Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) includes a group of heterogeneous tumors with generally invasive behavior. The PI3K/AKT pathway plays an important role in the pathogenesis of HNSCC.
Methods
In the current study, we investigated the expression of two negative feedback regulators of the PI3K pathway, namely PHLDA3 and GRHL3, in 45 paired samples of HNSCC and adjacent non-cancerous tissues (ANCTs).
Results
While expression of GRHL3 was down-regulated in tumoral tissues compared with ANCTs by the factor 4.21, PHLDA3 expression levels were up-regulated by 5.99-times. Gender-based analysis revealed a significant down-regulation of GRHL3 gene expression level in male patients compared with the control samples and significant up-regulation of PHLDA3 gene expression level in both sexes compared with the control samples. Differences in the expressions of both genes were significant in patients aged more than 60 years, but not in the younger patients. Expression of GRHL3 was only down-regulated in patients with positive smoking history. Expression of GRHL3 was decreased in grades 2 and 3 samples compared with controls. There was a significant increase in transcript levels of PHLDA3 in stages II and III HNSCC samples compared with the controls group. ROC curve analysis indicated that the expression level of PHLDA3 could be a promising marker for the diagnosis of HNSCC patients with a sensitivity and specificity of 0.666 and 0.688, respectively. In addition, sensitivity and specificity of GRHL3 were 0.755 and 0.577, respectively.
Discussion
The current study indicates dysregulation of regulators of PI3K pathway in HNSCC and their potential application as putative biomarkers for this cancer.
Keywords: GRHL3, PHLDA3, head and neck squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) includes a group of heterogeneous tumors that are generally invasive.1 Based on the GLOBOCAN statistics, more than 834,000 new cases of HNSCC have been diagnosed in 2018.2 This kind of cancer accounted for over 380,000 mortalities in 2018.2 High mortality rates and serious symptoms are the features that make management of HNSCC difficult. Half of patients die in the first 5 years after initial diagnosis and the rate of survival has not been improved significantly during recent decades.3,4 Regardless of recent advancements in surgery and radiotherapy, up to 50% of advanced local tumors relapse within 2 years after treatment.1,5 Thus, identification of the underlying mechanism for HNSCC development has practical significance.
The phosphatidylinositol 3-kinase (PI3K) pathway is one of the most common activated signaling pathways in human cancers. PI3K is a member of the family of 3ˈ-OH phosphatidylinositol phosphorylating enzymes that have important roles in cellular proliferation, differentiation, and survival. PI3Ks are activated by receptor tyrosine kinases such as EGFR and catalyze a number of enzymatic reactions that ultimately lead to phosphorylation of AKT. Activated AKT phosphorylates proteins that are involved in cell proliferation and survival.6,7 Dysregulation of the PI3K/AKT pathway is a common event in HNSCC.8 There are many pieces of evidence indicating that negative feedback regulators of the PI3K pathway act as tumor suppressors, and their loss of function leads to the initiation of cancer, proliferation, metastasis, and resistance to chemotherapy. As the PI3K/AKT pathway plays an important role in head and neck cancer, evaluation of transcriptome, proteome and methylation status of negative regulators of this pathway is important in the diagnosis, prognosis, and targeted treatment of HNSCC. Among negative regulators of this pathway are Grainyhead Like Transcription Factor 3 (GRHL3) and Pleckstrin Homology Like Domain Family A Member 3 (PHLDA3). Reduced expression of these negative regulators has been reported in different malignancies.9
GRHL3 is one of the very conserved and ancestral transcription factors which is crucial for the development and homeostasis of ectoderm in many species.10 GRHL3 may play a key role in prevention of HNSCC, as deletion of genomic region 1P36.11 containing this gene has been detected in human HNSCC. Evidence shows that GRHL3 evokes PTEN expression. Deletion of GRHL3 causes reduced expression of PTEN, so the PI3K/AKT/mTOR pathway is activated leading to metastasis of SCC. GHRL3 is a tumor suppressor.11 Reduced levels of GRHL3 and PTEN are also seen in human SCC and are accompanied by increased expression of miR-21. So the mir21/GRHL3/PTEN axis is defined as a tumor suppressor. Increased levels of miR-21 and reduced levels of PTEN and GRHL3 are seen in some human HNSCCs.12 GRHL3 also activates AKT and eNOS, two necessary factors for suppression of apoptosis.13 Loss of GRHL3 causes a loss of anti-apoptotic effect and may lead to faster tumor progression. Studies on GRHL3 expression and its protein in breast cancer have shown that stage I tumors have the highest expression levels and stage III has the lowest expression levels.14
PHLDA3 represses AKT activity by its PH domain. PHLDA3 was initially recognized as a DNA-damage-response induced protein by P53 and its expression is directly regulated by P53. PHLDA3 cooperates with P53 in activation of apoptosis. Its overexpression leads to increased apoptosis.15 P53 activation by PTEN and PHLDA3 suppresses PI3K/AKT and causes apoptosis. Inactivation of PHLDA3 leads to suppression of P53 by AKT.15,16 Loss of PHLDA3 has been frequently detected in early stages of cancer, so PHLDA3 is regarded as a tumor suppressor. Genomic loss of PHLDA3 locus has been seen in pancreatic neuroendocrine tumors.17
In the current study, we assessed expression of GRHL3 and PHLDA3 in HNSCC and adjacent non-cancerous tissues (ANCTs) to appraise their role in the pathogenesis of this cancer.
Patients and Methods
Study Participants
The current study was performed on tumor tissues from 45 patients with a definite diagnosis of HNSCC and their corresponding ANCTs as control samples. Tumor and ANCTs were separated from fresh samples by an expert pathologist. Patients were referred to Cancer, Institute Imam Khomeini Hospital during 2018. All the patients were Iranian and none of them received radiotherapy or chemotherapy before surgery. Tissue samples were transferred to the laboratory of the medical Genetics department in liquid nitrogen. The study protocol was approved by the ethical committee of Tehran University of Medical Sciences and all the methods were performed following the relevant guidelines and regulations. Informed written consent forms were signed by all patients.
Expression Study
Total RNA was extracted from cancerous tissues and ANCTs using the TRIzol™ Reagent (Invitrogen, Carlsbad, CA). The quality and quantity of RNA were evaluated using Nanodrop 2000C (Thermo Scientific, USA) and gel electrophoresis. cDNA was synthesized using SuperScript IV Reverse Transcriptase cDNA Synthesis Kit (Roche, Germany). Relative expressions of mRNAs were assessed in cancerous and non-cancerous tissues using SYBR Premix Ex Taq II (Tli RNaseH Plus) (TAKARA, Japan) in a LightCycler 96 Real-Time PCR System (Roche). SDHA gene was used as the reference gene. The sequences of primers and PCR product lengths are shown in Table 1.
Table 1.
Sequences of Primers Used in This Study
| Primer Name | Sequence | Primer Length (nt) | PCR Product Length (nt) |
|---|---|---|---|
| GRHL3-F | CTGCCTCTGAAGCGTACCTG | 20 | 117 |
| GRHL3-R | CTCAGTCTCCCTCCGCACAT | 20 | |
| PHLDA3-F | CGCACCATCTTTCCTTCATGCT | 22 | 121 |
| PHLDA3-R | CGTCCATGCCTTCCACCTTG | 20 | |
| SDHA-F | CTTGCCAGGACCTAGAGTTTGT | 22 | 86 |
| SDHA-R | CTCTCCACGACATCCTTCCG | 20 |
Statistical Analysis
Expressions of mRNAs in tumoral tissues compared with ANCTs were calculated using the Efficiency ^Ct normalizer Gene-Efficiency ^CT target gene method. To compare the expression level of two or multiple groups, an independent t-test or One-Way ANOVA was utilized. Pearson correlation coefficient was calculated to demonstrate the level of correlation between the variables. The receiver operating characteristic (ROC) curves were depicted to determine the diagnostic values of PHLDA3 and GRHL3 transcript levels for diagnosis of HNSCC. Two-sided P-values <0.05 were considered as statistically significant. The data analysis software used was GraphPad Prism 8.2.1 (CAA).
Results
General Data of Study Participants
The general data of the study participants are shown in Table 2.
Table 2.
Demographic and Pathological Characteristics of the Study Participants
| Parameters | Number (%) | |
|---|---|---|
| Age | <60 | 30 (67.7) |
| ≥60 | 15 (33.3) | |
| Gender | Male | 33 (73.3) |
| Female | 12 (26.7) | |
| Stage | I–II | 18 (40) |
| III–IV | 27 (60) | |
| Grade | G1 | 9 (20) |
| G2 | 15 (33.3) | |
| G3 | 21 (46.7) | |
| Smoking | Yes | 30 (66.7) |
| No | 15 (33.6) | |
| Alcohol consumption | Yes | 2 (4.4) |
| No | 43 (95.6) | |
Expression Analysis of GRHL3 and PHLDA3 in the HNSCC Patients
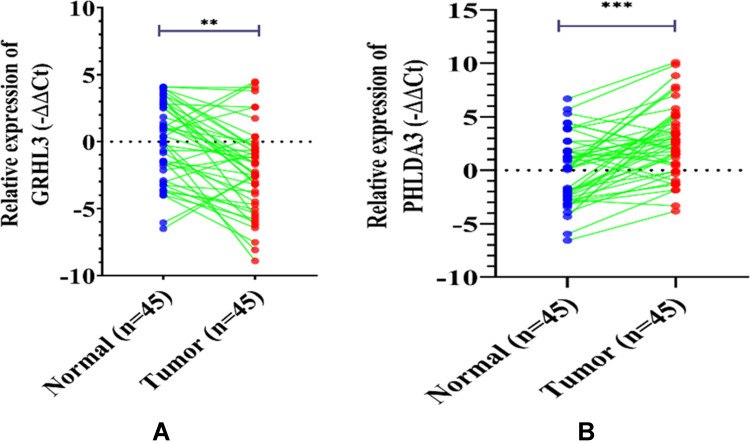
The results of GRHL3 and PHLDA3 gene expression in HNSCC tissues as compared with their ANCTs are depicted in Figure 1. While expression of GRHL3 was down-regulated in tumoral tissues compared with ANCTs by the factor 4.21 (P<0.0022, 95% CI=−10.439 to −1.698), PHLDA3 expression levels were up-regulated by 5.99-times (P<0.0003, 95% CI= 2.344 to −15.326).
Figure 1.
While GRHL3 expression was downregulated (A), the transcript level of PHLDA3 was increased (B) in HNSC tissues (n=45) compared with ANCTs (n=45). Expression was assessed by qRT-PCR and normalized to SDAH1 expression (shown as the -ΔΔCT method). The data represent the mean±SD of three replicates. **P<0.01; ***P<0.001.
Expression Analysis of GRHL3 and PHLDA3 in Association with Clinicopathological Features of Patients
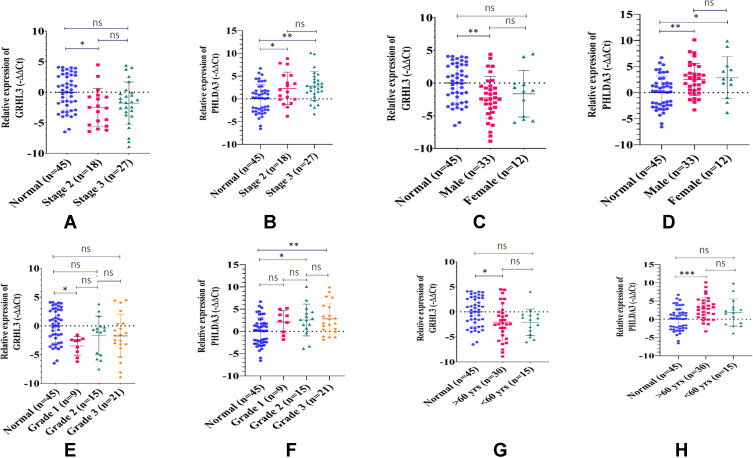
Data were also analyzed according to gender, age, smoking history, histologic grade, and pathologic stage of study participants (Tables 3 and 4). The gender-based analysis revealed significant down-regulation of GRHL3 gene expression level in male patients compared with the control samples (adjusted P=0.0073) and significant up-regulation of PHLDA3 gene expression level in both sexes compared with control samples (adjusted P-values of 0.0035 and 0.0223 for males and females, respectively). Moreover, the differences in the expressions of both genes were significant in patients aged more than 60 years but not in the younger patients. Expression of GRHL3 was only down-regulated in patients with positive smoking history. However, expression of PHLDA3 was up-regulated in patients with both positive and negative smoking history. Besides, expression of GRHL3 was decreased in grades 2 and 3 samples compared with controls. There was a significant increase in the transcript levels of PHLDA3 in stage II and III HNSCC samples when compared with the control group.
Table 3.
Relative Expression of GRHL3 in HNSCC Tissues and Adjacent Tissues Based on Clinicopathological Features
| Characteristics | Mean Difference | 95.00% CI of Difference | Adjusted P-values | Significance |
|---|---|---|---|---|
| Gender | ||||
| Normal (n=45) vs Male (n=33) | 2.230 | 0.514 to 3.945 | 0.007 | ** |
| Normal (n=45) vs Female (n=12) | 1.646 | −0.785 to 4.078 | 0.24 | ns |
| Male (n=33) vs Female (n=12) | −0.583 | −3.107 to 1.939 | 0.845 | ns |
| Age | ||||
| Normal (n=45) vs >60 years (n=30) | 2.06 | 0.292 to 3.827 | 0.018 | * |
| Normal (n=45) vs <60 years (n=15) | 2.104 | −0.131 to 4.34 | 0.069 | ns |
| >60 years (n=30) vs <60 years (n=15) | 0.044 | −2.326 to 2.416 | 0.998 | ns |
| Smoking history | ||||
| Normal (n=45) vs Yes (n=30) | 2.137 | 0.370 to 3.904 | 0.013 | * |
| Normal (n=45) vs No (n=15) | 1.949 | −0.286 to 4.184 | 0.100 | ns |
| Yes (n=30) vs No (n=15) | −0.188 | −2.559 to 2.182 | 0.980 | ns |
| Histologic grade | ||||
| Normal (n=45) vs Grade 1 (n=9) | −2.089 | −5.213 to 1.034 | 0.303 | ns |
| Normal (n=45) vs Grade 2 (n=15) | −2.561 | −5.111 to −0.010 | 0.048 | * |
| Normal (n=45) vs Grade 3 (n=21) | −2.811 | −5.072 to −0.550 | 0.008 | ** |
| Grade 1 (n=9) vs Grade 2 (n=15) | −0.471 | −4.079 to 3.135 | 0.986 | ns |
| Grade 1 (n=9) vs Grade 3 (n=21) | −0.721 | −4.130 to 2.686 | 0.94 | ns |
| Grade 2 (n=15) vs Grade 3 (n=21) | −0.250 | −3.142 to 2.642 | 0.995 | ns |
| Pathologic stage | ||||
| Normal (n=45) vs Stage 2 (n=18) | 2.502 | 0.417 to 4.586 | 0.014 | * |
| Normal (n=45) vs Stage 3 (n=27) | 1.789 | −0.030 to 3.609 | 0.054 | ns |
| Stage 2 (n=18) vs Stage 3 (n=27) | −0.712 | −2.987 to 1.561 | 0.736 | ns |
Notes: *P<0.05; **P< 0.01.
Abbreviation: ns, non-significant.
Table 4.
Relative Expression of PHLDA3 in HNSCC Tissues and Adjacent Tissues Based on Clinicopathological Features
| Characteristics | Mean Difference | 95.00% CI of Difference | Adjusted P-values | Significance |
|---|---|---|---|---|
| Gender | ||||
| Normal (n=45) vs Male (n=33) | −2.485 | −4.261 to −0.708 | 0.003 | ** |
| Normal (n=45) vs Female (n=12) | −2.854 | −5.372 to −0.336 | 0.022 | * |
| Male (n=33) vs Female (n=12) | −0.369 | −2.981 to 2.243 | 0.939 | ns |
| Age | ||||
| Normal (n=45) vs >60 years (n=30) | −2.936 | −4.753 to −1.120 | 0.000 | *** |
| Normal (n=45) vs <60 years (n=15) | −1.878 | −4.175 to 0.420 | 0.131 | ns |
| >60 years (n=30) vs <60 years (n=15) | 1.059 | −1.379 to 3.496 | 0.556 | ns |
| Smoking history | ||||
| Normal (n=45) vs Yes (n=30) | −2.494 | −4.321 to −0.666 | 0.004 | ** |
| Normal (n=45) vs No (n=15) | −2.762 | −5.073 to −0.451 | 0.014 | * |
| Yes (n=30) vs No (n=15) | −0.268 | −2.720 to 2.183 | 0.963 | ns |
| Histologic grade | ||||
| Normal (n=45) vs Grade 1 (n=9) | 3.482 | 0.496 to 6.468 | 0.015 | * |
| Normal (n=45) vs Grade 2 (n=15) | 1.683 | −0.755 to 4.121 | 0.276 | ns |
| Normal (n=45) vs Grade 3 (n=21) | 1.751 | −0.410 to 3.912 | 0.154 | ns |
| Grade 1 (n=9) vs Grade 2 (n=15) | −1.800 | −5.247 to 1.648 | 0.523 | ns |
| Grade 1 (n=9) vs Grade 3 (n=21) | −1.731 | −4.989 to 1.527 | 0.507 | ns |
| Grade 2 (n=15) vs Grade 3 (n=21) | 0.068 | −2.696 to 2.833 | >0.999 | ns |
| Pathologic stage | ||||
| Normal (n=45) vs Stage 2 (n=18) | −2.272 | −4.432 to −0.113 | 0.0368 | * |
| Normal (n=45) vs Stage 3 (n=27) | −2.791 | −4.675 to −0.905 | 0.001 | ** |
| Stage 2 (n=18) vs Stage 3 (n=27) | −0.518 | −2.874 to 1.838 | 0.859 | ns |
Notes: *P<0.05; **P<0.01; ***P<0.001.
Abbreviation: ns, non-significant.
Figure 2 shows the relative expression of the mentioned genes in different groups of tissues.
Figure 2.
Expression of GRHL3 and PHLDA3 in association with clinicopathological features of HNSCC. Expression levels of GRHL3 (A) and PHLDA3 (B) were determined by qRT-PCR in different clinical stages of HNSCC tissue samples. The gender-based analysis showed that the expression levels of GRHL3 (C) and PHLDA3 (D) were significantly different in males and females. High HNSCC grade was associated with significant reduction of GRHL3 (E) and up-regulation of PHLDA3 (F). Patients with age of greater than 60 presented a down-regulation in the expression of GRHL3 (G) and up-regulation of PHLDA3 (H) expression level. The data represent the mean±SD of two replicates. *P<0.05; **P<0.01; ***P<0.001, ns, not significant.
Co-Expression Analysis of GRHL3 and PHLDA3 in HNSCC Tissues and ANCTs
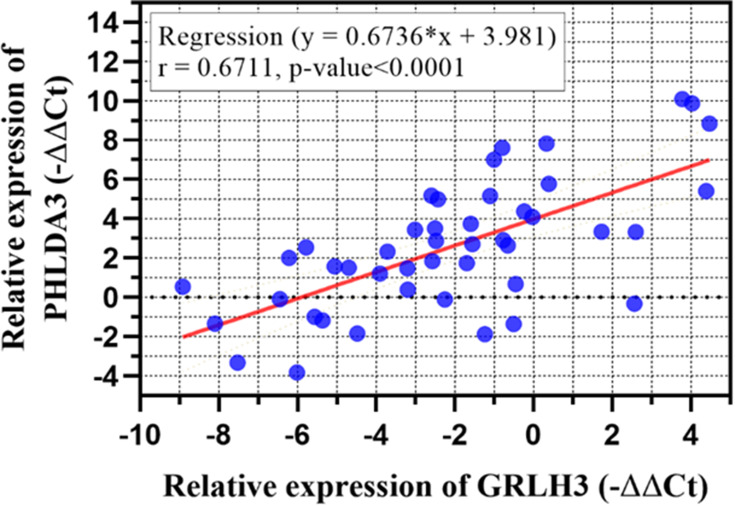
Pearson’s correlation analysis revealed a positive correlation between GRHL3 and PHLDA3 expression levels (Figure 3).
Figure 3.
Co-expression analysis between expression of genes in ANCTs and HNSCC tissues demonstrated a significant correlation between PHLDA3 and GRHL3 (r=0.6711, P<0.0001). Each dot indicates an individual sample.
ROC Curve Analysis
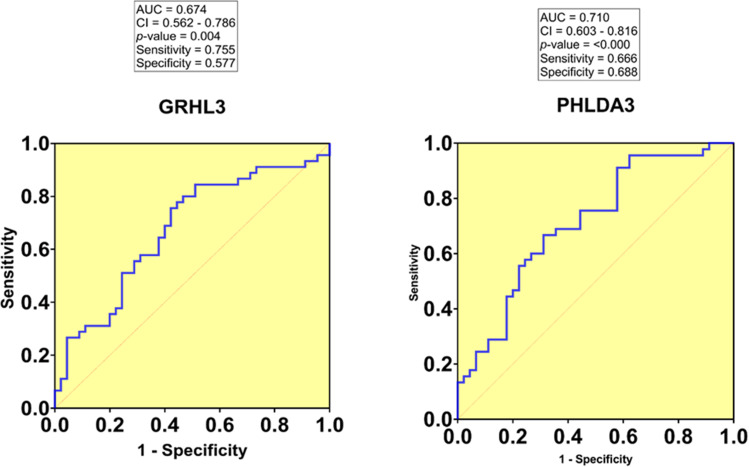
We evaluated the diagnostic value of the studied genes for HNSCC using the calculation of the area under the curve (AUC) in ROC curves. AUC close to 1.0 signifies that the test has almost perfect diagnostic value. The maximum Youden index was used as a cut-off point. ROC curve analysis indicated that the tissue PHLDA3 expression level could be considered as a promising marker for the diagnosis of HNSCC patients with a sensitivity and specificity of 0.666 and 0.688, respectively (AUC= 0.710, P-value=0.000, 95% CI=0.603–0.816). In addition, the sensitivity and specificity of GRHL3 were 0.755 and 0.577, respectively (AUC=0.674, P-value=0.004, 95% CI=0.562–0.786), demonstrating its ability to discriminate HNSCC patients from healthy controls (Figure 4).
Figure 4.
The ROC curve was constructed using GraphPad Prism. The area under the curve (AUC) for GRHL3 and PHLDA3 is 0.674 and 0.710, respectively.
TCGA Dataset Analysis
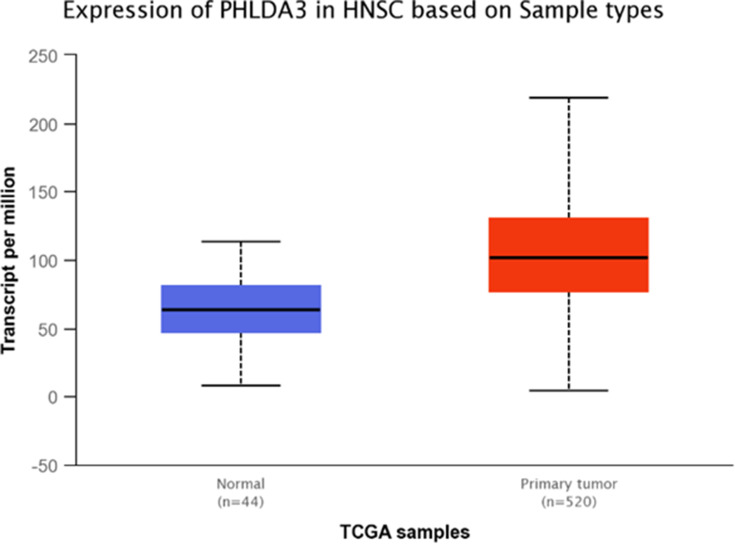
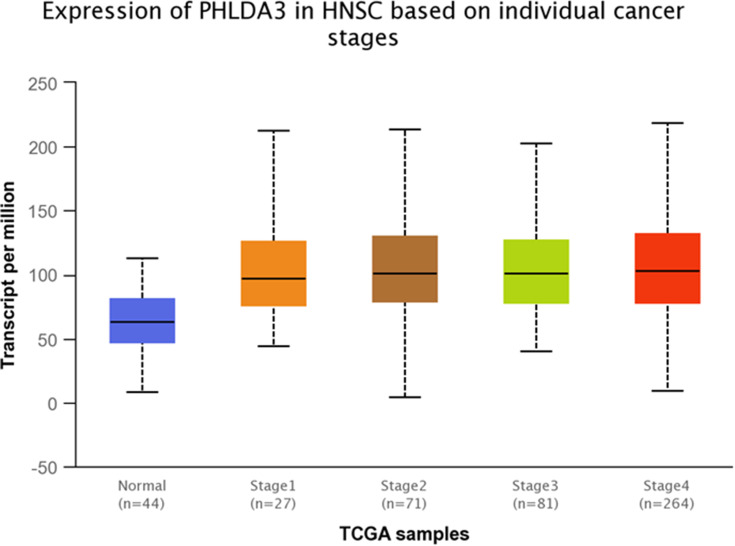
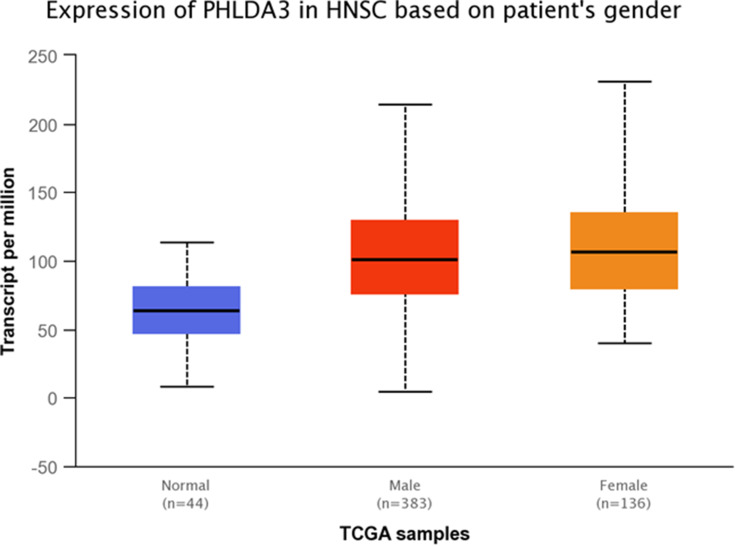
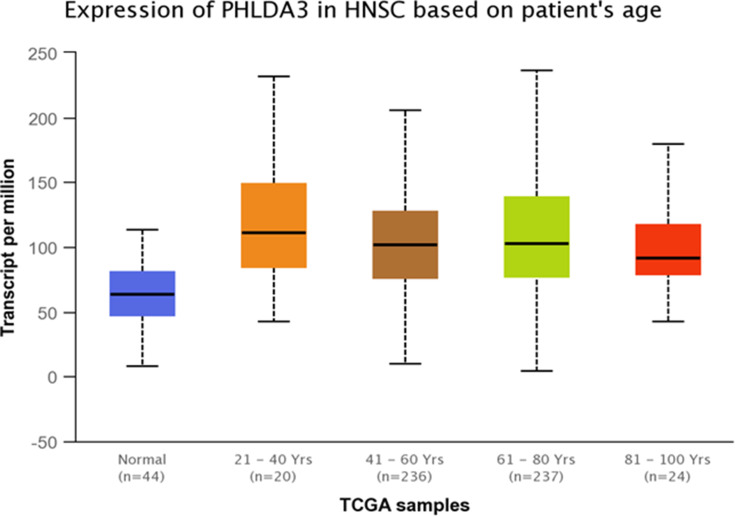
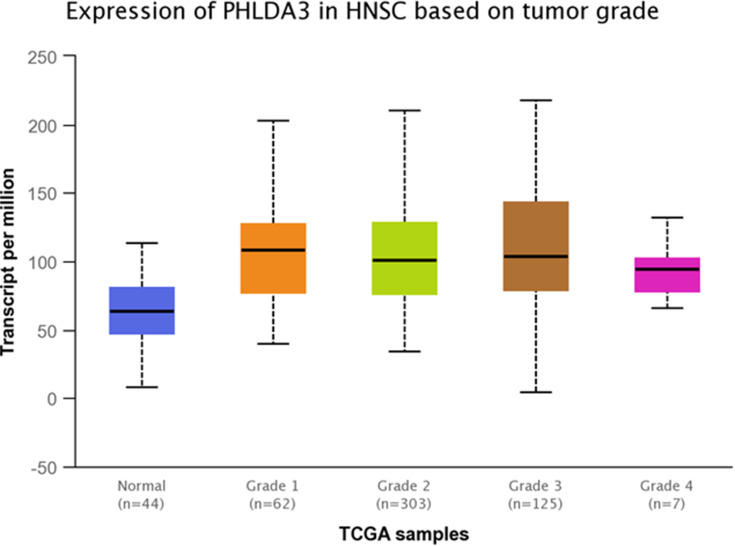
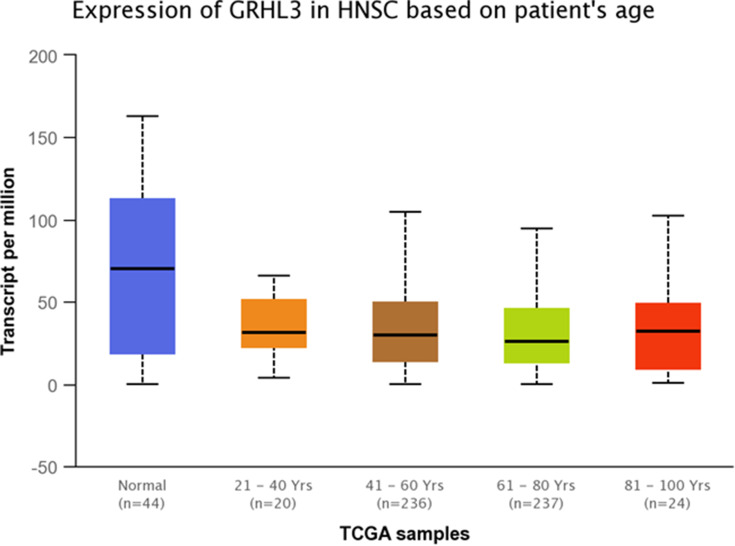
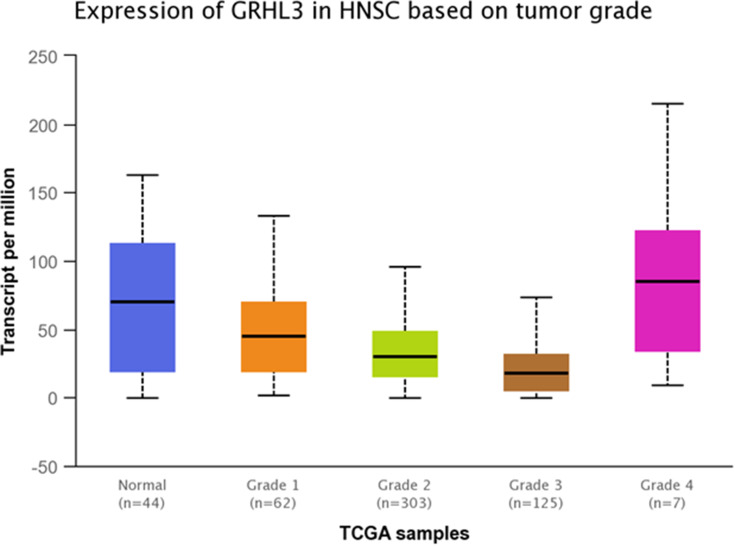
TCGA dataset analyses were performed to evaluate expressions of GRHL3 and PHLDA3 in HNSCC. The results showed that expression of PHLDA3 gene was significantly up-regulated in HNSCC tissues compared with normal tissues (Figure 5). Further assessment of data based on stages of HNSCC showed significant up-regulation of PHLDA3 in all stages of HNSCC compared with normal tissues (Figure 6 and Table 5). However, no difference was detected between different stages. We also assessed expression of genes based on their sex. Expression of PHLDA3 was higher in HNSCC tissues of both sexes compared with normal tissues. However, there was no difference between male and female patients (Figure 7 and Table 6). Assessment of expression of PHLDA3 in TCGA samples showed different levels of expression in tumors of all age-based subgroups compared with normal samples (Figure 8 and Table 7). Additional evaluation of data based on HNSCC grades showed significant up-regulation of PHLDA3 in all grades of HNSCC compared with normal tissues (Figure 9 and Table 8). However, no difference was detected between different grades except for comparison between grades 3 and 4.
Figure 5.
Relative expression of PHLDA3 in primary HNSCC tumors and normal tissues based on the data provided by TCGA (Statistical significance=1.0325074129014E-14).
Figure 6.
Relative expression of PHLDA3 in different stages of HNSCC tumors and normal tissues based on the data provided by TCGA.
Table 5.
Statistical Significance of Relative Expression of PHLDA3 in Different Stages of HNSCC Tumors and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Stage 1 | 4.967000E-04 |
| Normal vs Stage 2 | 9.33959998228318E-09 |
| Normal vs Stage 3 | 2.27800001084688E-09 |
| Normal vs Stage 4 | 1.68376423914651E-12 |
| Stage 1 vs Stage 2 | 7.347200E-01 |
| Stage 1 vs Stage 3 | 8.158600E-01 |
| Stage 1 vs Stage 4 | 9.008800E-01 |
| Stage 2 vs Stage 3 | 8.705000E-01 |
| Stage 2 vs Stage 4 | 6.441400E-01 |
| Stage 3 vs Stage 4 | 7.983600E-01 |
Note: Significance is shown by bold numbers.
Figure 7.
Relative expression of PHLDA3 in HNSCC tumors obtained from male and female patients and normal tissues based on the data provided by TCGA.
Table 6.
Statistical Significance of Relative Expression of PHLDA3 in HNSCC Samples of Different Sexes and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Male | 6.7612582199672E-14 |
| Normal vs Female | 5.14033260401447E-14 |
| Male vs Female | 2.862600E-01 |
Note: Significance is shown by bold numbers.
Figure 8.
Relative expression of PHLDA3 in HNSCC patients and normal tissues based on patients’ age.
Table 7.
Statistical Significance of Relative Expression of PHLDA3 in HNSCC Patients and Normal Tissues Based on Patients’ Age
| Comparison | Statistical Significance |
|---|---|
| Normal vs Age (21–40 years) | 8.019400E-04 |
| Normal vs Age (41–60 years) | 1.06492592522045E-12 |
| Normal vs Age (61–80 years) | 1.64968039229052E-12 |
| Normal vs Age (81–100 years) | 6.767000E-04 |
| Age (21–40 years) vs Age (41–60 years) | 1.880810E-01 |
| Age (21–40 years) vs Age (61–80 years) | 3.723800E-01 |
| Age (21–40 years) vs Age (81–100 years) | 5.286600E-01 |
| Age (41–60 years) vs Age (61–80 years) | 3.376200E-01 |
| Age (41–60 years) vs Age (81–100 years) | 7.099800E-01 |
| Age (61–80 years) vs Age (81–100 years) | 9.649600E-01 |
Note: Significance is shown by bold numbers.
Figure 9.
Relative expression of PHLDA3 in different grades of HNSCC tumors and normal tissues based on the data provided by TCGA.
Table 8.
Statistical Significance of Relative Expression of PHLDA3 in Different Grades of HNSCC Tumors and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Grade 1 | 5.96100000471722E-08 |
| Normal vs Grade 2 | 1.96609395430869E-12 |
| Normal vs Grade 3 | 1.91924254266951E-12 |
| Normal vs Grade 4 | 2.179900E-02 |
| Grade 1 vs Grade 2 | 4.443600E-01 |
| Grade 1 vs Grade 3 | 5.715600E-01 |
| Grade 1 vs Grade 4 | 6.253600E-02 |
| Grade 2 vs Grade 3 | 8.107700E-02 |
| Grade 2 vs Grade 4 | 3.607800E-01 |
| Grade 3 vs Grade 4 | 1.877650E-02 |
Note: Significance is shown by bold numbers.
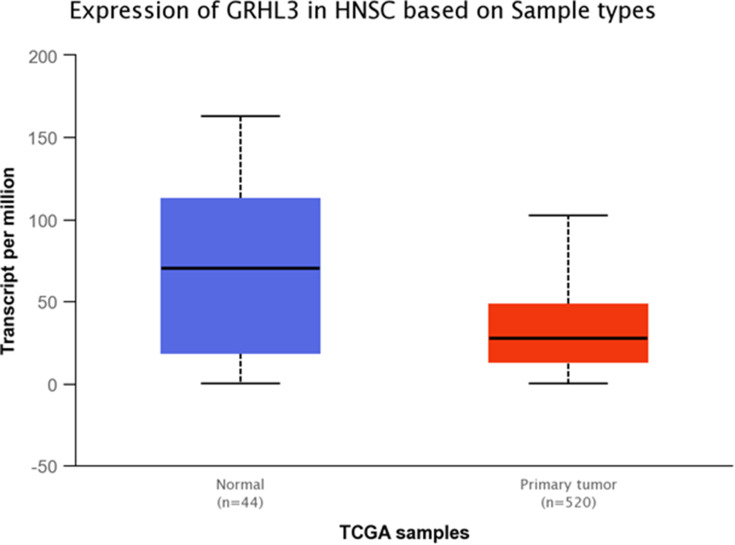
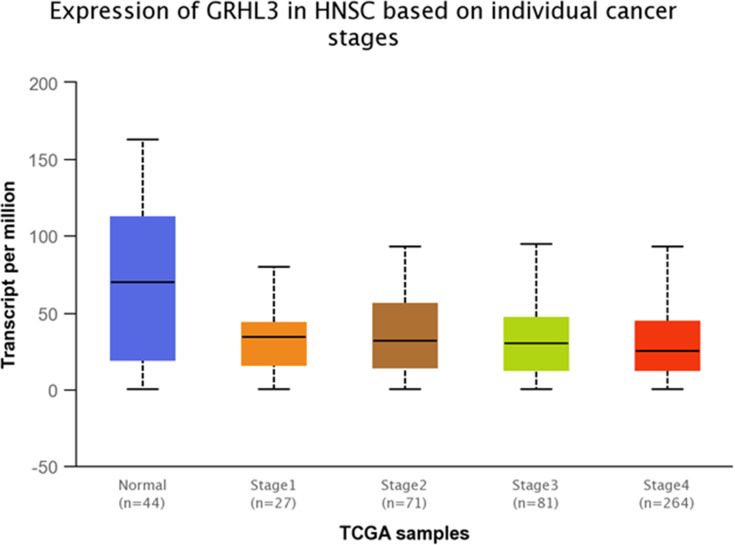
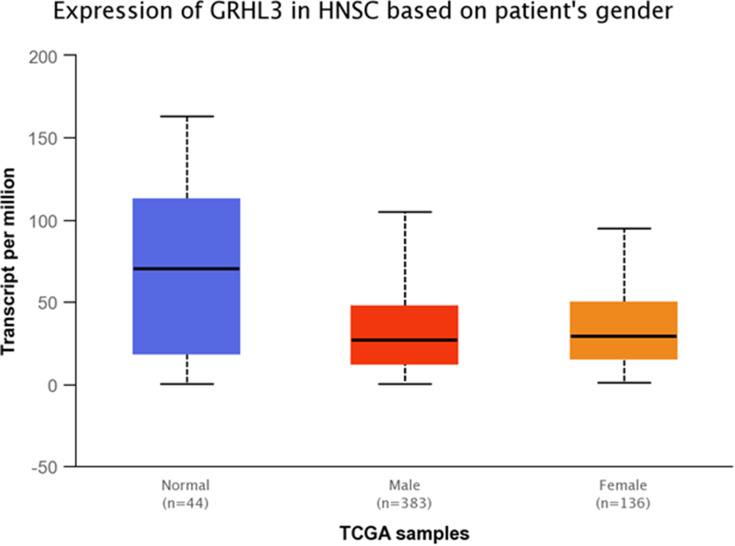
Expression of GRHL3 was significantly down-regulated in TCGA tumor samples compared with normal tissues (Figure 10). Additionally, GRHL3 was down-regulated in all stages of HNSCC compared with normal tissues (Figure 11 and Table 9). However, no difference was detected between different stages. Expression of GRHL3 was lower in HNSCC tissues of both sexes compared with normal tissues. However, there was no difference between male and female patients (Figure 12 and Table 10). Assessment of expression of GRHL3 in TCGA samples showed different levels of expression in tumors of all age-based subgroups compared with normal samples (Figure 13 and Table 11). Finally, evaluation of data based on HNSCC grades showed significant down-regulation of GRHL3 in grades 1–3 compared with normal tissues (Figure 14 and Table 12). However, no difference was detected between grade 4 and normal tissues.
Figure 10.
Relative expression of GRHL3 in primary HNSCC tumors and normal tissues based on the data provided by TCGA (Statistical significance=4.35819999999865E-05).
Figure 11.
Relative expression of GRHL3 in different stages of HNSCC tumors and normal tissues based on the data provided by TCGA.
Table 9.
Statistical Significance of Relative Expression of GRHL3 in Different Stages of HNSCC Tumors and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Stage 1 | 1.384340E-04 |
| Normal vs Stage 2 | 1.212600E-04 |
| Normal vs Stage 3 | 2.177500E-04 |
| Normal vs Stage 4 | 2.26539999998998E-05 |
| Stage 1 vs Stage 2 | 7.566200E-01 |
| Stage 1 s Stage 3 | 6.730800E-01 |
| Stage 1 vs Stage 4 | 8.224800E-01 |
| Stage 2 vs Stage 3 | 8.817200E-01 |
| Stage 2 vs Stage 4 | 4.218400E-01 |
| Stage 3 vs Stage 4 | 3.271000E-01 |
Note: Significance is shown by bold numbers.
Figure 12.
Relative expression of GRHL3 in HNSCC tumors obtained from male and female patients and normal tissues based on the data provided by TCGA.
Table 10.
Statistical Significance of Relative Expression of GRHL3 in HNSCC Samples of Different Sexes and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Male | 5.94999999999901E-05 |
| Normal vs Female | 3.90430000000164E-05 |
| Male vs Female | 5.796800E-01 |
Note: Significance is shown by bold numbers.
Figure 13.
Relative expression of GRHL3 in HNSCC patients and normal tissues based on patients’ age.
Table 11.
Statistical Significance of Relative Expression of GRHL3 in HNSCC Patients and Normal Tissues Based on Patients’ Age
| Comparison | Statistical Significance |
|---|---|
| Normal vs Age (21–40 yeas) | 1.606560E-04 |
| Normal vs Age (41–60 years) | 1.205040E-04 |
| Normal vs Age (61–80 years) | 2.27739999999477E-05 |
| Normal vs Age (81–100 years) | 6.153900E-04 |
| Age (21–40 years) vs Age (41–60 years) | 7.301600E-01 |
| Age (21–40 years) vs Age (61–80 years) | 6.165200E-01 |
| Age (21–40 years) vs Age (81–100 years) | 9.720000E-01 |
| Age (41–60 years) vs Age (61–80 years) | 1.568030E-01 |
| Age (41–60 years) vs Age (81–100 years) | 7.824200E-01 |
| Age (61–80 years) vs Age (81–100 years) | 7.384200E-01 |
Note: Significance is shown by bold numbers.
Figure 14.
Relative expression of GRHL3 in different grades of HNSCC tumors and normal tissues based on the data provided by TCGA.
Table 12.
Statistical Significance of Relative Expression of GRHL3 in Different Grades of HNSCC Tumors and Normal Tissues Based on the Data Provided by TCGA
| Comparison | Statistical Significance |
|---|---|
| Normal vs Grade 1 | 9.601700E-03 |
| Normal vs Grade 2 | 6.4151999999984E-05 |
| Normal vs Grade 3 | 4.95859999949388E-07 |
| Normal vs Grade 4 | 4.573400E-01 |
| Grade 1 vs Grade 2 | 8.861900E-03 |
| Grade 1 vs Grade 3 | 2.76570000001808E-06 |
| Grade 1 vs Grade 4 | 1.804570E-01 |
| Grade 2 vs Grade 3 | 6.20969999999277E-05 |
| Grade 2 vs Grade 4 | 9.618300E-02 |
| Grade 3 vs Grade 4 | 5.520000E-02 |
Note: Significance is shown by bold numbers.
Discussion
In the current study, we appraised the expression of two negative regulators of PI3K/AKT pathway, namely PHLDA3 and GRHL3, in 45 paired samples of HNSCC and ANCTs. We also compared our results with the results of TCGA data. Based on our results, expression of GRHL3 was down-regulated in tumoral tissues compared with ANCTs. However, expression of PHLDA3 was up-regulated in tumor tissues compared with ANCTs. Notably, analysis of their expression in TCGA samples verified our results.
GRHL3 has been identified as an important tumor suppressor which directly modulates PTEN and the PI3K/AKT/mTOR signaling pathway.12 Deletion of this gene has been commonly detected in HNSCC.11 Tumor suppressor function of Grhl3 in HNSCC has been verified in a conditional knockout mouse line. Although deletion of Grhl3 in oral epithelial cells did not affect the PTEN/PI3K/AKT/mTOR pathway, it caused significant down-regulation of GSK3B, leading to stabilization and accretion of c-MYC and development of aggressive HNSCC.11
PHLDA3 has been regarded as a p53-regulated suppressor of Akt.18 Assessment of genetic aberrations of the PHLDA3 locus in a large cohort of lung cancer samples revealed loss of this locus in large-cell neuroendocrine carcinoma and carcinoid samples, though no chromosomal aberration was detected in this locus in other histological types of lung cancer.18 Notably, Muroi et al19 have evaluated expression of PHLDA3 protein in esophageal SCC samples and detected a high level of its expression in the majority of samples, which is in accordance with our results in HNSCC. However, they found low PHLDA3 expression as a prognostic factor for tumor recurrence and low survival.19 Although PHLDA3 has been identified as a tumor suppressor which acts through suppression of Akt,18 Qiao et al.20 have shown that PHLDA3 inhibits somatic cell reprogramming through activation of the Akt-GSK3β pathway. This signaling pathway has a prominent role in attainment of the epithelial-mesenchymal transition (EMT) phenotype in the gefitinib-resistant HNSCC cells.21 Thus, it is possible that PHLDA3 exerts dual tumor suppressor and oncogenic roles based on the preferential mode of its regulatory effects on AKT downstream genes.
Based on the different directions of regulatory roles of PHLDA3 and GRHL3 on GSK3β, dysregulation of this downstream target of Akt is the possible mechanism for participation of PHLDA3 and GRHL3 in the pathogenesis of HNSCC.
The gender-based analysis revealed significant down-regulation of GRHL3 gene expression level in male patients compared with the control samples. However, the PHLDA3 gene was up-regulated in both sexes compared with control samples. Thus, one can consider a sex-based difference in the role of GRHL3 in HNSCC. Previous studies have noted sex-based differences in clinical manifestations and risk factors of HNSCC in terms of smoking and drinking habits and primary tumor site distribution.22 Moreover, the aneuploidy index has been different among HNSCC tumors of males and females.23 Consistent with these reports, Qadir et al.24 reported distinct molecular signatures in the HNSCC of males and females. However, assessment of expression of PHLDA3 and GRHL3 in TCGA samples revealed no difference in their expression between males and females.
Moreover, the differences in the expressions of both genes were significant in patients aged more than 60 years but not in the younger patients. Age has a remarkable influence on HNSCC prognosis and is associated with a distinctive epidemiology, hazards, and preferences.25 The distinct expression of these genes in elderly HNSCC patients adds to these unique patterns and might be associated with patients’ prognosis. However, assessment of TCGA data showed no difference in expression of genes between age-based subgroups. The inconsistency between TCGA data and our data is explained by the difference in determination of age subgroups.
Expression of GRHL3 was only down-regulated in patients with a positive smoking history. However, expression of PHLDA3 was up-regulated in patients with both positive and negative smoking history. Smoking might affect gene expression in HNSCC samples. Irimie et al.26 have identified a number of differentially expressed genes that distinguish HNSCC patients based on their smoking history. Most of these genes were enriched in cellular metabolism pathways. These observations indicate the prominence of the interaction between environmental risk factors and genetic elements in the pathogenesis of HNSCC.
Besides, expression of GRHL3 was decreased in grades 2 and 3 samples compared with controls. There was a significant increase in the transcript levels of PHLDA3 in stage II and III HNSCC samples when compared with the control group. Differences in the expression levels of these genes between distinct stages and grades of HNSCC have also been detected in TCGA samples. These data further support the participation of these genes in the pathogenesis of HNSCC.
ROC curve analysis indicated that the tissue expression level of PHLDA3 and GRHL3 could be considered as putative markers for assessment in larger cohorts of HNSCC patients. Taken together, the current study indicates dysregulation of regulators of the PI3K pathway in HNSCC and their potential application as putative biomarkers for this cancer.
Acknowledgment
The current study was supported by a grant from Tehran University of Medical Sciences.
Availability of Data
The data that support the findings of this study are available from the corresponding author (Dr. Abbas Shakoori) upon reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99(10):777–789. doi: 10.1093/jnci/djk179 [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550–4559. doi: 10.1200/JCO.2013.50.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pignon J-P, Le Maitre A, Maillard E, Bourhis J, Group M-NC. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): an update on 93 randomised trials and 17,346 patients. Radiother Oncol. 2009;92(1):4–14. doi: 10.1016/j.radonc.2009.04.014 [DOI] [PubMed] [Google Scholar]
- 6.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550. doi: 10.1038/nrc2664 [DOI] [PubMed] [Google Scholar]
- 7.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486. doi: 10.1038/onc.2008.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Dodhia S, Su GH. Dysregulations in the PI3K pathway and targeted therapies for head and neck squamous cell carcinoma. Oncotarget. 2017;8(13):22203. doi: 10.18632/oncotarget.14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729. doi: 10.1038/nrc1974 [DOI] [PubMed] [Google Scholar]
- 10.Wilanowski T, Cerruti L, Lin-Lin Z, Cunningham JM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370(3):953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgy SR, Cangkrama M, Srivastava S, et al. Identification of a novel proto-oncogenic network in head and neck squamous cell carcinoma. J Natl Cancer Inst. 2015;107(9). doi: 10.1093/jnci/djv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darido C, Georgy SR, Wilanowski T, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20(5):635–648. doi: 10.1016/j.ccr.2011.10.014 [DOI] [PubMed] [Google Scholar]
- 13.Lukosz M, Mlynek A, Czypiorski P, Altschmied J, Haendeler J. The transcription factor Grainyhead like 3 (GRHL3) affects endothelial cell apoptosis and migration in a NO-dependent manner. Biochem Biophys Res Commun. 2011;412(4):648–653. doi: 10.1016/j.bbrc.2011.08.018 [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Liu C, Zhao Z, et al. Clinical implications of GRHL3 protein expression in breast cancer. Tumor Biol. 2014;35(3):1827–1831. doi: 10.1007/s13277-013-1244-7 [DOI] [PubMed] [Google Scholar]
- 15.Ohki R PH domain-only protein PHLDA3; a p53-regulated repressor of Akt and a novel tumor suppressor of endocrine tumors. Paper presented at: GENES & GENETIC SYSTEMS2013. [Google Scholar]
- 16.Ohki R, Kawase T, Ohta T, Ichikawa H, Taya Y. Dissecting functional roles of p53 N‐terminal transactivation domains by microarray expression analysis. Cancer Sci. 2007;98(2):189–200. doi: 10.1111/j.1349-7006.2006.00375.x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Ohki R, Saito K, Chen Y, et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc Natl Acad Sci. 2014;111(23):E2404–E2413. doi: 10.1073/pnas.1319962111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawase T, Ohki R, Shibata T, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136(3):535–550. doi: 10.1016/j.cell.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 19.Muroi H, Nakajima M, Satomura H, et al. Low PHLDA3 expression in oesophageal squamous cell carcinomas is associated with poor prognosis. Anticancer Res. 2015;35(2):949–954. [PubMed] [Google Scholar]
- 20.Qiao M, Wu M, Shi R, Hu W. PHLDA3 impedes somatic cell reprogramming by activating Akt-GSK3β pathway. Sci Rep. 2017;7(1):2832. doi: 10.1038/s41598-017-02982-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maseki S, Ijichi K, Tanaka H, et al. Acquisition of EMT phenotype in the gefitinib-resistant cells of a head and neck squamous cell carcinoma cell line through Akt/GSK-3β/snail signalling pathway. Br J Cancer. 2012;106(6):1196–1204. doi: 10.1038/bjc.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang T, Zhang C, Geye HM, et al. Gender associations in head and neck squamous cell carcinoma (HNSCC). J Clin Oncol. 2012;30(15_suppl):e16010. [Google Scholar]
- 23.Hollows R, Wei W, Cazier JB, et al. Association between loss of Y chromosome and poor prognosis in male head and neck squamous cell carcinoma. Head Neck. 2019;41(4):993–1006. doi: 10.1002/hed.25537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadir F, Lalli A, Dar HH, et al. Clinical correlation of opposing molecular signatures in head and neck squamous cell carcinoma. BMC Cancer. 2019;19(1):830. doi: 10.1186/s12885-019-6059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massa ST, Cass LM, Challapalli S, et al. Demographic predictors of head and neck cancer survival differ in the elderly. Laryngoscope. 2019;129(1):146–153. doi: 10.1002/lary.27289 [DOI] [PubMed] [Google Scholar]
- 26.Irimie AI, Braicu C, Cojocneanu R, et al. Differential effect of smoking on gene expression in head and neck cancer patients. Int J Environ Res Public Health. 2018;15(7):1558. doi: 10.3390/ijerph15071558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (Dr. Abbas Shakoori) upon reasonable request.