocin signaling pathway | CACNA1C,CACNA1S,CALM1,EGFR,GNAQ,GNAS,MAP2K1,MAPK1,MAPK3,PTGS2,RYR1 |
| cAMP signaling pathway | ADORA1,ADORA2A,ADRB2,AKT1,CACNA1C,CACNA1S,CALM1,CFTR,CHRM2,F2R,GABBR1,GNAS,GRIA1,GRIA2,GRIA3,GRIA4,GRIN1,GRIN2A,GRIN2B,GRIN2D,HCN2,MAP2K1,MAPK1,MAPK3,MAPK8,RELA |
| Dopaminergic synapse | AKT1,CACNA1B,CACNA1C,CALM1,DRD3,GNAQ,GNAS,GRIA1,GRIA2,GRIA3,GRIA4,GRIN2A,GRIN2B,MAPK8,SLC6A3,TH |
| Ras signaling pathway | AKT1,CALM1,EGFR,GRIN1,GRIN2A,GRIN2B,HTR7,MAP2K1,MAPK1,MAPK3,MAPK8,NGFR,RELA |
| T cell receptor signaling pathway | AKT1,FYN,IL2,MAP2K1,MAPK1,MAPK3,RELA,TNF |
| Glutamatergic synapse | CACNA1C,DLG4,GNAQ,GNAS,GRIA1,GRIA2,GRIA3,GRIA4,GRIK1,GRIK5,GRIN1,GRIN2A,GRIN2B,GRIN2D,GRM1,GRM5,GRM7,MAPK1,MAPK3 |
| Neurotrophin signaling pathway | AKT1,CALM1,IRAK1,MAP2K1,MAPK1,MAPK3,MAPK8,NGFR,NTRK2,RELA |
| Estrogen signaling pathway | AKT1,CALM1,EGFR,GABBR1,GNAQ,GNAS,GRM1,MAP2K1,MAPK1,MAPK3 |
| Cholinergic synapse | AKT1,CACNA1B,CACNA1C,CACNA1S,CHRM2,CHRNA4,CHRNB2,FYN,GNAQ,JAK2,MAP2K1,MAPK1,MAPK3,SLC18A3 |
| Long-term depression | GNAQ,GNAS,GRIA1,GRIA2,GRIA3,GRM1,MAP2K1,MAPK1,MAPK3,NOS1,RYR1 |
| Long-term potentiation | CACNA1C,CALM1,GNAQ,GRIA1,GRIA2,GRIN1,GRIN2A,GRIN2B,GRIN2D,GRM1,GRM5,MAP2K1,MAPK1,MAPK3 |
| Apelin signaling pathway | AKT1,CALM1,GNAQ,MAP2K1,MAPK1,MAPK3,NOS1,RYR1,TGFBR1 |
| Nicotine addiction | CACNA1B,CHRNA4,CHRNB2,GABRA2,GRIA1,GRIA2,GRIA3,GRIA4,GRIN1,GRIN2A,GRIN2B,GRIN2D |
| Prolactin signaling pathway | AKT1,JAK2,MAP2K1,MAPK1,MAPK3,MAPK8,RELA,TH |
| Circadian entrainment | CACNA1C,CALM1,GNAQ,GNAS,GRIA1,GRIA2,GRIA3,GRIA4,GRIN1,GRIN2A,GRIN2B,GRIN2D,MAPK1,MAPK3,NOS1,RYR1 |
| Alzheimer’s disease | CACNA1C,CACNA1S,CALM1,FAS,GNAQ,GRIN1,GRIN2A,GRIN2B,GRIN2D,MAPK1,MAPK3,NOS1,TNF |
| TGF-beta signaling pathway | MAPK1,MAPK3,TGFB1,TGFB2,TGFB3,TGFBR1,TGFBR2,TNF |
| Cocaine addiction | DLG4,GNAS,GRIA2,GRIN1,GRIN2A,GRIN2B,GRIN2D,RELA,SLC6A3,TH |
| Alcoholism | ADORA2A,CALM1,GNAS,GRIN1,GRIN2A,GRIN2B,GRIN2D,MAP2K1,MAPK1,MAPK3,NTRK2,SLC6A3,TH |
| Amphetamine addiction | CACNA1C,CALM1,GNAS,GRIA1,GRIA2,GRIA3,GRIA4,GRIN1,GRIN2A,GRIN2B,GRIN2D,SLC6A3,TH |
| Retrograde endocannabinoid signaling | CACNA1B,CACNA1C,CACNA1S,GABRA2,GNAQ,GRIA1,GRIA2,GRIA3,GRIA4,GRM1,GRM5,MAPK1,MAPK3,MAPK8,PTGS2 |
| EGFR tyrosine kinase inhibitor resistance | AKT1,EGFR,JAK2,MAP2K1,MAPK1,MAPK3 |
| Natural killer cell mediated cytotoxicity | FAS,FYN,MAP2K1,MAPK1,MAPK3,TNF |
| Inflammatory mediator regulation of TRP channels | CALM1,GNAQ,GNAS,HTR2C,MAPK8 |
A network view of the gene sets and enriched pathways by training and prioritized candidate sets of genes is presented in Figure 2. The complex nature and inter-dependence of mechanisms contributing to acute and chronic pain phenotypes is evident from the shared gene networks. We also observed that there are potentially novel processes defined by the candidate genes that were not enriched by training genes. To further understand the relative contribution of unique and shared pathways in acute and chronic postsurgical pain, we present a heat map view based on hierarchical clustering of significantly enriched biological processes (-log p-values>1.3) for the pain phenotypes (Figure 3). The biological processes were condensed under broad headings based on similarities in mechanisms as described in Supplementary Table 2. The heat map shows clustering of several enriched processes, common to the pain phenotypes which indicates similar underlying pathophysiology. Our results point to various top molecular mechanisms like cAMP response element-binding protein (CREB) phosphorylation, ion channels, N-methyl-D-aspartate (NMDA) underlying both acute and chronic postsurgical pain. In addition, some immune/inflammatory (toll-like receptor signaling, interferon gamma signaling, cytokines, mitogen-activated protein kinase / extracellular signal-regulated protein kinase signaling (MAPK/ERK) and other neurotransmitter involved processes (purinergic, oxytocin, gamma amino butyric acid (GABA), glutaminergic, catecholaminergic and dopaminergic) are associated in differing degrees to acute and chronic postsurgical pain.
Figure 2:
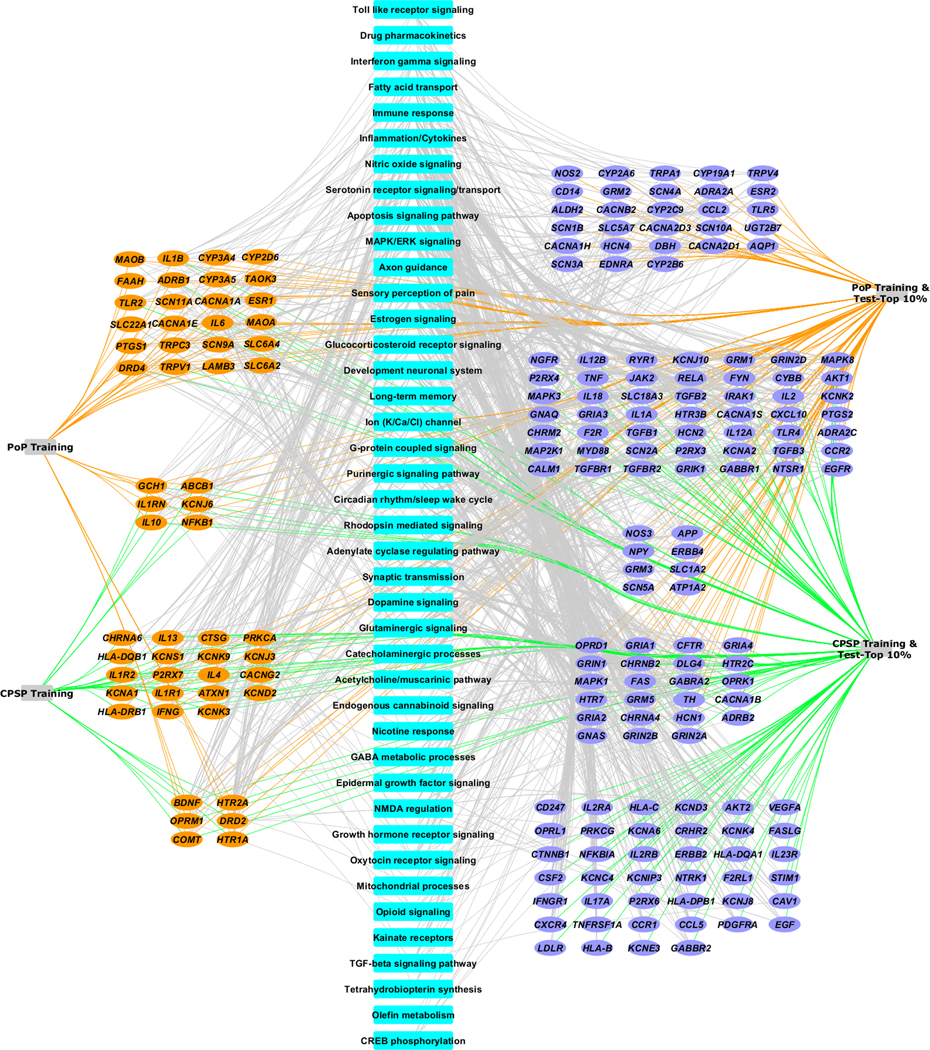
Network representation of genes (training and top candidate) and functionally enriched pathways in postoperative pain (PoP) and chronic postoperative pain (CPSP). Novel candidate genes for the phenotypes were identified using system biology-based approaches. Blue nodes represent biologic processes important to outcomes. Orange nodes represent training (known literature-curated) genes, and purple nodes represent novel candidate genes identified using system biology methodology likely to be associated with biologic process important to outcomes. This network is remarkable for the overlapping pathways underlining the complicated genetic architecture of acute postsurgical pain and its maintenance.
Figure 3:

Heat map showing hierarchical clustering of pathways and biological processes based on – log p-values for PoP: Postoperative pain and CPSP: chronic post-surgical pain. The key for the -log p-values is given above the heat map. A value of 1.3 signifies a statistically significant value of 0.05. Genes enriched for CPSP and PoP phenotypes are overrepresented in several statistically significant processes/pathways. Also, the clustering is intuitive in that mechanisms traditionally considered functionally similar are clustered together: for example, CREB (cAMP response element binding) phosphorylation and ion channels are together; MAPK/ERK (mitogen activated protein kinase/extracellular signal-regulated kinases), immune response and interferon gamma signaling. Other abbreviations: NMDA: N-methyl D-aspartate; GABA: gamma amino butyric acid; TGF: Transforming growth factor. In comparing p-values, immune processes seem to more significantly enriched for CPSP, while catecholaminergic, glutaminergic, tetrohydrobiopterin (GTP Cyclohydrolase 1) are among some pathways more significantly enriched for PoP compared to CPSP.
4. Discussion
This study demonstrates the utility of a systems biology and machine learning-based approach in identifying novel candidate genes and enriched pathways involved in acute and chronic post-surgical pain. Identification of overlapping biological processes involved in acute pain after surgery and its persistence helps understand the mechanisms that could be targeted to prevent CPSP. Molecular biology-based bioinformatics methods are thus useful in informing future research efforts focused on functional genomic pathways and novel molecular targets and could potentially lead to clinically translatable findings.
Systems biology assisted candidate gene prioritization helped identify 2610 genes that could be associated with postoperative pain and CPSP (top 10th percentile were further examined in this study), compared to 57 literature-curated genes with reported variant associations. Thus, many novel genes not previously reported to be associated with the specific phenotypes were identified. Consequently, novel biological processes/pathways were enriched by the newly identified sets of genes, compared to those enriched by training gene sets. The overlap of biological processes involving different neurotransmitters and molecular mechanisms contributing to acute postoperative pain and its persistence underlines the complex genetic architecture of pain, and the rationale for multimodal therapies targeted at different mechanisms. Since it is beyond the scope of this manuscript to discuss all the pathways identified, we will discuss key pathways, based on hierarchical clustering. Similar pathways are identified in the heatmap compared to those enriched by common training genes only - for example, adenylate cyclase regulating pathway (similar to cAMP signaling) and oxytocin/glutaminergic signaling, etc. (akin to neuroactive ligand receptor interaction are similarly enriched for both pain phenotypes, although in different orders of statistical significance.
Based on the heat map, the pathway that was most significantly over-represented for both pain outcomes was the cAMP response element-binding protein (CREB) pathway. This could be explained by the fact that CREB is a downstream molecule in the cAMP/Protein kinase A/CREB pathway, which is a potential molecular mechanism involved in pain memory.[36] Pain memory is pivotal for pain maintenance states, as it is involved in sensory-discriminative, emotional affective, and cognitive evaluative aspects of the pain experience. Nociceptive stimuli activate spinal cAMP pathways which phosphorylate CREB, a transcription factor that induces gene expression. Studies have shown reversal of mechanical hyperalgesia with the use of adenylate cyclase and protein kinase A (PKA) inhibitors with decreased CREB phosphorylation.[37] However, we did not identify significant over-representation in PKA signaling pathways. This might suggest relatively higher importance of certain molecular targets over others for targeting CPSP.
Ion channels are the next significant pathway over-represented in both pain phenotypes. This class encompasses processes involving calcium, potassium, chloride and sodium channels. This supports use of ion channel modulators like local anesthetics and drugs like gabapentin, which have been used in the treatment of pain for years.[38] Familial congenial insensitivity to pain as well as increased sensitivity from both loss and gain of function mutations in voltage gated sodium channel subtypes 1.3 and 1.7–1.9 mutations, have previously provided insight into the role of specific sodium channels in pain. [39, 40]
Neurotransmission pathways were significantly over-represented in both phenotypes. This finding is aligned with those of a comprehensive survey of up-to-date genetic association results with chronic pain conditions, which revealed that nearly half of identified genetic variants in migraine and musculoskeletal pain involve neurotransmission pathways.[17] We find that glutaminergic, NMDA regulation, catecholaminergic, dopaminergic, purinergic, glucocorticoid, oxytocin, and muscarinic pathways are involved to varying extent in pain outcomes, at a higher significance for CPSP compared to acute post-surgical pain. This may be explained by the fact that dopaminergic mechanisms modulate motivational salience of pain stimuli and influence affective sequelae of pain, thus contributing to chronic pain and disability.[41] Neurotransmitters and neuroendocrine processes are also involved in anxiety and psychological factors that play an important role in pain persistence.[42] Circadian rhythm regulation was also enriched for pain, as has been described in previous studies.[43, 44]
Surgical insult is known to trigger release of proinflammatory cytokines, so it is not surprising that inflammatory pathways are over-represented in post-surgical pain phenotypes. Previous studies have shown that G-protein coupled receptors (GPCRs) activated at nociceptors after surgical injury coupled with cytokines and other inflammatory mediators (such as interleukin-1 beta and tumor necrosis factor (TNF), evoke acute pain responses. When inflammatory processes are sustained by activation of ion channels (transient receptor TRP channels, sodium channels) mediated by protein kinases (MAP kinases, PKA, PKC), it leads to phenomena of peripheral sensitization and neuro-inflammation, which are key to maintenance of pain, and development of neuropathic pain.[45, 46] Our findings are aligned with enrichment of MAPK/ERK signaling in CPSP. These pathways also offer promising therapeutic targets for CPSP; for example, ERK/MAPK regulation [47] and anti-inflammatory treatment (TNFα antagonist etanercept) have been studied for amelioration of injury-induced neuropathic pain,[48] and post-amputation phantom limb pain.[49] Similarly, commercially available recombinant IL1r antagonist, anakinra, is already in use in rheumatoid arthritis; our study could provide a basis for perioperative trials for such medications in CPSP, as IL-1R, a cytokine receptor is also implicated in postoperative pain.[50] Another cytokine pathways (transforming growth factor-β (TGF-β) signaling pathway) is enriched for CPSP, but not acute postoperative pain. TGF-β1 belongs to another superfamily of cytokines, has protective effects against the development of chronic neuropathic pain by inhibiting neuroimmune responses and promoting the expression of endogenous opioids within the spinal cord.[51] Specific members of the TGF-β family and molecules in their signaling pathways may also be promising molecular targets for novel therapeutic agents for pain management.
The clustering of immune processes with inflammatory processes on the heat map with a higher enrichment for CPSP compared to acute postoperative pain also points to the role of neuroimmune reactions regulating inflammatory responses in especially chronic postsurgical pain.[6, 52]. One such immune process involves T-cell signaling pathways (regulation of CD4-positive, CD25-positive and alpha-beta regulatory T cell differentiation). T-cell infiltration in the dorsal horn in an animal model of spared nerve injury was previously found to be involved in neuropathic pain alongwith gamma interferon (IFNγ).[53] The role of immune processes involving T-cell receptor pathways in CPSP is aligned with reports of genetic variants in the T-cell receptor pathway associated with greater odds of pain in a chronic pain condition (temporomandibular disorder).[54] Similarly, another computational study of human gene polymorphisms modulating persistent pain phenotypes identified major roles for neuroimmune processes in addition to nitric oxide signaling across several chronic pain conditions.[21] We also found Toll like receptors (TLR 1–8) signaling pathways enriched for CPSP. This is aligned with other reports of TLR signaling involvement in models of nerve injury, cancer pain and opioid induced hyperalgesia.[55–57] TLR are a family of pattern recognition receptors located on macrophages. It is known that pain signals activating these receptors increases cytokine release from the macrophages.[58]
Similar to Kringel et. al’s observations,[21] nitric oxide (NO) signaling was significantly over represented in acute and chronic postsurgical pain. NO signaling plays a major role in pain sensitization and development of hyperalgesia; preclinical studies show that nitro oxidative stress leads to post-translational nitration of enzymes and transporters linked to glutamatergic neurotransmission, which may represent a novel mechanism of central sensitization. Further proof for this process comes from findings that pretreatment with peroxynitrate decomposition catalyst and NMDA antagonists reversed hyperalgesia following intraplantar injection of carrageenan in rats.[59]
In general, use of training sets based on candidate gene approaches limit the functional annotation-based prioritization discoveries of candidate genes. The annotations and analyses, as well as the prioritization, are as accurate as the underlying sources from which the annotations are retrieved. Moreover, underpinning studies, may be limited by sample size, varied outcome definitions, varying populations, multiple comparisons and lack of replication. Given the paucity of GWAS and replicated studies in this area, we included all reported genotype-phenotype associations for our analyses so as to allow for an extensive base of training genes. We could not assess the weight of individual genetic contributions. Hence, there is a risk of magnifying spurious findings. Despite the aforesaid limitations, we believe use of systems biology to generate novel gene sets and pathways is important for future targeted approaches to develop polygenic risk scores. The gene enrichment results from our study will provide a basis for future studies to overcome limitations posed by sample sizes and heterogenous surgical populations which make clinical studies using GWAS difficult to conduct. However, the strength of this approach is in combining all available bioinformatics data to complement available published data from clinical studies. We believe this is especially important given the lack of consistent genetic association data for these phenotypes from human studies thus far.
In conclusion, this is the first study to use systems biology to describe gene enriched pathways that play a role in postoperative pain and CPSP. Our findings help prioritize mechanisms that need to be studied further and identifies potential targets and focus areas for personalized prevention and treatment of CPSP.
Supplementary Material
Acknowledgements:
We would like to acknowledge Susan Glynn, CCRC IV, who independently checked the results of the literature review.
Funding: This project was supported by the National Institutes of Health ` EUNICE KENNEDY SHRIVER NATIONAL INSTITUTE OF CHILD HEALTH & HUMAN DEVELOPMENT (5K23HD08278 PI: Chidambaran)
Footnotes
Conflicts of interest disclosure: None of the authors have any conflicts of interest to disclose.
References
- [1].Weiser TG, Regenbogen SE, Thompson KD, Haynes AB, Lipsitz SR, Berry WR, et al. An estimation of the global volume of surgery: a modelling strategy based on available data. Lancet. 2008;372:139–44. [DOI] [PubMed] [Google Scholar]
- [2].Brennan F, Carr DB, Cousins M. Pain management: a fundamental human right. Anesth Analg. 2007;105:205–21. [DOI] [PubMed] [Google Scholar]
- [3].Chidambaran V, Ding L, Moore DL, Spruance K, Cudilo EM, Pilipenko V, et al. Predicting the pain continuum after adolescent idiopathic scoliosis surgery: A prospective cohort study. European journal of pain. 2017;21:1252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miech R, Johnston L, O’Malley PM, Keyes KM, Heard K. Prescription Opioids in Adolescence and Future Opioid Misuse. Pediatrics. 2015;136:e1169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Parsons B, Schaefer C, Mann R, Sadosky A, Daniel S, Nalamachu S, et al. Economic and humanistic burden of post-trauma and post-surgical neuropathic pain among adults in the United States. J Pain Res. 2013;6:459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pogatzki-Zahn EM, Segelcke D, Schug SA. Postoperative pain—from mechanisms to treatment. PAIN Reports. 2017;2:e588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].LaCroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152:1888–98. [DOI] [PubMed] [Google Scholar]
- [8].Argoff C Mechanisms of pain transmission and pharmacologic management. Curr Med Res Opin. 2011;27:2019–31. [DOI] [PubMed] [Google Scholar]
- [9].Angst MS, Phillips NG, Drover DR, Tingle M, Ray A, Swan GE, et al. Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain. 2012;153:1397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Norbury TA, MacGregor AJ, Urwin J, Spector TD, McMahon SB. Heritability of responses to painful stimuli in women: a classical twin study. Brain. 2007;130:3041–9. [DOI] [PubMed] [Google Scholar]
- [11].Hocking LJ, Generation S, Morris AD, Dominiczak AF, Porteous DJ, Smith BH. Heritability of chronic pain in 2195 extended families. European journal of pain. 2012;16:1053–63. [DOI] [PubMed] [Google Scholar]
- [12].Katz J One man’s risk factor is another man’s outcome: difference in risk factor profiles for chronic postsurgical pain maintenance vs transition. Pain. 2012;153:505–6. [DOI] [PubMed] [Google Scholar]
- [13].Katz J, Seltzer Z. Transition from acute to chronic postsurgical pain: risk factors and protective factors. Expert Rev Neurother. 2009;9:723–44. [DOI] [PubMed] [Google Scholar]
- [14].Kim H, Clark D, Dionne RA. Genetic contributions to clinical pain and analgesia: avoiding pitfalls in genetic research. J Pain. 2009;10:663–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sadhasivam S, Chidambaran V. Pharmacogenomics of opioids and perioperative pain management. Pharmacogenomics. 2012;13:1719–40. [DOI] [PubMed] [Google Scholar]
- [16].Mogil JS. The genetic mediation of individual differences in sensitivity to pain and its inhibition. Proc Natl Acad Sci U S A. 1999;96:7744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zorina-Lichtenwalter K, Meloto CB, Khoury S, Diatchenko L. Genetic predictors of human chronic pain conditions. Neuroscience. 2016;338:36–62. [DOI] [PubMed] [Google Scholar]
- [18].Radde NE, Hütt M-T. The Physics behind Systems Biology. EPJ Nonlinear Biomedical Physics. 2016;4:7. [Google Scholar]
- [19].Antunes-Martins A, Perkins JR, Lees J, Hildebrandt T, Orengo C, Bennett DL. Systems biology approaches to finding novel pain mediators. Wiley interdisciplinary reviews Systems biology and medicine. 2013;5:11–35. [DOI] [PubMed] [Google Scholar]
- [20].Slade GD, Smith SB, Zaykin DV, Tchivileva IE, Gibson DG, Yuryev A, et al. Facial pain with localized and widespread manifestations: separate pathways of vulnerability. Pain. 2013;154:2335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kringel D, Lippmann C, Parnham MJ, Kalso E, Ultsch A, Lotsch J. A machine-learned analysis of human gene polymorphisms modulating persisting pain points to major roles of neuroimmune processes. European journal of pain. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kurowski BG, Treble-Barna A, Pitzer AJ, Wade SL, Martin LJ, Chima RS, et al. Applying Systems Biology Methodology To Identify Genetic Factors Possibly Associated with Recovery after Traumatic Brain Injury. J Neurotrauma. 2017;34:2280–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big Data: Astronomical or Genomical? PLoS Biol. 2015;13:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research. 2009;37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Werner MU, Kongsgaard UE. I. Defining persistent post-surgical pain: is an update required? British journal of anaesthesia. 2014;113:1–4. [DOI] [PubMed] [Google Scholar]
- [26].Chidambaran V, Gang Y, Pilipenko V, Ashton M, Ding L. Systematic review and meta-analysis of genetic risk of developing chronic postsurgical pain. The journal of pain : official journal of the American Pain Society. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, et al. Canonical genetic signatures of the adult human brain. Nature neuroscience. 2015;18:1832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic acids research. 2004;32:D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic acids research. 2006;34:D354–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic acids research. 2005;33:6083–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Aoki Y, Nishi A, et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry. 2014;19:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cook-Sather SD, Li J, Goebel TK, Sussman EM, Rehman MA, Hakonarson H. TAOK3, a novel genome-wide association study locus associated with morphine requirement and postoperative pain in a retrospective pediatric day surgery population. Pain. 2014;155:1773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim H, Ramsay E, Lee H, Wahl S, Dionne RA. Genome-wide association study of acute post-surgical pain in humans. Pharmacogenomics. 2009;10:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mieda T, Nishizawa D, Nakagawa H, Tsujita M, Imanishi H, Terao K, et al. Genome-wide association study identifies candidate loci associated with postoperative fentanyl requirements after laparoscopic-assisted colectomy. Pharmacogenomics. 2016;17:133–45. [DOI] [PubMed] [Google Scholar]
- [35].Warner SC, van Meurs JB, Schiphof D, Bierma-Zeinstra SM, Hofman A, Uitterlinden AG, et al. Genome-wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein-kinase C gene. European journal of human genetics : EJHG. 2017;25:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Molecular Brain. 2012;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hoeger-Bement MK, Sluka KA. Phosphorylation of CREB and mechanical hyperalgesia is reversed by blockade of the cAMP pathway in a time-dependent manner after repeated intramuscular acid injections. J Neurosci. 2003;23:5437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Skerratt SE, West CW. Ion channel therapeutics for pain. Channels. 2015;9:344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52:767–74. [DOI] [PubMed] [Google Scholar]
- [41].Taylor AMW, Becker S, Schweinhardt P, Cahill C. Mesolimbic dopamine signaling in acute and chronic pain: implications for motivation, analgesia, and addiction. Pain. 2016;157:1194–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Martin EI, Ressler KJ, Binder E, Nemeroff CB. The Neurobiology of Anxiety Disorders: Brain Imaging, Genetics, and Psychoneuroendocrinology. The Psychiatric clinics of North America. 2009;32:549–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hagenauer MH, Crodelle JA, Piltz SH, Toporikova N, Ferguson P, Booth V. The Modulation of Pain by Circadian and Sleep-Dependent Processes: A Review of the Experimental Evidence. bioRxiv. 2017. [Google Scholar]
- [44].Gillin JC. Are sleep disturbances risk factors for anxiety, depressive and addictive disorders? Acta psychiatrica Scandinavica Supplementum. 1998;393:39–43. [DOI] [PubMed] [Google Scholar]
- [45].Zhang G, Yang P. Bioinformatics Genes and Pathway Analysis for Chronic Neuropathic Pain after Spinal Cord Injury. BioMed Research International. 2017;2017:6423021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Matsuda M, Huh Y, Ji R-R. Roles of inflammation, neurogenic inflammation, and neuroinflammation in pain. Journal of anesthesia. 2019;33:131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ma W, Quirion R. The ERK/MAPK pathway, as a target for the treatment of neuropathic pain. Expert Opinion on Therapeutic Targets. 2005;9:699–713. [DOI] [PubMed] [Google Scholar]
- [48].Iwatsuki K, Arai T, Ota H, Kato S, Natsume T, Kurimoto S, et al. Targeting anti-inflammatory treatment can ameliorate injury-induced neuropathic pain. PLoS One. 2013;8:e57721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dahl E, Cohen SP. Perineural injection of etanercept as a treatment for postamputation pain. Clin J Pain. 2008;24:172–5. [DOI] [PubMed] [Google Scholar]
- [50].Buvanendran A, Kroin JS. Does manipulating local surgical wound cytokines improve surgical outcomes? Anesthesia and analgesia. 2010;111:1335–6. [DOI] [PubMed] [Google Scholar]
- [51].Lantero A, Tramullas M, Diaz A, Hurle MA. Transforming growth factor-beta in normal nociceptive processing and pathological pain models. Mol Neurobiol. 2012;45:76–86. [DOI] [PubMed] [Google Scholar]
- [52].Ren K, Dubner R. Interactions between the immune and nervous systems in pain. Nature medicine. 2010;16:1267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Costigan M, Moss A, Latremoliere A, Johnston C, Verma-Gandhu M, Herbert TA, et al. T-Cell Infiltration and Signaling in the Adult Dorsal Spinal Cord Is a Major Contributor to Neuropathic Pain-Like Hypersensitivity. The Journal of Neuroscience. 2009;29:14415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Slade GD, Smith SB, Zaykin DV, Tchivileva IE, Gibson DG, Yuryev A, et al. Facial pain with localized and widespread manifestations: separate pathways of vulnerability. Pain. 2013;154: 10.1016/j.pain.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stokes JA, Cheung J, Eddinger K, Corr M, Yaksh TL. Toll-like receptor signaling adapter proteins govern spread of neuropathic pain and recovery following nerve injury in male mice. J Neuroinflammation. 2013;10:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lan LS, Ping YJ, Na WL, Miao J, Cheng QQ, Ni MZ, et al. Down-regulation of Toll-like receptor 4 gene expression by short interfering RNA attenuates bone cancer pain in a rat model. Molecular pain. 2010;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hutchinson MR, Zhang Y, Shridhar M, Evans JH, Buchanan MM, Zhao TX, et al. Evidence that opioids may have toll-like receptor 4 and MD-2 effects. Brain Behav Immun. 2010;24:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lacagnina MJ, Watkins LR, Grace PM. Toll-like receptors and their role in persistent pain. Pharmacology & therapeutics. 2018;184:145–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen Z, Muscoli C, Doyle T, Bryant L, Cuzzocrea S, Mollace V, et al. NMDA-receptor activation and nitroxidative regulation of the glutamatergic pathway during nociceptive processing. PAIN®. 2010;149:100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


