Abstract
The National Center for Advancing Translational Sciences (NCATS) has developed an online open science data portal for its COVID-19 drug repurposing campaign – named OpenData – with the goal of making data across a range of SARS-CoV-2 related assays available in real-time. The assays developed cover a wide spectrum of the SARS-CoV-2 life cycle, including both viral and human (host) targets. In total, over 10,000 compounds are being tested in full concentration-response ranges from across multiple annotated small molecule libraries, including approved drug, repurposing candidates and experimental therapeutics designed to modulate a wide range of cellular targets. The goal is to support research scientists, clinical investigators and public health officials through open data sharing and analysis tools to expedite the development of SARS-CoV-2 interventions, and to prioritize promising compounds and repurposed drugs for further development in treating COVID-19.
The pandemic of the atypical pneumonia coronavirus disease (COVID-19) caused by a novel beta coronavirus SARS-CoV-2 has significantly impacted global society, from both an economic and public health standpoint. As a result, biomedical scientists around the world – from academic and government laboratories to biotechnology companies and pharmaceutical corporations – have mobilized to understand the disease, to develop therapeutic interventions to mitigate its impact and to develop protective vaccines. This rapid research response has resulted in the initiation of repurposing clinical trials for a range of agents, including three that have received Emergency Use Authorizations (EUAs) from the Federal Drug Agency (FDA) in the United States: remdesivir1, chloroquine and hydroxychloroquine. These drugs, and others that have entered clinical trials, were prioritized based on clinical observations or a contemporary understanding of SARS-CoV-2 biology. In addition, numerous drug repurposing efforts have been undertaken, screening both approved and experimental agents2–10. Yet many published reports solely focus on active hits, and do not disclose the majority (usually >95%) of tested compounds that were inactive – information that is critical for understanding and validating disease and drug mechanism-of-action, and for nominating repurposed and novel clinical lead candidates. Rapid and open sharing of complete screening datasets, including negative results, will greatly accelerate the research and discovery process essential to the COVID-19 pandemic response.
To address this, the National Center for Advancing Translational Sciences (NCATS) has developed an online open science data portal for its COVID-19 drug repurposing campaign – named OpenData – with the goal of making data across a range of SARS-CoV-2 related assays available in real-time (Figure 1). This approach allows researchers rapid access to drug repurposing datasets that can support subsequent mechanistic study of compounds that perturb viral infectivity11. In this manner, open data sharing can facilitate important insight and associated publications towards the development of interventions against COVID-19. The OpenData dashboard, first shared publicly on May 25, 2020, makes quantitative high-throughput screening (HTS) data and detailed protocol information available for every assay screened. The goal is to provide clinicians and researchers with a user-friendly tool that allows direct comparison of compounds across multiple assays, with all primary concentration-response data made freely available, through direct download. https://opendata.ncats.nih.gov/covid19/
Figure 1. OpenData portal objective.
The goal of the OpenData portal is to provide actionable data and validated methodologies to the global research community to support rapid development of therapeutics against SARS-CoV-2.
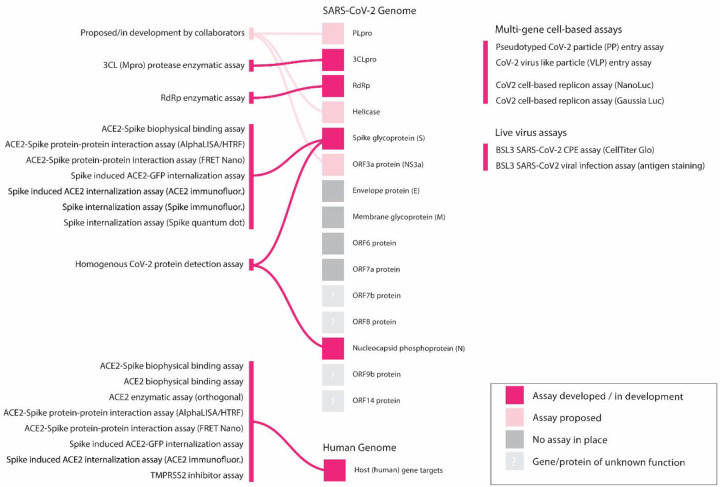
The assays that have been developed to date cover a wide spectrum of the SARS-CoV-2 life cycle (Figure 2), including both viral and human (host) targets, grouped into the following five categories based on different mechanisms of experimental design: viral entry, viral replication, in vitro infectivity, live virus infectivity, and counter-screens, which could flag false positives due to assay interference artifacts or cytotoxic effects. These assays encompass protein-based assays such as the SARS-CoV-2 spike protein-ACE2 interaction and viral enzyme activity assays, in addition to cell-based pseudotyped particle entry and live virus cytopathic effect assays. As additional assays are validated and screened, this list will be expanded and updated. Importantly, all assay documentation including assay overview, methodology and detailed assay protocols are available, and readily retrievable, on the OpenData site to facilitate adoption and ensure that other laboratories can replicate these assays (Table 1).
Figure 2. NCATS SARS-CoV-2 assays currently employed to address a range of both viral and host targets.
Overview of established assays and those in development. These include assays for specific viral targets, viral-host interaction assays, viral detection assays and viral cytopathic effect and replication assays, among others.
Table 1.
Validated OpenData portal assays and those under development.
| Assay Name | Assay Type | Target Category | Detection Strategy | Cell Line | Status |
|---|---|---|---|---|---|
| Spike-ACE2 protein-protein interaction (AlphaLISA) | Proximity | Viral Entry | AlphaLISA | n/a | Screening in progress |
| Spike-ACE2 protein-protein interaction (TruHit counterscreen) | Proximity | Counterscreen | AlphaLISA | n/a | Screening in progress |
| Spike-ACE2 protein-protein interaction (QD) | Proximity | Viral Entry | High-content imaging | n/a | Under development |
| Spike-ACE2 binding | Biophysical | Viral Entry | Bio-Layer Interferometry | n/a | Screening in progress |
| ACE2 binding | Biophysical | Viral Entry | Microscale thermophoresis | n/a | Screening in progress |
| ACE2 enzymatic activity | Biochemical | Viral Entry | Fluorescence | n/a | Screening in progress |
| ACE2 internalization | Cell-based | Viral Entry | High-content imaging | HEK293 | Under development |
| TMPRSS2 enzymatic activity | Biochemical | Viral Entry | Fluorescence | n/a | Under development |
| 3CL enzymatic activity | Biochemical | Viral replication | Fluorescence | n/a | Under development |
| RdRp enzymatic activity | Biochemical | Viral replication | Fluorescence | n/a | Under development |
| SARS-CoV-2 replicon | Cell-based | Viral replication | Luminescence | TBD | Under development |
| Pseudotyped particle entry | Cell-based | In vitro infectivity | Luminescence | TBD | Under development |
| Virus-like particle entry | Cell-based | In vitro infectivity | TBD | TBD | Under development |
| SARS-CoV-2 cytopathic effect (CPE) | Cell viability | Live virus infectivity | Luminescence | Vero E6 | Screening in progress |
| SARS-CoV-2 cytopathic effect (host toxicity counterscreen) | Cell viability | Counterscreen | Luminescence | Vero E6 | Screening in progress |
| SARS-CoV-2 Viral Infection | Cell-based | Live virus infectivity | TBD | TBD | Under development |
| SARS-CoV-2 Viral Infection (host toxicity counterscreen) | Cell-based | Counterscreen | TBD | TBD | Under development |
| Spike protein detection | Proximity | Live virus infectivity | AlphaLISA/HTRF | n/a | Under development |
| Nucleocapsid protein detection | Proximity | Live virus infectivity | AlphaLISA/HTRF | n/a | Under development |
QD, quantum dot; TBD, to be determined; HTRF, Homogeneous Time Resolved Fluorescence
In total, over 10,000 compounds are being tested in full concentration-response ranges from across multiple annotated small molecule libraries including: (1) The NCATS Pharmaceutical Collection (NPC), a library of 2,678 compounds approved for use by the Food and Drug Administration and related agencies in other countries12,13; (2) a collection of 739 anti-infective compounds with potential anti-coronavirus activity that have been reported in the literature as repurposing candidates; and (3) an annotated/bioactive collection of diverse small molecules with known mechanisms of action, including as small molecule probes and experimental therapeutics designed to modulate a wide range of targets, such as natural products, epigenetic-associated compounds or compounds developed for oncology indications14,15. Together, these libraries yield 9,958 compound responses, 8,624 of which represent structurally unique compounds, of which 1,820 (21%) are approved for use in humans with an additional 989 (11%) having entered human clinical evaluation (Phase 1–3 trials). Importantly, of these unique compounds, 5,224 (60%) have at least one annotated target, providing insight into potential mechanism-of-action underlying active compounds that perturb the SARS-CoV-2 life cycle in some manner.
A key goal of the OpenData portal is to allow users to view and inspect the concentration-response data for all compounds screened, not limited to only active compounds. These complete datasets, including full concentration-response information and compound annotations, can be directly downloaded, providing extensive information for further analysis and data mining. In addition, a user-friendly heatmap visualization has been implemented to allow direct and convenient comparison of compound activity across the panel of all assays tested including orthogonal and counter assays (Figure 3).
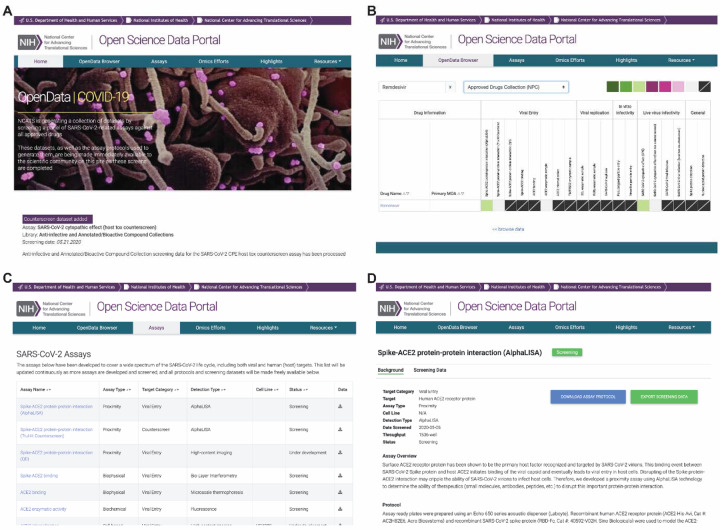
Figure 3. Representative screen captures from the OpenData portal.
A) OpenData portal home page with updates section. B) OpenData browser tab, with columns depicting activity across various protein-based, biochemical, and cell-based assays via heatmap visualization of compound activity to permit direct comparison across a panel of assays. The browser has both sort and search functionalities to facilitate interrogation of data. C) The portal assay page organizes access to all validated assays and those in development, including methodology, detection strategy and status. D) Detailed assay protocols are provided to allow independent evaluation and confirmation of method and compound activities.
Currently, the OpenData portal can be queried by a search term (e.g., drug name, primary mechanism-of-action, gene symbol) and ordered by any assay data field for increased interpretability. A high-level overview of compound activity across multiple different assays is displayed by heatmap, with sort functions based on curve class, efficacy, and potency. In the heatmap, darker colors indicate compounds that are more potent and efficacious (i.e., high-quality actives), lighter colors indicate less potent and efficacious compounds (low-quality actives), beige represents inactive compounds, and slashed boxes denote compounds/assays not yet tested.
In addition to the NCATS COVID-19 drug repurposing screening data described above, two regularly updated external resource sections have also been incorporated into the OpenData portal: a collection of publications focusing on small molecule drug repurposing screens, and a list of publicly available multiomics datasets generated to evaluate aberrant biochemical pathways or identify putative biomarkers for COVID-19.
In summary, the OpenData portal described herein has been designed to share NCATS SARS-CoV-2 complete datasets openly and without restriction, and, importantly, in real-time given the time urgency of COVID-19 pandemic response. In addition, we have constructed the portal such that HTS datasets from studies published by other investigators can be integrated into the portal to allow a global view of COVID-19 drug repurposing efforts, and enable comparison of screening hits across multiple centers. We envisage this site could act as a central portal and a unique resource to compare complete repurposing data generated in both the public and private sectors—the authors welcome contributions of external HTS data. We hope that this platform will help research scientists, clinical investigators and public health officials to facilitate open data sharing and analysis to expedite the development of SARS-CoV-2 interventions, and to prioritize promising compounds and repurposed drugs for further development in treating COVID-19.
Acknowledgments:
We would like to thank Tudor Oprea (UNM Health Sciences Center) for helpful suggestions and comments regarding the OpenData portal.
Funding: This research was supported in by the National Center for Advancing Translational Sciences (NCATS) Intramural Research Program of the NIH.
References
- 1.Eastman RT et al. (2020). Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Central Science. 6.5 (2020): 672–683. Web. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellinger B et al. (2020). Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Research Square. 10.21203/rs.3.rs-23951/v1 [DOI] [Google Scholar]
- 3.Fan H et al. (2020). Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin Med J. 10.1097/CM9.0000000000000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon DE et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon S et al. (2020). Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 10.1128/AAC.00819-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meehyun K et al. (2020). Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. bioRxiv. 2020.02.25.965582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riva L et al. (2020). A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. bioRxiv. 2020.04.16.044016. [Google Scholar]
- 8.Touret F et al. (2020). In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. bioRxiv. 2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weston S et al. (2020). Broad anti-coronaviral activity of FDA approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. bioRxiv. 2020.03.25.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing J et al. (2020). Reversal of Infected Host Gene Expression Identifies Repurposed Drug Candidates for COVID-19. bioRxiv. 2020.04.07.030734. [Google Scholar]
- 11.Gorshkov K et al. (2020). The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv, 2020.2005.2016.091520. [Google Scholar]
- 12.Huang R et al. (2019). The NCATS Pharmaceutical Collection: A 10-year Update. Drug Discov Today. Dec;24(12):2341–2349. doi: 10.1016/j.drudis.2019.09.019 [DOI] [PubMed] [Google Scholar]
- 13.Huang R et al. (2011). The NCGC Pharmaceutical Collection: A Comprehensive Resource of Clinically Approved Drugs Enabling Repurposing and Chemical Genomics. Sci Transl Med. Apr 27;3(80):80; doi: 10.1126/scitranslmed.3001862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews LA et al. (2012). A 1536-Well Quantitative High-Throughput Screen to Identify Compounds Targeting Cancer Stem Cells. J Biomol Screen. Oct;17(9):1231–42. doi: 10.1177/1087057112458152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coussens NP et al. (2018). High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem. Aug 31; 293(35): 13750–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]