Summary
Antibody responses develop following SARS-CoV-2 infection, but little is known about their epitope specificities, clonality, binding affinities, epitopes, and neutralizing activity. We isolated B cells specific for the SARS-CoV-2 envelope glycoprotein spike (S) from a COVID-19-infected subject 21 days after the onset of clinical disease. 45 S-specific monoclonal antibodies were generated. They had undergone minimal somatic mutation with limited clonal expansion, and three bound the receptor-binding domain (RBD). Two antibodies neutralized SARS-CoV-2. The most potent antibody bound the RBD and prevented binding to the ACE2 receptor, while the other bound outside the RBD. Thus, most anti-S antibodies that were generated in this patient during the first weeks of COVID-19 infection were non-neutralizing and target epitopes outside the RBD. Antibodies that disrupt the SARS-CoV-2 S-ACE2 interaction can potently neutralize the virus without undergoing extensive maturation. Such antibodies have potential preventive and/or therapeutic potential and can serve as templates for vaccine design.
Keywords: COVID-19, SARS, SARS-CoV-2, ACE2, antibodies, B cells, spike protein, receptor-binding domain, neutralization, MERS
Graphical Abstract
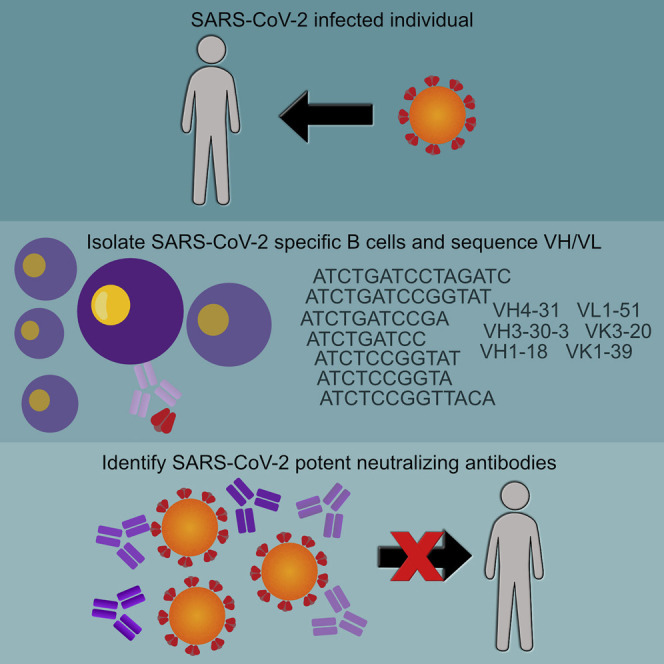
Highlights
-
•
Early B cell responses to SARS-CoV-2 spike protein are analyzed from a COVID-19 patient
-
•
Most antibodies target non-neutralizing epitopes outside the RBD
-
•
A potent neutralizing mAb blocks the interaction of the S protein with ACE2
-
•
Neutralizing antibodies are minimally mutated
Seydoux et al. analyze B cell responses in a COVID-19 patient and find that SARS-CoV-2 infection expands diverse B cell clones against the viral spike glycoprotein (S). Two neutralizing antibodies were identified that bind S with high affinity despite being minimally mutated. Thus, vaccine-induced neutralizing antibody responses may require activation of specific naive B cells without requiring extensive somatic mutation.
Introduction
The World Health Organization (WHO) declared the 2020 COVID-19 to be a global pandemic on March 11, 2020 (World Health Organization, 2020). According to data compiled from multiple local and government sources compiled by a team at Johns Hopkins University, as of June 12, 2020, there are currently 7.5 million documented cases of COVID-19 and over 420,000 deaths (Dong et al., 2020). The infection is caused by SARS-CoV-2, a beta coronavirus, closely related to SARS-CoV (Wan et al., 2020). Presently, the immune response to COVID-19 is not well understood and preventative measures, such as vaccines, are not available. It is also unclear which immune responses are required to prevent or control SARS-CoV-2 infection.
High-resolution structures of the SARS-CoV-2 prefusion-stabilized spike (S) ectodomain revealed that it adopts multiple conformations with either one receptor-binding domain (RBD) in the “up” or “open” conformation or all RBDs in the “down” or “closed” conformation, similar to previous reports on both SARS-CoV S and MERS-CoV S (Gui et al., 2017, Kirchdoerfer et al., 2018, Pallesen et al., 2017, Song et al., 2018, Walls et al., 2019, Walls et al., 2020, Wrapp et al., 2020, Yuan et al., 2017). Like SARS-CoV, SARS-CoV-2 utilizes angiotensin-converting enzyme 2 (ACE2) as an entry receptor binding with nM affinity (Li et al., 2003, Walls et al., 2020, Wrapp et al., 2020; Hoffmann et al., 2020, Letko et al., 2020, Ou et al., 2020). Indeed, the S proteins of the two viruses share a high degree of amino acid sequence homology, 76% overall and 74% in RBD (Wan et al., 2020).
Although binding and neutralizing antibody responses are known to develop following SARS-CoV-2 infection (Ni et al., 2020, Okba et al., 2020), no information is currently available on the epitope specificities, clonality, binding affinities, and neutralizing potentials of the antibody response.
Monoclonal antibodies (mAbs) isolated from SARS-CoV-infected subjects can recognize the SARS-CoV-2 S protein (Yuan et al., 2020), and immunization with SARS S protein can elicit anti-SARS-CoV-2 neutralizing antibodies in wild-type and humanized mice, as well as llamas (Walls et al., 2020, Wang et al., 2020, Wrapp et al., 2020). However, SARS-CoV-2 infection appears to not elicit strong anti-SARS-CoV neutralizing antibody responses and vice versa (Ou et al., 2020).
Here, we employed diverse but complementary approaches to investigate the serum binding and neutralizing antibody responses to a stabilized ectodomain variant of the SARS-CoV-2 S-protein (S2P) as well as the frequency and clonality of S2P-specific B cells in a SARS-CoV-2-infected individual 21 days following the onset of clinical disease. We isolated anti-SARS-CoV-2 S mAbs and characterized their binding properties and determined their neutralizing potencies. Among all B cells analyzed, no particular variable heavy (VH) or variable light (VL) gene family was expanded, and the isolated antibodies were minimally mutated. Our analysis reveals that only a small fraction of S2P-specific B cells recognized the RBD. Of the forty-five mAbs analyzed, only three displayed neutralizing activity. The most potent mAb, CV30, bound the RBD in a manner that disrupted the S-ACE2 interaction. The other two mAbs, CV1 and CV35, were clonal variants that bound to an epitope distinct from the RBD and were much less potent.
Results
A SARS-CoV-2 Infected Donor Displays Potent Neutralizing Activity within 3 Weeks of Clinical Disease Onset
Serum and peripheral blood mononuclear cells (PBMCs) were collected 21 days after the onset of clinical disease from one of the first patients infected with SARS-CoV-2 in the state of Washington. He was a 35-year-old male hospitalized for over 10 days with severe disease and received therapy with fluids, oxygen, and remdesivir.
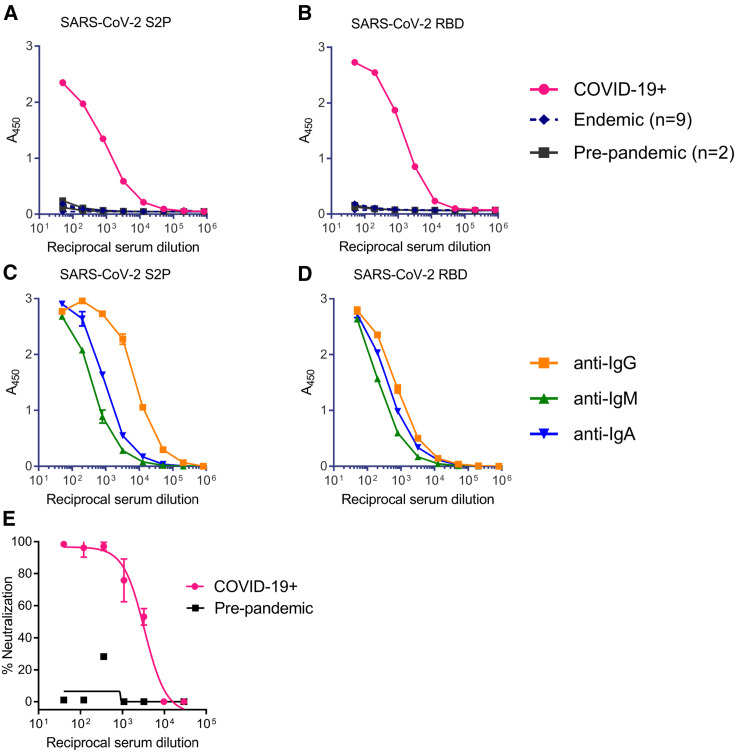
The serum contained high titers of antibodies to the SARS-CoV-2 S2P (Figure 1 A). The specificity of this response was confirmed by the absence of S2P reactivity by serum antibodies isolated from donors collected prior to the SARS-CoV-2 pandemic or donors with confirmed infection by endemic coronaviruses. We also measured the serum antibody response to RBD and again observed specific high titers of binding antibodies (Figure 1B). Isotype-specific ELISA revealed that the immunoglobulin G (IgG) titers were higher than the IgA and the IgM titers to both S2P and RBD, which suggested that a significant portion of the antibody responses to the ectodomain of the SARS-CoV-2-S were IgG (Figures 1C and 1D). The serum from the SARS-CoV-2 infected donor displayed potent neutralizing activity (reciprocal ID50 ∼3,000) against a pseudovirus expressing the S protein from SARS-CoV-2 isolate Wuhan-Hu-1 (Figure 1E). We concluded that this donor had developed strong binding and neutralizing antibody responses within 3 weeks of disease onset.
Figure 1.
SARS-CoV-2 Infection Elicits Binding and Neutralizing Antibodies Directed at the Spike Protein
(A–D) Total antibody binding in serum from a donor with confirmed SARS-CoV-2 infection (COVID-19+), from two donors collected prior to the COVID-19 pandemic with an unknown history of coronavirus infection (pre-pandemic), and from nine donors with confirmed infection by endemic corona viruses (endemic), was tested for binding to the SARS-CoV-2 S2P ectodomain (A) and the RBD (B) by ELISA. Serum from the donor in SARS-CoV-2 infection in (A) was tested for binding to the SARS-CoV-2 S2P ectodomain (C) and the RBD (D) using isotype-specific secondary antibodies by ELISA.
(E) Serum from donor with confirmed SARS-CoV-2 infection, and serum from a pre-pandemic donor were evaluated for their ability to neutralize a SARS-CoV-2 pseudovirus. Data points indicate the mean, and error bars represent standard deviation of 2–3 technical replicates. Due to the limited amount of serum available, these analyses were restricted to a single analysis.
Isolation of SARS-CoV-2 Spike Specific B Cells
To identify B cells specific to the SARS-CoV-2 S protein that were circulating at this time point, fluorescently labeled S2P and RBD probes were used as baits. S2P was labeled with either phycoerythrin (PE) or brilliant violet 711 (BV711) and used to stain B cells concurrently. This double labeling strategy helped to discriminate between bona fide S2P-specific B cells and non-specific background staining to the fluorophores. RBD was labeled with Alexa Fluor 647 to identify B cells specific for that domain.
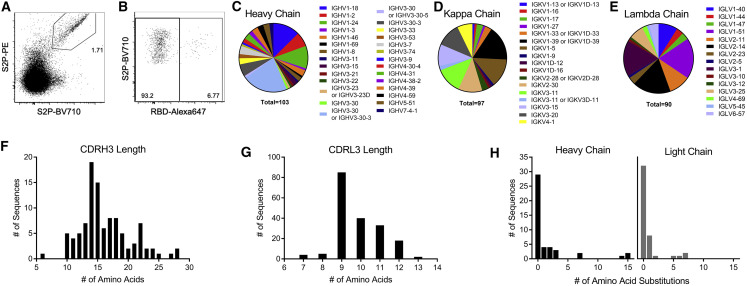
Approximately 0.65% of total CD19+ B cells were S2P+ compared to 0.07% of total B cells from a naive donor (Figures S1A and S1B). The dominant responding B cells were IgM+ IgD+ (49% of S2P+ B cells) (Figure S1C); 90% of which were CD27+ that suggested that although these B cells have not class-switched, they were antigen-experienced memory B cells. The second most prominent subset of S2P-specific B cells were class-switched IgG+ IgD− B cells (27% of S2P+ B cells) (Figure S1C). In fact, 1.7% of the IgG+ B cells stained with S2P (Figure 2 A) and of those ∼7% (or 0.12% of total IgG+ B cells) (Figure 2B) were also positive for the RBD.
Figure 2.
Early B Cell Response to SARS-CoV-2 Is Diverse and Largely Unmutated
(A) Class switched (IgM− IgG+) B cells were stained with SARS-CoV-2 S2P labeled with BV710 or PE.
(B) SARS-CoV-2 S2P+ IgG+ B cells were further analyzed for binding to Alexa Fluor 647-labeled SARS-CoV-2 RBD.
(C–E) Individual SARS-CoV-2 S2P+ IgG+ B cells were sorted into separate wells of a 96-well plate and sequenced using RT-PCR. VH (C), VK (D), and VL (E) gene usage of successfully sequenced S2P-specific B cells.
(F and G) CDRH3 (F) and CDRL3 (G) length distributions of successfully sequenced S2P-specific B cells.
(H) Number of amino acid substitutions from germline in S2P-specific heavy and light chains.
We hypothesized that the class-switched SARS-CoV-2-specific B cells were more likely to have undergone some affinity maturation and contain antibodies capable of neutralizing the virus. Thus, we focused on B cell receptor (BCR) sequencing of S2P+ IgG+ B cells. 576 S2P+ B cells were single-cell sorted into individual wells of 96-well plates and the variable heavy and light chain regions of B cell receptor transcripts were sequenced using nested RT-PCR. We successfully recovered 103 VH sequences, and 187 VL sequences, 97 of which were kappa and 90 were lambda. B cells specific for S2P were derived from diverse antibody heavy and light chain genes (Figures 2C–2E) and had normal distributions of CDRH3 and CDRL3 lengths (Figures 2F and 2G). Consistent with the relatively short time of infection, the majority of BCR sequences showed low levels of somatic mutation (Figure 2H). We conclude that the early B cell response to the SARS-CoV-2 S protein is polyclonal and directed at epitopes mostly outside of the RBD.
Determining the Binding Specificities of Anti-S mAbs
Among all successfully sequenced VH and VL transcripts, we obtained paired sequences from forty-five. These were produced as recombinant mAbs of the IgG1 isotype and tested for binding to recombinant S-derived proteins (Figure 3 ).
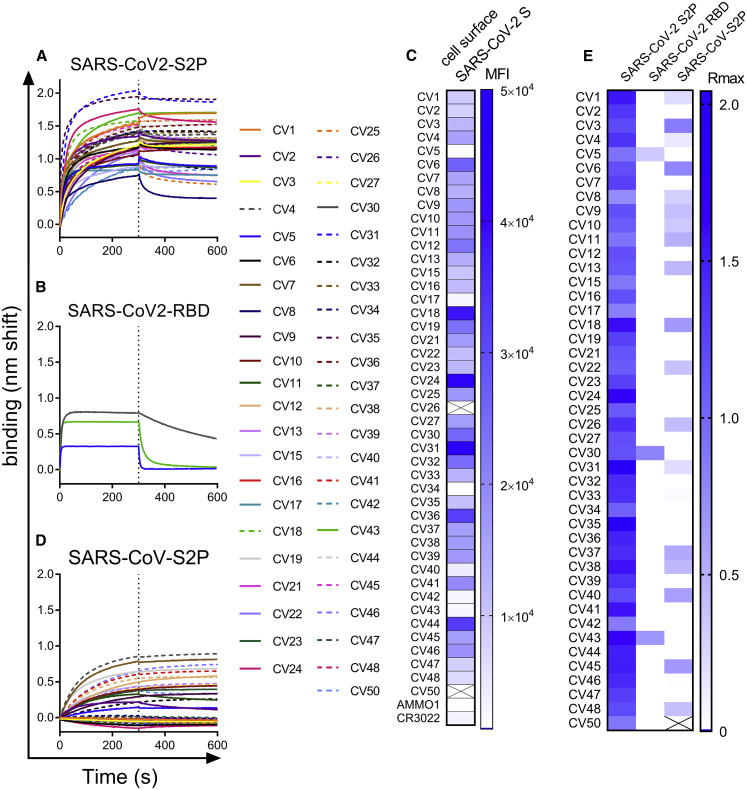
Figure 3.
Recombinant Anti-spike mAbs Bind to a Stabilized SARS-CoV-2 Ectodomain Trimer and a Subset Cross-React with SARS-CoV S
(A and B) mAbs isolated from SARS-CoV-2 S2P-specific B cells were tested for binding to SARS-CoV-2 S2P (A) and to SARS-CoV-2 RBD (B) using BLI.
(C) mAbs were labeled with phycoerythrin (PE) and used to stain 293 cells transfected with wild-type SARS-Cov-2 S by flow cytometry. Heatmap shows mean fluorescence intensity of PE+ cells at 2.5 μg/mL. Titration curves are shown in Figure S2. Boxes with an X through them indicate antibodies that were not tested.
(D) mAbs were tested for binding to SARS-CoV S2P by BLI (D).
(E) Heatmap shows maximum binding response (average nm shift of the last 5 s of association phase) of binding data in (A), (B), and (D). Data presented here are representative of 2 independent experiments.
See also Figure S2.
Most of the mAbs represented unique rearrangements; however, we identified at least 4 expanded clones. CV4 and CV7 were identical and unmutated. CV1 and CV35 differed by one synonymous LC mutation, CV40 and CV47 differed by 5 amino acids (Table S1). CV12, CV39, and CV46 were much more mutated, up to 15 heavy chain and 6 light chain mutations for CV12 (Table S1). Due to the relatively short window of time between SARS-CoV-2 infection and PBMC collection, these mAbs may represent an expansion of a memory B cell from a previous infection with an endemic coronavirus.
All the mAbs bound to the stabilized SARS-CoV-2 ectodomain, S2P (Figures 3A and 3E). Consistent with the B cell staining that revealed very few RBD-specific B cells (Figure 2B), only three mAbs, CV5, CV30, and CV43, also bound the SARS-CoV-2 RBD (Figures 3B and 3E). The majority of S2P-specific mAbs also bound to full-length membrane-bound wild-type SARS-CoV-2 S on the surface of 293 cells (Figures 3C and S2). The observation that some S2P-specific mAbs failed to bind to cell surface S indicated that there may be conformational differences between the stabilized soluble ectodomain and cell surface S. The fact that a subset of the mAbs bound to a stabilized ectodomain variant of the closely related SARS-CoV S protein (Figures 3D and 3E) demonstrated that there are conserved epitopes among the two viruses. Consistent with the lower degree of conservation of the S1 subunit between SARS-CoV and SARS-CoV-2, the anti-RBD mAbs CV30 and CV43 did not cross react with SARS-CoV S2P, whereas CV5 showed weak binding. These results confirmed that the antibody responses to the S-protein are largely outside of the RBD, and the conformation of the stabilized ectodomain variant may differ from the membrane anchored SARS-CoV-2 S.
The Early Antibody Response to SARS-CoV-2 Infection Is Largely Non-neutralizing
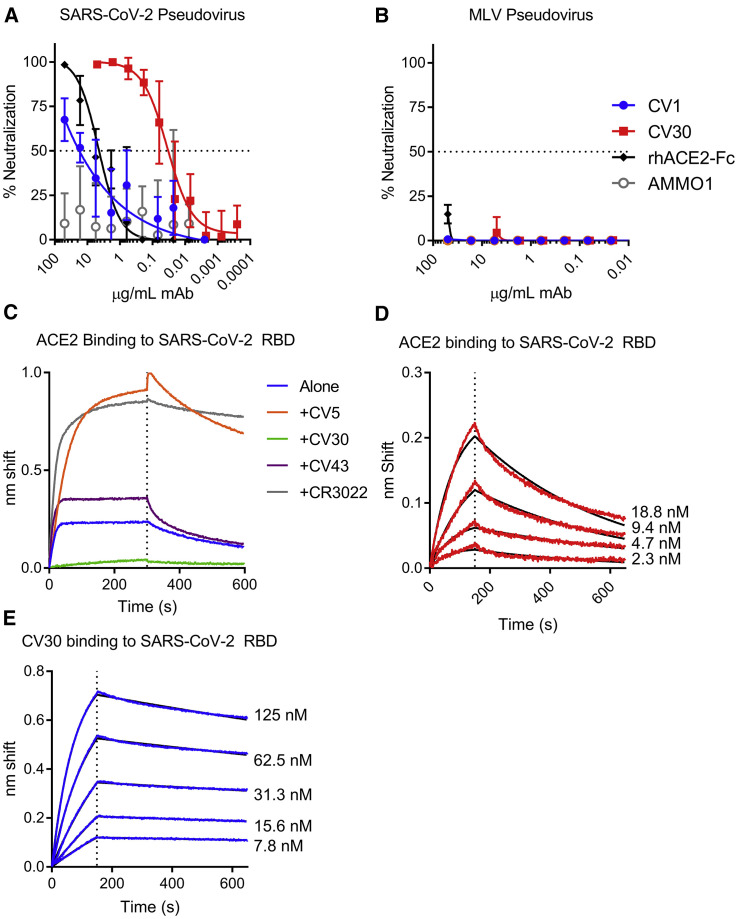
The S2P-binding mAbs were evaluated for their ability to neutralize SARS CoV-2 pseudovirus infection of 293T cells stably expressing ACE2. All but three of the mAbs were non-neutralizing (Figure 4 A; Table S1). Although they did not achieve 100% neutralization at the highest concentration, CV1 and the clonal variant CV35, which bound to an epitope outside the RBD, were weakly neutralizing (IC50 = 15 μg/mL) (Figure 4A; Table S1). CV1 and CV35 neutralized less potently than a rhesus ACE2-Fc fusion protein that acts as a soluble competitor for the interaction between S and the cell surface-expressed ACE2 (IC50 = 2.2 μg/mL). In contrast, CV30 achieved 100% neutralization and was ∼530 times more potent than CV1 and CV35 (IC50 = 0.03 μg/mL) (Figure 4A; Table S1). CV1, CV30, and the ACE2-Fc fusion did not neutralize a murine leukemia virus pseudovirus demonstrating their specificity for the SARS-CoV-2 S protein (Figure 4B). CV30 is derived from a heavy chain utilizing an IGHV3-53∗01 VH gene and an IGKV3-20∗01 VL gene. CV1 and CV35 bind an epitope outside the RBD and are derived from an IGHV4-38∗02 VH gene and an IGLV1-44∗01 VL gene.
Figure 4.
The RBD-Specific mAb CV30 Neutralizes SARS CoV-2 by Blocking the ACE2- SARS-CoV-2 S Interaction
(A) CV1 and CV30 were serially diluted and tested for their ability to neutralize SARS-CoV-2 pseudovirus infection of 293T cells stably expressing ACE2. An ACE2-FC fusion and the anti-EBV mAb AMMO1 were included as positive and negative controls. Data points represent the mean and, error bars indicate the standard deviation of quadruplicate replicates. Data are representative of 6 independent experiments (see Table S1 for details).
(B) The same mAbs were tested for neutralization of an MLV pseudovirus. Data points represent the mean, and error bars indicate the standard deviation of quadruplicate replicates.
(C) Biotinylated ACE2-Fc was immobilized on streptavidin biosensors and then tested for binding to SARS-CoV-2 RBD in the absence and presence of the indicated mAbs using BLI.
(D) ACE2-Fc was immobilized Protein A biosensors and binding to the indicated serial dilutions of SARS-CoV-2 RBD were measured by BLI and used to determine the dissociation constant (KD). Red lines represent the measured data and black lines indicate the theoretical fit.
(E) CV30 was immobilized onto anti-human Fc biosensors and binding to the indicated serial dilutions of SARS-CoV-2 RBD were measured by BLI and used to determine the binding constant (kD). Blue lines represent the measured data and black lines indicate the theoretical fit.
Kinetic measurements from (D) and (E) are summarized in Table S2. BLI analyses are representative of 2–3 independent experiments.
Based on the observations that CV30 is potently neutralizing and it binds the RBD, we investigated whether it would block the interaction between the SARS-CoV-2 S protein and the ACE2 receptor. To this end, we set up binding competition experiments using BLI. Indeed, CV30 inhibited the RBD-ACE2 interaction. In contrast, CV5 and CV43, the other two anti-RBD mAbs and the CR3022 mAb, which binds the RBD outside of the ACE2 binding site, did not (Figure 4C) (ter Meulen et al., 2006, Yuan et al., 2020). We also measured the relative binding affinities of CV30 and ACE2 to the SARS-CoV-2 RBD. ACE2 and CV30 bound the RBD with comparable affinities, 4.4 nM and 3.6 nM (Figures 4D and 4E; Table S2). However, the kinetics of the interactions were notably different. ACE2 had both a faster association and dissociation rate than CV30 (Figures 4D and 4E; Table S2). Collectively these results indicate that CV30 neutralizes SARS-CoV-2 infection by blocking the S-ACE2 interaction through an interaction that is slightly higher affinity.
Discussion
The development of therapeutic interventions, of immunoprophylaxis, and of an effective vaccine against SARS-CoV-2 will benefit from understanding the protective immune responses elicited during infection. A recent report indicates that neutralizing antibodies are present in the sera collected from convalescent COVID-19 patients (Ni et al., 2020, Okba et al., 2020). However, the kinetics of neutralizing antibody development as well as the characteristics and epitope specificities of neutralizing antibodies generated during SARS-CoV-2 infection are presently poorly understood.
Serological analysis revealed that the SARS-CoV-2-infected patient studied here developed high titers of binding and neutralizing antibody responses 21 days following the onset of clinical disease. The development of neutralizing antibody titers at this early time point has been reported for other COVID-19 patients (Ni et al., 2020, Okba et al., 2020) and is consistent with the rapid development of neutralizing responses to SARS-CoV infection (Corti et al., 2011). At this time point, IgG constituted the major fraction of anti-S2P and anti-RBD serum antibodies, although both IgM and IgA antibodies against these viral antigens were detected in the serum as well. The S2P-specific, class-switched B cells circulating at this time point were not dominated by any particular clone. Rather, they were derived from a diverse VH/VL gene repertoire, with frequencies similar to those reported in healthy uninfected individuals (Briney et al., 2019, DeKosky et al., 2016, Soto et al., 2019, Vázquez Bernat et al., 2019).
Although anti-S2P antibodies isolated at this time point could bind the S protein, the majority lacked neutralizing activity. However, two unique mAbs, CV1/CV35 and CV30, were able to neutralize SARS-CoV-2. Thus, although diverse B cell clones became activated during infection, the serum neutralizing activity is likely directed against a relatively small subset of epitope specificities.
CV1/CV35 binds to an unknown epitope region outside of the RBD, but the more potent CV30 recognizes the RBD and likely neutralizes infection by directly inhibiting SARS-CoV-2 S binding to the ACE2 receptor. The RBD is a major target of neutralizing antibodies in SARS-CoV infection (Cao et al., 2010). Several neutralizing mAbs that block the interaction of SARS-CoV with the ACE2 receptor have been described (Hwang et al., 2006, Prabakaran et al., 2006, Rockx et al., 2008, Sui et al., 2004, Walls et al., 2019). Moreover, the neutralizing potency correlated with the degree of S-ACE2 inhibition (Rockx et al., 2008). The RBD of MERS-CoV is also a target of potent neutralizing antibodies (Jiang et al., 2014, Niu et al., 2018, Tang et al., 2014, Ying et al., 2014), highlighting the importance of receptor blocking antibodies for coronavirus vaccine development. Although SARS-CoV and SARS-CoV-2 share extensive amino acid sequence in the RBD (74%), and both viruses utilize human ACE2 for entry, the amino acid identity in the receptor binding motif is only ∼50% (Wan et al., 2020). In line with this, potent anti-SARS-CoV neutralizing mAbs that bind RBD fail to cross react with SARS-CoV-2 (Wrapp et al., 2020), similarly the anti-RBD mAb, CV30 described herein fails to cross react with the SARS-CoV S-protein.
Consistent with the short time period post-infection, the majority of S-specific BCRs from individual B cells was unmutated or had only accumulated very few mutations. This was true for the neutralizing antibodies as well. The V-gene encoded amino acid sequences of CV1 and CV35 were identical to the chromosomally encoded sequence, while CV30 had 2 amino acid mutations in VH and none in VL. Largely unmutated antibodies against SARS-CoV S (Prabakaran et al., 2006, Sui et al., 2004) and MERS-CoV S (Jiang et al., 2014, Tang et al., 2014, Ying et al., 2014, Ying et al., 2015) have been isolated from phage display libraries created from uninfected donors.
Potent anti-SARS-CoV neutralizing mAbs that bind RBD are derived from different VH genes (VH1-18, VH1-69, or VH3-30) than CV30 (Prabakaran et al., 2006, Sui et al., 2004, Traggiai et al., 2004, Walls et al., 2019). Anti-RBD antibodies that neutralize MERS are derived from diverse gene families (Jiang et al., 2014, Niu et al., 2018, Tang et al., 2014).
Collectively, these results indicate that some high-affinity coronavirus-neutralizing antibodies require a short developmental pathway. This suggests that a vaccine against this virus may only need to activate a subset of B cells for potent neutralizing antibody responses to be developed, and potent neutralizing antibodies may not be V-gene restricted.
In sum, we provide information on the characteristics of early antibody and B cell responses to the SARS-CoV-2 S-protein during infection. Moreover, the neutralizing antibodies discussed here can serve as templates for the design of immunogens and potentially have utility as therapeutic and prophylactic agents to combat the SARS-CoV2 pandemic.
Limitations of Study
We acknowledge that our analysis is limited to one SARS-CoV-2-infected donor analyzed at an early time point. Comparable analyses of the humoral response from several SARS-CoV-2 infected donors with varying degrees of clinical symptoms, ideally with longitudinal sampling, will be required to determine whether the findings presented here are generalizable.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4 BB515 | BD Biosciences | Cat# 565996; RRID:AB_2739447 |
| CD38 BB700 | BD Biosciences | Cat# 566445; RRID:AB_2744375 |
| IgM PE/Dazzle 594 | BioLegend | Cat# 314530; RRID:AB_2566483 |
| CD14 PE-Cy5 | eBioscience | Cat# 15-0149-42; RRID:AB_2573058 |
| CD56 PE-Cy5 | BioLegend | Cat# 318308; RRID:AB_604105 |
| CD19 PE-Cy7 | BD Biosciences | Cat# 557835; RRID:AB_396893 |
| CD8 Fluor 700 | BD Biosciences | Cat# 557945; RRID:AB_396953 |
| CD69 APC/Fire750 | BioLegend | Cat# 310946; RRID:AB_396953 |
| CD3 BV510 | BioLegend | Cat# 317332; RRID:AB_396953 |
| CD27 BV605 | BioLegend | Cat# 302830; RRID:AB_2561450 |
| IgD BV650 | BD Biosciences | Cat# 740594; RRID:AB_2740295 |
| IgG BV786 | BD Biosciences | Cat# 564230; RRID:AB_2738684 |
| Peroxidase-conjugated AffiniPure Donkey Anti-Human IgG, Fcγ fragment specific | Jackson ImmunoResearch | Cat#709-035-098; RRID:AB_2340494 |
| Mouse anti-Human IgM-HRP | Southern Biotech | Cat# 9022-05; RRID:AB_2796584 |
| Mouse anti-Human IgA-HRP | Southern Biotech | Cat# 9130-05; RRID:AB_2796654 |
| PE-conjugated AffiniPure Fab fragment goat anti-human IgG | Jackson ImmunoResearch | Cat #109-117-008; RRID:AB_2632442 |
| CR3022 | ter Meulen et al., 2006 | N/A |
| AMMO1 | Snijder et al., 2018 | N/A |
| Biological Samples | ||
| PMBC from SARS-CoV-2-infected donor | This study | N/A |
| PMBC from pre-pandemic donors | This study | N/A |
| PBMC from donors with confirmed endemic CoV infection | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Streptavidin-Phycoerythrin (PE) | Invitrogen | Cat# S21388 |
| Streptavidin-Alexa Fluor 647 | Invitrogen | Cat# S32357 |
| Streptavidin-BV711 | BioLegend | Cat# 405241 |
| 7-AAD (7-Aminoactinomycin D) | Invitrogen | Cat# A1310 |
| HisTrap FF affinity column | GE Healthcare | Cat# 17-5255-01 |
| Strep-Tactin Sepharose | IBA Lifesciences | Cat# 2-1201-010 |
| Strep-Tactin Purification Buffer Set | IBA Lifesciences | Cat# 2-1201-001 |
| Superose 6 10/300 GL column | GE Healthcare | Cat# 17-5172-01 |
| HiLoad 16/600 Superdex 200 pg column | GE Healthcare | Cat# 28-9893-35 |
| Enrich SEC 650 10X300 column | Bio-Rad | Cat# 7801650 |
| Protein A agarose resin | GoldBio | Cat# P-400 |
| SureBlue Reserve TMB Peroxidase Substrate | Seracare KPL | Cat# 5120-0080 |
| Easylink NHS-PEG4-biotin | ThermoFisher | Cat# 21330 |
| RNase Out | ThermoFisher | Cat# 10777019 |
| 293 Free transfection reagent | EMD Millipore | Cat# 72181 |
| Critical Commercial Assays | ||
| HotStarTaq DNA Polymerase | QIAGEN | Cat# 203205 |
| iScript | Bio-Rad | Cat# 1708891 |
| inFusion HD cloning kit | TakaraBio | Cat# 639650 |
| polyethylenimine | PolySciences | Cat# 24765 |
| Deposited Data | ||
| Antibody variable gene sequences | This study | GenBank: MT462477 -MT462570 |
| Experimental Models: Cell Lines | ||
| HEK293T-hACE2 | BEI resources | Cat# NR-5251 |
| HEK293T | ATCC | Cat# CRL-3216 |
| 293-6E | National Research Council, Canada | N/A |
| Oligonucleotides | ||
| Primers for antibody nested PCR and sequencing | Tiller et al., 2008 | N/A |
| Recombinant DNA | ||
| pαH-SARS-CoV-2 S2P | Wrapp et al., 2020 | N/A |
| pαH-SARS-CoV S2P | Pallesen et al., 2017 | N/A |
| pαH- SARS-CoV-RBD-Fc | Wrapp et al., 2020 | N/A |
| pαH-ACE2-His-strep | Wrapp et al., 2020 | N/A |
| pTT3-SARS-CoV-2-S | This Study | N/A |
| pHDM-Hgpm2 | BEI resources | Cat# NR-52517 |
| pRC-CMV-rev1b | BEI resources | Cat# NR-52519 |
| pHDM-tat1b | BEI resources | Cat# NR-52518 |
| pHDM-SARS-CoV-2 Spike | BEI resources | Cat# NR-52514 |
| pHAGE-CMV-Luc2-IRES-ZsGreen-W | BEI resources | Cat# NR-52516 |
| Software and Algorithms | ||
| Flow Jo version 9.9.4 | Tree Star | https://www.flowjo.com |
| ForteBio data analysis software | ForteBio | N/A |
| R Version 3.4.1 | R Development Core Team, 2017 | https://www.r-project.org/ |
| Biostrings | Pages et al., 2018 | https://bioconductor.org/packages/release/bioc/html/Biostrings.html |
| seqinr | Charif and Lobry, 2007 | http://seqinr.r-forge.r-project.org/ |
| Tidyverse | Wickham, 2017 | https://www.tidyverse.org |
| Prism | Graphpad Software | https://www.graphpad.com |
| Other | ||
| SpectraMax M2 plate reader | Molecular Devices | https://www.moleculardevices.com/ |
| FACS Aria II | Becton, Dickinson and Company | N/A |
| Octet RED96e | ForteBio | N/A |
| Anti-Human IgG Fc capture (AHC) biosensors | ForteBio | Cat#18-5060 |
| Streptavidin biosensors | ForteBio | Cat#18-5019 |
| Symphony flow cytometer | Becton, Dickinson and Company | N/A |
| Fluoroskan Ascent FL | Thermofisher | Cat# 2805630 |
Resource Availability
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, Leonidas Stamatatos (lstamata@fredhutch.org).
Materials Availability
All reagents generated in this study are available upon request through Material Transfer Agreements. pTT3-derived plasmids and 293-6E cells require a license from the National Research Council (Canada)
Data and Code Availability
The sequences of codon-optimized monoclonal antibodies have been deposited in GenBank: MT462477 - MT462570.
Experimental Models and Subject Details
Human Subjects
Peripheral blood mononuclear cells (PBMCs) and serum were collected from a SARS-CoV-2 positive donor as part of the Hospitalized and Ambulatory Adults with Respiratory Viral Infections (HAARVI) study. The subject was a 35-year-old male hospitalized for over 10 days with severe disease, and received therapy with fluids, oxygen and remdesivir. The participant signed informed consent, and the following institutional human subjects review committee approved the protocol prior to study initiation: University of Washington IRB (Seattle, Washington, USA).
PBMCs and serum from pre-pandemic controls and from patients infected with endemic coronoaviruses were blindly selected at random with no considerations were made for age, or sex. These samples from pre-pandemic donors were recruited at the Seattle HIV Vaccine Trials Unit (Seattle, Washington, USA) as part of the study “Establishing Immunologic Assays for Determining HIV-1 Prevention and Control.” Samples from patients infected with endemic corona viruses were collected under FH 1829. All participants signed informed consent, and the following institutional human subjects review committee approved the protocol prior to study initiation: Fred Hutchinson Cancer Research Center IRB (Seattle, Washington, USA). PBMC samples from donors were blindly selected at random with no considerations were made for age, or sex.
Cell Lines
All cell lines were incubated at 37°C in the presence of 5% CO2 and were not tested for mycoplasma contamination. 293-6E (human female) and 293T cells (human female) cells were maintained in Freestyle 293 media with gentle shaking. HEK293T-hACE2 (human female) were maintained in DMEM containing 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (cDMEM).
Method Details
Recombinant Protein Expression and Purification
pαH-derived plasmids encoding a stabilized His- and strep-tagged SARS-CoV-2 ectodomain (pαH-SARS-CoV-2 S2P), SARS-CoV S2P (pαH-SARS-CoV S2P), and the SARS-CoV-2 receptor binding domain fused to a monomeric Fc (pαH- SARS-CoV-RBD-Fc) as well as a plasmid encoding the human ACE2 ectodomain (amino acids 1-615) with c-terminal His- and strep-tags (pαH-ACE2-His-strep) were a kind gift from Dr. Jason McLellan (Pallesen et al., 2017, Wrapp et al., 2020).
1L of 293-6E cells were cultured to a density of 1 million cells/ml and were transfected with 500μg of pαH-SARS-CoV-2 S2P, pαH-SARS-CoV S2P, pαH-SARS-CoV-2-RBD-Fc, or pαH-ACE2-His-strepusing 2 mg polyethylenimine. 6 days after transfection, supernatants were harvested by centrifugation and passed through a 0.22μm filter. Supernatant from cells transfected with SARS-CoV-2 S2P, SARS-CoV S2P, the ACE2 ectodomain was passed over a HisTrap FF affinity column pre-equilibrated in HisTrap binding buffer (20mM sodium Phosphate, 0.5M NaCl, 10mM Imidazole HCl, pH 7.4) and then washed with HisTrap binding buffer until a baseline A280 absorbance was reached and then eluted with 20mM sodium Phosphate, 0.5M NaCl, 500mM Imidazole HCl, pH 7.4). SARS-CoV S2P was further purified using a 2ml Strep-Tactin Sepharose column and Strep-Tactin Purification Buffer Set according to the manufacturer’s instructions. The S2P variants were then further purified using a Superose 6 10/300 GL column pre-equilibrated in 1XPBS or 2mM Tris 200mM NaCl, pH 8.0. The ACE2 ectodomain was further purified using an Enrich SEC 650 10X300 column (Bio-Rad) equilibrated in PBS.
Supernatant containing RBD-Fc was purified over Protein A agarose resin, cleaved with HRV3C protease (made in house) on-column. The eluate containing the RBD was further purified by SEC using HiLoad 16/600 Superdex 200 pg column (GE Healthcare) pre-equilibrated in 2mM Tris-HCl, 200mM NaCl, pH 8.0. Proteins were directly used for subsequent assays or aliquoted, flash frozen and kept at −80C until further use.
Protein biotinylation
Purified recombinant SARS-CoV or SARS-CoV-2 S2P, SARS-CoV-2 RBD, or human ACE2-Fc were biotinylated at a theoretical 1:1 ratio using the Easylink NHS-PEG4-biotin kit (Thermofisher) according to the manufacturer’s instructions. Excess biotin was removed via size exclusion chromatography using an ENrich SEC 650 10 × 300 mm column (Bio-Rad).
ELISA
Immulon 2HB microtiter plates (Thermo Scientific) were coated with 50ng/well of RBD or S2P overnight at room temperature. Plates were washed 4X with PBS with 0.02% Tween-20 (wash buffer). Plates were blocked with 250 μL of 10% non-fat milk and 0.02% Tween-20 in PBS (blocking buffer) for 1 hr at 37°C. After washing 4X with wash buffer, plasma was prepared at 1:50 dilution in blocking buffer and diluted in three-, four-, or fivefold serial dilutions in plate and incubated for 1 hr at 37°C. Plates were washed 4X in wash buffer and the secondary antibody Goat anti-Human Ig-HRP (Southern Biotech, Cat# 2010-05), Peroxidase-conjugated AffiniPure Donkey Anti-Human IgG, Fcγ fragment specific Mouse anti-Human IgM-HRP, or Mouse anti-Human IgA-HRP, was added and incubated at 37°C for 1 hr. After a final 4X wash, 50 μL of SureBlue Reserve TMB Peroxidase Substrate was added and incubated for 4 min followed by addition of 100 μL of 1 N H2SO4 to stop the reaction. The optical density at 450nm was measured using a SpectraMax M2 plate reader (Molecular Devices). All wash steps were performed using a BioTek 405 Select Microplate Washer.
B cell sorting
Fluorescent SARS-CoV-2-specific S2P and RBD probes were made by combining biotinylated protein with fluorescently labeled streptavidin (SA). The S2P probes were made at a ratio of 2 moles of trimer to 1 mole SA. Two S2P probes, one labeled with phycoerythrin (PE), one labeled with brilliant violet (BV) 711, were used in this panel in order to increase specificity of the detection of SARS-CoV-2-specific B cells. The RBD probe was prepared at a molar ratio of 4 to 1 of protein to SA, labeled with Alexa Fluor 647. Cryopreserved PBMC from the SARS-CoV-2-infected participant and a SARS-naive donor were thawed at 37°C and stained for SARS-CoV-2-specific memory B cells with a flow cytometry panel consisting of: a viability dye (7AAD), CD14 PE-Cy5, CD69 APC-Fire750, CD8a Alexa Fluor 700, CD3 BV510, CD27 BV605, IgM PE-Dazzle594, CD4 BB515, IgD BV650, IgG BV786, CD56 PE-Cy5, CD19 PE-Cy7, and CD38 BB700 (. Cells were stained first with the cocktail of the three SARS-CoV-2 probes for 30 min at 4°C, then washed with 2% FBS/PBS and stained with the remaining antibody panel and incubated for 30 min at 4°C. The cells were washed two times and resuspended for sorting in 10% FBS/RPMI media containing 7AAD. The sample was sorted on a FACS Aria II instrument using the following gating strategy: singlets, lymphocytes, live, CD3-, CD14-, CD4-, CD19+, IgD-, IgG+, S2P-PE+ and S2P-BV711+. Two plates of S2P double positive IgD- B cells were single-cell index-sorted into 96-well plates containing 16μl lysis buffer ((3.90% IGEPAL, 7.81mM DTT, 1250 units/ml RNase Out). 4 additional plates of the S2P double positive IgD- IgG+ B cell population were single-cell index-sorted into dry 96-well plates and flash frozen on dry ice. The RBD+ frequency of sorted B cells was analyzed post-sort using the index file data in Flow Jo version 9.9.4.
B cell sequencing
cDNA was generated from sorted B cells by adding 4μl of iScript and cycling according to the manufacturer’s instructions. The VH and VL sequences were recovered using gene specific primers and cycling conditions previously described (Tiller et al., 2008). VH or VL amplicons were sanger sequenced (Genewiz). The antibody gene usage was assigned using IMGT/V-QUEST (Brochet et al., 2008). Sequences were included in sequence analysis if a V and J gene identity could be assigned and the sequence contained an in-frame CDR3. Paired VH and VL sequences from S2P positive B cells were codon optimized for human expression using the Integrated DNA Technologies (IDT) codon optimization tool, synthesized as eBlocks (IDT) and cloned into full-length pTT3 derived IgL and IgK expression vectors (Snijder et al., 2018) or subcloned into the pT4-341 HC vector (Mouquet et al., 2010) using inFusion cloning.
Antibody purification
Antibody expression plasmids were co-transfected into 293-6E cells at a density of 1X 106 cells/ml in Freestyle 293 media using the 293Free transfection reagent according to the manufacturer’s instructions. Expression was carried out in Freestyle 293 media for 6 days, after which cells and cellular debris were removed by centrifugation at 4,000 × g followed by filtration through a 0.22 μm filter. Clarified cell supernatant containing recombinant antibodies was passed over Protein A Agarose, followed by extensive washing with PBS, and then eluted with 1 mL of Pierce IgG Elution Buffer, pH 2.0, into 0.1 mL of Tris HCl, pH 8.0. Purified antibodies were then dialyzed overnight into PBS, passed through a 0.2μM filter under sterile conditions and stored at −80°C until use.
Biolayer Interferometry (BLI)
BLI assays were performed on the Octet Red instrument at 30°C with shaking at 500-1,000 RPM.
mAb binding screen
mAbs were diluted in PBS to a concentration of 20μg/ml and captured using Anti-Human IgG Fc capture (AHC) biosensors for 240 s. After loading, the baseline signal was then recorded for 60 s in KB. The sensors were then immersed in PBS containing 0.5-2μM of purified SARS CoV-2 S2P, SARS CoV-2 RBD, or SARS-CoV S2P for a 300 s association step. The dissociation was then measured for 300 s by immersing sensors in kinetics buffer (KB: 1X PBS, 0.01% BSA, 0.02% Tween 20, and 0.005% NaN3, pH 7.4). As a control for non-specific binding the background signal of VRC01 binding to S2P or RBD was subtracted at each time point.
Kinetic analyses
For kinetic analyses CV30 was captured on anti-Human IgG Fc capture (AHC) sensors, and the biotinylated ACE2 ectodomain was captured on streptavidin biosensors. ligands were diluted to 10 μg/ml in PBS and loaded for 100 s. After loading, the baseline signal was then recorded for 1min in KB. The sensors were immersed into wells containing serial dilutions of purified SARS-CoV-2 RBD in KB for 150 s (association phase), followed by immersion in KB for an additional 600 s (dissociation phase). The background signal from each analyte-containing well was measured using empty reference sensors and subtracted from the signal obtained with each corresponding mAb loaded sensor. Kinetic analyses were performed at least twice with an independently prepared analyte dilution series. Curve fitting was performed using a 1:1 binding model and the ForteBio data analysis software. Mean k on, k off values were determined by averaging all binding curves that matched the theoretical fit with an R2 value of ≥ 0.95.
Antibody competition binding assays
Biotinylated ACE2 ectodomain was captured onto streptavidin biosensors (ForteBio) for 240 s. The baseline interference was then read for 60 s in KB buffer, followed by immersion in a 150nM solution of recombinant SARS CoV-2 RBD or a 150nM solution of recombinant SARS CoV-2 RBD pre-incubated with 150nM of mAb for the 300 s association phase. The dissociation was then measured for 300 s by immersing sensors in KB. As a control for non-specific binding the background signal of binding of RBD and mAb to uncoated biosensors was subtracted at each time point.
Cell surface SARS-CoV-2 S binding assay
cDNA for the full-length SARS CoV-2 S isolate USA-WA1/2020 was codon optimized and synthesized by Twist Biosciences and cloned into the pTT3 vector using InFusion cloning. pTT3-SARS-CoV-2-S was transfected into 293-6E cells using 293 Free transfection reagent according to the manufacturer’s instructions. Transfected cells were incubated for 24h at 37°C with shaking.
The next day, 1μg of each mAb was complexed with 3μg of PE-conjugated AffiniPure Fab fragment goat anti-human IgG, and the labeled mAb was incubated for 30 min at RT prior to dilution to 5μg/mL in Freestyle medium containing 10% FBS and 1% Pen/Strep. mAbs were then diluted 2-fold over 8 points in 96 well round bottom plates, and an equal volume containing 5x105 293E cells expressing SARS-CoV-2 spike proteins was added to each well. The mAb-cells mixture was incubated for 30 min at 37°C. Controls included cells treated with CR3022, an anti-SARS mAb that cross-reacts with SARS-CoV-2, with an unrelated mAb (AMMO1, specific for EBV), and untreated with mAb (cells only). The plates were then washed with FACS buffer (PBS + 2% FBS + 1mM EDTA) and fixed with 10% formalin. The mean fluorescence intensity (MFI) for PE+ cells was measured on an X-50 flow cytometer and the data analyzed using Flow Jo version 9.9.4. .
Neutralization Assay
HIV-1 derived viral particles were pseudotyped with full length wild-type SARS CoV-2 S (Crawford et al., 2020). Briefly, plasmids expressing the HIV-1 Gag and pol (pHDM-Hgpm2), HIV-1Rev (pRC-CMV-rev1b), HIV-1 Tat (pHDM-tat1b), the SARS CoV2 spike (pHDM-SARS-CoV-2 Spike) and a luciferase/GFP reporter (pHAGE-CMV-Luc2-IRES-ZsGreen-W) were co-transfected into 293T cells at a 1:1:1:1.6:4.6 ratio using 293 Free transfection reagent according to the manufacturer’s instructions. 72 hours later the culture supernatant was harvested, clarified by centrifugation and frozen at −80°C.
293 cells stably expressing ACE2 (HEK293T-hACE2) were seeded at a density of 4 X103 cells/well in a 100μl volume in 96 well flat bottom tissue culture plates. The next day, mAbs were initially diluted to 100μg/ml in 30μl of cDMEM in 96 well round bottom plates in triplicate. An equal volume of viral supernatant diluted to result in 2 × 105 luciferase units was added to each well and incubated for 60 min at 37°C. Meanwhile 50ul of cDMEM containing 6μg/ml polybrene was added to each well of 293T-ACE2 cells (2μg/ml final concentration) and incubated for 30 min. The media was aspirated from 293T-ACE2 cells and 100μl of the virus-antibody mixture was added. The plates were incubated at 37°C for 72 hours. The supernatant was aspirated and replaced with 100ul of Steadyglo luciferase reagent (Promega). 75μl was then transferred to an opaque, white bottom plate and read on a Fluorskan Ascent Fluorimeter. Control wells containing virus but no antibody (cells + virus) and no virus or antibody (cells only) were included on each plate.
% neutralization for each well was calculated as the RLU of the average of the cells + virus wells, minus test wells (cells +mAb + virus), and dividing this result difference by the average RLU between virus control (cells+ virus) and average RLU between wells containing cells alone, multiplied by 100.
mAbs that showed > 50% neutralization at 50μg/ml, or sera were further analyzed to determine neutralizing potency by preparing serial dilutions and conducting the neutralization assay as described above. The antibody concentration or plasma dilution that neutralized 50% of infectivity (IC50 or ID50, respectively) was interpolated from the neutralization curves determined using the log(inhibitor) versus response–Variable slope (four parameters) fit using automatic outlier detection in Graphpad Prism Software. As a control for specificity SARS CoV2-mAbs were tested for neutralizing activity against HIV-1 derived virions pseudotyped with murine leukemia virus envelope (MLV).
Quantification and Statistical Analysis
Amino acid mutations were identified by aligning the VH/VL gene sequences to the corresponding germline genes (IMGT Repertoire) using the Geneious Software (Version 8.1.9). Mutations were counted beginning at the 5′ end of the V-gene to the 3′ end of the FW3. To quantify the number of amino acid mutations, the sequence alignments were exported from Geneious and imported into R (Version 3.4.1) for analysis (R Development Core Team, 2017). This analysis uses the packages Biostrings (Pages et al., 2018), seqinr (Charif and Lobry, 2007), and tidyverse (Wickham, 2017) in R and GraphPad Prism version 7 were used to create graphs.
Acknowledgments
We thank Dr. McLellan for providing the SARS-CoV and SARS-CoV-2 S2P and RBD plasmids and Dr. Neil King for providing the rhesus ACE2-Fc fusion protein and the CR3022 mAb. We thank Todd Haight and Melinda Akoto for specimen processing. The repository of samples collected from patients infected with endemic corona viruses is supported by the Vaccine and Infectious Disease Division. We thank Sarah Ameny for assistance with B cell sorting. This work was supported by generous donations to Fred Hutch COVID-19 Research Fund.
Authors Contributions
Conceptualization, L.S., A.T.M., and M.P.; Investigation, E.S., L.J.H., A.J.M., K.R.P., N.K.H., M.F.J., N.R.A., A.B.S., Y.-H.W., J.F., R.E.W., S.S., and K.W.C.; Writing – Original Draft, L.S., A.T.M., and M.P.; Writing – Review & Editing, L.S., A.T.M., M.P., J.M., K.R.P., K.W.C., H.Y.C., and J.E; Funding Acquisition, L.S.; Resources, M.B., H.Y.C., and J.E.; Supervision, L.S., A.T.M., and M.P.
Declaration of Interests
The authors declare no competing financial interests. A provisional patent application (U.S. Provisional Application No. 63/016268) has been filed on the SARS-CoV-2 specific monoclonal antibodies isolated herein. H.Y.C. receives personal fees from Merck (consultant), personal fees from Glaxo Smith Kline (consultant), grants from Sanofi-Pasteur, non-financial support from Cepheid, non-financial support from Ellume, and non-financial support from Genentech. The content of these consultancies and support are unrelated to the work performed in this manuscript.
Published: June 8, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.06.001.
Contributor Information
Marie Pancera, Email: mpancera@fredhutch.org.
Andrew T. McGuire, Email: amcguire@fredhutch.org.
Leonidas Stamatatos, Email: lstamata@fredhutch.org.
Supplemental Information
References
- Briney B., Inderbitzin A., Joyce C., Burton D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503-8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Liu L., Du L., Zhang C., Jiang S., Li T., He Y. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol. J. 2010;7:299. doi: 10.1186/1743-422X-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D., Lobry J. SeqinR 1.0-2: a contributed package to the R Project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U., Porto M., Roman H.E., Vendruscolo M., editors. Structural Approaches to Sequence Evolution. Springer Verlag; 2007. pp. 207–232. [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 Spike protein for neutralization assays. Viruses. 2020;12:E513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., Georgiou G. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W.A., Liddington R.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang N., Zuo T., Shi X., Poon K.M., Wu Y., Gao F., Li D., Wang R., Guo J. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6:234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020 doi: 10.1016/j.immuni.2020.04.023. Published online May 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu P., Zhang S., Zhou P., Huang B., Deng Y., Qin K., Wang P., Wang W., Wang X., Zhou J. Ultrapotent Human Neutralizing Antibody Repertoires Against Middle East Respiratory Syndrome Coronavirus From a Recovered Patient. J. Infect. Dis. 2018;218:1249–1260. doi: 10.1093/infdis/jiy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.200841. Published online April 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages H., Aboyoun P., Gentleman R., DebRoy S. Biostrings: efficient manipulation of biological strings. 2018. https://rdrr.io/bioc/Biostrings/
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2017. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rockx B., Corti D., Donaldson E., Sheahan T., Stadler K., Lanzavecchia A., Baric R. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 2008;82:3220–3235. doi: 10.1128/JVI.02377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., McGuire A.T. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity. 2018;48:799–811. doi: 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Bombardi R.G., Branchizio A., Kose N., Matta P., Sevy A.M., Sinkovits R.S., Gilchuk P., Finn J.A., Crowe J.E., Jr. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. 2019;566:398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez Bernat N., Corcoran M., Hardt U., Kaduk M., Phad G.E., Martin M., Karlsson Hedestam G.B. High-Quality Library Preparation for NGS-Based Immunoglobulin Germline Gene Inference and Repertoire Expression Analysis. Front. Immunol. 2019;10:660. doi: 10.3389/fimmu.2019.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. Tidyverse., 2017. https://www.tidyverse.org.
- World Health Organization WHO announces COVID-19 outbreak a pandemic. 2020. http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L., Liu Q., Wang L., Feng Y., Wang Y. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D.S., Zhou T. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences of codon-optimized monoclonal antibodies have been deposited in GenBank: MT462477 - MT462570.