Abstract
Quantitative proteomic workflow based on mass spectrometry (MS) is recently developed by the researchers to screen for biomarkers in periodontal diseases comprising periodontitis. Periodontitis is known for chronic inflammatory disease characterized by progressive destruction of the tooth-supporting apparatus, yet has a lack of clear pathobiology based on a discrepancy between specified categories and diagnostic vagueness. The objective of this review was to outlined the accessible information related to proteomics studies on periodontitis. The Preferred Reporting Items for Systematical Reviews and Meta-Analysis (PRISMA) statement guides to acquaint proteomic analysis on periodontal diseases was applied. Three databases were used in this study, such as Pubmed, ScienceDirect and Biomed Central from 2009 up to November 2019. Proteomics analysis platforms that used in the studies were outlined. Upregulated and downregulated proteins findings data were found, in which could be suitable as candidate biomarkers for this disease.
Keywords: Cell biology, Proteins, Biochemistry, Molecular biology, Dentistry, Proteomics, Periodontitis, Periodontal disease, Biomarker
Cell biology; Proteins; Biochemistry; Molecular Biology; Dentistry; proteomics; Periodontitis; Periodontal disease; Biomarker
1. Introduction
Periodontal disease encompassing periodontitis and gingivitis, is highly prevalent and could impact on 90% of the worldwide population [1]. Gingivitis is a nonspecific inflammatory reaction to a nonspecific accreation of plaque which restrained to the gingival tissue, without causal destruction of periodontal tissue [2]. Meanwhile, periodontitis is a chronic multifactorial inflammatory disease that related to microfloral plaque biofilm, host-mediated inflammation that results in loss of periodontal attachment which characterized by gradual destruction of periodontal tissue support [3, 4].
Clinical considerations are efficacious instruments for determining the health or periodontal disease conditions in most patients. However, some individuals are more susceptible to developing periodontitis and severe generalized periodontitis and also fewer responsive to standard periodontitis treatment [3]. Periodontal tissues have a complex structure, therefore better understanding of a total set of cellular and matrix proteins in periodontal tissue is a necessity for futuristic advances [5]. Biomarkers are expected to supplement the information stipulated by the criterion clinical parameters and can also to improving diagnostic precision in early detection of periodontitis, which is expected to make an important contribution for a better assessment of periodontitis [3].
Recently, scientists have sought to uncover biomarkers for periodontitis. In fact, a number of genes, transcriptions and proteins that related to periodontitis have been identified [6]. Mass spectrometry (MS)-based proteomics studies allow the discovery of proteome and clinical condition correlations. It is also suitable for the study of complex multifactorial diseases including periodontitis [7]. Integration of several data platforms from clinical, radiographic, and proteomics in the study of periodontitis is expected to increase a better diagnosis of periodontitis [4]. The current review aims an in-depth analysis of proteomics approaches for biomarkers and diagnosis of periodontitis.
2. Methods
2.1. Literature search
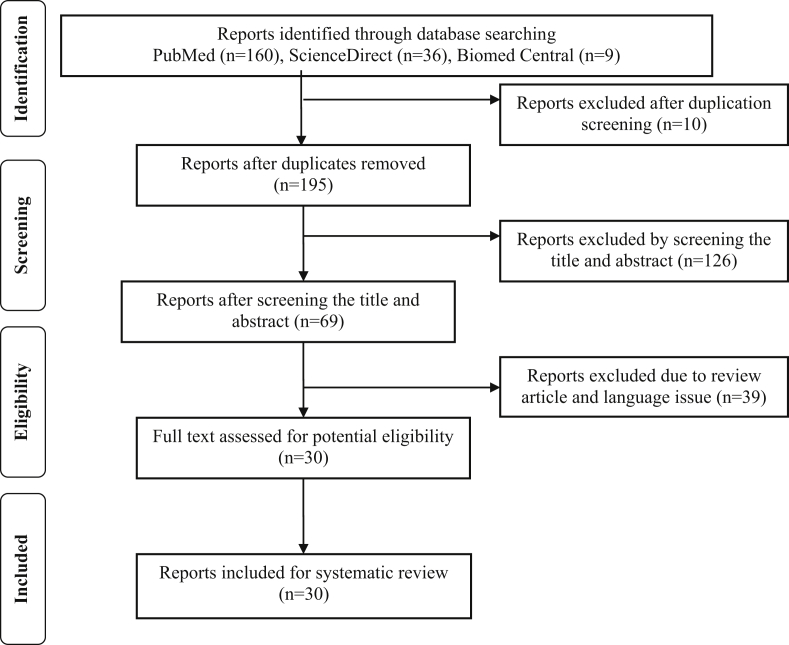
This review is applying The Preferred Reporting Items for Systematical Reviews and Meta-Analysis (PRISMA) statement guidelines to acquaint proteomic analysis on periodontal diseases. Full text manuscripts written in English from three databases were used in this study, including Pubmed, ScienceDirect and Biomed Central which published from 2009 up to November 2019. The following search terms by means of Boolean operators were “periodontitis”, “periodontal disease”, “proteomics”, “proteome”, and “protein”. Authors decided an article that achieve the inclusion criteria by following the guidelines (Figure 1).
Figure 1.
PRISMA flowchart for screening protocol.
We independently screened all titles and abstract. There are two types of studies that found in this study, they were observational and experiment in vitro studies. The articles which selected for full-text reading were read and assessed by individual reviewers independently to regain relevant data for review.
2.2. Inclusion and exclusion criteria
The inclusion criteria for this analysis are: (1) Observational and experiment studies, (2) Studies that analyzing proteomic profiles of periodontal diseases, (3) Methods of sample analysis were: Liquid chromatography tandem mass spectrometry (LC-MS/MS), two-dimensional electrophoresis (2DE), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and enzyme-linked immunosorbent assay (ELISA). Meanwhile, the exclusion criteria are: (1) A case reports studies, (2) Animal studies and (3) Duplicate publications.
2.3. Quality of evidence
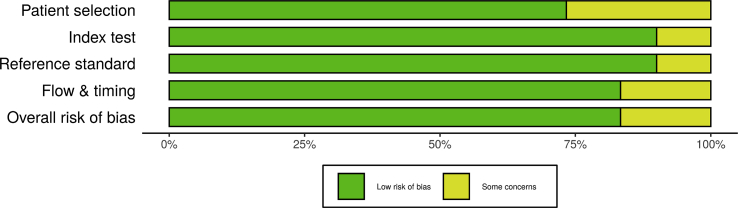
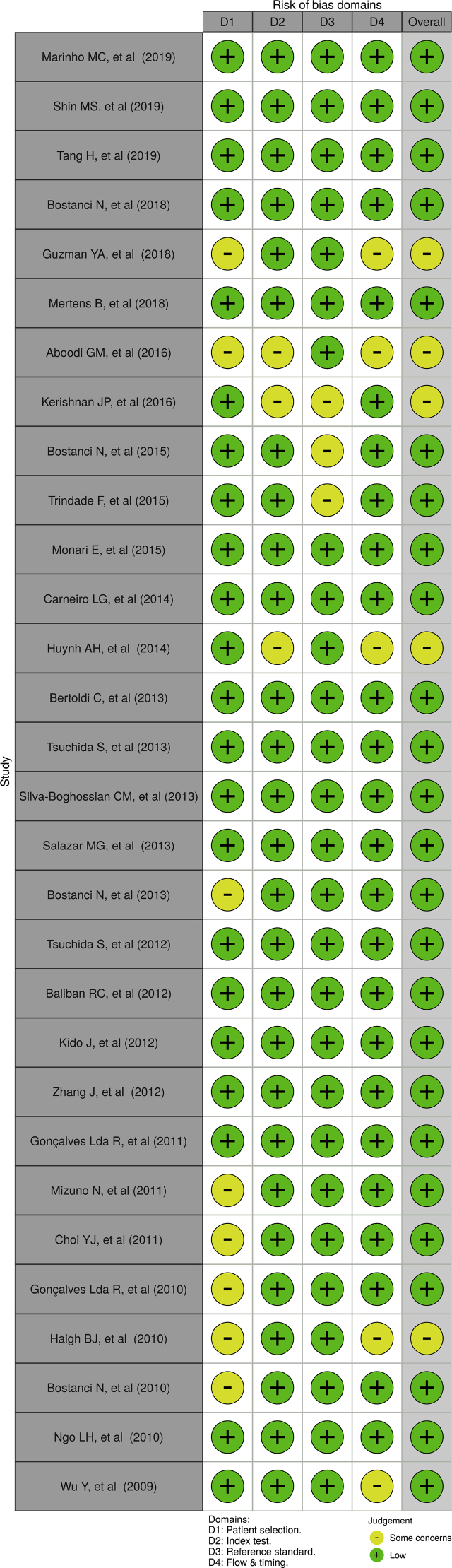
Quality evaluation from all appointed reports were accomplished using risk of bias assessment QUADAS-2 tool for diagnostic test accuracy studies [8]. Risk of bias of each individual study was assessed by two reviewers, independently. Risk of bias plots and graph were created using the robvis application (Figures 2 and 3). [9] The criteria of this assessment were four domains: patient selection, index test, reference criterion, along flow and timing. The additional signaling questions are included to assist judgments. They were answered as “yes,” “no,” or “unclear” and are phrased such that “yes” indicates low risk of bias. Risk of bias is judged with “low,” “high,” or “some concern.”
Figure 2.
The distribution of risk-of-bias judgments within each bias domain.
Figure 3.
Risk of bias plots of the domain-level judgements for each individual study.
3. Results
3.1. Type of study
Of the 30 articles that met the criteria, there were 29 observational studies, and one in vitro trial (Table 1) In observational studies, there is one study that uses systemic disease (Diabetes Mellitus type 2) subjects, and most of the studies utilize the 1999 classification of periodontal diseases for diagnosis. In this review, there were no proteomic studies using the latest classification of periodontal diseases. In the new classification for periodontal diseases, periodontitis was characterized based on staging and grading system [10].
Table 1.
Proteomics studies of periodontal diseases.
| Author (Year) | Diagnosis Criteria | Sample | Number of Subjects | Protein Analysis | Main Findings |
|---|---|---|---|---|---|
| Marinho MC, et al. (2019) [11] | CP with DM; PH with DM; CP without DM; PH without DM | GCF | 20 subjects (5 for each group) | iTRAQ labeling; LC-MS/MS |
- There were 104 proteins exhibiting significant differences between the controlled and the diseased groups |
| Shin MS, et al. (2019) [12] | P; H | WUS | 207 subjects (107 P, 100 H) | LC MS/MS; ELISA | - There were 744 proteins identified - S100A8 and S100A9 were the highest total relative abundance proteins |
| Tang H, et al. (2019) [13] | CP; G; PH | WUS GCF BS |
50 subjects (17 CP; 17 G; 16 PH) | MALDI-TOF; Nano-LC/ESI-MS/MS | - There were 91 peptides detected, seven of which showed significant differences between the CP and the PH group - Fifty-eight peptide peaks were found in BC, 13 of which showed significant differences between the CP and the PH groups, 10 peaks exhibited significantly upregulated in G group - Forty-eight peptide peaks were found in GCF, four of them were significantly higher in the CP group than in the PH group - One hundred and thirteen salivary peptide peaks were detected in G groups, three of which were significantly different from the PH group |
| Bostanci N, et al. (2018) [14] | CP; gAP; G; PH | WUS | 67 subjects in study group phase 1 (17 CP; 17 gAP, 17 gingivitis; 16 PH) and 82 subjects in study group phase 2 (21 CP; 21 gAP; 20 G; 20 PH) | LFQ LC-MS/MS; LC-SRM-MS |
- One hundred an nineteen proteins significant difference between PH and disease subjects - Five proteins with high predictive value for CP and gAP group (AUC >0.97); |
| Guzman YA, et al. (2018) [15] | CP | GCF | 10 subjects before and after treatment | LC-MS/MS | - Azurocidin, lysozyme C, and myosin-9 as biomarkers candidate at baseline - α-smooth muscle actin as biomarker candidate 13 weeks after treatment |
| Mertens B, et al. (2018) [16] | CP; gAP; PH | WUS | 33 subjects (10 CP; 11 gAP, 12 PH) | SDS-PAGE; LC-MRM |
- Hemopexin, plasminogen, and α-fibrinogen related to the presence of periodontitis compare with healthy subjects - Apolipoprotein H was found higher in gAP compare with CP |
| Aboodi GM, et al. (2016) [17] | Plaque-induced gingivitis | WUS | 5 healthy (experimental model) | Oral neutrophil quantification; LC-ESI-MS/MS | - Eighty-nine proteins showed significant level changes during experiment on gingivitis - Neutrophil count in G has a moderate correlation with salivary β-globin, thioredoxin, and albumin - Neutrophil count in G has a strong correlation with collagen α-1 and G protein–coupled receptor 98 |
| Kerishnan JP, et al. (2016) [18] | PH; mild CP; moderate CP; severe CP; gAP | BS | 90 subjects (42 PH; 9 mild CP; 12 moderate CP; 19 severe CP; 8 gAP) | 2DE; MALDI-TOF/TOF; WB | - Fourteen protein clusters identified - α1-antitrypsin, haptoglobin, Ig κ chain C region, kininogen significantly expressed in all stages of periodontitis compare to normal subjects. |
| Bostanci N, et al. (2015) [19] | Biofilm with red complex; Biofilm without red complex | Gingival epithelial culture secretome | Gingival epithelial culture | LC MS/MS | - One hundred and ninety-two proteins were quantified - The red complex bacteria in the biofilm was responsible for down-regulated effect - The upregulated proteins were associated with inflammation and apoptosis - The down regulated ones were associated with the alteration of epithelial tissue robustness and disablement of tissue turnover |
| Trindade F, et al. (2015) [20] | CP; PH | WUS | 9 CP; 10 PH | Nano-HPLC-MALDI-TOF/TOF; Protease prediction carried out in silico with Proteasix; Slot blot | - Protease prediction showed a different protease profiles in CP and H subjects - Histatin-1 increased in CP subjects compare to PH subjects - Eight peptides showed a specific association with CP |
| Monari E, et al. (2015) [21] | CP; PH | Periodontal pocket tissue | 15 CP; 15 PH | 2DE; LC MS/MS; Western blot | - Thirty-two proteins identified - Four proteins (S100A9, heat shock protein β1, galectin-7, and 14-3-3) were over-expressed in CP compared with PH subjects |
| Carneiro LG, et al. (2014) [22] | PH; Moderate- severe P | GCF | 40 PH; 40 moderate-severe P | LC-ESI-MS/MS; ICAT labeling; mTRAQ labeling; SDS-PAGE; ELISA | - One hundred and eighty proteins were quantified in both groups - Twenty six and 32 proteins were found only in PH and moderate-severe P subjects respectively - Immunoglobulin A2, lactotransferrin, neutrophil defensin-1, myeloperoxidase, and S100A8 proteins were upregulated in moderate-severe P - Fifty host proteins and 16 bacterial proteins were upregulated |
| Huynh AH, et al. (2014) [23] | PH; G; CP | GCF | 15 PH; 15 G; 15 CP | SDS-PAGE; LC-ESI-MS/MS | - One hundred and twentyone proteins were detected, two-thirds of which were identified in all three groups - Forty-two proteins were significant different according to specified criteria |
| Bertoldi C, et al. (2013) [24] | Moderate-advance CP | Pocket-associated and healthy tissues | 25 subjects before and after osseous resective surgery | 2DE; LC–MS/MS | - Fifteen proteins were differently expressed between pathological and healthy tissues - Annexin A2, actin cytoplasmic 1, carbonic anhydrase 1 & 2; Immunoglobulin κ chain C region and flavin reductase were upregulated in pathological condition - Heat shock protein β-1, 14-3-3 proteins sigma and zeta/delta, triosephosphate isomerase, peroxiredoxin-1, fatty acid-binding protein-epidermal, and galectin-7 were downregulated in pathological |
| Tsuchida S, et al. (2013) [25] | CP; PH | GCF | 31 CP; 16 PH | LC–MS/MS | Six hundred and nineteen proteins were identified |
| Silva-Boghossian CM, et al. (2013) [26] | CP; PH | GCF | 5 CP; 5 PH | LC–MS/MS | PH subjects (145 proteins) and CP subjects consist of three probing depth sites: P (deep probing depth sites: 214 proteins), G (shallow probing depth sites with bleeding on probing: 154 proteins), and H (shallow sites without bleeding on probing: 133 proteins) |
| Salazar MG, et al. (2013) [27] | P; H | WSS | 20 P; 20 H | LC–MS/MS | Three hundred and forty-four proteins identified |
| Bostanci N, et al. (2013) [28] | G | GCF | 20 H (experimental model) | LC–MS/MS | Two hundred and fifty-four human proteins, 18 bacterial proteins |
| Tsuchida S, et al. (2012) [29] | H; mild P; moderate P; severe P | GCF | 5 H; 3 mild P; 3 moderate P; 5 severe P | 2DE; SDS-PAGE; LC–MS/MS | Three hundred and twenty-seven proteins, including superoxide dismutase 1, apolipoprotein A-I and dermcidin were identified |
| Baliban RC, et al. (2012) [30] | CP; PH | GCF | 12 CP; 12 PH | LC-MS/MS | - Four hundred and thirty-two human and 30 bacterial proteins were detected - Angiotensinogen, clusterin, and thymidine phosphorylase found only in PH group - Neutrophil defensin-1, carbonic anhydrase-1, and elongation factor-1 gamma were associated with CP |
| Kido J, et al. (2012) [31] | PH; P | GCF | 1 PH; 8 P | SDS-PAGE; LC-MS/MS |
Two hundred and thirty-one proteins detected; 64 proteins were found only in PH sites and 63 proteins were founded only in P sites. |
| Zhang J, et al. (2012) [32] | OT-H; OT-P; P | WUS | 24 subjects (8 OT-H; 8 OT-P; 8 P) | MALDI-TOF MS combined with magnetic bead; nano-LC/ESI-MS/MS | - One hundred and nine protein mass peak detected - There are nine intensities peak differ among three groups |
| Gonçalves Lda R, et al. (2011) [33] | G; H | WUS | 10 G; 10 H | 2DE; MALDI-TOF; LC-MS/MS | - Ten proteins detected from 2DE and MALDI-TOF - Twenty-four proteins detected from LC-MS/MS |
| Mizuno N, et al. (2011) [34] | gAP | Neutrophil | 10 gAP with chemotaxis dysfunction; 10 gAP without dysfunction; 15 CP; 15 PH | 2DE; MALDI-TOF | - Lactoferrin, caldesmon, heat shock protein 70, and STAC showed a higher protein expression level in gAP with neutrophil dysfunction group than in the control group - The caldesmon mRNA levels in neutrophils from gAP with chemotaxis dysfunction were upregulated compared with other groups |
| Choi YJ, et al. (2011) [35] | CP | GCF | 12 CP; 11 PH | SDS-PAGE; LC-MS/MS | Three hundred and five proteins identified |
| Gonçalves Lda R, et al. (2010) [36] | CP | WUS | 10 CP; 10 PH | 2DE; MALDI-TOF; LC-MS/MS | - Four proteins detected from 2DE and MALDI-TOF with different abundance among groups - Twenty-seven proteins detected from LC-MS/MS |
| Haigh BJ, et al (2010) [37] | Severe P | WUS | 9 severe P before and after | 2DE; MALDI-TOF; LC-MS/MS | - One hundred and twenty-eight proteins identified across all saliva samples - Fifteen protein spots with altered abundance |
| Bostanci N, et al. (2010) [38] | gAP; PH | GCF | 5 gAP; 5 PH | LC-MS | - One hundred and fifteen proteins identified in gAP and 88 proteins in PH group - Four sources of origin were identified: human, bacterial, yeast, and virus - L-plastin detected only in gAP and annexin-A1 was upregulated 5-fold in PH group |
| Ngo LH, et al. (2010) [39] | CP; PH | GCF | 12 CP | SDS-PAGE; MALDI-TOF; LC-MS/MS | Sixty-six proteins were positively identified |
| Wu Y, et al. (2009) [40] | gAP; PH | WUS | 5 gAP; 5 PH | 2DE; LC-MS/MS | Eleven proteins exhibited a different level between gAP and PH subjects |
2DE: two dimensional electrophoresis; DM: diabetes mellitus; AUC: area under curve; BS: blood serum; CP: chronic periodontitis; ELISA: enzyme-linked immunosorbent assay; ESI: electrospray ionization; gAP: generalized aggressive periodontitis; GCF: gingival crevicular fluid; G: gingivitis; H: healthy; HPLC: high performance liquid chromatography; ICAT: isotope coded affinity tag; LC: liquid chromatography; LFQ: label-free quantitative; MALDI: matrix-assisted laser desorption/ionization; MRM: multiple-reaction monitoring; MS/MS: tandem mass spectrometry; mTRAQ: mass differential tags for relative and absolute quantification; P: periodontitis; PH: periodontally healthy; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SRM: selected reaction monitoring; OT: orthodontic treatment; WB: western blot; WUS: whole unstimulated saliva; WSS: whole stimulated saliva.
3.2. Sample of study
Gingival crevicular fluid (GCF) and saliva have become main body fluids used to study the molecular profile of periodontal diseases. It is because their ability to represent the local oral and systemic related to health condition [7]. Moreover, GCF and saliva are very beneficial because of their non-invasive and painless nature. In this review, there were 13 articles using GCF, 11 articles using saliva, one article using blood serum, one article using neutrophil extracted from blood, two articles using pocket tissue, one article using gingival epithelial culture secretome which were preconditioned with pooled human saliva to form a surface pellicle, and one article using three different samples, namely saliva, GCF and blood serum.
3.3. Quality appraisal
There was a high precentage of “low” over “some concern” risk of bias and there were no “high” risk of bias (Figures 2 and 3). We concluded that each of the studies that included, have a good quality based on the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.
4. Discussion
Assessment of complete protein profiles in health compare to periodontitis has been the goal of the proteomic approach [6]. Tandem mass spectrometry (LC-MS/MS) has been widely performed for proteomics analysis, such as bottom-up or shotgun methods. Using this method, proteins were extracted from cells, tissues or body fluids and digested into peptides. Trypsin was typically used for proteins cleavages. The peptides were then apart using liquid chromatography (LC) prior to mass spectrometry (MS) analysis. Electrospray ionization (ESI) was performed to ionize the eluted peptides. Peptides were subsequently transferred and fragmented into the mass spectrometer, and its mass-to-charge ratio (m/z) was found out. MS/MS analysis was implemented using the automatic acquisition approach. After acquisition, a list of fragment masses was created under the script control and yield to database for protein identification[41].
Protein quantification can also be performed using MS, either with stable isotope labeling or label-free quantification. Stable isotope labeling are used to generate mass difference between peptides, proteins, or reporter ions from different samples. This method can be achieved using stable-isotope labels such as isobaric tags for absolute and relative quantification (iTRAQ), isotope coded affinity tag (ICAT), mass differential tags for relative and absolute quantifications (mTRAQ), and also tandem mass tag (TMT) [11, 22, 25]. In brief, heavy and light proteins can only be distinguished by MS. Subsequently, the quantitation is achieved by determining the relative intensities of the heavy and light versions of protein or peptide in the MS or MS/MS spectra. These methods are accurate, nevertheless they can not be applied in a large scale, even on small scale they tend to time, labor and cost consuming [7].
On the other hand, label-free techniques are alternatively applied for protein quantification. Label-free quantitative methods in MS aim to measure peptides and proteins without using stable isotope labels. One of the approaches for the quantification of peptides and proteins from LC-MS data is extorting the LC-MS/MS amounts of peptide fragment ions of peptides either from DDA or from selected reaction monitoring experiments (SRM) that also called multiple reaction monitoring (MRM) [41].
In MS/MS typically only the highest-abundance peptide are able to identify, while low-abundance may never be detected. Those which discovered in one group but not on the other group are not essentially absent, but possibly down regulated. However, MRM/SRM assay would constantly detect low-abundance peptides [7]. In this review, there were two studies using this method. Bostanci et al. carried out both proteomic platforms; open ended label-free quantitative (LFQ) proteomics followed by SRM resulting five proteins of high predictive value for periodontal diseases. Meanwhile, Merten et al. applied MRM ensuing four candidate biomarkers for periodontitis [14, 16].
Table 2 shows discoveries of identified proteins that were up and downregulated in periodontal diseases from numerous studies. The S100 proteins are family of calcium-binding cytosolic proteins. They have vast extracelullar and intracellular functions such as regulating calcium balance, cell apoptosis, migration, proliferation, differentiation, energy metabolism, and also inflammation [42]. Some of its member, like S100A6, S100A7, S100A8, S100A9 and S100A11 were reported upregulated in periodontitis [11, 12, 21, 22, 26, 37, 43]. In fact, Gao et al. found that Calprotectin (heterodimer of S100A8 and S100A9) exerts the expression of proinflammatory cytokines in human gingival fibroblasts through S100A9-TLR4 interactions [44].
Table 2.
Protein alteration in periodontal disease.
| Protein | Authors (Year) |
|---|---|
| Upregulated | |
| Albumin | Wu Y et al. (2009), Ngo LH et al. (2009), Goncalves L da R et al. (2010), Goncalves L da R et al. (2011), Bostanci N et al. (2012), Silva-Boghossian CM et al. (2013), Carneiro LG et al. (2014), Aboodi GM et al. (2016) |
| S100A6 | Haigh BJ et al. (2010) |
| S100A7 | Kido et al. (2012) |
| S100A8; S100A9 | Haigh BJ et al. (2010), Kido et al. (2012), Silva-Boghossian CM et al. (2013), Carneiro LG et al. (2014), Monari et al. (2015), Shin M-S et al. (2018), Marinho MC et al. (2018), |
| S100A11 | Kido et al. (2012) |
| Matrix metalloproteinase 9 | Tsuchida S et al. (2013), Salazar MG et al. (2013), Trindade F et al. (2015), Bostanci N et al. (2018) |
| Immunoglobulins | Goncalves L da R et al. (2010), Goncalves L da R et al. (2011), Marinho MC et al. (2018) |
| Immunoglobulin G | Ngo LH et al. (2009) |
| Immunoglobulin γ 2 chain C region | Wu Y et al. (2009) |
| Immunoglobulin κ chain C region | Bertoldi C et al. (2013), Kerishnan JP et al. (2016) |
| Hemoglobin | Ngo LH et al. (2009), Goncalves L da R et al. (2010), Goncalves L da R et al. (2011) |
| Transferrin | Ngo LH et al. (2009) |
| Lactotransferrin | Mizuno N et al. (2011), Kido et al. (2012), Salazar MG et al. (2013) |
| Azurocidin | Guzman YA et al. (2018), Choi YJ et al. (2011) |
| Keratins | Goncalves L da R et al. (2010), Goncalves L da R et al. (2011), Silva-Boghossian CM et al. (2013) |
| Ceruloplasmin | Kido et al. (2012), Salazar MG et al. (2013) |
| L-plastin | Bostanci N et al. (2009) |
| α-amylase | Wu Y et al. (2009), Goncalves L da R et al. (2010) |
| Neutrophil defensin-1 | Bostanci N et al. (2012), Baliban CR et al. (2012) |
| Haptoglobin | Davis IJ et al. (2016) |
| Apolipoprotein | Ngo LH et al. (2009) |
| Apolipoprotein A-I | Tsuchida S et al. (2012), Silva-Boghossian CM et al. (2013) |
| Rap guanine nucleotide exchange factor | Marinho MC et al. (2018) |
| Hexokinase; legumain | Yang W et al (2017) |
| Carbonic anhydrase-1; elongation factor-1 γ | Baliban CR et al. (2012) |
| Superoxide dismutase 1; dermcidin | Tsuchida S et al. (2012) |
| Vitamin D-binding protein; zinc-α 2 glycoprotein | Wu Y et al. (2009) |
| α1-antitrypsin; haptoglobin; kininogen | Kerishnan JP et al. (2016) |
| Heat shock protein β-1; galectin-7; 14-3-3 | Monari et al. (2015) |
| Caldesmon; heat shock protein 70; STAC | Mizuno N et al. (2011) |
| Actins; histones; annexins | Silva-Boghossian CM et al. (2013) |
| Hemopexin; plasminogen; α-fibrinogen; Apolipoprotein H | Mertens B et al. (2017) |
| Histone 1.4; keratin type II cytoskeletal 6E; cDNA FLJ53910 (keratin type II cytoskeletal 6A) | Bostanci N et al. (2012) |
| Annexin A2; actin cytoplasmic 1; carbonic anhydrase 1 and 2; flavin reductase | Bertoldi C et al. (2013) |
| Salivary β-globin; thioredoxin; collagen α-1, and G-protein coupled receptor 98 | Aboodi GM et al. (2016) |
| Ras-related protein-1; actin-related protein 2/3 complex subunit 5 | Bostanci N et al. (2018) |
| Blood-, cytoskeleton-, immunity-, inflammation- and lipid-related proteins; glycogen phosphorylase, glutathione S-transferase; phosphoglycerate mutase; psoriasin; resistin; A1-antitrypsin; lipocalin; cathelicidin | Kido et al. (2012) |
| Leukotriene A-4 hydrolase; adenylyl cyclase-associated protein 1; catalase; rho GDP-dissociation inhibitor 2; S100-P; neutrophil collagenase; neutrophil defensin; peptidoglycan recognition protein 1 gelsolin; profilin-1; calreticulin; plastin-2; fibrinogen chain; α-2-macroglobulin; complement C3; α-2-HS-glycoprotein | Salazar MG et al. (2013) |
| Downregulated | |
| Cystatin | Goncalves L da R et al. (2010) |
| Cystatin B; cystatin S | Huynh AHS et al. (2014) |
| Cytokeratin | Yang W et al (2017) |
| 14-3-3 sigma | Wu Y et al. (2009), Bertoldi C et al. (2013) |
| 14-3-3 zeta/delta; heat shock protein β-1; triosephosphate isomerase; peroxiredoxin-1; fatty acid–binding protein-epidermal; galectin-7 | Bertoldi C et al. (2013) |
| Actin; myristoylated alanine-rich C-kinase substrate; glutathione S-transferase; cathelicidin | Marinho MC et al. (2018) |
| Lactotransferrin; elongation factor 2; short palate-, lung- and nasal-epithelium carcinoma-associated protein 2 precursor; carbonic anhydrate 6 | Wu Y et al. (2009) |
Bostanci et al. states that of all the proteins found, albumin has the highest quantity, followed by immunoglobulin and keratin. In the study, cytokines were not identified. This is likely due to its very small concentration and is covered by the quantity of albumin and immunoglobulin [38]. Albumin is in accordance with other studies that found to be upregulated in periodontitis [22, 26, 36, 39, 40] and gingivitis [17, 28, 33] in a large number. In contrast to cytokines, several enzymes associated with polymorphonuclear cells (PMN), such as matrix metalloproteinase-8 (MMP-8), cathepsin G, and myeloperoxidase are found. Matrix metalloproteinase-8 and cathepsin G were not even found in healthy subjects, whereas myeloperoxidase was still found in healthy subjects, even though only one in five subjects [38].
Matrix metalloproteinases are enzymes that zinc-dependent. It is defined as crucial tissue damaging enzymes in periodontitis. Other matrix metalloproteinase that also upregulated in periodontitis was MMP-9 [14, 20, 25, 27]. Furthermore, according to Bostanci et al., MMP-9 is one of the five proteins that have high predictive value for periodontal disease as well as Ras related protein-1, Actin-related protein 2/3 complex subunit, Clusterin, and Deleted in Malignant Brain Tumors-1 [14].
Group of actin proteins as a main component of microfilaments, uncovered to associate with periodontal disease [7]. Upregulated level of actins and actin-related proteins were detected in GCF and saliva of periodontitis subjects [14, 24, 26]. However, Guzman et al. consider smooth muscle actin as a protein biomarkers candidate for health condition due to up-regulated 13 weeks after periodontal treatment [15]. Another actin related protein in the spotlight is L-plastin (lymphocyte cytosolic protein-1). This protein was not identified in five healthy subjects but was found in four of the five aggressive periodontitis subjects. The role of L-plastin in the regulation of leukocyte adhesion and PMN signal transduction has been suggested. High concentration of L-plastin in GCF is assumed to facilitate PMN in the inflammatory area, so PMN hype reaction occurs in periodontal disease [38].
Antimicrobial protein azurocidin was likewise found upregulated and consider as protein biomarkers candidate in chronic periodontitis [15, 35]. It is known to be secreted by neutrophils and suggested that increased level of azurocidin at the early stage may have protective role in osteoclast-activated alveolar bone destruction due to its effect on osteoclast differentiation [35]. Recent study by Mertens et al. discovered a protein which can distinguish between chronic and aggressive periodontitis. The protein was apolipoprotein H (ApoH) that significantly increased in subjects with aggressive periodontitis when compared to chronic periodontitis and healthy subjects. ApoH, also known as β2-glycoprotein I, is a plasma glycoprotein that may circulate as a free protein or related with lipoprotein [16]. Noteworthy correlation between lipid regulators and periodontal disease is found by proteomic studies [7].
Among all upregulated proteins, there were also downregulated proteins that found in periodontitis. Cystatin has a protease inhibitory property that can inhibit proteases involved in periodontal tissue destruction, such as lysosomal cathepsins B, H and L [36]. Lower abundance of cystatin found in periodontitis group, and more generally exhibit in control group [23, 36]. Herein review, contradictory research results also were found. Heat shock protein beta 1 and 14-3-3 protein sigma were attained underexpressed in pathological tissue and saliva but increased in periodontal pocket tissue [21, 24, 40]. Another limitation of this study is lack of sensitivity and specificity information of diagnosis ability of protein biomarkers. However, Baliban, et al. states that using a mixed-integer linear optimization (MILP) model that was developed to identify the optimal combination of biomarkers which could clearly distinguish a blind subject sample as healthy or diseased. Overall cross validation of the capabilities of the MILP model is carried out on a training set consisting of 55 samples and consistently more than 99% accuracy is achieved when interpreting the test set sample as healthy or diseased [45].
5. Conclusion
Comprehensive analysis with a proteomic approach is expected to understand the pathogenesis of periodontal diseases, also to facilitate the development of more precise biomarkers for diagnostic and prognostic. The current task upstretched is how to combine and utilize the resulting data sets to benefit patient. The reviewed studies mainly focused on expression proteomics or differential display proteomics between disease and healty condition. In further studies, the assessment of diagnostic ability of biomarkers should be improved. Further research is also needed to overcome proteomic profiles based on the latest classification scheme for periodontal diseases.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This work was partly supported by Universitas Indonesia (PUTI Doktor 2020). No. NKB-595/UN2.RST/HKP. 05.00/2020.
Competing interest statement
The authors declare no conflict of interest.
Additional information
MIR is currently enrolled in a PhD program at Faculty of Dentistry Universitas Indonesia.
References
- 1.Pihlstrom B.L., Michalowicz B.S., Johnson N.W. Periodontal diseases. Lancet. 2005;366(9499):1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [pii]∖r10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 2.Lang N.P., Bartold P.M. Periodontal health. J. Clin. Periodontol. 2018;45(April 2017):S9–S16. doi: 10.1111/jcpe.12936. [DOI] [PubMed] [Google Scholar]
- 3.Tonetti M.S., Greenwell H., Kornman K.S. Staging and grading of periodontitis: framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018;45(February):S149–S161. doi: 10.1111/jcpe.12945. [DOI] [PubMed] [Google Scholar]
- 4.Papapanou P.N., Sanz M., Buduneli N. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 2018;89(Suppl 1):S173–S182. doi: 10.1002/JPER.17-0721. [DOI] [PubMed] [Google Scholar]
- 5.Gupta A., Govila V., Saini A. Proteomics - the research frontier in periodontics. J. Oral Biol. Craniofacial Res. 2015;5(1):46–52. doi: 10.1016/j.jobcr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trindade F., Oppenheim F.G., Helmerhorst E.J., Amado F., Gomes P.S., Vitorino R. Uncovering the molecular networks in periodontitis. Proteonomics Clin. Appl. 2014 doi: 10.1002/prca.201400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman Y.A., Sakellari D., Arsenakis M., Floudas C.A. Proteomics for the discovery of biomarkers and diagnosis of periodontitis: a critical review. Expert Rev. Proteomics. 2014;11(1):31–41. doi: 10.1586/14789450.2014.864953. [DOI] [PubMed] [Google Scholar]
- 8.Whiting P.F., Rutjes A.W.S., Westwood M.E. Quadas-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011 doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 9.McGuinness L.A. 2019. Robvis: an R Package and Web Application for Visualising Risk-Of-Bias Assessments.https://github.com/mcguinlu/robvis [DOI] [PubMed] [Google Scholar]
- 10.Caton J G., Armitage G., Berglundh T. A new classification scheme for periodontal and peri-implant diseases and conditions – introduction and key changes from the 1999 classification. J. Clin. Periodontol. 2018;45(March):S1–S8. doi: 10.1111/jcpe.12935. [DOI] [PubMed] [Google Scholar]
- 11.Marinho M.C., Pacheco A.B.F., Costa G.C.V., Ortiz N.D., Zajdenverg L., Sansone C. Quantitative gingival crevicular fluid proteome in type 2 diabetes mellitus and chronic periodontitis. Oral Dis. 2019;25(2):588–595. doi: 10.1111/odi.12996. [DOI] [PubMed] [Google Scholar]
- 12.Shin M.S., Kim Y.G., Shin Y.J., Ko B.J., Kim S., Kim H.D. Deep sequencing salivary proteins for periodontitis using proteomics. Clin. Oral Invest. 2019;23(9):3571–3580. doi: 10.1007/s00784-018-2779-1. [DOI] [PubMed] [Google Scholar]
- 13.Tang H., Yuan C., Ma Z. The potentiality of salivary peptide biomarkers for screening patients with periodontal diseases by mass spectrometry. Clin. Chim. Acta. 2019;495(March):278–286. doi: 10.1016/j.cca.2019.04.076. [DOI] [PubMed] [Google Scholar]
- 14.Bostanci N., Selevsek N., Wolski W. Targeted proteomics guided by label-free quantitative proteome analysis in saliva reveal transition signatures from health to periodontal disease. Mol. Cell. Proteomics. 2018;17(7):1392–1409. doi: 10.1074/mcp.RA118.000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman Y.A., Sakellari D., Papadimitriou K., Floudas C.A. High-throughput proteomic analysis of candidate biomarker changes in gingival crevicular fluid after treatment of chronic periodontitis. J. Periodontal. Res. 2018 doi: 10.1111/jre.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertens B., Orti V., Vialaret J. Assessing a multiplex-targeted proteomics approach for the clinical diagnosis of periodontitis using saliva samples. Bioanalysis. 2018;10(1):35–45. doi: 10.4155/bio-2017-0218. [DOI] [PubMed] [Google Scholar]
- 17.Aboodi G.M., Sima C., Moffa E.B. Salivary cytoprotective proteins in inflammation and resolution during experimental gingivitis-A pilot study. Front. Cell Infect. Microbiol. 2016;5(JAN):1–12. doi: 10.3389/fcimb.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerishnan J.P., Mohammad S., Alias M.S. Identification of biomarkers for periodontal disease using the immunoproteomics approach. PeerJ. 2016 doi: 10.7717/peerj.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bostanci N., Bao K., Wahlander A., Grossmann J., Thurnheer T., Belibasakis G.N. Secretome of gingival epithelium in response to subgingival biofilms. Mol. Oral Microbiol. 2015;30(4):323–335. doi: 10.1111/omi.12096. [DOI] [PubMed] [Google Scholar]
- 20.Trindade F., Amado F., Oliveira-Silva R.P. Toward the definition of a peptidome signature and protease profile in chronic periodontitis. Proteonomics Clin. Appl. 2015;9(9-10):917–927. doi: 10.1002/prca.201400191. [DOI] [PubMed] [Google Scholar]
- 21.Monari E., Cuoghi A., Bellei E. Analysis of protein expression in periodontal pocket tissue: a preliminary study. Proteome Sci. 2015;13(1):1–11. doi: 10.1186/s12953-015-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carneiro L.G., Nouh H., Salih E. Quantitative gingival crevicular fluid proteome in health and periodontal disease using stable isotope chemistries and mass spectrometry. J. Clin. Periodontol. 2014 doi: 10.1111/jcpe.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh A.H.S., Veith P.D., Mcgregor N.R. Gingival crevicular fluid proteomes in health, gingivitis and chronic periodontitis. J. Periodontal. Res. 2014;50(5):637–649. doi: 10.1111/jre.12244. [DOI] [PubMed] [Google Scholar]
- 24.Bertoldi C., Bellei E., Pellacani C. Non-bacterial protein expression in periodontal pockets by proteome analysis. J. Clin. Periodontol. 2013;40(6):573–582. doi: 10.1111/jcpe.12050. [DOI] [PubMed] [Google Scholar]
- 25.Tsuchida S., Satoh M., Kawashima Y. Application of quantitative proteomic analysis using tandem mass tags for discovery and identification of novel biomarkers in periodontal disease. Proteomics. 2013;13(15):2339–2350. doi: 10.1002/pmic.201200510. [DOI] [PubMed] [Google Scholar]
- 26.Silva-Boghossian C.M., Colombo A.P.V., Tanaka M., Rayo C., Xiao Y., Siqueira W.L. Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS One. 2013;8(10):1–15. doi: 10.1371/journal.pone.0075898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar M.G., Jehmlich N., Murr A. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J. Clin. Periodontol. 2013;40(9):825–832. doi: 10.1111/jcpe.12130. [DOI] [PubMed] [Google Scholar]
- 28.Bostanci N., Ramberg P., Wahlander Å. Label-free quantitative proteomics reveals differentially regulated proteins in experimental gingivitis. J. Proteome Res. 2013;12(2):657–678. doi: 10.1021/pr300761e. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchida S., Satoh M., Umemura H. Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics. 2012;12(13):2190–2202. doi: 10.1002/pmic.201100655. [DOI] [PubMed] [Google Scholar]
- 30.Baliban R.C., Sakellari D., Li Z., DiMaggio P.A., Garcia B.A., Floudas C.A. Novel protein identification methods for biomarker discovery via a proteomic analysis of periodontally healthy and diseased gingival crevicular fluid samples. J. Clin. Periodontol. 2012;39(3):203–212. doi: 10.1111/j.1600-051X.2011.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kido J., Bando M., Hiroshima Y. Analysis of proteins in human gingival crevicular fluid by mass spectrometry. J. Periodontal. Res. 2012 doi: 10.1111/j.1600-0765.2011.01458.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Zhou S., Li R. Magnetic bead-based salivary peptidome profiling for periodontal-orthodontic treatment. Proteome Sci. 2012 doi: 10.1186/1477-5956-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonçalves L.D.R., Soares M.R., Nogueira F.C.S. Analysis of the salivary proteome in gingivitis patients. J. Periodontal. Res. 2011;46(5):599–606. doi: 10.1111/j.1600-0765.2011.01378.x. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno N., Niitani M., Shiba H. Proteome analysis of proteins related to aggressive periodontitis combined with neutrophil chemotaxis dysfunction. J. Clin. Periodontol. 2011;38(4):310–317. doi: 10.1111/j.1600-051X.2010.01693.x. [DOI] [PubMed] [Google Scholar]
- 35.Choi Y.J., Heo S.H., Lee J.M., Cho J.Y. Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci. 2011;9 doi: 10.1186/1477-5956-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonçalves L.D.R., Soares M.R., Nogueira F.C.S. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J Proteomics. 2010;73(7):1334–1341. doi: 10.1016/j.jprot.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Haigh B.J., Stewart K.W., Whelan J.R.K., Barnett M.P.G., Smolenski G.A., Wheeler T.T. Alterations in the salivary proteome associated with periodontitis. J. Clin. Periodontol. 2010;37(3):241–247. doi: 10.1111/j.1600-051X.2009.01525.x. [DOI] [PubMed] [Google Scholar]
- 38.Bostanci N., Heywood W., Mills K., Parkar M., Nibali L., Donos N. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MS ( gingival exudatome ) 2010;9:2191–2199. doi: 10.1021/pr900941z. [DOI] [PubMed] [Google Scholar]
- 39.Ngo L.H., Veith P.D., Chen Y.Y., Chen D., Darby I.B., Reynolds E.C. Mass spectrometric analyses of peptides and proteins in human gingival crevicular fluid. J. Proteome Res. 2010;9(4):1683–1693. doi: 10.1021/pr900775s. [DOI] [PubMed] [Google Scholar]
- 40.Wu Y., Shu R., Luo L.J., Ge L.H., Xie Y.F. Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J. Periodontal. Res. 2009;44(5):636–644. doi: 10.1111/j.1600-0765.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- 41.Bantscheff M., Lemeer S., Savitski M.M., Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012;404(4):939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 42.Xia C., Braunstein Z., Toomey A.C., Zhong J., Rao X. S100 proteins as an important regulator of macrophage inflammation. Front. Immunol. 2018;8(JAN):1–11. doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kido J., Bando M., Hiroshima Y. Analysis of proteins in human gingival crevicular fluid by mass spectrometry. J. Periodontal. Res. 2012;47(4):488–499. doi: 10.1111/j.1600-0765.2011.01458.x. [DOI] [PubMed] [Google Scholar]
- 44.Gao H., Hou J., Meng H., Zhang X., Zheng Y., Peng L. Proinflammatory effects and mechanisms of calprotectin on human gingival fibroblasts. J. Periodontal. Res. 2017;52(6):975–983. doi: 10.1111/jre.12465. [DOI] [PubMed] [Google Scholar]
- 45.Baliban R.C., Sakellari D., Li Z., Guzman Y.A., Garcia B.A., Floudas C.A. Discovery of biomarker combinations that predict periodontal health or disease with high accuracy from GCF samples based on high-throughput proteomic analysis and mixed-integer linear optimization. J. Clin. Periodontol. 2013 doi: 10.1111/jcpe.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]