Abstract
Sunless (chemical) tanning is widely regarded as a safe alternative to solar UV-induced skin tanning known to be associated with epidermal genotoxic stress, but the cutaneous biology impacted by chemical tanning remains largely unexplored. Chemical tanning is based on the formation of melanin-mimetic cutaneous pigments (‘melanoidins’) from spontaneous amino-carbonyl (‘glycation’) reactions between epidermal amino acid/protein components and reactive sugars including the glycolytic ketose dihydroxyacetone (DHA). Here, we have examined the cutaneous effects of acute DHA-exposure on cultured human HaCaT keratinocytes and epidermal reconstructs, profiled by gene expression array analysis and immunodetection. In keratinocytes, DHA-exposure performed at low millimolar concentrations did not impair viability while causing a pronounced cellular stress response as obvious from rapid activation of phospho-protein signal transduction [p-p38, p-Hsp27(S15/S78), p-eIF2α] and gene expression changes (HSPA6, HMOX1, CRYAB, CCL3), not observable upon exposure to the non-ketose, tanning-inactive DHA-control glycerol. Formation of advanced glycation end products (AGEs) from posttranslational protein-adduction was confirmed by quantitative mass spectrometric detection of N-ε-(carboxyethyl)-l-lysine (CEL) and N7-carboxyethyl-l-arginine, and skin cells with CRISPR-Cas9-based elimination of the carbonyl stress response gene GLO1 (encoding glyoxalase 1) displayed hypersensitivity to DHA-cytotoxicity. In human epidermal reconstructs a topical use-relevant DHA-dose regimen elicited a comparable stress response as revealed by gene expression array (HSPA1A, HSPA6, HSPD1, IL6, DDIT3, EGR1) and immunohistochemical analysis (CEL, HO-1, p-Hsp27-S78). In DHA-treated SKH-1 hairless mouse skin IHC-detection revealed epidermal occurrence of CEL- and p-Hsp27-epitopes. For comparison, stress response gene expression array analysis was performed in epidermis exposed to a supra-erythemal dose of solar simulated UV (2 MEDs), identifying genes equally or differentially sensitive to either one of these cutaneous stimuli [DHA (‘sunless tanning’) versus solar UV (‘sun-induced tanning’)]. Given the worldwide use of chemical tanners in consumer products, these prototype data documenting a DHA-induced specific cutaneous stress response deserve further molecular exploration in living human skin.
Keywords: Sunless tanning, Dihydroxyacetone, Glycation, Reconstructed human epidermis, Stress response gene expression, Phosphoprotein signaling
1. Introduction
Solar ultraviolet (UV)-induced skin tanning is known to be associated with epidermal genotoxic stress [[1], [2], [3]]. After initial genomic photodamage associated with cyclobutane pyrimidine dimer (CPD) formation, melanogenesis occurs downstream of p53-dependent proopiomelanocortin (POMC) expression and paracrine signaling through α-MSH that activates melanocortin (MC1R) on melanocytes, followed by melanin production and redistribution to solar UV-exposed keratinocytes [1,4,5]. Chemical tanning is widely regarded as a safe alternative to solar UV-induced skin tanning, but the cutaneous biology impacted by sunless tanning remains largely unexplored [[6], [7], [8]]. Discovered in the context of nutritional interventions examining pediatric glycogen storage disease with systemic administration of triose sugars, chemical tanning is now a standard cosmetic intervention used by large numbers of consumers worldwide [[8], [9], [10]].
Chemical tanning is based on the formation of melanin-mimetic cutaneous pigments (referred to as ‘melanoidins’) from spontaneous amino-carbonyl (‘glycation’) reactions between epidermal amino acid/protein components and reactive sugars of the ketose family including the glycolytic triose DHA and the tetrose erythrulose [[7], [8], [9], [10], [11], [12]]. Interestingly, previous research has investigated glycation and metabolic impact of DHA and its glycolytic phospho-metabolite dihydroxyacetone phosphate [13,14]. Glycation reactions are associated with the formation of posttranslational protein modifications referred to as advanced glycation end products (AGEs) [15,16]. Moreover, the chemistry leading to AGE formation involves reactive intermediates such as reactive carbonyl (e.g. glyoxal and methylglyoxal) and oxygen species (ROS) [17,18]. Previous research has investigated the involvement of glycation reactions and formation of cutaneous AGEs in the context of diabetic wound healing, solar photodamage, photocarcinogenesis, and chronological aging [[19], [20], [21], [22]]. Moreover, chemical crosslinking of skin extracellular matrix proteins and photosensitization activity of specific AGE-chromophores have been observed in human skin, and specific protein-bound AGEs [e.g. Nε-carboxymethyl-l-lysine (CML) and Nε-carboxyethyl-l-lysine (CEL)] stimulate signaling through specialized receptors including receptor for advanced glycation end products (RAGE) and toll-like receptor 4 (TLR4) that trigger inflammatory signaling upon ligand binding [20,[23], [24], [25], [26]].
After topical application, chemical tanning agents are thought to be confined to the stratum corneum without affecting structure and function of viable epidermal layers [27]. However, safety concerns have been raised based on pharmacokinetic data demonstrating (i) DHA skin penetration after topical application, (ii) detection of glycation-associated formation of free radicals and AGEs, and (iii) induction of genotoxic stress [6,12].
Here, we have examined the cutaneous effects of acute DHA exposure employing cultured human HaCaT keratinocytes, organotypic epidermal reconstructs, and SKH-1 hairless mouse skin, profiled by gene expression array analysis and immunodetection. Taken together, our prototype data document that topical DHA application induces cutaneous stress response signaling and gene expression that occur within minutes of exposure. Given the worldwide use of chemical tanners including DHA in consumer products these findings deserve further molecular exploration in relevant model systems and live human skin.
2. Materials and methods
Chemicals: All chemicals were purchased from Sigma Aldrich (St. Louis, MO, USA) including dihydroxyacetone (D107204) and glycerol (G5516).
Human skin cell culture: Human immortalized keratinocytes (HaCaT) and malignant A375 melanoma cells were purchased from ATCC (Manassas, VA, USA) and maintained according to the manufacturer's instructions. In brief, both HaCaT and A375 cells were cultured in DMEM medium (Corning, Manassas, VA) or RPMI (ATCC), respectively, supplemented with 10% bovine calf serum (HyClone™ Laboratories, Logan, UT). Unless specified otherwise, DHA exposure was performed in PBS (instead of complete medium) in order to minimize the impact of indirect glycation reactions (e.g. DHA-modification of serum proteins that might confound cellular effects).
CRISPR/Cas9-based engineering of GLO1_KO A375 malignant melanoma cells: Homozygous GLO1 gene knock-out in human malignant A375 melanoma cells was performed using genetic engineering as published elsewhere [28]. Briefly, double strand breaks were generated on both sides of exon 2 (chromosome 6, positions: 38,687,313 bp; 38,685,738 bp) with guide CRISPR RNAs (5′-ACCCTCATGGACCAATCAGT-3′ and 5′-TGATCATAGGTGTATACGAG-3′). Parental cells were transfected with Cas9 protein, crRNAs, and trans-activating crRNA (Integrated DNA Technologies, San Diego, CA) using the Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA). Next, single cells were deposited in 96-well plates and once single cell colonies expanded (after approximately three weeks), individual clones were screened by PCR. Clones that were negative for a sequence inside the targeted deletion and negative for the undeleted chromosomal sequences but positive for ligation-junction fragment were scored as potentially homozygous for GLO1 exon 2 deletion. Absence of GLO1 expression was confirmed by single RT-qPCR, immunoblot, and enzymatic activity assays [29].
Human epidermal reconstructs: Before treatment, refrigerated epidermal reconstructs (EPI-200, 9 mm diameter; MatTek, Corp., Ashland, MA) were equilibrated in fresh growth medium (0.9 mL; EPI-200-ASY media per well, 1 h), following our standard procedures for maintenance and treatment as published before [[30], [31], [32]]. Briefly, stratum corneum of air exposed reconstructs was treated with topical DHA [100 μL; 10% in Vanicream™ (Pharmaceutical Specialties, Inc., Rochester, MN) or Vanicream™ carrier only; 6 h exposure time, 37 °C; 5% CO2]. Following exposure, carrier (with or without DHA) was removed using a cotton swab. Digital colorimetry assessing DHA-induced tanning was performed using the Image Studio™ Lite quantification software version 5.2 (LI-COR Biosciences, Lincoln, NE). The epidermal reconstruct was then processed for (i) RNA extraction using the RNeasy Mini kit (Qiagen, Germantown, MD) or (ii) immunohistochemical analysis as described before [[30], [31], [32]].
Cell viability analysis by flow cytometry: Cells were treated with DHA (≤50 mM; 1 h in PBS), followed by culture in fresh growth medium (≤24 h). Cell viability was then determined using flow cytometric analysis of annexinV (AV)-propidium iodide (PI) stained cells using an apoptosis detection kit (APO-AF, Sigma, St. Louis, MO) according to the manufacturer's specifications as published before [33].
Cell proliferation assay: Cells (5000 per 35 mm dish) were seeded; the following day (d 0), cells were treated with DHA (≤50 mM; 1 h in PBS), followed by culture in fresh growth medium (72 h). Cells were counted (d0 and d3) using a Z2 analyzer (Beckman Coulter, Fullerton, CA, USA).
Cell cycle analysis: Cell cycle analysis was performed treating cells as detailed for proliferation analysis. After 48 h, cells were harvested and fixed in ethanol (100%). Cell pellets were then incubated [37 °C, 30 min; Ribonuclease A (1 mg/mL) with propidium iodide (4 mg/mL) in PBS] and then analyzed by flow cytometry. Cellular DNA content was analyzed using ModFit LT software, version 5.0 (Verity, Topsham, ME) [34].
Human Stress & Toxicity PathwayFinder RT2 ProfilerTM gene expression array analysis: After DHA treatment, total mRNA from cultured HaCaT keratinocytes (200,000 in 35 mm dish format) or epidermal reconstructs was prepared using the RNeasy Mini kit (Qiagen, Valencia, CA) following our published standard procedures. Reverse transcription was then performed using the RT2 First Strand kit (Qiagen) from 500 ng total RNA. For gene expression array analysis, the human Stress & Toxicity PathwayFinder RT2 Profiler™ technology (Qiagen), assessing expression of 84 stress response-related genes, was used as published before [[30], [31], [32], [33]]. Quantitative PCR was run using the following conditions: 95 °C (10 min), followed by 40 cycles at 95 °C (15 s) alternating with 60 °C (1 min) (Applied Biosystems, Carlsbad, CA). Gene-specific products were normalized to a group of 5 housekeeping genes (ACTB, B2M, GAPDH, HPRT1, RPLP0) and quantified using the comparative ΔΔCt method (ABI Prism 7500 sequence detection system user guide). Expression values were averaged across at least three independent array experiments, and standard deviation was calculated for graphing and statistical analysis as published before.
Individual RT-qPCR analysis: Total cellular mRNA was isolated using the Qiagen RNeasy Mini Kit (Qiagen, Gaithersburg, MD) according to the manufacturer's protocol. Human primer probes [GLO1 (Hs_02861567_m1), HMOX1 (Hs00157965_m1), HSPA1A (Hs00359163_s1), HSPA6 (Hs00275682_s1), RSP18 (housekeeper; Hs_01375212_g1)], were obtained from Thermo Fisher Scientific, Waltham, MA. After cDNA synthesis, quantitative PCR reactions were performed as follows: 10 min (95 °C) followed by 15 s (95 °C), 1 min (60 °C), 40 cycles, using the ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplification plots were generated, and Ct values were recorded as published before [33].
Immunoblot analysis: Detection of proteins by immunoblot analysis was conducted using the following primary antibodies: Nrf2 (13032, Santa Cruz Biotechnology, Santa Cruz, CA), HO-1 (5853, Cell Signaling, Danvers, MA), αβ-crystallin (45844, Cell Signaling), HSP70B’ (ADI-SPA-754, Enzo Life Sciences, Farmingdale, NY), β-actin (A4700, Sigma), phospho-p38 (9211, Cell Signaling), total p38 (9212, Cell Signaling), phospho-eIf2α (9721, Cell Signaling), total eIF2α (9722, Cell Signaling), phospho-HSP27 [serine 15 (ADI-SPA-525), serine 78 (ADI-SPA-523), serine 82 (ADI-SPA-524)], total HSP27 (ADI-SPA-803, Enzo Life Sciences), phospho-ERK1/2 (5726, Cell Signaling), total ERK1/2 (4696, Cell Signaling) and GLO1 (ab96032, Abcam, Cambridge, MA). The secondary antibodies used were goat anti-mouse (115-035-146, Jackson Immunological Research, West Grove, PA) or anti-rabbit (111-035-144, Jackson Immunological Research), followed by enhanced chemiluminescent detection (32106, ThermoFisher Scientific, Waltham, MA). For quantification of immunoblots, digital image analysis was performed using Image Studio™ Lite quantification software (LI-COR Biosciences, Lincoln, NE).
Detection of intracellular oxidative stress: HaCaT cells were exposed to DHA (≤40 mM; < 2 h in PBS). Fresh medium was supplemented with 2′,7′-dichlorodihydrofluorescein-diacetate (DCF) according to a previously published method [[30], [31], [32], [33]]. Cells were then harvested and analyzed by flow cytometric detection of DCF-stained cells.
Comet assay (alkaline single cell gel electrophoresis): HaCaT cells were treated with DHA (20 mM; 1 h in PBS) followed by cell lysis and alkaline comet assay (Trevigen, Gaithersburg, MD) with DAPI fluorescence microscopy as published before. In addition, the Fpg-FLARE assay kit (Trevigen, Gaithersburg, MD) was utilized for detection of oxidative DNA base damage as published before [32].
Detection of γ-H2AX (S139): Following a published standard procedure, HaCaTs were incubated with DHA (≤40 mM; 1 h in PBS). Cells were then washed with PBS and fixed (4% formalin, 15 min, room temperature). After permeabilization (90% methanol in PBS on ice), cells were stained using γ-H2AX Alexa 488-conjugate antibody (1:50; 0.5% BSA in PBS, Cell Signaling, Danvers, MA) followed by PBS-wash and flow cytometric analysis [34].
QuARKMod (Quantitative Analysis and Discovery of Lysine and Arginine Modifications) analysis for posttranslational modification quantitation via LC-MS/MS: Cells were exposed to DHA (≤20 mM, 1 h in PBS; n = 6) and replenished/cultured under growth medium for an additional 5 h. After cell harvest, total protein extraction, and BCA-assay based protein quantification, lysine and arginine glycation-adducts were quantified (per 100 μg) as described earlier [35]. Briefly, 50 mM ammonium bicarbonate, pH 8.0 (60 μL), was added to each protein pellet followed by the addition of an internal standard mix (10 μL, see table below) and 1 μg of sequencing-grade trypsin (Promega, 10 μL). Proteins were then digested overnight (37 °C). After incubation, trypsin was denatured via heating (95 °C; 10 min). After the samples returned to room temperature, 15 μg of aminopeptidase M (Millipore, 10 μL) was added to each sample and incubated overnight at 37 °C. Following digestion, 10 μL of a solution containing heptafluorobutyric acid (HFBA) and water (1:1) was added to each sample. Insoluble debris was removed via centrifugation at 14,000×g for 10 min 10 μL of the supernatant was then chromatographed [Shimadzu LC system; 150 × 2.1 mm, 3.5 μm particle diameter Eclipse XDB-C8 column (Agilent, Santa Clara, CA); flow rate: 0.425 mL/min; mobile phase A: 10 mM HFBA in water; mobile phase B: 10 mM HFBA in acetonitrile]. The following gradient was used: 0.5 min, 5% B; 8 min, 50% B; 8.5 min, 80% B; 9 min 80% B; 9.5 min, 5% B. Scheduled multiple reaction monitoring (MRM) was conducted in positive-ion mode using an AB SCIEX 4500 QTRAP. MRM detection window was 50 s with a target scan time of 0.75 s. The following parameters were used for detection:
| Species | Q1 (m/z) | Q3 (m/z) | Time (min) | CE (V) |
|---|---|---|---|---|
| Lys | 147.1 | 84.1 | 4.7 | 29 |
| 13C615N2 Lys | 155.1 | 90.1 | 4.7 | 29 |
| Arg | 175.1 | 70.1 | 5.0 | 47 |
| 13C615N4 Arg | 185.1 | 75.1 | 5.0 | 47 |
| Leu | 132.1 | 86.1 | 5.4 | 17 |
| 13C615N Leu | 139.1 | 93.1 | 5.4 | 17 |
| MG-H1 | 229.2 | 70.1 | 5.5 | 53 |
| 13C-MG-H1 | 230.2 | 70.1 | 5.5 | 53 |
| CEA | 247.2 | 70.1 | 5.4 | 55 |
| 13C-CEA | 248.2 | 70.1 | 5.4 | 55 |
| LactoylLys | 219.2 | 84.1 | 3.3 | 41 |
| CEL | 219.2 | 84.1 | 4.7 | 41 |
| CEL-d4 | 223.2 | 88.1 | 4.7 | 41 |
Analytes were quantified using their respective internal standards. LactoylLys was quantified against CEL-d4. Lysine and arginine analytes were measured to ensure consistency and minimal changes of amino acids between samples. All samples were corrected with enzyme controls and normalized to leucine [35].
GLO1 enzymatic activity assay: Glyoxalase I specific enzymatic activity in melanoma A375 cell cytosolic fractions was analyzed using a colorimetric assay kit (Abcam, Cambridge, UK) according to manufacturer's instructions [29]. Briefly, pelleted cells (approximately 2 × 106) were homogenized with 300 μL of ice-cold Glo I Assay Buffer containing protease inhibitor PMSF and centrifuged (12,000 g; 4 °C; 10 min). Supernatant cytosolic fractions were analyzed for protein content (Pierce™ BCA Protein Assay Kit, Thermo Fisher Scientific, Waltham, MA); equal amounts (10 μg protein) were mixed with substrate and then examined for enzymatic activity by measuring absorbance at 240 nm in kinetic mode (room temperature; 10–20 min).
Irradiation with solar simulated UV light (SSL): A KW large area light source solar simulator, model 91293, from Oriel Corp. (Stratford, CT) was used, equipped with a 1000 W xenon arc lamp power supply, model 68920, and a VIS-IR band pass blocking filter plus either an atmospheric attenuation filter (output 290–400 nm plus residual 650–800 nm for solar simulated light) [[30], [31], [32], [33],36]. The output was quantified using a dosimeter from International Light Inc. (Newburyport, MA), model I L1700, with an SED240 detector for UVB (range 265–310 nm, peak at 285 nm) or a SED033 detector for UVA (range 315–390 nm peak 365 nm) at a distance of 365 mm from the source, which was used for all experiments. At 365 mm from the source, total solar UV intensity was 7.60 mJ/cm2 s (UVA) and 0.41 mJ/cm2 s (UVB).
Immunohistochemistry: Following DHA treatment of mouse skin and epidermal reconstructs, immunohistochemical detection was performed as follows: Tissues were collected, fixed in NBF (10%), and embedded in paraffin. Following deparaffinization, hydration, and antigen retrieval the following primary antibodies were used: HO-1 (1:250, 13248, abcam), total HSP27 (1:500, ADI-SPA-803, Enzo Life Sciences), phospho-HSP27 (S78) (1:100, ADI-SPA-523, Enzo Life Sciences). Moreover, for detection of Nε-carboxyethyl-l-lysine (CEL; 1:3000; 30917, abcam), an antibody known to equally recognize Nε-carboxymethyl-l-lysine (CML) and CEL was used [37,38]. After overnight incubation, secondary antibody was applied followed by streptavidin/horseradish peroxidase incubation (RTU PK7200, Vector Laboratories, Burlingame, CA), development using diaminobenzidine/hydrogen peroxide (Vectastain ABC, SK-4103, Vector Laboratories), and hematoxylin counterstaining. Negative controls were performed on each run substituting the primary antibody with mouse IgG1 (X0931, Agilent/DAKO, Santa Clara, CA). Images were obtained with an Olympus microscope (BX50 L98-029) and camera (DP72, Center Valley, PA) using the cellSens program.
Mouse experiment: SKH-1 Elite™ mice (6–8 weeks old; n ≥ 3 per group) were obtained from Charles River Laboratories (Stain code 477) and maintained at the University of Arizona, according to an approved protocol by the Institutional Animal Care and Use Committee (IACUC). 72 h after topical treatment [DHA (10% in Vanicream™) or carrier only; 200 μL], skin was processed for immunohistochemical analysis according to published procedures [39].
Statistical analysis: For every data point presented, at least three independent experiments were conducted and analyzed [39]. Unless indicated otherwise, statistical significance was calculated employing the Student's two-tailed t-test, utilizing Excel (Microsoft®, Redmond, WA). The level of statistical significance was marked as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
3. Results
Dose response relationship of DHA-induced impairment of cell viability, proliferation, and cell cycle progression in human HaCaT keratinocytes.
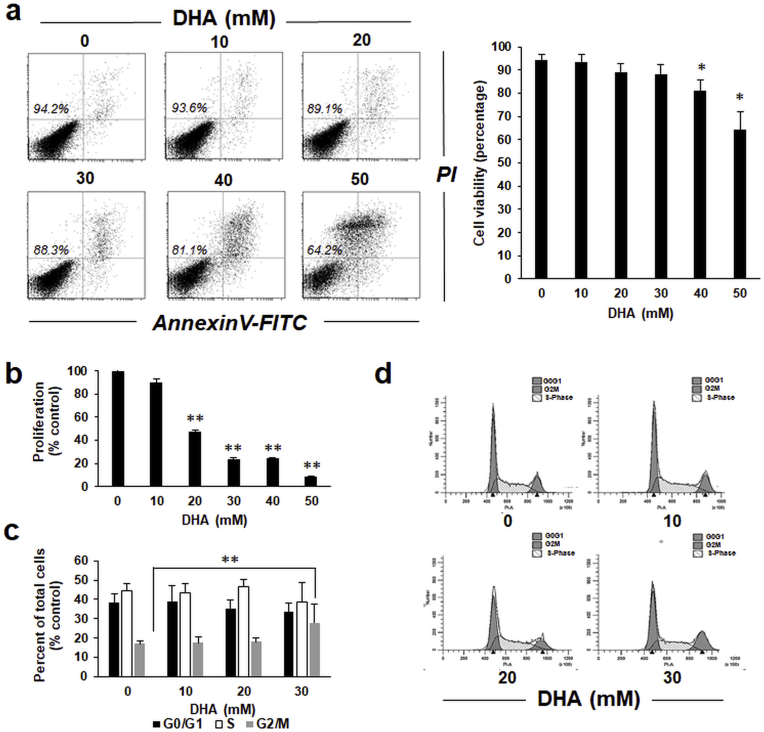
Following previous studies that have addressed the potential cytotoxicity of DHA on human skin, we first determined the dose response relationship of DHA-induced impairment of cell viability, proliferation, and cell cycle progression as assessed in HaCaT keratinocytes [12,40]. Indeed, significant impairment of cell viability was observed only upon exposure to high DHA concentrations (≥40 mM; Fig. 1a). Likewise, inhibition of cell proliferation occurred as a result of exposure to high DHA concentrations (≥20 mM; Fig. 1b). At 20 mM DHA, a concentration that did not interfere with cell viability, half-maximal inhibition of proliferation was observed. Next, in order to further explore the role of cell cycle in the causation of inhibited proliferation, we performed flow cytometric analysis of cell cycle progression as a function of DHA exposure (Fig. 1c and d). Strikingly, only at higher concentrations (≥30 mM) DHA treatment caused a significant increase in cells in G2/M phase of the cell cycle. Taken together, these initial dose response relationship experiments indicate a range of DHA concentration (≤20 mM) that does not interfere with cell viability and is devoid of adverse effects on cell cycle progression. Thus, this concentration range was chosen for execution of our subsequent experiments to define the early cellular stress response displayed by cultured human keratinocytes subjected to short term DHA exposure.
Fig. 1.
DHA attenuates cell viability, proliferation and cell cycle progression in human keratinocytes. (a) Impairment of cellular viability in response to acute DHA exposure (≤50 mM; 1 h in PBS followed by 24 h in growth medium) as assessed by flow cytometry (annexin V-PI staining). Numbers in quadrants indicate viable cells (AV-negative, PI-negative) in percent of total gated cells; bar graph summarizes numerical values (mean ± SD). (b) DHA-induced impairment of cellular proliferation [≤ 50 mM, continuous exposure (72 h) in growth medium; n = 3]. (c) DHA-induced cell cycle alteration [≤ 30 mM, continuous exposure (48 h) in growth medium] as assessed by flow cytometry of PI-stained cells; bar graph summarizes numerical values (mean ± SEM, n = 3). (d) Representative cell cycle histograms per treatment group.
Expression array analysis reveals stress response gene expression in human HaCaT keratinocytes exposed to DHA.
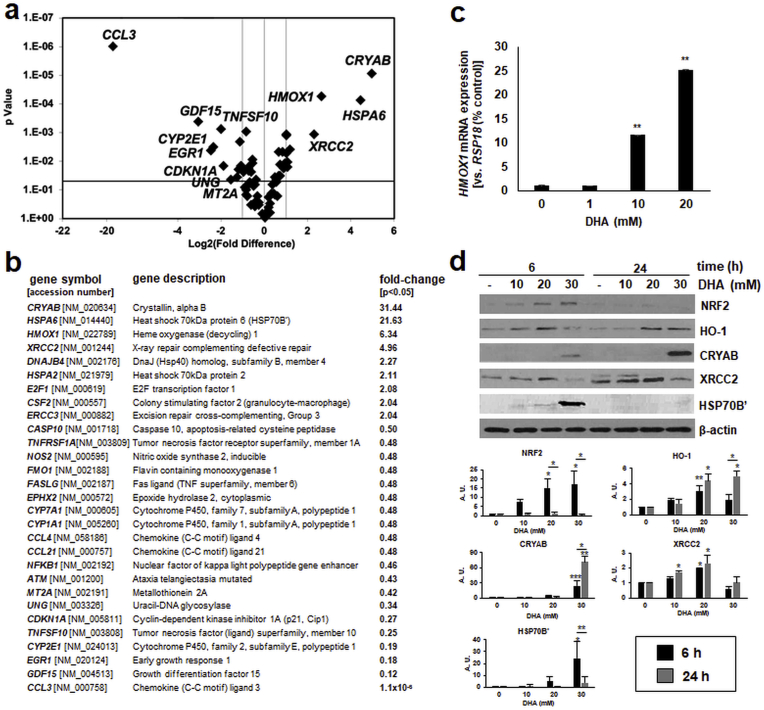
Next, to determine keratinocyte stress response gene expression elicited by DHA exposure (performed at concentrations that do not impair cell viability), we employed the Human Stress and Toxicity PathwayFinder™ PCR Array technology. Strikingly, acute exposure to DHA (1 h in PBS followed by 5 h culture in growth medium) caused a pronounced upregulation of stress response gene expression including genes encoding specific heat shock [CRYAB (31-fold), HSPA6 (22-fold), DNAJB4 (2-fold)], oxidative stress [HMOX1 (6-fold)], and DNA damage [XRCC2 (5-fold), ERCC3 (2-fold)] response factors (Fig. 2a and b). Moreover, DHA treatment downregulated expression of genes encoding inflammatory chemokines [CCL3, CCL4, CCL21] and other mediators of inflammation and tissue remodeling [NOS2, NFKB1, GDF15] as well as members of the cytochrome P450 class [CYP2E1, CYP1A1, CYP7A1]. Also, independent RT-qPCR analysis confirmed DHA-induced upregulation of HMOX1 encoding the antioxidant enzyme heme oxygenase-1 detectable as concentrations as low as 10 mM (Fig. 2c).
Fig. 2.
Array analysis reveals early stress response gene expression in human keratinocytes receiving acute DHA exposure. (a) HaCaT keratinocytes underwent short term DHA exposure (20 mM; 1 h in PBS followed by 5 h in growth medium). Stress response gene expression was then assessed by RT2 Profiler™ Gene Expression Array analysis (volcano blot; p < 0.05). (b) Table summarizes numerical values (n = 3, p < 0.05). (c) Confirmatory single RT-qPCR analysis (HMOX1, mean ± SEM). (d) Immunoblot analysis profiling stress response protein expression (≤30 mM DHA, ≤ 24 h); bar graphs summarize densitometric analysis of antigens (mean ± SEM).
Next, in order to determine if DHA-induced gene expression changes were observable at the protein level, immunoblot analysis was performed (Fig. 2d). Indeed, upregulated cellular levels of heat shock proteins [crystallin alpha B (encoded by CRYAB); HSP70B’ encoded by HSPA6], the DNA damage repair enzyme XRCC2 (encoded by XRCC2), and oxidative stress response factors including the transcription factor NRF2 and its target heme oxygenase-1 (HO-1 encoded by HMOX1) were detectable within 6 h of DHA exposure. Moreover, sustained upregulation of HO-1, CRYAB, and XRCC2 levels was confirmed by immunoblot analysis performed at 24 h after DHA treatment.
Taken together, these data indicate that acute DHA exposure at sublethal concentrations causes pronounced stress response gene expression observable at the mRNA and protein levels in cultured HaCaT keratinocytes.
DHA induces phosphoprotein-associated stress signaling in human HaCaT keratinocytes.
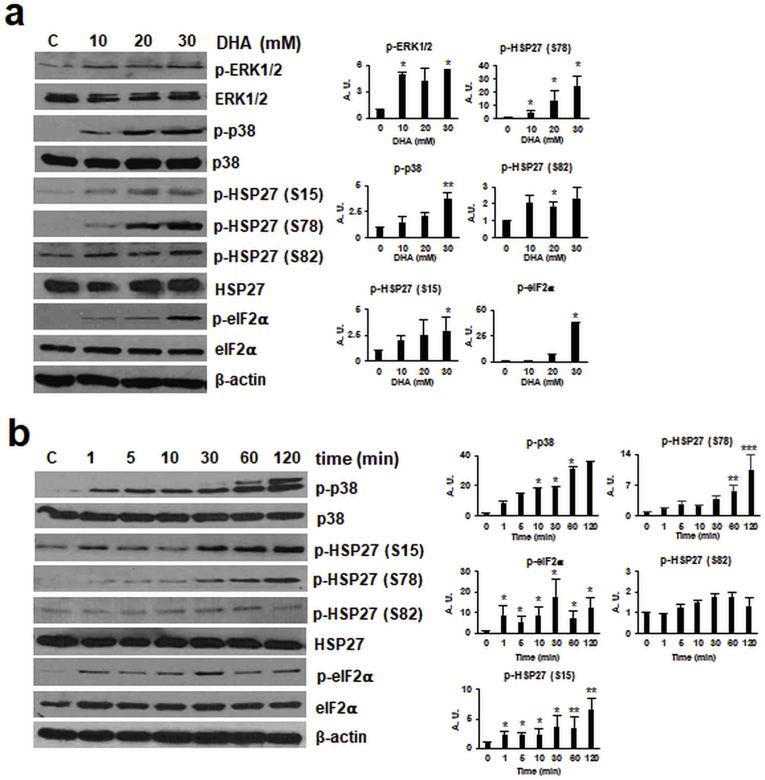
After analysis of DHA-induced gene expression changes detectable at the mRNA and protein levels, we then focused on immunodetection of DHA-induced phospho-protein signaling (Fig. 3). To this end, HaCaT keratinocytes were DHA-exposed (0–30 mM; 1 h in PBS), followed by immunoblot detection of stress protein phosphorylation, focusing on crucial components of (i) MAPK-, (ii) ER-, and (iii) heat shock-stress response pathways. Indeed, phosphorylation of ERK1/2-MAPK (Fig. 3a) was found to be responsive to DHA exposure. Likewise, p38-MAPK displayed a pronounced, dose-dependent increase in activational phosphorylation in response to DHA exposure (Fig. 3a), and time course analysis revealed p38 phosphorylation within 1 min exposure time (Fig. 3b). In the context of UV-induced cell stress, p38 is known to become activated and then phosphorylate small heat shock proteins (HSP27, αβ-crystallin, etc.) [41,42]. Consistent with the phosphorylation of the established p38-MAPK downstream target HSP27 at three distinct sites (serine 15, 78, and 82)], documented earlier to be responsive to environmental stressors including solar UV, we observed that HaCaT exposure to DHA (10 mM) caused pronounced HSP27-phosphorylation (S15, S78) detectable within 1 min exposure time (Fig. 3a and b). In addition, it was observed that DHA exposure causes pronounced induction of ER-stress response signaling as indicated by phosphorylation of eIF2α detectable within 1 min DHA exposure, an observation consistent with rapid induction of DHA induced protein translation blockade (Fig. 3a and b) [43].
Fig. 3.
Acute DHA exposure induces dose- and time-dependent phospho-protein stress signaling in human keratinocytes. Protein phosphorylation in response to acute DHA exposure was profiled by immunoblot analysis: (a) dose-response relationship (≤30 mM DHA in PBS; 1 h); (b) time course (20 mM DHA in PBS; ≤ 2 h); bar graphs summarize densitometric analysis of antigens (mean ± SEM).
Taken together, these data indicate that acute DHA exposure performed at dose-regimens that do not impair viability induce rapid and pronounced MAPK-, ER-, and heat shock-stress response signaling detectable at the mRNA and (phospho)-protein levels.
DHA induces AGE formation and cytotoxic glycation stress, antagonized by expression of the glycation defense enzyme glyoxalase 1 (encoded by GLO1).
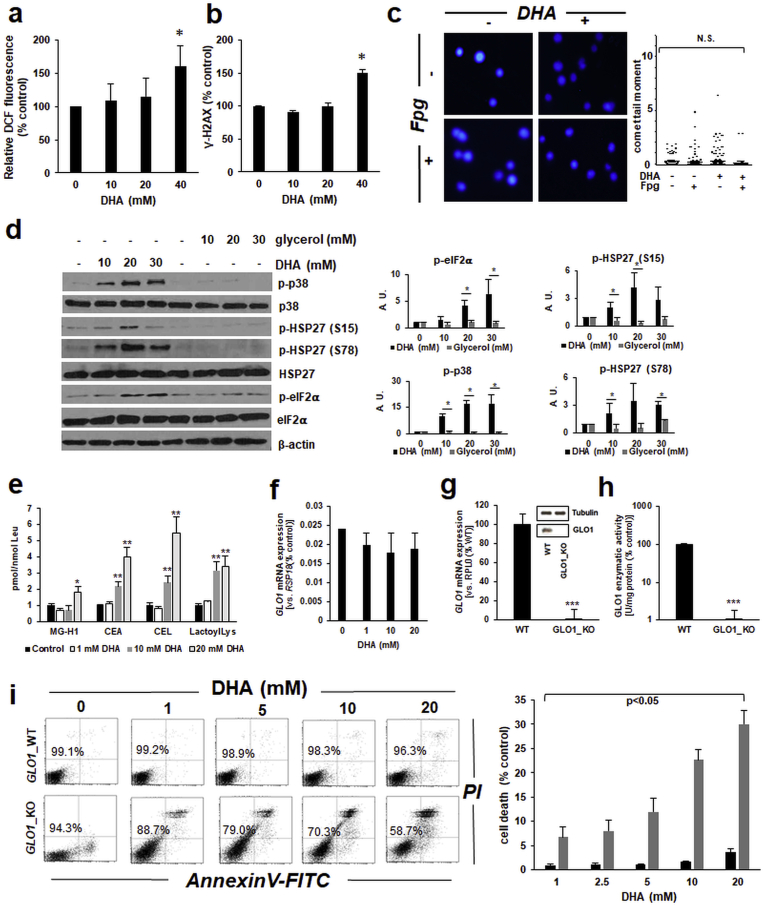
Next, in order to explore the molecular nature of cellular stress imposed by DHA treatment in HaCaT keratinocytes (Fig. 2, Fig. 3), we examined the occurrence of DHA-induced oxidative stress, as already suggested by DHA-upregulated expression of the antioxidant defense gene HMOX1 (Fig. 2). Significant elevation of intracellular ROS levels (as assessed by DCF fluorescence-based flow cytometry) was observed only when HaCaT keratinocytes were exposed to high DHA concentrations (≥40 mM) (Fig. 4a), and similar results were observed when the reduced glutathione pool was examined using a luminescent luciferase-coupled assay (data not shown). Next, examination of DHA-induced genotoxic effects (as assessed by detection of γ-H2AX, an established marker of electrophilic DNA damage and double strand breaks) indicated significant elevation of γ-H2AX levels as detected by flow cytometry, again observable only at high DHA concentrations (Fig. 4b). Interestingly, alkaline single cell gel electrophoresis (comet analysis) assessing genotoxic consequences of DHA exposure did not reveal the occurrence of DHA-induced DNA damage, even if performed using the N-formylpyrimidine-glycosylase (Fpg) digestion method detecting oxidative DNA damage through cleavage at 8-oxo-dG sites (Fig. 4c).
Fig. 4.
Glycation stress and AGE-formation in DHA-exposed human keratinocytes. (a) DHA-induced (≤40 mM, 2 h; mean ± SEM) intracellular oxidative stress as assessed by flow cytometric analysis of DCF-stained cells. (b) DHA-induced activation of DNA damage response as detected by flow cytometric analysis of γ-H2AX staining. (c) DHA-induced (20 mM) oxidative DNA damage as detected by comet assay with FPG-digestion for 8-oxo-dG detection [mean comet tail moment ± SEM; representative images (left) with dot blot depiction of quantitative analysis (right)]. (d) Glycerol-induced (≤30 mM; 1 h in PBS) phosphoprotein signaling as assessed by immunoblot analysis; bar graphs summarize densitometric analysis of antigens (mean ± SEM). (e) AGE-type posttranslational modification detected in total protein extracts from DHA-treated keratinocytes (≤20 mM, 1 h in PBS, followed by 5 h in growth medium, n = 6, mean ± SEM) by LC-MS/MS analysis. (f) DHA-modulation (≤20 mM) of GLO1 mRNA expression in HaCaT keratinocytes determined by RT-qPCR. (g) GLO1 mRNA expression in A375_WT and GLO1_KO cells as determined by RT-qPCR; insert depicts representative immunoblot analysis of GLO1 protein levels. (h) GLO1 enzymatic activity compared between A375_WT and GLO1_KO cells. (i) DHA-induced impairment of cell viability (A375_WT versus A375-GLO1_KO) as determined in Fig. 1a; bar graph summarizes numerical values (mean ± SD).
Next, in order to explore the possibility that signaling responses observed in HaCaT keratinocytes may be attributable to compound-induced osmotic stress (as expected to occur as a result of exposure to millimolar sugar concentrations), analogous experimentation was performed using glycerol, the trihydroxy-alcohol analogue of the keto-triose sugar DHA, serving as molecular control devoid of tanning activity due to the absence of a glycation-active α-hydroxy-carbonyl moiety (Fig. 4d). Strikingly, it was observed that glycerol (employed at equimolar concentrations) did not elicit stress response signaling characteristic of DHA (Fig. 3), as evident from analogous phospho-protein immunoblot analysis examining responsiveness of p38, Hsp27, and eIF2α phospho-signaling (Fig. 4d).
Next, we examined the occurrence of glycation damage as a result of DHA exposure [19,44]. To this end, mass spectrometric analysis of AGE-protein adducts from DHA-treated keratinocytes was performed (Fig. 4e) [35]. Indeed, it was observed that DHA treatment induced posttranslational protein adduction as evident from up to five-fold elevated levels of specific AGEs known to result from triose-adduction of protein targets. Specifically, DHA-dependent formation of N7-(carboxyethyl)-l-arginine (CEA), (carboxyethyl)-l-lysine (CEL), and lactoyl-l-lysine was detectable in HaCaT keratinocytes exposed to concentrations as low as 10 mM, and the hydroimidazolone-type AGE MG-H1, derived from the DHA-oxidation product methylglyoxal, was detectable upon exposure to higher DHA concentrations (20 mM) [45,46].
After substantiating the occurrence of DHA-induced glycation stress as assessed by AGE-formation, we explored the role of GLO1 expression in determining DHA sensitivity displayed by cultured skin cells. GLO1 is an essential glycation stress response gene encoding glyoxalase 1, an enzyme involved in the detoxification of the triose-derived glycolytic byproduct methylglyoxal. However, we noticed that acute DHA exposure [performed in analogy to the mRNA expression array analysis as depicted above (Fig. 2)] did not cause significant changes in GLO1 expression levels detected in HaCaT keratinocytes (Fig. 4f). Next, to further explore the potential role of GLO1 expression in DHA sensitivity, we compared DHA cytotoxicity between isogenic cultured skin cells displaying differential GLO1 expression levels as achieved by CRISPR/Cas9-engineering (Fig. 4g–i). To this end, human malignant melanoma (A375-GLO1_WT versus KO) cells, generated and validated recently in our laboratory to study the impact of GLO1 expression on melanoma invasiveness, were used to examine whether GLO1 status would determine cellular DHA sensitivity. First, validation of KO status of these cells indicated that A375-GLO1_KO (as compared to A375-GLO1_WT) cells display loss of GLO1 mRNA (Fig. 4g), immunoreactivity (Fig. 4g; insert), and enzymatic activity (Fig. 4h). It was then observed that A375-GLO1_KO as compared to wildtype control cells displayed hypersensitivity to DHA-induced loss of viability as assessed by flow cytometric analysis (Fig. 4i). This differential hypersensitivity towards DHA exposure potentially attributable to GLO1 deletion in these isogenic cell line variants was evident from detection of increased induction of cell death in response to DHA concentrations as low as 1 mM. Taken together, these data indicate that DHA treatment induces skin cell glycation stress as evident from AGE-formation in HaCaT keratinocytes and hypersensitivity to DHA-induced loss of viability displayed by genetically-engineered human malignant A375 melanoma cells that do not express the glycation stress defense gene GLO1.
In human epidermal reconstructs topical DHA induces a distinct stress response gene expression profile (different from that induced by exposure to solar UV) and AGE-formation with upregulation of stress response protein-phosphorylation (p-Hsp27).
Given the pronounced skin cell stress response induced by DHA detectable in cultured human keratinocytes, further experiments were performed that would test the possibility of similar responses in reconstructed fully differentiated human epidermis, an accepted organotypic model of human skin used widely for the toxicological assessment of topical treatments [19,[30], [31], [32]]. Following exposure [10% DHA in carrier (Vanicream™) versus carrier only], it was observed that topical DHA caused pronounced epidermal yellowing (i.e. melanoidin-type pigmentation) in epidermal reconstructs detectable (within 24 h exposure) by visible inspection and colorimetric image analysis (Fig. 5a). Next, heat shock stress response gene expression [as observed earlier in HaCaT keratinocytes exposed to DHA (Fig. 2)] was first examined by single RT-qPCR analysis indicating significant upregulation of HSPA6 and HSPA1A observable at a 10 mM exposure level (Fig. 5b). Subsequently, epidermal stress response gene expression array profiling was executed (Fig. 5c and Table 1), performed in direct analogy to the analysis of DHA-exposed cultured human keratinocytes (Fig. 2a–c). Moreover, for comparison, epidermal stress response gene expression array analysis was performed in response to exposure to a supra-erythemal dose of solar simulated UV [2 minimal erythemal doses (MEDs): 240 mJ/cm2 (UVB); 4.6 J/cm2 (UVA)], an experimental design that would allow the identification of genes equally or differentially sensitive to either one of these relevant cutaneous stimuli, [i.e, DHA (‘sunless tanning’) versus solar UV (‘sun exposure-induced tanning’)] (Fig. 5d and Table 1) [36]. These comparative gene expression data (DHA-versus UV-exposed epidermal reconstructs) are displayed in Venn-diagram depiction and also in numerical table format (Fig. 5e and Table 1, respectively).
Fig. 5.
Differential stress response gene expression analysis in human reconstructed epidermis exposed to topical DHA or acute solar simulated UV. (a) After treatment [DHA (10% in Vanicream™) versus carrier control; 24 h], organotypic human epidermal reconstructs were imaged for colorimetric analysis; left: representative images; right: bar graph summarizing colorimetric values (mean ± SD). (b–c) After treatment [DHA (10% in Vanicream™) versus carrier control; 6 h], organotypic human epidermal reconstructs were analyzed for gene expression changes: (b) Individual RT-qPCR analysis (HSPA1A, HSPA6; mean ± SD; DHA: 1 or 10%). (c) Gene expression changes as determined using the Human Stress and Toxicity PathwayFinder™ PCR Array technology comparing DHA- and carrier-exposed epidermal reconstructs depicted as a volcano plot and summarized numerically in Table 1 (d-e) Comparative gene expression array analysis of solar simulated UV-exposed epidermal reconstructs [240 mJ/cm2 UVB versus unexposed carrier control; 6 h after UV) displayed as a volcano plot (d), versus DHA (in Venn diagram depiction with total number of genes per group) (e), and summarized numerically in Table 1. (f) Immunohistochemical analysis of epidermal reconstructs [prepared as in (a)]. (g) Immunohistochemical analysis of SKH-1 mouse skin 3 d after treatment [DHA (10% in Vanicream™) versus carrier control].
Table 1.
Comparative gene expression changes in human epidermal reconstructs (DHA versus UV) as performed and detected by Human Stress and Toxicity PathwayFinder™ PCR Array analysis specified in Fig. 5; all statistically significant changes detected in at least one treatment group are depicted (p < 0.05).
| Gene symbol |
gene description |
fold-change |
|
|---|---|---|---|
| [accession number] | UV | DHA | |
| CYP1A1 [NM_005260] | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 48.58 | ns |
| CYP2E1 [NM_024013] | Cytochrome P450, family 2, subfamily E, polypeptide 1 | 7.19 | ns |
| HSPA1A [NM_013371] | Heat shock 70 kDa protein 1A | 7.06 | 4.98 |
| CCL21 [NM_000757] | Chemokine (C–C motif) ligand 21 | 6.99 | ns |
| CYP7A1 [NM_000605] | Cytochrome P450, family 7, subfamily A, polypeptide 1 | 6.99 | ns |
| FASLG [NM_002187] | Fas ligand (TNF superfamily, member 6) | 6.99 | 1.95 |
| IL6 [NM_000600] | Interleukin 6 (interferon, beta 2) | ns | 6.35 |
| CCL4 [NM_058186] | Chemokine (C–C motif) ligand 4 | 6.13 | ns |
| NOS2 [NM_000595] | Nitric oxide synthase 2, inducible | 5.68 | ns |
| UGT1A4 [NM_003807] | UDP glucuronosyltransferase 1 family, polypeptide A4 | 5.64 | ns |
| IL1B [NM_000879] | Interleukin 1, beta | 4.67 | ns |
| HMOX1 [NM_022789] | Heme oxygenase (decycling) 1 | 4.60 | ns |
| CSF2 [NM_000557] | Colony stimulating factor 2 (granulocyte-macrophage) | 1.90 | 3.85 |
| HSPA5 [NM_012275] | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 3.76 | ns |
| DNAJA1 [NM_002170] | DnaJ (Hsp40) homolog, subfamily A, member 1 | 3.64 | ns |
| FMO1 [NM_002188] | Flavin containing monooxygenase 1 | 3.38 | ns |
| LTA [NM_000880] | Lymphotoxin alpha (TNF superfamily, member 1) | 3.28 | ns |
| DNAJB4 [NM_002176] | DnaJ (Hsp40) homolog, subfamily B, member 4 | 3.02 | ns |
| HSPD1 [NM_018724] | Heat shock 60 kDa protein 1 (chaperonin) | 2.20 | 3.00 |
| PCNA [NM_002341] | Proliferating cell nuclear antigen | 2.77 | 5.25 |
| DDIT3 [NM_002169] | DNA-damage-inducible transcript 3 | ns | 2.82 |
| GSTM3 [NM_013278] | Glutathione S-transferase mu 3 (brain) | 2.75 | ns |
| HSPA2 [NM_000576] | Heat shock 70 kDa protein 2 | 2.61 | ns |
| HSPA6 [NM_014440] | Heat shock 70 kDa protein 6 (HSP70B′) | 2.46 | 5.30 |
| HSPA8 [NM_173205] | Heat shock 70 kDa protein 8 | 2.44 | ns |
| TNFRSF1A [NM_003809] | Tumor necrosis factor receptor superfamily, member 1A | 2.30 | 2.87 |
| UNG [NM_003326] | Uracil-DNA glycosylase | 2.27 | ns |
| MDM2 [NM_000584] | Mdm2 p53 binding protein homolog (mouse) | 2.25 | ns |
| CDKN1A [NM_005811] | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 2.12 | ns |
| XRCC2 [NM_001244] | X-ray repair complementing defective repair | 2.11 | 2.90 |
| EGR1 [NM_020124] | Early growth response 1 | ns | 2.13 |
| CHEK2 [NM_016204] | CHK2 checkpoint homolog (S. pombe) | 2.11 | 0.5 |
| NFKB1 [NM_003240] | Nuclear factor of kappa light polypeptide gene enhancer | 2.07 | ns |
| GADD45A [NM_000585] | Growth arrest and DNA-damage-inducible, alpha | 1.98 | ns |
| HSPE1 [NM_021803] | Heat shock 10 kDa protein 1 (chaperonin 10) | 2.00 | 1.98 |
| ERCC1 [NM_000641] | Excision repair cross-complementing, complementation group 1 | 2.00 | ns |
| CASP10 [NM_001718] | Caspase 10, apoptosis-related cysteine peptidase | 0.58 | 0.5 |
| ATM [NM_001200] | Ataxia telangiectasia mutated | 0.50 | ns |
| HSP90AA2P [NM_019618] | Heat shock protein 90 kDa alpha, class A member 2 | 0.49 | ns |
| HSPA1L [NM_000575] | Heat shock 70 kDa protein 1-like | 0.49 | ns |
| CCNC [NM_000639] | Cyclin C | 0.46 | 3.35 |
| ANXA5 [NM_006129] | Annexin A5 | 0.44 | ns |
| CASP8 [NM_001719] | Caspase 8, apoptosis-related cysteine peptidase | 0.42 | ns |
| SERPINE1 [NM_000594] | Serpin peptidase inhibitor, clade E, member 1 | 0.41 | ns |
| TNFSF10 [NM_003808] | Tumor necrosis factor (ligand) superfamily, member 10 | 0.40 | ns |
| TNF [NM_003701] | Tumor necrosis factor | 0.32 | 3.40 |
| RAD50 [NM_003239] | RAD50 homolog (S. cerevisiae) | 0.22 | ns |
| CCL3 [NM_000758] | Chemokine (C–C motif) ligand 3 | 0.03 | ns |
| BCL2L1 [NM_130851] | BCL2-like 1 | 0.01 | ns |
Strikingly, comparative gene expression array analysis revealed that only thirteen genes were jointly responsive to UV- and DHA-exposure (Fig. 5e and Table 1). The number of genes modulated exclusively by DHA exposure (a total of three only: IL6, DDIT3, EGR1) was much lower than the number of genes responsive exclusively responsive to UV (a total of thirty three) which included those encoding specific members of the cytochrome P450 family (e.g. CYP1A1, CYP2E1, CYP7A1) and also affecting inflammatory (CCL21, CCL3, CCL4, IL1B, LTA, NOS2, TNFSF10), heat shock (DNAJA1, DNAJB4, HSPA2, HSPA5, HSPA8, HSPA1L, HSP90AA2P), antioxidant (GSTM3, HMOX1), DNA damage (ATM, ERCC1, GADD45A, RAD50), proliferative (CCNC, CDKN1A), and cell survival (ANXA5, BCL2L1, CASP8, FASLG) control. These data allow to draw the conclusion that (at specific exposure levels examined in this limited prototype study), the transcriptional consequences and epidermal impact of solar UV-exposure far outweigh those imposed by DHA-exposure.
Interestingly, among the thirteen genes (HSPA1A, FASLG, CSF2, HSPD1, PCNA, HSPA6, TNFRSF1A, XRCC2, CHEK2, HSPE1, CASP10, CCNC, TNF) jointly responsive to either UV- or DHA-exposure, some expression changes imposed by UV (fold changes) quantitatively outweighed those imposed by DHA [such as: HSPA1A (7- versus 5-fold upregulation), FASLG (7- versus 2-fold upregulation), respectively (Table 1)]. Vice versa, among these expression changes (responsive to both UV- or DHA-exposure), some DHA-induced alterations significantly outweighed those imposed by UV [such as: CSF2 (2- versus 4-fold upregulation), PCNA (3- versus 5-fold upregulation), HSPA6 (3- versus 5-fold upregulation), TNF (0.3- versus 3-fold downregulation), respectively (Table 1)].
Next, immunohistochemical analysis was used to explore the consequences of DHA-treatment in epidermal reconstructs (Fig. 5f). Strikingly, as observed before in human keratinocytes (Figs. 2d and 3), pronounced upregulation of DHA-induced staining for CEL, HO-1, total HSP27, and phospho-HSP27 (S78) was detected, indicative of glycation-stress-associated AGE-epitope formation and heat shock-related stress response signaling.
Likewise, to explore the possibility that similar responses are detectable in an in vivo exposure model, prototype experimentation was performed in hairless SKH-1 mouse skin treated with DHA (Fig. 5g). Indeed, immunohistochemical analysis revealed that DHA-exposed mouse skin displayed prominent epidermal staining for AGE-epitopes (CML/CEL), antioxidant markers (HO-1), and heat shock response [total HSP27 and phospho-HSP27 (S78)], changes reminiscent of immunohistochemical alterations observed in human epidermal reconstruct exposed to topical DHA. However, future experiments performed at the transcriptional and protein expression levels will have to be performed (using mouse and human skin) in order to further substantiate these prototype findings obtained in live murine skin exposed topically to DHA.
4. Discussion
Since its accidental discovery almost 50 years ago, chemical tanning is widely regarded as a safe alternative to solar UV-induced skin tanning known to be associated with epidermal genotoxic stress, but the cutaneous biology impacted by sunless tanning that involves chemical reactions remains largely unexplored [[8], [9], [10], [11]]. Here, we have examined the cutaneous effects of acute DHA exposure employing cultured human keratinocytes and epidermal reconstructs, profiled by gene expression array analysis and immunodetection. In human HaCaT keratinocytes, DHA exposure (up to 20 mM; 6 h) did not impair viability while causing a pronounced cellular stress response as obvious from activation of phospho-protein signal transduction [p-p38, p-Hsp27(S15/S78), p-eIF2α] and gene expression changes (HSPA6, HMOX1, CRYAB, CCL3), both of which were not observed upon exposure to the non-ketose, tanning-inactive DHA-control glycerol. Likewise, a cosmetic use-relevant topical DHA dose-regimen elicited a pronounced transcriptional stress response observable in human epidermal reconstructs as revealed by gene expression array (HSPA1A, HSPA6, HSPD1, IL6, DDIT3, EGR1) and immunohistochemical analysis (CEL, HO-1, p-Hsp27-S78). Strikingly, in human epidermal reconstructs, DHA treatment, compared directly to the consequences of acute exposure to solar simulated light (delivered at approximately two MEDs), caused modulation of thirteen genes also responsive to solar UV. Strikingly, expression of only three genes was exclusively responsive to this chemical tanning agent, including IL6 (encoding the pro-inflammatory cytokine ‘interleukin-6’), DDIT3 [also known as GADD153, encoding the proteotoxic stress-response transcription factor ‘C/EBP homologous protein’ (CHOP)], and EGR1 (encoding the tumor protein p53-responsive stress transcription factor ‘early growth response protein 1’), whereas a much higher number of genes (a total of thirty three, as assessed by array analysis) was responsive exclusively to solar UV, indicating that DHA treatment causes a more focused and limited gene expression pattern as compared to solar UV exposure.
Obviously, functional consequences of acute cellular and tissue responses elicited by topical DHA exposure, profiled for the first time in this prototype study in cultured human keratinocytes and epidermal reconstructs, remain largely unexplored and must be validated and expanded in adequate follow up studies using relevant exposure regimens, preferably performed in live human skin ex vivo and in vivo [6,8,12,14]. Likewise, more detailed cutaneous pharmacokinetic and pharmacodynamic exploration must be performed, examining effects of DHA and related chemical tanning agents (applied in commercially relevant formulations and vehicles including spray tan applications) as encountered in consumer products used worldwide, with inclusion of adequate vehicle controls that represent these often complex formulations optimized for epidermal delivery and residence time [8,12].
Given the significant epidermal changes that occur in response to DHA exposure impacting stress response gene expression and phosphoprotein signaling, it is tempting to speculate that these changes might also engage in molecular cross-talk with established environmental cutaneous stressors (including solar UV, ozone, arsenic, particulate matter etc.), potentially synergizing with or even attenuating inflammatory, genotoxic, glycoxidative, redox, and proteotoxic impact of these environmental stressors, relevant to skin barrier function, aging, carcinogenesis, and other determinants of skin health [6,15,19,44,[47], [48], [49], [50]]. Moreover, sunless skin tanning through glycation reactions is also known to provide a moderate sun protection factor thought to originate from UV-absorptivity of melanoidin pigments, and it will be interesting to probe if DHA-induced photoprotection might also be attributable to modulation of stress response pathways as reported here for the first time [51].
The role of glycation stress associated with sunless tanning using reactive triose and tetrose sugars (including DHA and erythrulose, respectively), imposed by enabling epidermal glycation reactions that cause formation of melanoidin-pigments mimicking solar UV-induced melanogenesis, remains to be explored in much detail [20]. In our prototype study, formation of DHA-derived AGEs resulting from posttranslational protein adduction was confirmed by quantitative mass spectrometric detection of Nε-(carboxyethyl)-l-lysine (CEL) and N7-carboxyethyl-l-arginine (CEA) in HaCaT keratinocytes as well as CEL-immunodetection performed in human epidermal reconstructs and mouse skin. In this context, it will be interesting to explore the specific identity of molecular targets undergoing glycation by DHA upstream of stress response signaling and gene expression. Moreover, it might be speculated that glycation-active glycolytic intermediates formed from cellular energy metabolism, such as dihydroxyacetone phosphate (and methylglyoxal derived thereof), might elicit cellular stress responses similar to those detected by us in response to exogenous DHA [13,46,52]. Likewise, the role of the intrinsic cellular glycation defense attenuating glycation damage deserves further examination, as evidenced by our observation that A375 melanoma cells with genetic GLO1 deletion display hypersensitivity towards DHA, focusing on the potential cytoprotective role of GLO1 expression in cutaneous keratinocytes and intact human skin [29,40,53]. Interestingly, DHA-dependent induction of cell death observable in cultured malignant melanoma cells has been reported before, but the potential therapeutic implication of this observation remains to be substantiated [40]. Importantly, glycation stress has long been known to impact structural integrity of major skin components such as collagens and elastins, relevant to the occurrence of increased glycoxidative skin damage observable in diabetic patients, but the occurrence of similar changes in response to chemical tanning through topical application of glycation-active ingredients might largely depend on specific skin pharmacokinetic profile of these agents and formulations [15,19,44]. It has now been demonstrated that AGEs, serving as damage-associated molecular patterns (DAMPs) that form in response to electrophilic environmental stressors, are also recognized by specialized receptors including RAGE and TLR4, widely expressed throughout skin and known to be involved in innate immunity, inflammatory remodeling, and even tumorigenic progression (as demonstrated for RAGE-dependent melanoma cell invasion and metastasis) [20,[24], [25], [26]]. Finally, a role of AGEs as cutaneous photosensitizers and mediators of skin photooxidative stress, activated by solar UVA and blue light excitation, has recently been substantiated, and it remains to be seen if DHA-derived cutaneous AGEs might serve a similar function [17,23,47].
Taken together, these prototype data profile for the first time a comprehensive DHA-induced acute epidermal stress response at the transcriptional level that profoundly differs from that elicited by solar UV exposure, a finding that deserves further molecular exploration in living human skin [5]. Molecular pathways that may facilitate sunless tanning and skin pigmentation are of much interest in the context of both, therapeutic intervention targeting dyspigmentation pathologies such as vitiligo and cosmetic intervention facilitating skin tanning in the absence of UV-induced genotoxic stress [1,5,6,8,54]. Given the worldwide use of DHA in consumer products accentuated by a growing interest in safer molecular approaches that allow sunless tanning without compromising skin barrier and function, these prototype data may provide a first framework for future mechanistic studies.
Declaration of competing interest
The authors state no conflict of interest.
Acknowledgements
Supported in part by grants from the National Institutes of Health (1R01CA229418, 1R03CA230949, 1R21ES029579, 1P01CA229112, ES007091, ES006694, and UA Cancer Center Support Grant CA023074). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.D'Orazio J.A., Nobuhisa T., Cui R., Arya M., Spry M., Wakamatsu K., Igras V., Kunisada T., Granter S.R., Nishimura E.K., Ito S., Fisher D.E. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Malek Z.A., Knittel J., Kadekaro A.L., Swope V.B., Starner R. The melanocortin 1 receptor and the UV response of human melanocytes--a shift in paradigm. Photochem. Photobiol. 2008;84:501–508. doi: 10.1111/j.1751-1097.2008.00294.x. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J., Douki T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018;17:1816–1841. doi: 10.1039/c7pp00395a. [DOI] [PubMed] [Google Scholar]
- 4.Cui R.T., Widlund H.R., Feige E., Lin J.Y., Wilensky D.L., Igras V.E., D'Orazio J., Fung C.Y., Schanbacher C.F., Granter S.R., Fisher D.E. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 5.Chen H.X., Weng Q.Y., Fisher D.E. UV signaling pathways within the skin. J. Invest. Dermatol. 2014;134:2080–2085. doi: 10.1038/jid.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akin F.J., Marlowe E. Non-carcinogenicity of dihydroxyacetone by skin painting. J. Environ. Pathol. Toxicol. Oncol. 1984;5:349–351. [PubMed] [Google Scholar]
- 7.Huang A., Brody N., Liebman T.N. Dihydroxyacetone and sunless tanning: knowledge, myths, and current understanding. J. Am. Acad. Dermatol. 2017;77:991–992. doi: 10.1016/j.jaad.2017.04.1117. [DOI] [PubMed] [Google Scholar]
- 8.Braunberger T.L., Nahhas A.F., Katz L.M., Sadrieh N., Lim H.W. Dihydroxyacetone: a review. J. Drugs Dermatol. JDD. 2018;17:387–391. [PubMed] [Google Scholar]
- 9.Wittgenstein E., Berry H.K. Staining of skin with dihydroxyacetone. Science. 1960;132:894–895. doi: 10.1126/science.132.3431.894. [DOI] [PubMed] [Google Scholar]
- 10.Wittgenstein E., Guest G.M. Biochemical effects of dihydroxyacetone. J. Invest. Dermatol. 1961;37:421–426. doi: 10.1038/jid.1961.137. [DOI] [PubMed] [Google Scholar]
- 11.Tressl R., Wondrak G.T., Garbe L.A., Kruger R.P., Rewicki D. Pentoses and hexoses as sources of new melanoidin-like Maillard polymers. J. Agric. Food Chem. 1998;46:1765–1776. doi: 10.1021/jf970657c. [DOI] [PubMed] [Google Scholar]
- 12.Petersen A.B., Wulf H.C., Gniadecki R., Gajkowska B. Dihydroxyacetone, the active browning ingredient in sunless tanning lotions, induces DNA damage, cell-cycle block and apoptosis in cultured HaCaT keratinocytes. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2004;560:173–186. doi: 10.1016/j.mrgentox.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Seneviratne C., Dombi G.W., Liu W., Dain J.A. In vitro glycation of human serum albumin by dihydroxyacetone and dihydroxyacetone phosphate. Biochem. Biophys. Res. Commun. 2012;417:817–823. doi: 10.1016/j.bbrc.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 14.Smith K.R., Hayat F., Andrews J.F., Migaud M.E., Gassman N.R. Dihydroxyacetone exposure alters NAD(P)H and induces mitochondrial stress and autophagy in HEK293T cells. Chem. Res. Toxicol. 2019;32:1722–1731. doi: 10.1021/acs.chemrestox.9b00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournet M., Bonte F., Desmouliere A. Glycation damage: a possible hub for major pathophysiological disorders and aging. Aging Dis. 2018;9:880–900. doi: 10.14336/AD.2017.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrone A., Giovino A., Benny J., Martinelli F. Advanced glycation end products (AGEs): biochemistry, signaling, analytical methods, and epigenetic effects. Oxid Med Cell Longev. 2020;2020:3818196. doi: 10.1155/2020/3818196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wondrak G.T., Roberts M.J., Jacobson M.K., Jacobson E.L. Photosensitized growth inhibition of cultured human skin cells: mechanism and suppression of oxidative stress from solar irradiation of glycated proteins. J. Invest. Dermatol. 2002;119:489–498. doi: 10.1046/j.1523-1747.2002.01788.x. [DOI] [PubMed] [Google Scholar]
- 18.Roberts M.J., Wondrak G.T., Laurean D.C., Jacobson M.K., Jacobson E.L. DNA damage by carbonyl stress in human skin cells. Mutat. Res. Fund Mol. Mech. Mutagen. 2003;522:45–56. doi: 10.1016/s0027-5107(02)00232-4. [DOI] [PubMed] [Google Scholar]
- 19.Pageon H., Zucchi H., Rousset F., Monnier V.M., Asselineau D. Skin aging by glycation: lessons from the reconstructed skin model. Clin. Chem. Lab. Med. 2014;52:169–174. doi: 10.1515/cclm-2013-0091. [DOI] [PubMed] [Google Scholar]
- 20.Lee E.J., Kim J.Y., Oh S.H. Advanced glycation end products (AGEs) promote melanogenesis through receptor for AGEs. Sci. Rep. 2016;6 doi: 10.1038/srep27848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Putte L., De Schrijver S., Moortgat P. The effects of advanced glycation end products (AGEs) on dermal wound healing and scar formation: a systematic review. Scars Burn Heal. 2016;2 doi: 10.1177/2059513116676828. 2059513116676828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen A.H., Detty S.Q., Agrawal D.K. Clinical implications of high-mobility group box-1 (HMGB1) and the receptor for advanced glycation end-products (RAGE) in cutaneous malignancy: a systematic review. Anticancer Res. 2017;37:1–7. doi: 10.21873/anticanres.11282. [DOI] [PubMed] [Google Scholar]
- 23.Wondrak G.T., Jacobson M.K., Jacobson E.L. Endogenous UVA-photosensitizers: mediators of skin photodamage and novel targets for skin photoprotection. Photochem. Photobiol. Sci. 2006;5:215–237. doi: 10.1039/b504573h. [DOI] [PubMed] [Google Scholar]
- 24.Lamore S.D., Cabello C.M., Wondrak G.T. HMGB1-directed drug discovery targeting cutaneous inflammatory dysregulation. Curr. Drug Metabol. 2010;11:250–265. doi: 10.2174/138920010791196337. [DOI] [PubMed] [Google Scholar]
- 25.Pandolfi F., Altamura S., Frosali S., Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin. Therapeut. 2016;38:1017–1028. doi: 10.1016/j.clinthera.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Dickinson S.E., Wondrak G.T. TLR4 in skin cancer: from molecular mechanisms to clinical interventions. Mol. Carcinog. 2019;58:1086–1093. doi: 10.1002/mc.23016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Younick J.J., Koenig M.L., Yourick D.L., Bronaugh R.L. Fate of chemicals in skin after dermal application: does the in vitro skin reservoir affect the estimate of systemic absorption? Toxicol. Appl. Pharmacol. 2004;195:309–320. doi: 10.1016/j.taap.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Jandova J., Perer J., Hua A., Snell J.A., Wondrak G.T. Genetic target modulation employing CRISPR/Cas9 identifies glyoxalase 1 as a novel molecular determinant of invasion and metastasis in A375 human malignant melanoma cells in vitro and in vivo. Cancers (Basel) 2020 May 26;12(6):E1369. doi: 10.3390/cancers12061369. PMID: 32466621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bair W.B., 3rd, Cabello C.M., Uchida K., Bause A.S., Wondrak G.T. GLO1 overexpression in human malignant melanoma. Melanoma Res. 2010;20:85–96. doi: 10.1097/CMR.0b013e3283364903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamore S.D., Wondrak G.T. Zinc pyrithione impairs zinc homeostasis and upregulates stress response gene expression in reconstructed human epidermis. Biometals. 2011;24:875–890. doi: 10.1007/s10534-011-9441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao S., Justiniano R., Zhang D.D., Wondrak G.T. The Nrf2-inducers tanshinone I and dihydrotanshinone protect human skin cells and reconstructed human skin against solar simulated UV. Redox Biol. 2013;1:532–541. doi: 10.1016/j.redox.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S.L., Justiniano R., Williams J.D., Cabello C.M., Qiao S., Wondrak G.T. The tryptophan-derived endogenous aryl hydrocarbon receptor ligand 6-Formylindolo[3,2-b]Carbazole is a nanomolar UVA photosensitizer in epidermal keratinocytes. J. Invest. Dermatol. 2015;135:1649–1658. doi: 10.1038/jid.2014.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis A.L., Qiao S., Lesson J.L., Rojo de la Vega M., Park S.L., Seanez C.M., Gokhale V., Cabello C.M., Wondrak G.T. The quinone methide aurin is a heat shock response inducer that causes proteotoxic stress and Noxa-dependent apoptosis in malignant melanoma cells. J. Biol. Chem. 2015;290:1623–1638. doi: 10.1074/jbc.M114.592626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabello C.M., Bair W.B., 3rd, Ley S., Lamore S.D., Azimian S., Wondrak G.T. The experimental chemotherapeutic N6-furfuryladenosine (kinetin-riboside) induces rapid ATP depletion, genotoxic stress, and CDKN1A(p21) upregulation in human cancer cell lines. Biochem. Pharmacol. 2009;77:1125–1138. doi: 10.1016/j.bcp.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galligan J.J., Kingsley P.J., Wauchope O.R., Mitchener M.M., Camarillo J.M., Wepy J.A., Harris P.S., Fritz K.S., Marnett L.J. Quantitative analysis and discovery of lysine and arginine modifications. Anal. Chem. 2017;89:1299–1306. doi: 10.1021/acs.analchem.6b04105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams J.D., Bermudez Y., Park S.L., Stratton S.P., Uchida K., Hurst C.A., Wondrak G.T. Malondialdehyde-derived epitopes in human skin result from acute exposure to solar UV and occur in nonmelanoma skin cancer tissue. J. Photochem. Photobiol., B. 2014;132:56–65. doi: 10.1016/j.jphotobiol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai R., Araki T., Horiuchi S. Preparation of specific antibody against CML, one of major AGE structures. Maillard Reaction in Food Chemistry and Medical Science: Update for the Postgenomic Era. 2002;1245:479–480. [Google Scholar]
- 38.Koito W., Araki T., Horiuchi S., Nagai R. Conventional antibody against N-epsilon-(carboxymethyl)lysine (CML) shows cross-reaction to N-epsilon-(carboxyethyl)lysine (CEL): immunochemical quantification of CML with a specific antibody. J. Biochem. 2004;136:831–837. doi: 10.1093/jb/mvh193. [DOI] [PubMed] [Google Scholar]
- 39.Justiniano R., Perer J., Hua A., Fazel M., Krajisnik A., Cabello C.M., Wondrak G.T. A topical zinc ionophore blocks tumorigenic progression in UV-exposed SKH-1 high-risk mouse skin. Photochem. Photobiol. 2017;93:1472–1482. doi: 10.1111/php.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith K.R., Granberry M., Tan M.C.B., Daniel C.L., Gassman N.R. Dihydroxyacetone induces G2/M arrest and apoptotic cell death in A375P melanoma cells. Environ. Toxicol. 2018;33:333–342. doi: 10.1002/tox.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong J.W., Shi B., Farboud B., McClaren M., Shibamoto T., Cross C.E., Isseroff R.R. Ultraviolet B-mediated phosphorylation of the small heat shock protein HSP27 in human keratinocytes. J. Invest. Dermatol. 2000;115:427–434. doi: 10.1046/j.1523-1747.2000.00077.x. [DOI] [PubMed] [Google Scholar]
- 42.Jonak C., Mildner M., Klosner G., Paulitschke V., Kunstfeld R., Pehamberger H., Tschachler E., Trautinger F. The hsp27kD heat shock protein and p38-MAPK signaling are required for regular epidermal differentiation. J. Dermatol. Sci. 2011;61:32–37. doi: 10.1016/j.jdermsci.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Baird T.D., Wek R.C. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pageon H., Zucchi H., Dai Z., Sell D.R., Strauch C.M., Monnier V.M., Asselineau D. Biological effects induced by specific advanced glycation end products in the reconstructed skin model of aging. Biores Open Access. 2015;4:54–64. doi: 10.1089/biores.2014.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galligan J.J., Wepy J.A., Streeter M.D., Kingsley P.J., Mitchener M.M., Wauchope O.R., Beavers W.N., Rose K.L., Wang T., Spiegel D.A., Marnett L.J. Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U. S. A. 2018;115:9228–9233. doi: 10.1073/pnas.1802901115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaffney D.O., Jennings E.Q., Anderson C.C., Marentette J.O., Shi T., Schou Oxvig A.M., Streeter M.D., Johannsen M., Spiegel D.A., Chapman E., Roede J.R., Galligan J.J. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem Biol. 2020;27:206–213. doi: 10.1016/j.chembiol.2019.11.005. e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wondrak G.T., Jacobson E.L., Jacobson M.K. Photosensitization of DNA damage by glycated proteins. Photochem. Photobiol. Sci. 2002;1:355–363. doi: 10.1039/b202732c. [DOI] [PubMed] [Google Scholar]
- 48.Wondrak G.T. Let the sun shine in: mechanisms and potential for therapeutics in skin photodamage. Curr. Opin. Invest. Drugs. 2007;8:390–400. [PubMed] [Google Scholar]
- 49.Weinmuellner R., Kryeziu K., Zbiral B., Tav K., Schoenhacker-Alte B., Groza D., Wimmer L., Schosserer M., Nagelreiter F., Rosinger S., Mildner M., Tschachler E., Grusch M., Grillari J., Heffeter P. Long-term exposure of immortalized keratinocytes to arsenic induces EMT, impairs differentiation in organotypic skin models and mimics aspects of human skin derangements. Arch. Toxicol. 2018;92:181–194. doi: 10.1007/s00204-017-2034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrara F., Pambianchi E., Pecorelli A., Woodby B., Messano N., Therrien J.P., Lila M.A., Valacchi G. Redox regulation of cutaneous inflammasome by ozone exposure. Free Radic. Biol. Med. 2019 Nov 26;S0891-5849(19):31662–31664. doi: 10.1016/j.freeradbiomed.2019.11.031. [DOI] [PubMed] [Google Scholar]
- 51.Fusaro R.M., Rice E.G. The Maillard reaction for sunlight protection. Maillard Reaction: Chemistry at the Interface of Nutrition, Aging, and Disease. 2005;1043:174–183. doi: 10.1196/annals.1333.022. [DOI] [PubMed] [Google Scholar]
- 52.Bollong M.J., Lee G., Coukos J.S., Yun H., Zambaldo C., Chang J.W., Chin E.N., Ahmad I., Chatterjee A.K., Lairson L.L., Schultz P.G., Moellering R.E. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature. 2018;562:600–604. doi: 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou X.Y., Ding D., Zhan N., Liu X.M., Pan C., Xia Y.M. Glyoxalase I is differentially expressed in cutaneous neoplasms and contributes to the progression of squamous cell carcinoma. J. Invest. Dermatol. 2015;135:589–598. doi: 10.1038/jid.2014.377. [DOI] [PubMed] [Google Scholar]
- 54.Passeron T. Indications and limitations of afamelanotide for treating vitiligo. JAMA Dermatol. 2015;151:349–350. doi: 10.1001/jamadermatol.2014.4848. [DOI] [PubMed] [Google Scholar]