Abstract
Background
DNAs released from tumor cells into blood (circulating tumor DNAs, ctDNAs) carry tumor-specific genomic aberrations, providing a non-invasive means for cancer detection. In this study, we aimed to leverage somatic copy number aberration (SCNA) in ctDNA to develop assays to detect early-stage HCCs.
Methods
We conducted low-depth whole-genome sequencing (WGS) to profile SCNAs in 384 plasma samples of hepatitis B virus (HBV)-related HCC and cancer-free HBV patients, using one discovery and two validation cohorts. To fully capture the robust signals of WGS data from the complete genome, we developed a machine learning-based statistical model that is focused on detection accuracy in early-stage HCC.
Findings
We built the model using a discovery cohort of 209 patients, achieving an overall area under curve (AUC) of 0.893, with 0.874 for early-stage (Barcelona clinical liver cancer [BCLC] stage 0-A) and 0.933 for advanced-stage (BCLC stage B-D). The performance of the model was then assessed in two validation cohorts (76 and 99 patients) that only consisted of patients with stage 0-A HCC. Our model exhibited a robust predictive performance, with an AUC of 0.920 and 0.812 for the two validation cohorts. Further analyses showed the impact of tumor sample heterogeneity in model training on detecting early-stage tumors, and a refined model addressing the heterogeneity in the discovery cohort significantly increased model performance in validation.
Interpretation
We developed an SCNA-based, machine learning-driven model in the non-invasive detection of early-stage HCC in HBV patients and demonstrated its performance through strict independent validations.
Keywords: Copy number aberration (CNA), Hepatocellular carcinoma (HCC), Early detection, Machine learning
Abbreviations: AFP, alpha fetoprotein; AUC, area under curve; BCLC, Barcelona clinic liver cancer; ctDNAs, circulating tumor DNAs; ccfDNA, circulating cell free DNA; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; SCNA, somatic copy number aberration; TCGA, The Cancer Genome Atlas
Research in context.
Evidence before this study
Hepatocellular carcinoma (HCC) is a leading cause of cancer death. Currently, the only clinically available blood-based biomarker for HCC screening is alpha fetoprotein (AFP); however, the clinic utility of AFP is severely limited due to its low sensitivity and specificity. Liquid biopsy using circulating tumor DNA (ctDNA) has been found to hold potential as a noninvasive mean to detect cancers. ctDNA, a small portion of circulating cell-free DNA (ccfDNA), carries genomic aberrations in various forms such as DNA methylations, point mutations, and somatic copy number aberrations (SCNAs). It has been increasingly recognized that SCNAs are likely to be a superior approach for ctDNA-based cancer early detection because, compared to point mutations, SCNAs contributes a much larger number of ctDNA fragments to the overall pool of cfDNA, and span a much larger genomic regions, thus offering higher statistical confidence in detection. However, none of the studies is conducted in early-stage HCC patients with a strict study design that focusing on SCNAs. Thus, it is of great significance to leverage SCNAs in ctDNA to develop assays to detect early-stage HCC
Added value of this study
In the present study, we conducted low-depth whole-genome sequencing (WGS) to profile SCNAs in 384 plasma samples of hepatitis B virus (HBV)-related HCC and cancer-free HBV patients, using one discovery and two validation cohorts. We developed a novel weighted Random Forest-driver model to take advantage of the genome-wide SCNA profiles in ccfDNA, as well as prior knowledge from large-scale external data to boost diagnostic accuracy.
Implications of all the available evidence
Our findings suggest the promise of machine learning-based genome-wide interrogation of SCNAs in ctDNA for HCC early detection. This is an important step towards a more comprehensive understanding of ctDNA, which will lay the groundwork for early cancer detection by using whole-genome SCNA profiling coupled with innovative machine learning.
Alt-text: Unlabelled box
1. Introduction
Hepatocellular carcinoma (HCC) is a leading cause of cancer death. HCC patients diagnosed at early stages have a much better prognosis compared to those diagnosed at advanced-stages [1]. When mass lesions suspicious for HCC are found, diagnosis may be confirmed by liver biopsy, an invasive procedure with limitations that include incomplete tumor capture due to intratumoral heterogeneity, and with associated risk of cancer cell dissemination through the needle track. Currently, the only clinically available blood-based biomarker for HCC screening is alpha fetoprotein (AFP); however, the clinic utility of AFP is severely limited due to its low sensitivity and specificity [2], [3], [4]. Therefore, the development of novel non-invasive and clinically applicable HCC markers remains an urgent unmet clinical need.
Liquid biopsy using circulating tumor DNA has been found to hold potential as a noninvasive mean to detect cancers. ctDNA, a small portion of circulating cell-free DNA (ccfDNA), carries genomic aberrations in various forms such as DNA methylations, point mutations, and somatic copy number aberrations (SCNAs) [5], [6], [7], [8], [9], [10]. Methylation-based liquid biopsy assay has shown promise in diagnosing cancer patients [5,11,12]. For example, Oussalah et al. [12] demonstrated a high diagnostic accuracy for HCC based on a novel ccfDNA-based epigenetic biomarker, SEPT9 promoter methylation. However, most of these studies have largely enrolled late-stage tumors, the majority of which can be detected by other currently available assays. ctDNA point mutation detection methods developed to date likely lack either the scope or analytical sensitivity necessary to be useful for cancer screening, due to the low (<1%) ctDNA fractions derived from early stage tumors [13]. Moreover, it has been increasingly recognized that point mutations might not be ideal markers for early cancer detection due to clonal hematopoiesis in which somatic mutations are identified in cfDNAs derived from normal cells [14]. In comparison, SCNAs are likely to be a superior approach for ctDNA-based cancer early detection because, compared to point mutations, SCNAs contributes a much larger number of ctDNA fragments to the overall pool of cfDNA [13], and span a much larger genomic regions, thus offering higher statistical confidence in detection. Moreover, compared to methylation, SCNAs are much less affected by confounders such as age, diet, and life style [15]. A seminal study by Chan et al demonstrated that the concordant copy number variants could be detected in cfDNA and the corresponding resected tumor tissues, and indicated that the analysis of plasma CNA was specific for differentiating between cancer patients and individuals without a cancer [16]. Another elaborate in silico assessment suggested the potential of ctDNA CNA-based screening in selected cancer types [13]. However, none of the studies is conducted in early-stage HCC patients with a strict study design that includes independent validations.
In this study, we aimed to develop a blood-based non-invasive assay, coupled with innovative machine learning approaches, to systematically evaluate its potential for the detection of early-stage HCC, through SCNA profiling derived from low-depth whole genome sequencing (WGS) data [5].
2. Materials and Methods
2.1. Study design and patient enrollment
All the enrolled subjects in this study were HBV-infected patients, including one discovery cohort for investigating the characteristics of early- and late-stage HCC detection, and two independent validation cohorts for testing the model performance in early-stage HCC. The discovery cohort included HCC patients at all stages, whereas the validation cohorts selectively enrolled HCC patients at early stages (Barcelona-Clinic-Liver-Cancer (BCLC) stage 0 or A). HBV controls who were cancer free for at least 6 months of follow-up were recruited in each cohort. More details for patient enrollment can be found in Supplementary Materials. The study was approved by the Ethics Committees of the involved hospitals and written informed consent was obtained from all participants.
2.2. Sample collection, processing and sequencing library construction
Whole blood collected in EDTAK2 tubes was processed within 3 hours. Plasma DNA was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Sequencing library for Illumina was constructed using approximately 20ng ccfDNA. Please see Supplementary Materials for details.
2.3. WGS experiments and data processing
WGS was performed using the Illumina HiSeq X10 platform based on a paired-end 150 bp protocol [17]. The raw paired-end reads were aligned to the reference human genome GRCh37 using BWA-0.7.4 [18] and the BAM files were marked duplicate reads by picard-tools-1.92 (http://broadinstitute.github.io/picard/). The genome-level statistics for quality control assessment was analyzed by QPLOT [19]. Tumor burden was estimated by quantifying tumor fraction (TFx) in ccfDNA via low-coverage WGS of ccfDNA using ichorCNA, an algorithm without the need for prior knowledge of tumor mutations [20].
2.4. Development of a machine learning-based statistical model to analyze genome-wide SCNAs
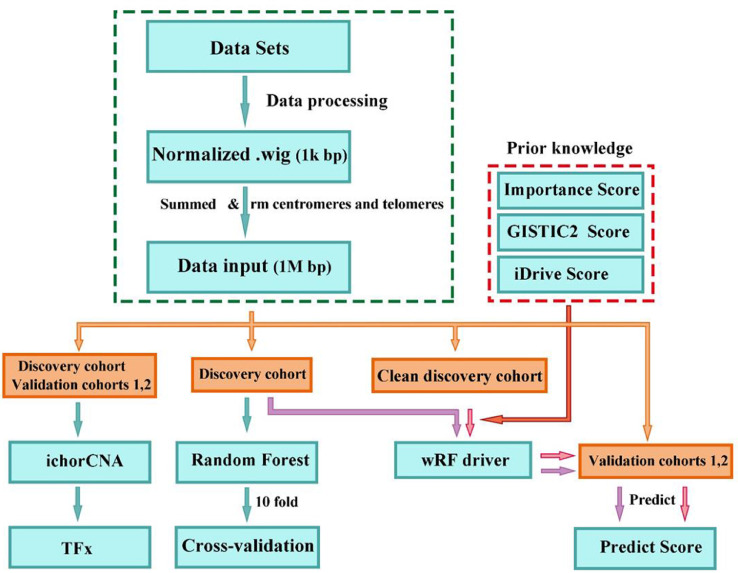
The flowchart of the analysis procedure is summarized in Fig. 1. After alignment, we divided aligned reads into non-overlapping windows of 1 Kb on the genome and obtained normalized read depth in each window by correcting for GC content and mappability biases (http://bioconductor.org/packages/2.11/bioc/html/HMMcopy.html). We then summed all copy numbers (Cor.map column) in all windows in each bin of 1 Mb. The summed copy numbers in each 1-Mb bin represents the copy number information in that bin. We filtered regions in the centromeres and telomeres as well as the boundaries or connecting bins. The normalized read count in each bin along the genome represents the genome-wide SCNA profiling, and constitutes the data input for machine learning algorithms. We used the R implication of Random Forest (RF) (version 4.6-12) for the initial data exploration. Briefly, we applied a classical RF algorithm to train a classification model based on the genome-wide SCNA profiles of the samples in the discovery cohort. We then performed 10-fold cross-validation 300 times by randomly shuffling the samples in the discovery cohort for each of the 10-fold cross-validation, and reported the average accuracy across the 300 replicates to obtain robust estimates of the model's performance.
Fig. 1.
Flowchart of the analysis procedure
2.5. Development of a novel weighted model to incorporate prior knowledge
We assume that external data such as that from The Cancer Genome Atlas (TCGA) [21], [22], [23], [24] and the International Cancer Genome Consortium (ICGC) [25] can be leveraged to increase accuracy of HCC early detection. We further developed a novel weighted model (weighted random forest-driver, wRF-driver) to incorporate such prior knowledge to further augment detection performance. In the classical RF, the Gini impurity index was used to select the best features for the split of sub-nodes in each tree of the forest. In the wRF-driver, we incorporated prior knowledge as weights in the process of dividing a node into two sub-nodes for each tree (Supplementary Fig. S1). Specifically, we adjusted the Gini impurity index using the penalty scores penalty, , where D and A denote datasets and a feature respectively. A higher penalty corresponds to a lower weight or a weaker importance. In the decision tree construction, when each node is split into two sub-nodes, the best splitting is selected on all available variables. We added a penalty into the Gini index score such that important variables, which were reflected as prior knowledge, should be more likely to be selected as the splitting variable, while the noisy variables are suppressed (Supplementary Fig. S1). For a two-class problem, the Gini impurity index is defined as:
where and , and pij is the probability of Dj belonging to class i, and n is the number of classes in Dj.
In this study we incorporated three sources of prior knowledge: 1) the GISTIC2 [26] scores from TCGA [21], [22], [23], [24] representing the evidence of driver SCNAs, 2) the iDriver [27] scores that were calculated from multi-omics data from TCGA, representing the evidence of driver genes when considering all of the –omics data, and 3) the importance scores derived from the discovery cohort of the current study representing the important features directly obtained from the SCNA profiling in ccfDNA of the discovery cohort using random forest. More details regarding the algorithm, model evaluation, and the investigation of the weighting schemes, are available in Supplementary Materials, Supplementary Fig. S2 and S3. Model performance was assessed by the area under the curve (AUC) after constructing receiver operating characteristic (ROC) curves.
2.6. Data deposition
The raw sequencing data are available in BIG Sub with access number subCRA002891 (https://bigd.big.ac.cn/gsub/).
3. Results
3.1. Patient cohorts
Table 1 summarizes characteristics of patients. We enrolled 73 (67.6%) early-stage (6 stage 0 and 67 stage A) and 35 (32.4%) stage B-D patients in the discovery cohort. All the HCC patients in the validation cohorts are early-stage patients with most having stage A tumor (92.1% for validation cohort 1 and 82.4% for validation cohort 2). There were 41.6%, 57.9%, and 45.8% cirrhotic HBV controls, and 76.9%, 68.4%, and 62.7% cirrhotic HCC patients in the discovery, validation 1 and validation 2 cohorts, respectively. In addition, HCC patients had higher AFP values and elevated level of liver enzymes (such as alanine aminotransferase and aspartate aminotransferase) than HBV patients in these cohorts.
Table 1.
Patient characteristics
| Characteristics | Discovery cohort | Validation cohort 1 | Validation cohort 2 | |||
|---|---|---|---|---|---|---|
| (N=209) | (N=76) | (N=99) | ||||
| HBV controls | HCC patients | HBV controls | HCC patients | HBV controls | HCC patients | |
| (N=101) | (N=108) | (N=38) | (N=38) | (N=48) | (N=51) | |
| Gender | ||||||
| Female, N (%) | 27 (26.7) | 14 (13.0) | 8 (21.1) | 7 (18.4) | 11 (22.9) | 14 (27.5) |
| Male, N (%) | 74 (73.3) | 94 (87.0) | 30 (78.9) | 31 (81.6) | 37 (77.1) | 37 (72.5) |
| Age (years), mean (SD) | 49.9 (9.0) | 53.3 (10.2) | 46.2 (7.9) | 53.6 (7.3) | 45.8 (12.3) | 55.0 (10.7) |
| AFP | ||||||
| Negative, < 25 ng/mL, N (%) | 89 (88.1) | 48 (44.4) | 36 (94.7) | 15 (39.5) | 43 (89.6) | 24 (47.1) |
| Positive, ≥ 25 ng/mL, N (%) | 4 (4.0) | 54 (50.0) | 0 (0) | 22 (57.9) | 5 (10.4) | 27 (52.9) |
| NA, N (%) | 8 (7.9) | 6 (5.6) | 2 (5.3) | 1 (2.6) | 0 (0) | 0 (0) |
| ALT | ||||||
| Normal, ≤ 40 U/L, N (%) | 57 (56.4) | 63 (58.3) | 29 (76.3) | 22 (57.9) | 35 (72.9) | 32 (62.7) |
| Elevated, > 40 U/L, N (%) | 41 (40.6) | 45 (41.7) | 9 (23.7) | 16 (42.1) | 12 (25.0) | 19 (37.3) |
| NA, N (%) | 3 (3.0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.1) | 0 (0) |
| AST | ||||||
| Normal, ≤ 37 U/L, N (%) | 70 (69.3) | 52 (48.2) | 30 (78.9) | 29 (76.3) | 36 (72.9) | 38 (74.5) |
| Elevated, > 37 U/L, N (%) | 28 (27.7) | 56 (51.8) | 8 (21.1) | 9 (23.7) | 12 (25.0) | 13 (25.5) |
| NA, N (%) | 3 (3.0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.1) | 0 (0) |
| ALP | ||||||
| Normal, ≤ 117 U/L, N (%) | 59 (58.4) | 66 (61.1) | 23 (60.5) | 27 (71.1) | 30 (62.5) | 44 (86.3) |
| Elevated, > 117 U/L, N (%) | 13 (12.9) | 42 (38.9) | 5 (13.2) | 11 (28.9) | 15 (31.3) | 7 (13.7) |
| NA, N (%) | 29 (28.7) | 0 (0) | 10 (26.3) | 0 (0) | 3 (6.2) | 0 (0) |
| HBV DNA loading | ||||||
| Undetectable, N (%) | 9 (8.9) | 0 (0) | 0 (0) | 0 (0) | 28 (58.3) | 23 (45.1) |
| Low, ≤ 2000 IU/mL, N (%) | 77 (76.2) | 38 (35.2) | 34 (89.5) | 12 (31.6) | 10 (20.8) | 14 (27.5) |
| High, > 2000 IU/ml, N (%) | 13 (12.9) | 16 (14.8) | 4 (10.5) | 2 (5.3) | 10 (20.8) | 12 (23.5) |
| NA, N (%) | 2 (2.0) | 54 (50.0) | 0 (0) | 24 (63.1) | 0 (0) | 2 (3.9) |
| Cirrhosis status | ||||||
| No, N (%) | 59 (58.4) | 25 (23.1) | 16 (42.1) | 12 (31.6) | 26 (54.2) | 19 (37.3) |
| Yes, N (%) | 42 (41.6) | 83 (76.9) | 22 (57.9) | 26 (68.4) | 22 (45.8) | 32 (62.7) |
| BCLC stage | ||||||
| 0, N (%) | - | 6 (5.6) | - | 3 (7.9) | - | 9 (17.6) |
| A, N (%) | - | 67 (62.0) | - | 35 (92.1) | - | 42 (82.4) |
| B, N (%) | - | 22 (20.4) | - | 0 (0) | - | 0 (0) |
| C, N (%) | - | 12 (11.1) | - | 0 (0) | - | 0 (0) |
| D, N (%) | - | 1 (0.9) | - | 0 (0) | - | 0 (0) |
| Tumor grade | ||||||
| Well differentiated | - | 8 (7.4) | - | 4 (10.5) | - | 0 (0) |
| Moderately differentiated | - | 45 (41.7) | - | 18 (47.4) | - | 31 (60.8) |
| Poorly differentiated | - | 6 (5.5) | - | 7 (18.4) | - | 17 (33.3) |
| Unknown | - | 49 (45.4) | - | 9 (23.7) | - | 3 (5.9) |
SD: standard deviation; AFP: alpha fetoprotein; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; NC: not available; BCLC stage: Barcelona-Clinic-Liver-Cancer stage; ALT: alanine aminotransferase; AST: aspartate aminotransferase; ALP: alkaline phosphatase.
3.2. Sequencing depth-based profiling of SCNAs across the genome
The average sequencing depth of cleaned WGS data after removing duplicates was ∼5X, and the most dominant insert size of paired-end sequencing reads was at ∼167 bp. The normalized sequencing depth across all bins in the genome of a patient represents the SCNA profile of that patient. Supplementary Fig. S4A, S4B and S4C show the SCNA profiles of typical HBV, early-stage (stage A) HCC, and advanced-stage (stage C) HCC patients, respectively. There were 24 out of 35 advanced-stage samples, and 40 out of 73 early-stage samples, exhibiting pronounced and visually detectable aberrations, similar to Supplementary Fig. S4C. On the other hand, HBV samples rarely show such visually detectable aberrations, with a few exceptions of 5 cirrhotic patients that showed similar visually detectable aberrations, for which the possibility cannot be ruled out that such cirrhotic HBV patients may actually be undiagnosed early-stage HCC patients. For most early-stage patients, the SCNA profiles are largely similar to HBV patients, making it impossible to visually separate them from HBV patients.
3.3. Applicability of ctDNA burden in HCC detection
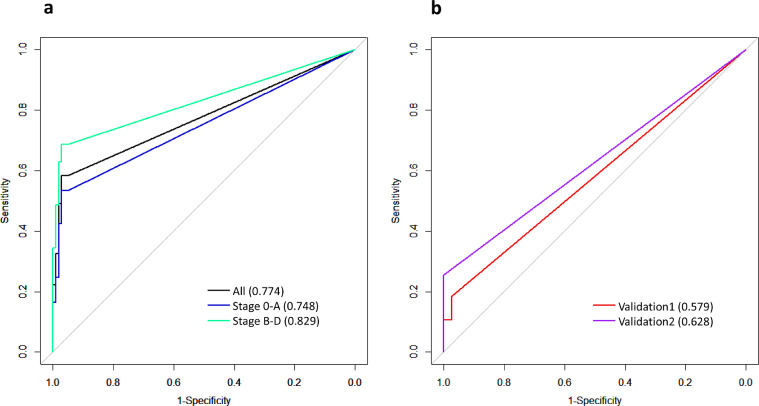
We aimed to leverage SCNA profiles to infer tumor burden and to explore the utility of the estimated tumor burden for HCC detection. Toward that end, we applied ichorCNA [20] to estimate SCNAs and quantify TFx using the WGS data. The distributions of the TFx statistics for the discovery and validation cohorts are in Supplementary Fig. S5. Using TFx>0 as the cutoff, we achieved a sensitivity of 0.583 and a specificity of 0.950 when considering all samples in the cohort. When we separated the analysis by stage 0-A and stage B-D, at a specificity of 0.95, the sensitivity was 0.534 and 0.686 to detect stage 0-A and stage B-D patients. In the two validation cohorts, the proportions of samples with detectable tumor burden (i.e., TFx>0) are drastically lower than that in the discovery cohort, with 18% and 29% of HCC samples having TFx>0 in validation cohort 1 and 2 (Supplementary Fig. S5), respectively, confirming the dominance of early-stage HCC patients in the two validation cohorts and demonstrating the poor performance of TFx in detecting early-stage HCC. Although having low sensitivity, TFx statistics were highly specific, with remarkable specificity of 97.4% and 95.6% for the two cohorts, respectively. By constructing ROC curves, we found that TFx values alone resulted in an AUC of 0.774 when all patients in the discovery cohort were considered, and an AUC of 0.748 and 0.829 for stage 0-A and stage B-D HCC, respectively (Fig. 2A). For the validation cohorts, given the low ctDNA burden, the TFx-derived AUC statistics have limited predictive utility, with an AUC of 0.579 and 0.628, for validation cohort 1 and 2, respectively (Fig. 2B). Altogether, although ctDNA burden has remarkable specificity when ctDNA fraction can be estimated from the WGS data, regardless of tumor stages, it suffers from markedly reduced sensitivity in early-stage patients, indicating that a large proportion of HCC patients do not have considerably elevated ctDNA fractions in their blood readily detectable by the TFx statistics.
Fig. 2.
ROC curves and AUC statistics based on the TFx statistics estimated by ichorCNA in the discovery cohort (A) and two validation cohorts (B).
3.4. An RF-based machine learning model leveraging genome-wide SCNA profiles for HCC detection
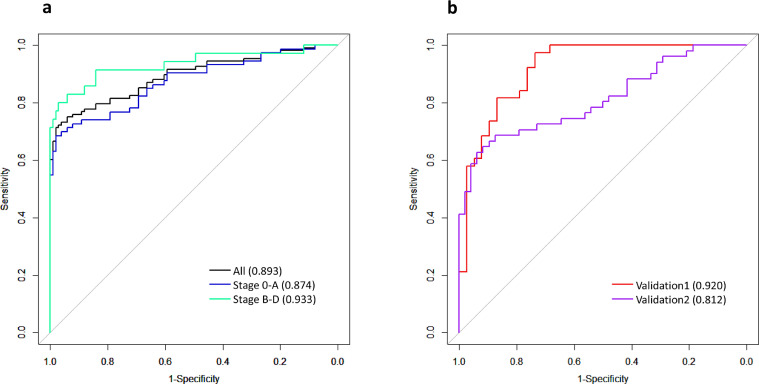
One limitation of the TFx approach is that it ignores the sequencing depth information among the whole genome. To address this limitation, we employed an RF-based machine learning algorithm to distinguish HCC from HBV patients based on the sequencing depth-derived SCNA profiles across the genome. When considering all patients in the cohort, RF achieved an AUC of 0.893 (Fig. 3A), a significant improvement over TFx. The model exhibited a much higher accuracy in stage B-D than stage 0-A patients (AUC 0.933 vs. 0.874) (Fig. 3A). At specificity of 0.95, the model was able to achieve sensitivity of 0.699 for stage 0-A HCC patients and 0.829 for stage B-D patients. Compared with the TFx statistics, the RF model universally achieved improved accuracy, and the relative strength of RF vs. TFx is particularly pronounced for early-stage HCC patients (AUC=0.874 vs. 0.748), due largely to RF's ability in capturing and amplifying weak signals in SCNA profiles across the genome compared to the TFx approach.
Fig. 3.
(A) The cross-validation ROC curves and AUC statistics of the random forest model based on SCNA profiles data in the discovery cohort. (B) ROC curves and AUC statistics of the wRF-driver model in the two validation cohorts based on the model trained using genome-wide SCNA profiles data in the discovery cohort.
3.5. Application of a novel weighted machine learning model in validation cohorts
As our primary goal is for the detection of early-stage HCC, we selectively collected early-stage HCC patients in the two independent validation cohorts. Moreover, there were a higher number of HCC patients with very early stage (stage 0) tumor in the validation cohort 2 than in the validation cohort 1 (17.6% vs. 7.9%, Table 1). Given the much lower ctDNA burden observed in the two validation cohorts (Fig. 2 and Supplementary Fig. S5), it is inevitably much more challenging to detect HCC in the validation cohorts. Thus, to increase the detection accuracy for early-stage HCC, we developed a novel framework to improve the model performance, by incorporating prior knowledge derived from external genomics data. The rationale is that genome aberrations in early-stage patients are enriched for driver events, and genome regions harboring driver events play a more prominent role in discriminating early-stage HCC from HBV patients. To test the hypothesis, we used GISTIC2 [26] and iDriver [27] scores as the external evidence for driver events, and observed a significant enrichment of driver events in the top important genomics features, with p values of 2.5 × 10−14 and 5.9 × 10−4, respectively, for GISTIC2 and iDriver scores. Motivated by the findings, we developed a novel framework, wRF-driver, denoting a weighted Random Forest model incorporating driver evidence derived from external data.
We applied the wRF-driver model to the two validation cohorts, and obtained an AUC of 0.920 and 0.812 in validation cohort 1 and 2 (Fig. 3B). The accuracy in validation cohort 2, noticeably lower than that in validation cohort 1, was likely due to more patients with very early stage tumor in validation cohort 2. When we chose a cut-off of predictive probability of 0.5, we obtained a sensitivity (SE) of 0.56 and a specificity (SP) of 0.9 for the validation cohort 1, and a SE of 0.53 and a SP of 0.96 for the validation cohort 2. Our results showed that the performance of the wRF-drive model in the validation cohort 1 was higher than that in the early-stage patients in the discovery cohort (cross-validation AUC=0.874, Fig. 3A), although the tumor burden in the validation cohort 1 was considerably lower than that in the discovery cohort (Fig. 2, Supplementary Fig. S5).
3.6. Impact of heterogeneity of tumor samples on model performance
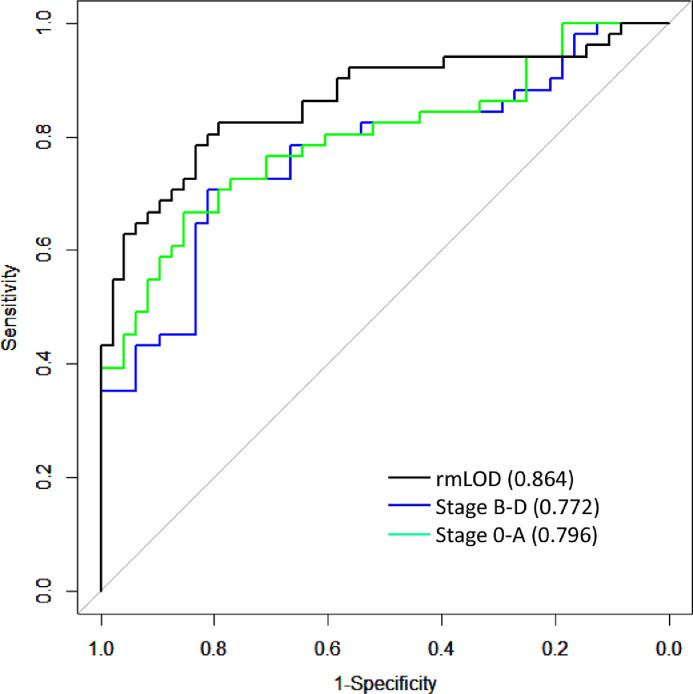
We noticed that the predictive performance of the model differed for the two validation cohorts, with the validation cohort 2 having appreciably lower accuracy than validation cohort 1. One of the key differences between the two validation cohorts was that validation cohort 2 was collected in a different hospital in a different city (Supplementary Materials), thus could serve as a better (i.e., more independent) validation cohort. Moreover, because our interest is early-stage HCC detection, and there are more early stage HCC patients in the validation cohort 2 (Table 1), we therefore, used the validation cohort 2 for this evaluation to achieve our goals. We first constructed three categories of HCC samples, each with 75 patients, from the discovery cohort, and then built three wRF-driver models using HCC cases in each of the three categories as the positives and randomly chosen 75 HBV patients as controls. We used the same number of samples in each model to make the performance comparable across models. These three categories were: 1) stage 0-A HCC patients, 2) stage B-D HCC patients, and 3) HCC patients with ctDNA lower than the limit of detection (LOD) removed. Specifically, category 1 was enriched in stage 0-A HCC samples, category 2 enriched in stage B-D HCC samples, and category 3 enriched in “cleaned” HCC samples after removing HCC samples with ctDNA level below LOD. Technically, samples with signals below LOD can be considered as “mislabeled” training set, and including such “mislabeled” samples in the training set will reduce the model performance [28,29]. The performance of the three models on the validation cohort 2 is shown in Fig. 4. The model trained using stage 0-A patients in the discovery cohort has better accuracy than that trained using stage B-D patients, for predicting HCC patients in the validation cohort 2, suggesting that matching characteristics of discovery and validation samples is critical to improve model performance. Among the three models, the largest performance improvement was observed for the model trained using category 3 samples (AUC=0.864, Fig. 4), the AUC increased considerably compared to the original model (AUC=0.812, Fig. 3B), suggesting the pronounced impact of HCC samples with low ctDNA burden on the model performance. Additional details are available in Supplementary Materials, Supplementary Fig. S6 and S7.
Fig. 4.
The performance (ROC and AUC statistics) of three wRF-driver models evaluated on the validation cohort 2. The three models were trained based on data of three categories of samples with distinct stages and levels of ctDNA burden. The blue, green and black lines represent the performance of the model trained on stage B-D samples, stage 0-A samples, and samples after filtering LOD, respectively.
3.7. Effect of sequencing depth on model performance
The above analyses were based on sequencing data of ∼5X. We were interested in learning the effect of reducing sequencing depth on model performance. To this end, we evaluated the performance of the wRF-driver model at reduced depths of 1.0X and 0.1X, by pooling all of the patients in the three cohorts. Compared to 5.0X (AUC=0.910), 1.0X depth resulted in a negligible reduction in performance (AUC=0.894), whereas 0.1X resulted in a more dramatic reduction (AUC=0.858) (Supplementary Fig. S8A). When separating by stages, we observed similar patterns for stage B-D tumors, with remarkably close performance for data with 5.0X vs. 1.0X (Supplementary Fig. S8C), indicating that 1.0X is probably adequate for detection of stage B-D HCC patients. For stage 0-A patients, however, the adequate sequencing coverage for effective detection is less clear, as the performance of the model was noticeably reduced with 1.0X compared to 5.0X, and the performance was further reduced for 0.1X (Supplementary Fig. S8B).
4. Discussion
In the current study, our extensive analyses demonstrated the potential for early cancer detection by using whole-genome SCNA profiling coupled with innovative machine learning. It is worth noting that our study is unique in focusing on detecting early-stage HCC, whereas the vast majority of previous studies [5,7,11] focused on late-stage patients that are generally more easily to be detected by other routine technologies already available in clinics. A key promise of liquid biopsy for tumor detection lies in its ability to identify tumors at early stages. Detecting early-stage tumors is inevitably much mor challenging, as these tumors tend to release much less DNA into blood, thus resulting in low ctDNA burden and requiring an unrealistic amount of blood to analyze ctDNA mutations [30]. This is consistent with the lower accuracy of our validation chorts, which are predominalty early stage tumors, compared to the discovery cohort. For SCNAs, the weak signals in early-stage tumors are embedded in the WGS-derived genome-wide data, and machine learning approaches are able to more effectively capture these weak signals to distinguish cancer from non-cancer patients. To boost accuracy of HCC early detection, we developed the wRF-driver algorithm to take advantage of both the genome-wide SCNA profiles as well as prior knowledge gained from external data, and demonstraded its robust performance in the two validation cohorts.
Previous studies have reported higher levels of ccfDNA in patients with HCC than in those with HBV/HCV-related chronic hepatitis and healthy subjects [31], [32], [33]. In recent studies conducted in patients with various cancer types including liver cancer, the concentration of plasma ctDNA increases with tumor stage, and discrimination accuracy between cancer patients and healthy controls decreases from stage III to stage I tumor [34,35]. The use of high-dimensional machine learning approach offers an opportunity to improve diagnostic sensitivity and specificity [36]. Studies such as that of Panagopoulou et al [37] did not observe the correlation, possibly due to study population, quantification method, sample size, and cancer type. Furthermore, in this study, we used cancer-free HBV patients as controls. Previous studies on HCC early detection used HBV-free healthy subjects as normal controls, which may potentially inflate the detection accuracy of HCC, in that it may not be able to differentiate the genomic aberrations derived from HCC or cancer-free HBV patients. Our study design, although more technically and analytically challenging, has greater clinical implications because HBV infection is a primary risk factor for HCC.
A tumor is considered to be a constantly evolving system in which genomic aberrations cumulate stochastically during evolution [38]. As a result, there is pervasive and extensive heterogeneity among individual tumors [38]. For early cancer detection, how to effectively capture genomics features that are predictive of early-stage tumors is critically important. It is reasonable to assume that, in late-stage tumors, a considerable proportion of genomic aberrations are passenger events, which are likely to be distinct among individual tumors, as well as different from early-stage tumors [39]. Therefore, we speculated that the composition of the heterogeneous samples in the discovery cohort has impact on the accuracy of our model. Indeed, we observed that matching tumor stages in the discovery and validation samples is potentially a key to improve the model performance. Tumor burden as well as biological factors affect the release of tumor DNA [36]. Based on currently available technologies, ctDNA could be harvested and analyzed to signify cancer only when the tumor weight is in the range of 100 mg to 1 g or has an approximate diameter of ≥1 cm [30]. The released ctDNA may be undetectable in a typical blood draw of 10 mL if early stage cancer patients have a very small tumor. In situation where LOD exists, the empirical ROC curve may fail to provide a valid estimate [40]. When conducting analyses, the samples with ctDNA level below LOD were considered as non-cancerous samples for the model training purpose, even if they were collected from HCC patients, which introduced misclassification and decreased diagnostic accuracy. A noteworthy finding of this study was that removing these samples from the discovery cohort remarkably improved predictive accuracy (Fig. 4).
Currently most liquid biopsy studies for tumor detection utilize only one aspect of genomic aberrations, such as mutation [9,41,42], methylation [7,11,12] or SCNA [13,43]. Since tumor genomes often harbor all such aberrations simultaneously, integrating multiple sources of data holds great promise for increasing detection accuracy, especially for early-stage tumors. The rationale lies in that distinct aberrations provide complementary signals, thus possibly leading to a higher prediction performance [9,35,44]. Given the extensive inter- and intra-heterogeneity of tumor genomes, genome-wide profiling is likely the most effective strategy to comprehensively survey genomics aberrations. Machine learning effectively integrates all sources of data and indispensably captures the complex dependency among the high-dimension multi-omics data; in doing so, it fully utilizes the complementary information carried in individual -omics data. In the long run, integrating other types of information, such as genomics, genetics, molecular, clinical, and environmental data when available, should further boost the accuracy of early cancer detection.
One limitation of this study is the limited sample size. Given the extensive tumor heterogeneity, the sample size required to adequately capture the heterogeneous genome aberrations in blood-based non-invasive assay is essentially hard to determine but should be expected to far outnumber the sample size used in the current study. Thus, multi-site independent validations become essential to ensure the robustness of the model developed in discovery cohorts to minimize model overfitting [45]. As the model performance is critically dependent on the sample size, it is reasonable to anticipate further improvement when the sample size increases. We focused on HBV patients to control confounding from disease etiology. Thus, it remains to be tested whether this model may be applicable to other high-risk populations such as those with HCV infection, alchoholic liver, or non-alcoholic fatty liver diseases. The current study was based on retrospectively collected blood samples, and the clinical utility of our developed model needs to be further evaluated in prospective cohorts of noncancerous patient to fully assess the predictive accuracy.
Acknowledgments
Acknowledgements
We thank the patients for their participation in the study.
Funding Sources
This work was supported by funding from the National Natural Science Foundation of China (81320108021) (J. Xing), the Science & Technology Co-ordination and Innovation Project of Shaanxi Province, China (2016KTZDS-F01-04) (J. Xing), the Key Research and Development Program of Shaanxi Province, China (2018ZDXM-SF-061), and the National Science and Technology major projects of China (2018ZX10302205-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ Disclosures or Potential Conflicts of Interest
Q. Zhang and C. Yang are employees of Oriomics Inc. No other disclosures were reported.
Authors' contributions
JX and CY designed the study. KT and ZB performed most of the experiments, wrote and revised the manuscript. QZ, XG and CY performed part of the experiments. YW, KZ, SW and MS analyzed and interpreted the data. All authors read and approved the final manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102811.
Contributor Information
Chuhu Yang, Email: chuhu.yang@oriomics.com.
Jinliang Xing, Email: xingjl@fmmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Singal A.G., Pillai A., Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS medicine. 2014;11(4) doi: 10.1371/journal.pmed.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hann H.W., Fu X., Myers R.E., Hann R.S., Wan S., Kim S.H. Predictive value of alpha-fetoprotein in the long-term risk of developing hepatocellular carcinoma in patients with hepatitis B virus infection–results from a clinic-based longitudinal cohort. Eur J Cancer. 2012;48(15):2319–2327. doi: 10.1016/j.ejca.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson P.J. Role of alpha-fetoprotein in the diagnosis and management of hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14(Suppl):S32–S36. doi: 10.1046/j.1440-1746.1999.01873.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnson P.J. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5(1):145–159. doi: 10.1016/s1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- 5.Chan K.C., Jiang P., Chan C.W., Sun K., Wong J., Hui E.P. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A. 2013;110(47):18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douville C., Springer S., Kinde I., Cohen J.D., Hruban R.H., Lennon A.M. Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs) Proc Natl Acad Sci U S A. 2018;115(8):1871–1876. doi: 10.1073/pnas.1717846115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo S., Diep D., Plongthongkum N., Fung H.L., Zhang K., Zhang K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet. 2017;49(4):635–642. doi: 10.1038/ng.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer S.U., Chen C.H., Rodriguez Pena M.D.C., Li L., Douville C., Wang Y. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife. 2018;7:e32143. doi: 10.7554/eLife.32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Li L., Douville C., Cohen J.D., Yen T.T., Kinde I. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018;10(433) doi: 10.1126/scitranslmed.aap8793. eaap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang P., Chan K.C.A., Lo Y.M.D. Liver-derived cell-free nucleic acids in plasma: Biology and applications in liquid biopsies. Journal of hepatology. 2019;71(2):409–421. doi: 10.1016/j.jhep.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 12.Oussalah A., Rischer S., Bensenane M., Conroy G., Filhine-Tresarrieu P., Debard R. Plasma mSEPT9: A Novel Circulating Cell-free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. EBioMedicine. 2018;30:138–147. doi: 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molparia B., Nichani E., Torkamani A. Assessment of circulating copy number variant detection for cancer screening. PloS one. 2017;12(7) doi: 10.1371/journal.pone.0180647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayrhofer M., De Laere B., Whitington T., Van Oyen P., Ghysel C., Ampe J. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome medicine. 2018;10(1):85. doi: 10.1186/s13073-018-0595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dor Y., Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 16.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clinical chemistry. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 17.Mouliere F., Chandrananda D., Piskorz A.M., Moore E.K., Morris J., Ahlborn L.B. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466) doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Zhan X., Wing M.K., Anderson P., Kang H.M., Abecasis G.R. QPLOT: a quality assessment tool for next generation sequencing data. Biomed Res Int. 2013;2013 doi: 10.1155/2013/865181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adalsteinsson V.A., Ha G., Freeman S.S., Choudhury A.D., Stover D.G., Parsons H.A. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N., Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7) doi: 10.1016/j.cell.2017.05.046. 1327-41 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Baran J., Cros A., Guberman J.M., Haider S., Hsu J. International Cancer Genome Consortium Data Portal–a one-stop shop for cancer genomics data. Database (Oxford) 2011:2011. doi: 10.1093/database/bar026. bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Wei Q., Zhong X., Yang H., Li B. Cancer driver gene discovery through an integrative genomics approach in a non-parametric Bayesian framework. Bioinformatics. 2017;33(4):483–490. doi: 10.1093/bioinformatics/btw662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlenbach F., Lallich S., Zighed D.A. Identifying and handling mislabelled instances. J Intell Inf Syst. 2004;22:89–109. [Google Scholar]
- 29.Zeng X., Martinez T.R. An algorithm for correcting mislabeled data. Intelligent Data Analysis. 2001;5:491–502. [Google Scholar]
- 30.Fiala C., Diamandis E.P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC medicine. 2018;16(1):166. doi: 10.1186/s12916-018-1157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piciocchi M., Cardin R., Vitale A., Vanin V., Giacomin A., Pozzan C. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatology international. 2013;7(4):1050–1057. doi: 10.1007/s12072-013-9481-9. [DOI] [PubMed] [Google Scholar]
- 32.Pezzuto F., Buonaguro L., Buonaguro F.M., Tornesello M.L. The Role of Circulating Free DNA and MicroRNA in Non-Invasive Diagnosis of HBV- and HCV-Related Hepatocellular Carcinoma. International journal of molecular sciences. 2018;19(4) doi: 10.3390/ijms19041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D., Hu X., Long G., Xiao L., Wang Z.M., Zhou L.D. The clinical value of total plasma cell-free DNA in hepatitis B virus-related hepatocellular carcinoma. Annals of translational medicine. 2019;7(22):650. doi: 10.21037/atm.2019.10.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224) doi: 10.1126/scitranslmed.3007094. 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heitzer E., Haque I.S., Roberts C.E.S., Speicher M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nature reviews Genetics. 2019;20(2):71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 37.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., Karamitrousis E. Circulating cell-free DNA in breast cancer: size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38(18):3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 38.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 39.McGranahan N., Favero F., de Bruin E.C., Birkbak N.J., Szallasi Z., Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283) doi: 10.1126/scitranslmed.aaa1408. 283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bantis L.E., Yan Q., Tsimikas J.V., Feng Z. Estimation of smooth ROC curves for biomarkers with limits of detection. Statistics in medicine. 2017;36(24):3830–3843. doi: 10.1002/sim.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng C.K.Y., Di Costanzo G.G., Tosti N., Paradiso V., Coto-Llerena M., Roscigno G. Genetic profiling using plasma-derived cell-free DNA in therapy-naive hepatocellular carcinoma patients: a pilot study. Annals of oncology: official journal of the European Society for Medical Oncology. 2018;29(5):1286–1291. doi: 10.1093/annonc/mdy083. [DOI] [PubMed] [Google Scholar]
- 42.Labgaa I., Villacorta-Martin C., D'Avola D., Craig A.J., von Felden J., Martins-Filho S.N. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene. 2018;37(27):3740–3752. doi: 10.1038/s41388-018-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schutz E., Akbari M.R., Beck J., Urnovitz H., Zhang W.W., Bornemann-Kolatzki K. Chromosomal instability in cell-free DNA is a serum biomarker for prostate cancer. Clinical chemistry. 2015;61(1):239–248. doi: 10.1373/clinchem.2014.226571. [DOI] [PubMed] [Google Scholar]
- 44.Haque I.S., Elemento O. Challenges in Using ctDNA to Achieve Early Detection of Cancer. bioRxiv. 2017 [Google Scholar]
- 45.Dietterich T. Overfitting and undercomputing in machine learning. ACM computing surveys (CSUR) 1995;27(3):326–327. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.