Abstract
Background
Accurate lymph nodes (LNs) assessment is important for rectal cancer (RC) staging in multiparametric magnetic resonance imaging (mpMRI). However, it is incredibly time-consumming to identify all the LNs in scan region. This study aims to develop and validate a deep-learning-based, fully-automated lymph node detection and segmentation (auto-LNDS) model based on mpMRI.
Methods
In total, 5789 annotated LNs (diameter ≥ 3 mm) in mpMRI from 293 patients with RC in a single center were enrolled. Fused T2-weighted images (T2WI) and diffusion-weighted images (DWI) provided input for the deep learning framework Mask R-CNN through transfer learning to generate the auto-LNDS model. The model was then validated both on the internal and external datasets consisting of 935 LNs and 1198 LNs, respectively. The performance for LNs detection was evaluated using sensitivity, positive predictive value (PPV), and false positive rate per case (FP/vol), and segmentation performance was evaluated using the Dice similarity coefficient (DSC).
Findings
For LNs detection, auto-LNDS achieved sensitivity, PPV, and FP/vol of 80.0%, 73.5% and 8.6 in internal testing, and 62.6%, 64.5%, and 8.2 in external testing, respectively, significantly better than the performance of junior radiologists. The time taken for model detection and segmentation was 1.3 s/case, compared with 200 s/case for the radiologists. For LNs segmentation, the DSC of the model was in the range of 0.81–0.82.
Interpretation
This deep learning–based auto-LNDS model can achieve pelvic LNseffectively based on mpMRI for RC, and holds great potential for facilitating N-staging in clinical practice.
Keywords: Lymph node, Deep learning, Detection and segmentation
Research in context.
Evidence before this study
Accurate LN assessment is critical for RC staging based on mpMRI, and smaller LNs can be challenging to be detected in limited time.
The auto-LNDS model based on deep learning proposed in this paper enables fast and accurate nodal localization and delineation based on mpMRI. This auto-LNDS model outperformed junior radiologists, and can help to eliminate inter-observer differences and reduce the workload for radiologist potentially.
Added value of this study
Based on the data from multiple clinical centers, we present an auto-LNDS for the detection and segmentation of LNs and the model were significantly faster and better than the junior radiologist performance.
Implications of all the available evidence
The proposed method can help to increase the efficiency of the clinical workflow, and also has the potential to assist physicians in identifying the distribution of LNs.
Alt-text: Unlabelled box
1. Introduction
Lymph nodes (LNs) are the most common metastatic site for rectal cancer (RC), and nodal status is critical for treatment decisions and prognosis. According to the National Comprehensive Cancer Network (NCCN) guidelines and the American Joint Committee on Cancer (AJCC) staging criteria, both the location and number of metastatic LNs should be evaluated pre-treatment for guiding treatment decisions [1,2]. Accurate identification and removal of the metastatic LNs at surgery are crucial for reducing tumor recurrence, especially for lateral LNs. Some studies have demonstrated that enlarged lateral lymph nodes (LLNs) may have a close relationship with local recurrence [3], and suggested lateral lymph-node dissection (LLND) for patients with metastatic LNs in these regions to improve the prognosis and reduce the local recurrence rate in low RC patients [4,5]. Whereas LLND is another procedure independent of routine total mesorectal excision (TME) and always has a higher incidence of surgery complications, including operative mortality and long-term sexual and urinary dysfunction [6]. Therefore, accurate detection and identification of the number and location of metastatic LNs before surgery is of great importance to inform the treatment decision [7,8].
Multiparametric magnetic resonance imaging (mpMRI) has been accepted as the first choice for RC examination, and N-staging is necessary for all MR reports, for which accurate detection and segmentation of LNs is the first step. Then the morphology and signal of the LNs on each MRI sequence are assessed to determine whether they are metastatic. In a recent report Gröne et al. [9] reported an unsatisfactory result using short-diameter of 5 mm as the cutoff value for metastatis, in which the sensitivity, specificity and accuracy of MRI diagnosis for RC N-staging is 72%, 45.7% and 56.7%, respectively. In another study, Langman et al. reported that even LNs ≤ 3 mm in short-diameter still have a high probability of being malignant (28%, 95/334), suggesting that small LNs (≤ 3 mm in short-diameter) should not be overlooked [10]. Therefore, it is important to identify all the LNs in the scan area as much as possible. However, it is a highly challenging task. In practice, even LNs ≥ 3 mm in short-diameter may be missed by both inexperienced and experienced radiologists as a result of their small size, despite rigourous works. For a radiologist, finding tiny LNs from hundreds or thousands of images in a limited time is a difficult and monotonous task, which directly relates to the efficiency of the subsequent diagnosis of metastatic LNs.
Therefore, fully automated LNs detection and segmentation is desirable. This work is challenging due to the lack of comparison between LNs and the surrounding structures and the individual anatomic variation. To date, a limited number of studies have been published on automated LNs detection and segmentation. To the best of our knowledge, morphological-based blob detectors [[11],12], learning-based methods combining spatial prior map [13], [14], [15], [16], and graph-based and fast-marching methods [17], [18], [19] have been used to analyzed CT data for LNs detection or segmentation. However, all these semiautomatic LN algorithms require substantial time-consuming manual interaction. Furthermore, these algorithms are generally applied for a nodal size of 8 mm or larger. For MRI data, some researchers have utilized T1-weighted imaging (T1WI) and/or T2-weighted imaging (T2WI) for LNs detection and segmentation [20,21]. However, T2WI and diffusion-weighted imaging (DWI) are the most important sequences for nodal identification in clinical practice [22].
In recent years, deep learning techniques have simulated great interest for tackling challenging computer vision tasks in medical imaging, such as tumor segmentation [23], [24], [25] and pulmonary nodule detection [26,27]. However, due to the considerable individual differences in the location and size of LNs, the detection of LNs is even more complicated and the capability of convolutional neural networks (CNN) is inadequate for this task. The object detection framework—Mask R-CNN (regional convolutional neural network) proposed by He et al. [28] has shown great promise in object detection. We hypothesize that using the fusion of T2WI and DWI of mpMRI images as input to the Mask R-CNN may improve the performance of MR-based LNs detection and segmentation, especially including all LNs ≥ 3 mm. In this work, we sought to develop and validate the feasibility of an automated LNs detection and segmentation (Auto-LNDS) model using deep learning techniques on multivendor and multicentre mpMRI datasets.
2. Materials and methods
This study was approved by the institutional review boards of the participating centres. The need for signed informed consent was waived because of the retrospective nature of our study.
2.1. Dataset
MpMRI data from 293 patients with rectal adenocarcinoma, confirmed by surgical pathology between July 2013 and June 2016 at the Sixth Affiliated Hospital of Sun Yat-sen University (Guangzhou, China), was collected and used as the training dataset in this study. All scans were generated on the 1.5T GE Optima MR360 scanner (General Electric Medical Systems, Milwaukee, WI, USA) using an eight-channel phased-array body coil in the supine position. Data from another 31 patients collected from the same center were utilized as the internal testing dataset. An external testing dataset consisting of 50 patients was collected from three other medical centres (the First Affiliated Hospital of Soochow University; Beijing Hospital; and Guizhou Province Hospital of Traditional Chinese Medicine). The rectal MR protocol of each center is shown in Supplementary Table S1.
To ensure all the LNs were annotated correctly, the ground truths were generated based on the decisions of three radiologists with varying seniority (35, 25 and 24 years, respectively). All LNs ≥ 3 mm in the short-diameter were annotated on the axial T2WI images by the two senior radiologists with 25 and 24 years’ experience using Medical Imaging Interaction Toolkit (MITK) software (version 2013.12.0; http://www.mitk.org/). If there was a difference, the third senior radiologist (35 years’ experience) was involved to provide a decision on LN presence. A total of 5789 LNs were annotated in the training dataset and were used to develop the auto-LNDS model, and another 2133 LNs were annotated in the internal and external testing datasets for model evaluation.
2.2. Preprocessing
DWI volumes were aligned to the T2WI volumes using a rigid registration with trilinear interpolation based on open-source Insight Segmentation and Registration Toolkit (ITK, version 4.7.2; https://itk.org/), to obtain the same resolution, spacing, and origin [29]. The DWI images with high b value were used in this study, as the higher the b value, the stronger the diffusion effects. On the high b-value DWI images the signal of the background tissue is well suppressed, so the high or slightly high signal intensity LNs can be clearly displayed and easily identified. In addition, Mask R-CNN requires three-channels of image input. In order to obtain the best combination mode of T2WI and DWI for training the auto-LNDS model, four kinds of combinations modes, including three channels set T2WI, three channels set DWI, two channels set DWI + one set T2WI and two channels set T2WI + one channel set DWI, were tested and compared for their performance both in the internal and external testing datasets. Images were cropped manually with 256 × 256 matrix as showed in Fig. 1, and perirectal and lateral LNs were included in this region for detection and segmentation. Finally, a total of 5694 processed images (each case has approximately 15 to 25 slices) of each combination mode were used as training dataset, and another 1192 and 2572 images were used as the internal and external testing datasets, respectively. The results of statistical analysis of the size of the LNs in internal and external testing datasets are shown in Fig. 2.
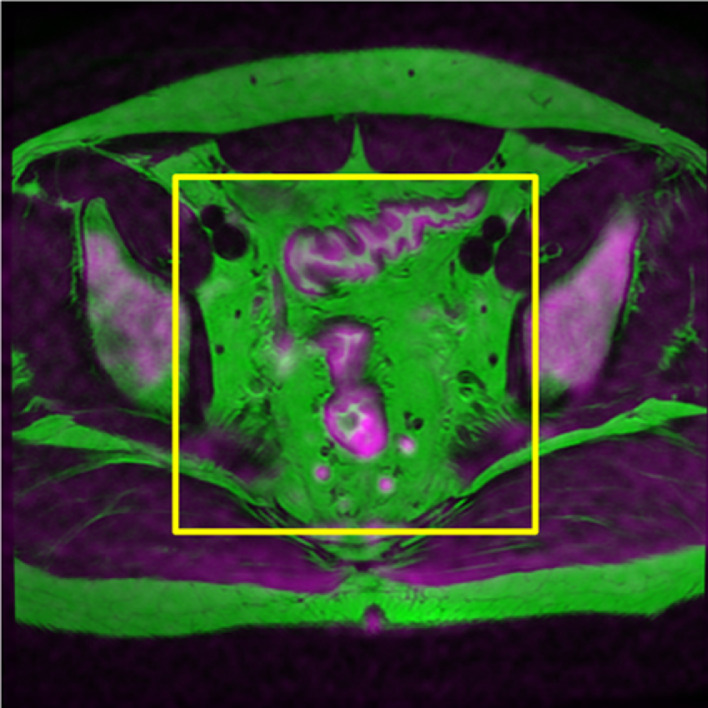
Fig. 1.
This image is a three-channel image obtained by the fusion of DWI and T2WI images. Both the perirectal and lateral lymph nodes are included in the cropping range (yellow box). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
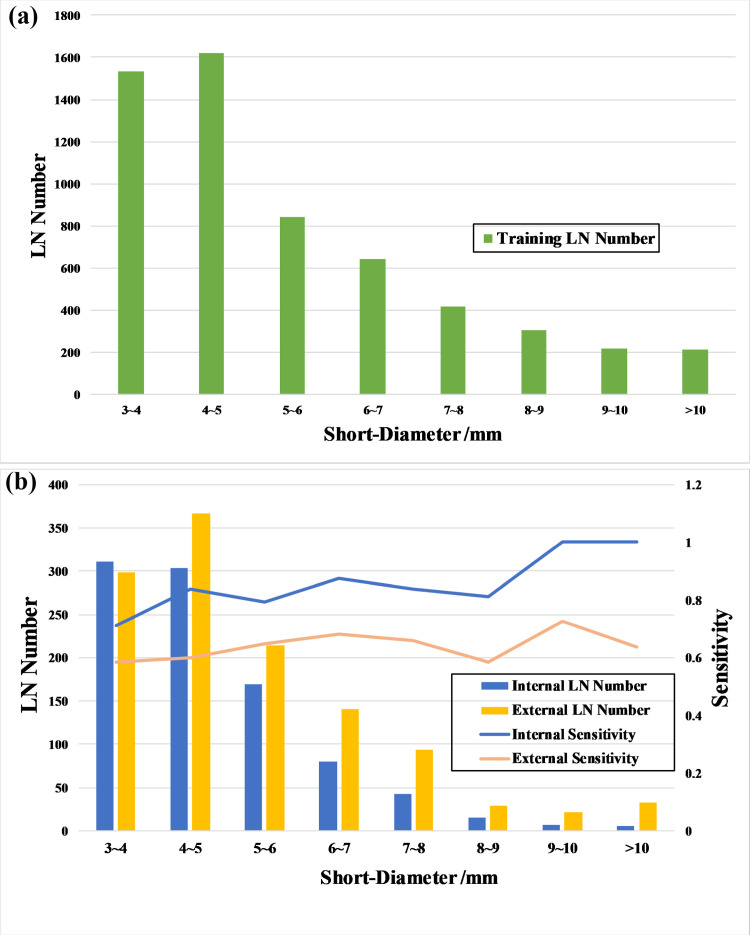
Fig. 2.
(a) The distribution of lymph nodes short-diameters in the training dataset; (b) The distribution of lymph nodes short-diameters in the testing datasets; The Sensitivity Curves of the auto-LNDS model for lymph nodes with different short-diameters in the internal and external testing datasets.
2.3. Development of auto-LNDS model
2.3.1. Data augmentation
Artificial data augmentation is a common procedure for generating sufficient training data in the context of CNN. It can also teach the network the desired invariances and robustness properties when the data set is insufficient [30]. In this study, we utilized the data augmentation package of python—imgaug (https://github.com/aleju/imgaug) to extend the training dataset. We adopted image cropping, affine transformations, flipping horizontally or vertically, adding noise and blur on image, and changing the contrast and brightness of image. Our training data set was augmented during the training by generating new images through 0 to 2 kinds of transformations randomly chosen from those mentioned above. Details are shown in the Supplementary Material and Methods.
2.3.2. Training model
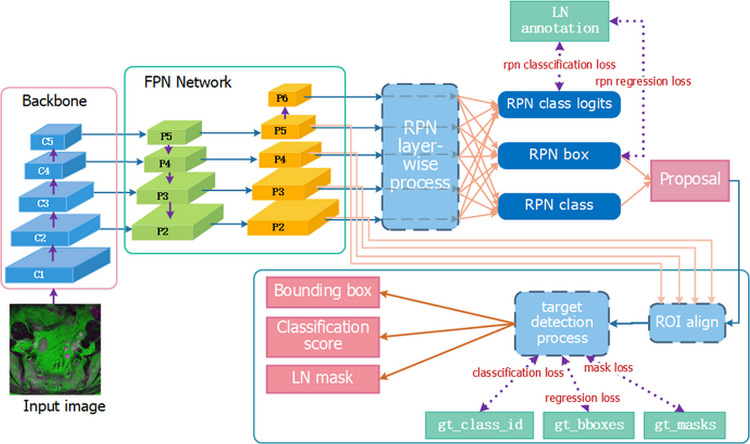
The framework of Mask R-CNN [28] can efficiently detect objects in an image while simultaneously generating a high-quality segmentation mask for each instance. Mask R-CNN is composed of the backbone network, Feature Pyramid Network (FPN) [31], the Region Proposal Network (RPN) [32], and the head network. The Resnet-101 [33] was chosen to be the backbone of Mask R-CNN, in which the identity mapping block was used as a shortcut to solve the degradation problem and make it possible to train the deeper network. Details are shown in the Supplementary Material and Methods and Fig. S1. The FPN inside Mask R-CNN is capable of detecting multi-scale objects so as to improve the detection of small targets, and the RPN shares the convolutional features of the full image with the head network, and can generate the candidates effectively, which broke a bottleneck for target detection [34,35]. Considering that the number and size of LNs vary from patient to patient, we adopted Mask R-CNN for the nodal detection and segmentation. To achieve a quick convergence of our network, a pre-trained Mask R-CNN (initialized on ImageNet dataset) was applied for LN detection and segmentation (as illustrated in Fig. 3).
Fig. 3.
Architecture of Mask RCNN. The gt_class_id, gt_bboxes, and gt_masks represent the nodal ground truth of class, position, and segmentation.
Relying on the high-level neural networks API—Keras, using the tensorflow as backend, the Mask R-CNN model was trained on an Ubuntu 16.04 computer with 1 Intel Xeon CPU, using a NVIDIA GTX 1080 Ti 11Gb GPU for training and testing, with 32 Gb available in RAM memory. Training of layers was performed by stochastic gradient descent in batches of four images per step using an Adam Optimizer [36] with the default value (β1 = 0.9, β2 = 0.999). The training hyper parameters are shown in Table 1 and illustrated in more detail in the Supplementary Material and Methods. Among the training dataset images, one-tenth randomly selected from the 5694 images were used to evaluate the learning effect of deep-learning model, and the remaining images were used to train the model. During testing, our model took less than 1.5 s to complete LN detection and segmentation per volume.
Table 1.
Training Hyper-parameters of Mask R-CNN.
| Hyper-parameters | Value |
|---|---|
| Iteration | 100 |
| Batch size | 4 |
| Learning rate | 1.e-6 |
| Optimizer | Adam |
| Weight decay | 1.e-4 |
| Scale of anchor | [8, 16, 32, 64, 128] |
| Aspect ratio of anchor | [0.5, 1, 2] |
| RPN NMS threshold | 0.8 |
2.4. Evaluation of the auto-LNDS model
2.4.1. Evaluation criteria of LN detection
We evaluated the LN detection results according to the method used in the Ref. [16], described as following: A true positive (TP) means that there exists a detection with the center inside the manually annotated LN bounding box, and a false negative (FN) means there is no center of any detections inside the box. A detection is considered to be false positive (FP) if its center is not inside any annotated LN box. We used the sensitivity and positive predictive value (PPV) to evaluate the model's performance. The higher the value of both, the better the performance of the algorithm.
Sensitivity is the proportion of the true LNs detected by auto-LNDS to total true LNs, being defined as:
| (1) |
PPV is the proportion of the true LNs identified by auto-LNDS to all the LNs identified by auto-LNDS, defined as:
| (2) |
The false positive per volume (FP/vol) is a measure of the average number of FPs per each case, defined as:
| (3) |
2.4.2. Evaluation criteria of LN segmentation
The Dice similarity coefficient (DSC) quantitatively evaluates the degree of similarity between the segmentation results of auto-LNDS and the ground truth. The DSC ranges from 0 to 1, and a larger value indicates a higher segmentation accuracy. The DSC was defined as Eq. (4), as follows:
| (4) |
Where P denotes the segmentation result given by the segmentation algorithm, G is the ground truth and N represents the number of pixels in the corresponding set.
2.5. Comparison of the auto-LNDS model with radiologist performance and other models
Four radiologists with varying experience in imaging diagnosis of abdominal diseases (1.5, 4, 7 and 9 years, respectively), were assigned to read the MR images from the internal and external testing datasets and marked all the LNs ≥ 3 mm including the perirectal and lateral LNs. A detection is considered TP if there exists a detection marked by the radiologist inside the segmented LN of the ground truth; a detection is considered FP if the marker is not inside any annotated LN; FN means a ground truth is not detected but marked by the radiologist. The sensitivity, PPV and FP/vol were used to evaluate the radiologists’ performance. In addition, the results were compared with the auto-LNDS model results to analyze sensitivity, PPV, FP/vol and length of time taken.
2.6. Statistical analysis
Statistical analysis was performed by using R software (version 3.5.1, https://www.r-project.org/). Two-side, one sample t-test was applied to assess the differences of the performance between the radiologists and auto-LNDS model. A p value smaller than 0.05 was considered significant.
3. Results
3.1. LN detection performance of the auto-LNDS model
The performances of the auto-LNDS model trained with the four kinds of T2WI and DWI combination modes are shown in Table 2. The auto-LNDS model with two channels set DWI and one channel set T2WI has the best detection performance, with the sensitivity of 80.0% (95%CI, 76.9%−82.2%), PPV of 73.5% (95%CI, 70.7%−76.2%) and FP/vol of 8.6 (95%CI, 6.9–10.3) in the internal testing dataset; and the sensitivity of 62.6% (95%CI, 59.5%−65.1%), PPV of 64.5% (95%CI, 61.7%−67.3%) and FP/vol of 8.2 (95%CI, 7.0–9.5) in the external testing dataset.
Table 2.
The performance of the auto-LNDS model trained with four combination modes of T2WI and DWI for lymph nodes detection.
| Combination Mode | Sens (95%CI) | PPV (95%CI) | FP/vol (95%CI) | DSC (95%CI) | |
|---|---|---|---|---|---|
| Internal Dataset | 3 T2WI | 63.0% (59.7%−65.9%) | 54.7% (51.7%−57.7%) | 15.7 (13.5−18.0) | 0.85 (0.84−0.86) |
| 3 DWI | 52.0% (48.7%−55.2%) | 66.7% (63.1%−70.1%) | 7.8 (6.2−9.5) | 0.63 (0.62−0.65) | |
| 2 T2WI+1 DWI | 81.3% (78.6%−83.7%) | 59.7% (56.9%−62.4%) | 16.5 (14.1−19.0) | 0.83 (0.82−0.84) | |
| 2 DWI+1 T2WI | 80.0% (76.9%−82.2%) | 73.5% (70.7%−76.2%) | 8.6 (6.9−10.3) | 0.82 (0.82−0.83) | |
| External Dataset | 3 T2WI | 45.5% (42.7%−48.4%) | 44.2% (41.4%−47.0%) | 13.8 (12.2−15.4) | 0.85 (0.85−0.86) |
| 3 DWI | 36.0% (33.4%−38.7%) | 44.7% (41.7%−47.7%) | 11.9 (9.9−13.8) | 0.56 (0.54−0.57) | |
| 2 T2WI+1 DWI | 58.1% (55.2%−60.9%) | 56.0% (53.2%−58.7%) | 11.0 (9.1−12.9) | 0.84 (0.84−0.85) | |
| 2 DWI+1 T2WI | 62.6% ( 59.5%−65.1%) | 64.5% (61.7%−67.3%) | 8.2 (7.0−9.5) | 0.81 (0.80−0.82) |
Table 3 lists the results of the comparison of this auto-LNDS model with the previously reported LN detection methods. The sensitivity of the auto-LNDS model for LN detection in the internal testing dataset (80.0%) is close to Barbu's (80%) [16] and Feuerstein's (82.1%) [11] results, and higher than Kitasaka's (57%) [12] and Feulner's [14] (65.4%) results. However, only Barbu's research [16] focused on the pelvic and abdominal LNs and was limited to the LNs > 10.0 mm. The PPV of the auto-LNDS model for LN detection in the internal testing dataset (73.5%) was much higher than Feuerstein's (13.3%) [11], Kitasaka's (30.3%) [12] and Feulner's (52.6%) [14] results, and close to Barbu's (72.6%) [16] results. Though the performance of the auto-LNDS model declined for the external testing dataset, its PPV is still higher than Feuerstein, Kitasaka and Feulner's results, and its sensitivity is a little higher than or close to Kitasaka's and Feulner's results which focused on the LNs > 5 mm in short-diameter [[11],12,14]. To our knowledge, Feuerstein's study [11] enrolled the smallest LNs with a short-diameter > 1.5 mm in mediastinum for automatic LN detection, and the sensitivity was generally satisfactory but the FP/vol was too large to exceed the acceptable range. In this research, we focused on the LNs with a short-diameter ≥ 3 mm and obtained a relatively acceptable FP/vol, which can meet clinical needs well. The performance of the auto-LNDS model for the external dataset from three centres is shown in Table 4. Besides these, the algorithm of this auto-LNDS model is more than ten times faster than the previous fastest algorithm according to our knowledges [16].
Table 3.
The performance of the current auto-LNDS model and others in the literatures for lymph nodes detection. thods.
| Method | Target area | Scan type | #cases | Nodal Size | #Nodes | #FP | #TP | #FN | Sens (95%CI) | PPV (95%CI) | FP/vol (95%CI) | Time/Vol |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Current-IT | Pelvic | MRI | 31 | ≥ 3.0mm | 935 | 268 | 745 | 190 | 80.0% (76.9%−82.2%) | 73.5% (70.7%−76.2%) | 8.6 (6.9–10.3) | 1.37sec |
| Current-ET | Pelvic | MRI | 50 | ≥3.0mm | 1198 | 412 | 750 | 448 | 62.6% ( 59.5%−65.1%) | 64.5% (61.7%−67.3%) | 8.2 (7.0–9.5) | 1.43sec |
| Barbu[16] | Pelvic+Aebden | CT | 54 | >10.0mm | 569 | 172 | 455 | 114 | 80.0% | 72.6% | 3.2 | 15–40sec |
| Feuerstein[11] | Mediastinum | CT | 5 | >1.5mm | 106 | 567 | 87 | 19 | 82.1% | 13.3% | 113.4 | 1–6min |
| Kitasaka[12] | Abdomen | CT | 5 | >5.0mm | 221 | 290 | 126 | 95 | 57.0% | 30.3% | 58 | 2–3h |
| Feulner[14] | Mediastinum | CT | 54 | >10.0mm | 266 | 157 | 174 | 92 | 65.4% | 52.6% | 2.9 | 135sec |
IT: Internal testing dataset; ET: External testing dataset.
Table 4.
The performance of the auto-LNDS model for lymph nodes detection in three external datasets.
| center | Sens (95%CI) | PPV (95%CI) | FP/vol (95%CI) | DSC (95%CI) |
|---|---|---|---|---|
| Beijing Hospital | 67.0% (62.8%−71.0%) | 68.9% ( 64.6%−72.8%) | 8.0 (5.9−10.0) | 0.82 (0.81−0.83) |
| the First Affiliated Hospital of Soochow University | 60.0% (50.4%−68.9%) | 62.2% (52.4%−71.0%) | 6.0 (2.99.1) | 0.83 (0.81−0.85) |
| Guizhou Province Hospital of Traditional Chinese Medicine | 58.4% (54.2%−62.5%) | 60.9% (56.7%−65.0%) | 9.2 (7.5−10.8) | 0.79 (0.78−0.81) |
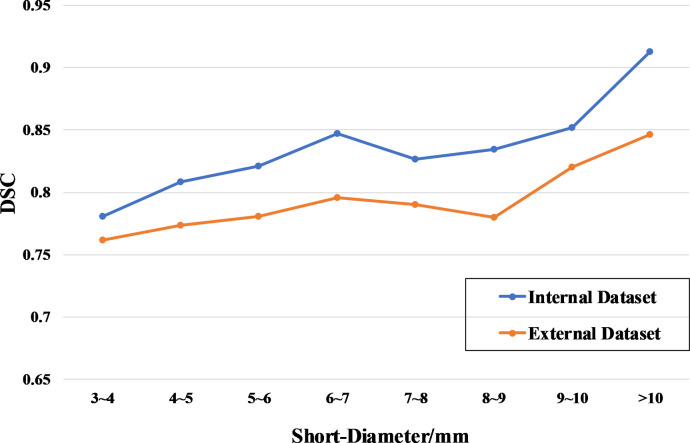
The Sensitivity Curves of the auto-LNDS model for detecting the LNs with different short-diameters in the internal and external test datasets are shown in Fig. 2(b), which shows that the sensitivity of the auto-LNDS model increases with the size (short-diameter) increase of the LNs.
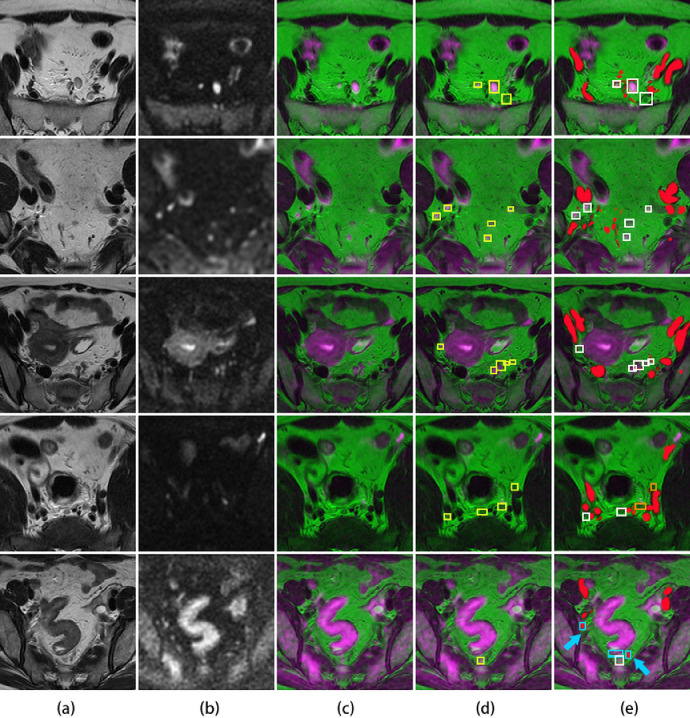
Some examples of the performance of the auto-LNDS model for LNs detection are shown in Fig. 4. The cases from the first row to the third row show the LNs correctly detected by the auto-LNDS model both in large and small size, and in discrete and clustered distribution. Besides these, both of the cases in the first row and the forth row show the correctly detected right lateral LNs by the auto-LNDS model. Whereas, the case in the forth row shows two missed LNs by the auto-LNDS model: one perirectal LN is missed due to insufficient image registration and inconspicuous display on the fusion image (c); one left lateral LN is missed due to adjacent to the branches of iliac vessels and iso-intensity on DWI. And the case in the fifth row shows three misdiagnosed LNs by the auto-LNDS model: two of them are cross section of small vessels; one is a small part of intestinal wall. Therefore, it does exhibit some false positive and false negative detections as indicated in the Fig. 4, which might be due to insufficient image registration, the iso-intensity of LN on DWI, and the overlap between the LNs and small vessels or intestinal wall as a result of partial volume effects.
Fig. 4.
Lymph node detection. (a): the original T2WI. (b): the original DWI. (c): the fusion image. (d): the ground truth of annotated lymph nodes with yellow boxes on the fusion image. (e): the detected results of auto-LNDS displayed on the fusion images. The white boxes represent the true positives, the cyan boxes represent the false positives and the orange boxes represent the false negatives. Vessels were filled with red. The case in the fourth row shows two missed lymph nodes by the auto-LNDS model. In the case of the fifth row, two cyan boxes with red color inside are small vessels misdiagnosed as lymph nodes by the auto-LNDS model (cyan arrow), and the other cyan box is intestinal wall misdiagnosed as a lymph node. See main text for additional details (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Detection performance of the radiologists for the internal and external testing datasets are shown in Tables 5 and 6, respectively. In both of the internal and external testing datasets, the sensitivity and PPV of the auto-LNDS model were higher than those of all the junior radiologists with less than ten-years experience (p < 0.05, t-test). The average time consumed by the radiologists was more than 200 s per case, compared with only 1–2 s per case for the auto-LNDS model (p < 0.05, t-test).
Table 5.
Results of radiologists vs. Auto-LNDS model in internal testing dataset.
| Doctor | Sens (95%CI) | PPV (95%CI) | FP/vol (95%CI) | Time/sec |
|---|---|---|---|---|
| D1(1.5y) | 43.2% (38.0%−48.4%) | 42.0% (38.7%−45.3%) | 19.4 (16.7–22.1) | 345.6 |
| D2 ( 4y ) | 31.1% (25.7%−36.5%) | 48.7% (44.0%−53.4%) | 10.8 (8.8–12.8) | 133.8 |
| D3 ( 7y ) | 37.7% (32.6%−42.8%) | 43.4% (39.7%−47.1%) | 16.7 (13.8–19.6) | 199.2 |
| D4 ( 9y ) | 40.6% (34.9%−46.3%) | 41.1% (36.1%−46.1%) | 18.3 (16.1–20.5) | 147.0 |
| Mean | 38.2% (33.1%−43.3%) | 43.8% (40.5%−47.1%) | 16.3 (12.6–20.0) | 206.4 |
| Auto-LNDS | 80.0% (76.9%−82.2%) | 73.5% (70.7%−76.2%) | 8.6 (6.9–10.3) | 1.37 |
| p value | 0.0004 | 0.0002 | 0.0138 | 0.0121 |
P values were derived from the t-test of comparing each metrics between the radiologists and the auto-LNDS model.
Table 6.
Results of radiologists vs. Auto-LNDS model in external testing dataset.
| Doctor | Sens (95%CI) | PPV (95%CI) | FP/vol (95%CI) | Time/sec |
|---|---|---|---|---|
| D1(1.5y) | 39.2% (33.6%−44.8%) | 24.4% (20.7%−28.1%) | 27.5 (25.2–29.8) | 350.4 |
| D2 ( 4y ) | 27.3% (22.7%−31.9%) | 43.6% (38.1%−49.1%) | 7.5 (6.3–8.7) | 118.8 |
| D3 ( 7y ) | 34.6% (30.9%−38.3% ) | 36.0% (32.0%−40.0%) | 14.3 (12.4–16.2) | 224.4 |
| D4 ( 9y ) | 45.6% (32.6%−58.6%) | 39.5% ( 25.6%−43.4%) | 15.4 (13.8–17.0) | 134.4 |
| Mean | 36.7% (29.1%−44.3%) | 35.9% (27.8%−44.0%) | 16.2 (8.0–24.4) | 207.0 |
| Auto-LNDS | 62.6% (59.5%−65.1%) | 64.5% (61.7%−67.3%) | 8.2 (7.0–9.5) | 1.43 |
| p value | 0.0033 | 0.0025 | 0.0755 | 0.0153 |
P values were derived from the t-test of comparing each metrics between the radiologists and the auto-LNDS model.
3.2. LN segmentation performance of the auto-LNDS model
Our auto-LNDS model was evaluated on 745 detected LNs in the internal testing dataset and 750 detected LNs in the external testing dataset. DSC is 0.82 (95%CI, 0.82–0.83) and 0.81 (95%CI, 0.80–0.82) for the internal and external testing datasets, respectively.
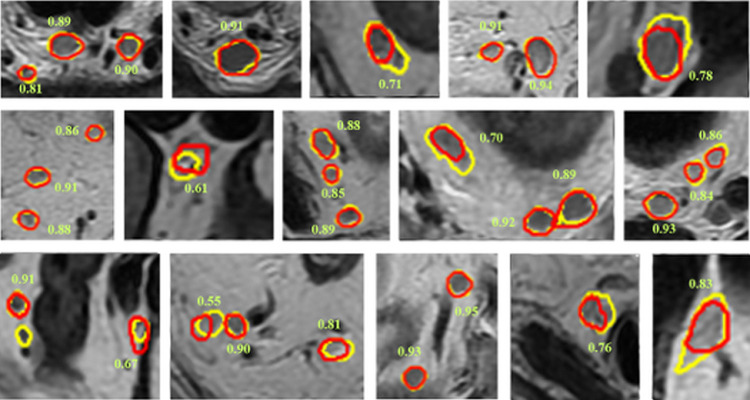
The examples of LNs segmentation are shown in Fig. 5. The DSC distribution of LNs segmentation in internal and external datasets are shown in Fig. 6. We find that the segmentation boundaries of the larger LNs have better overlap with the ground truth than those of the smaller LNs.
Fig. 5.
Nodal segmentation examples displayed on T2WI. Ground truth results are shown in yellow, and segmentation results of the auto-LNDS model are shown in red. The number besides the lymph node is the corresponding DSC (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 6.
DSC distribution of lymph node with different short-diameters in internal and external testing datasets.
The loss function values of the total networks, the detection network, mask network and the region proposal network (RPN) in the training process were output as shown in Supplementary Fig. S2.
4. Discussion
It is well known that, for patients with RC, detecting all the LNs and distinguishing the malignant from the benign LNs is an important and challenging job for the radiologists. N-staging is one of the key factors affecting treatment decisions and patient prognosis. The first step in this process is detecting all the LNs, and it is a monotonous and time-consuming job. In this study, we proposed an innovative deep learning approach (auto-LNDS model) which enables rapid and accurate detection and segmentation for LNs on MR examination (T2WI and DWI) in the setting of rectal cancer N staging. By this method, a map of LNs could be rapidly acquired in less than 2 s to support the real-time diagnostic interpretation, which can greatly save the search time for the radiologists. According to our knowledge, this is the first attempt to automatically detect and segment LNs simultaneously based on MRI data. Most of the prior attempts with other methods have focused on the LNs with the short-diameter sizes > 5 mm [11,12,14,16], whereas our study expands the range of detection to LNs>3 mm in the short-diameter. Barbu's method [16] obtained good results with a PPV of 72.6% and a DSC of 0.76, but that research focused on LNs with a short-diameter > 10 mm, which means that all the metastatic LNs with a short diameter ≤ 10 mm will be missed and these are very common in daily work. Feuerstein's team [11] tried to automatically detect LNs <5 mm, but the PPV of 13.3% was too low for clinical relevance. In addition, our algorithm is highly integrated and the output displays the detection and segmentation results, whereas other algorithms [16,18] depend on complicated cascaded detectors with an additional segmentation algorithm or some manual initialization. Our method aimed at LNs with a short-diameter ≥ 3 mm and achieved great performance in the internal testing dataset and good generalization performance on the external testing dataset. To date, very few studies provide both LNs detection and segmentation as we do. Meanwhile, we found that the size of the LNs is an important factor to influence the performance of the model for LN detection, and the sensitivity of the auto-LNDS model increases with the size increase in both of the internal and external testing datasets as shown in Fig. 2(b), which means that the large LNs are easier to be detected than the small ones. Although the criterion of 3 mm will reduce the performance of the model, we believe that it is more meaningful and this setting will better meet the future clinical needs.
In this study, we tested and compared the performance of the auto-LNDS model with different T2WI and DWI combination modes both in the internal and external testing datasets (Table 2). We can conclude that both of the image type and the algorithm have an impact on the detection performance. The auto-LNDS models with the combination of T2WI and DWI achieved better performance than the models with single sequence on the detection of LNs, which indicates that the information in T2WI and DWI are mutually complementary. For LNs detection tasks, the performance of the auto-LNDS model with two channels of DWI is better than that with one channel of DWI in external testing dataset, which indicates that this combination mode is more robust.
In addition, as shown in Table 3, our auto-LNDS model obtained acceptable LNs detection results both in the internal and external testing datasets. More meaningfully, as listed in Table 4, the performance of the auto-LNDS model for external datasets from other three hospitals with different MR parameters also achieved good results, which confirmed the generalization of this model. As shown in Tables 4 and 5, the performance of this auto-LNDS model was much higher than those of the junior radiologists with less than ten-years experience (p < 0.05, t-test) in both of the internal and external testing datasets, which means this auto-LNDS method could significantly improve the LNs detection ability of junior and inexperienced radiologists, and could even directly supply LNs maps to surgeons for intraoperative reference as shown in Fig. 5. Meanwhile, we found inter-observer variability among the radiologists, which might be attributed to subjectivity, fatigue, or degree and experience, but the auto-LNDS model can minimize these differences. So, we believe that this auto-LNDS model could help to improve the accuracy and shorten the time of the radiologists for LNs detection. We consider the above is the most important contribution of this research. Finally, as shown in Fig. 4, we believe that we can use this auto-LNDS model to automatically detect LNs, including lateral LNs, and it can be predicted that our auto-LNDS model will provide more favourable help for lateral LNs detection and the decision of LLND in the future.
For RC N-staging, after detection and segmentation of the LNs, the next step is to assess the LNs for metastatic involvement. Recently, a deep learning based automated diagnosis model for LNs was reported [37], in which the reference standard for metastatic LNs was made by the subjective impression of radiologists based on imaging criteria (ie, short-diameter ≥ 5 mm, indistinct borders, irregular morphology, or high signal intensity on DWI images). Whereas, without direct LNs mapping to pathology results, the true metastatic status of each LN is still uncertain, and these subjective reference standards had been proved to be unsufficient to be used as the ground truths [38,39]. Therefore, in this study, we did not further distinguish between benign and malignant LNs. However, our study has taken an important first step towardsautomatic nodal staging, and in the future assessment using carefully matched one-to-one MR-pathological confirmed datasets to prompt the final step of identifying malignant LNs will be necessary for the final step.
We acknowledge limitations to our research. Firstly, our dataset size is smaller than the natural detection task dataset, which could be a reason that errors are made in this automatic system. In this study, both the training and the internal testing datasets were all generated by the same MR vendor from one medical center, which contained limited variances. However, the external testing dataset was collected from different medical centres. This may account in part for the better results acquired from the internal testing dataset, while the results acquired from the external testing dataset are decreased. In the future, extending the training dataset to multivendor and multicentre platforms may further promote the performance of the auto-LNDS model. Secondly, in this study, there are still some false positive and false negative results, and the reasons may be related to the following factors: insufficient image registration due to DWI image distortion and respiratory movement, some overlap between the LN and vessel or small intestinal wall due to partial volume effect, and not included the dynamic contrast-enhance MRI (DCE-MRI) sequences in the datasets. As we know, some LNs show isosignal intensity on high b-value DWI, and may be missed by the auto-LNDS as the lateral LN shown in the fourth row in Fig. 4. The thin-layer DCE-MRI was not included in the dataset in this study, although it is an effective method to further observe the process of LNs enhancement, which may be helpful for distinguishing the vessels and LNs and further identifying the benign and the malignant LNs. Meanwhile, it would be accurate if one-to-one MR-surgical pathological LN confirmation could be acquired, but it is really difficult in clinical practice. In this study we use the common opinion of three senior radiologists to establish the ground truth. In the future, inviting more reputable senior radiologists from well-known clinical centres to join the study may help to obtain more representative results. To evaluate the effectiveness of auto-LNDS, we compared its results with those of four junior radiologists. Although the four radiologists can not adequately represent the general level of all junior radiologists under ten years experience, all of them come from the first-rate hospitals specialized on gastro-intestinal disease in China, so we suppose that their ability will not be lower than the average of all junior radiologists. The low sensitivity and PPV of their results may be related to the fact that, in order to save time, they neglected part of the small (3–5 mm) LNs and oblong LNs because they believe that these LNs are less likely to be malignant, or some small LNs are too small to be noticed.In the future, to invite more junior radiologists to participate in this test may give better representation. In this study, the Mask R-CNN we used is a 2D network, and most of the LNs (diameter, 3–6 mm) appeared on only one slice (slice thickness of T2WI and DWI: 3–6 mm), which means axis images can cover most of the information of LNs, and so the 2D network should be adequate. However, those LNs with relatively large sizeare likely to be shown on two or more adjacent images, and a 3D network may be expected to fully display the overall shape of them. In addition, the performance of LN segmentation was acceptable but perhaps suboptimal. This likely relates to the inclusion of very small LNs (often less than 5 pixels per image), which will continues to pose a challenge.
In conclusion, based on Mask R-CNN, we developed an auto-LNDS modeland evaluated it both on the internal and external testing datasets, which shows this deep-learning auto-LNDS model can accurately detect and segment LNs on mpMRI with relatively high performance compared with the junior radiologists and existing studies. So we believe that this auto-LNDS model could help to quickly detect and segment LNs, improve clinical efficiency, and minimize the differences among the radiologists with different experiences.
Acknowledgments
Author contributions
Xin Gao and Xiaochun Meng are co-corresponding authors. Xin Gao supervised the model designing, building and manuscript editing. Xiaochun Meng supervised the study designing, data acquisition, LNs annotation and manuscript editing.
Xingyu Zhao made important contributions to the model designing, analysis and manuscript drafting. Peiyi Xie conducted data acquisition, collection and LNs annotation. Mengmeng Wang made contribution of image processing and model building. Wenru Li made contribution in data acquisition and LNs annotation. Perry J. Pickhardt made contributions in manuscript drafting. Wei Xia made contribution to the image processing and manuscript editing. Fei Xiong, Rui Zhang and Yao Xie made contribute in the data acquisition and LNs annotation. Junming Jian and Honglin Bai made statistical analysis. Caifang Ni and Jinhui Gu performed patient samples and LNs annotation. Tao Yu performed patient samples. Yuguo Tang made statistical analysis.
Funding sources
This work was supported by National Natural Science Foundation of China [grant number: 81871439, 61801474]; Key Research and Development Program of Jiangsu [grant number: BE2017671]; Suzhou science and technology plan project [grant number: SYG201908]; CAS-VPST Slik Road Science Fund 2018 [grant number: GJHZ1857]; Science and Technology Plan Projects of Tianjin [grant number: 19YDYGHZ00030]
Acknowledgements
The funding source had no involvement in study design, analysis, writing of the report, or in the decision to submit the paper for publication. The corresponding authors confirm to have full access to all the data and have final responsibility for the decision to submit for publication. The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102780.
Contributor Information
Xin Gao, Email: xingaosam@yahoo.com, gaox@sibet.ac.cn.
Xiaochun Meng, Email: mengxch3@mail.sysu.edu.cn, mengxch1972@163.com.
Appendix. Supplementary materials
References
- 1.Amin M.B., Edge S.B. Springer; 2017. AJCC cancer staging manual. [Google Scholar]
- 2.Harisinghani M.G., Weissleder R. Sensitive, Noninvasive Detection of Lymph Node Metastases. PLoS Med. 2004;1:e66. doi: 10.1371/journal.pmed.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T.H., Jeong S.-.Y., Choi D.H., Kim D.Y., Jung K.H., Moon S.H. Lateral lymph node metastasis is a major cause of Locoregional recurrence in rectal cancer treated with preoperative chemoradiotherapy and curative resection. Ann Surg Oncol. 2008;15:729–737. doi: 10.1245/s10434-007-9696-x. [DOI] [PubMed] [Google Scholar]
- 4.Ishihara S., Kawai K., Tanaka T., Hata K., Nozawa H. Correlations between the sizes of lateral pelvic lymph nodes and metastases in rectal cancer patients treated with preoperative chemoradiotherapy. ANZ J Surg. 2018;88:1306–1310. doi: 10.1111/ans.14717. [DOI] [PubMed] [Google Scholar]
- 5.Ogura A., Konishi T., Cunningham C., Garcia-Aguilar J., Iversen H., Toda S. Neoadjuvant (Chemo)radiotherapy with total mesorectal excision only is not sufficient to prevent lateral local recurrence in enlarged nodes: results of the multicenter lateral node study of patients with Low cT3/4 Rectal Cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2019;37:33–43. doi: 10.1200/JCO.18.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiou P., Tan E., Gouvas N., Antoniou A., Brown G., Nicholls R.J. Extended lymphadenectomy versus conventional surgery for rectal cancer: a meta-analysis. Lancet Oncol. 2009;10:1053–1062. doi: 10.1016/S1470-2045(09)70224-4. [DOI] [PubMed] [Google Scholar]
- 7.Arii K., Takifuji K., Yokoyama S., Matsuda K., Higashiguchi T., Tominaga T. Preoperative evaluation of pelvic lateral lymph node of patients with lower rectal cancer: comparison study of MR imaging and CT in 53 patients. Langenbecks Arch Surg. 2006;391:449–454. doi: 10.1007/s00423-006-0066-0. [DOI] [PubMed] [Google Scholar]
- 8.Ishibe A., Ota M., Watanabe J., Suwa Y., Suzuki S., Kanazawa A. Prediction of lateral pelvic lymph-node metastasis in low rectal cancer by magnetic resonance imaging. World J Surg. 2016;40:995–1001. doi: 10.1007/s00268-015-3299-7. [DOI] [PubMed] [Google Scholar]
- 9.Gröne J., Loch F.N., Taupitz M., Schmidt C., Kreis M.E. Accuracy of various lymph node staging criteria in rectal cancer with magnetic resonance imaging. J Gastrointest Surg Off J Soc Surg Aliment Tract. 2018;22:146–153. doi: 10.1007/s11605-017-3568-x. [DOI] [PubMed] [Google Scholar]
- 10.Langman G., Patel A., Bowley D.M. Size and distribution of lymph nodes in rectal cancer resection specimens. Dis Colon Rectum. 2015;58:406–414. doi: 10.1097/DCR.0000000000000321. [DOI] [PubMed] [Google Scholar]
- 11.Feuerstein M., Deguchi D., Kitasaka T., Iwano S., Imaizumi K., Hasegawa Y. nternational Society for Optics and Photonics. 2009. Automatic mediastinal lymph node detection in chest CT. Med. Imaging 2009 Comput.-Aided Diagn; p. 72600V. [DOI] [Google Scholar]
- 12.Kitasaka T., Tsujimura Y., Nakamura Y., Mori K., Suenaga Y., Ito M. Automated Extraction of Lymph Nodes from 3-D Abdominal CT Images Using 3-D Minimum Directional Difference Filter. In: Ayache N., Ourselin S., Maeder A., editors. Vol. 4792. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. pp. 336–343. (Med. image comput. comput.-assist. interv. – MICCAI 2007). [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Hoffman J., Zhao J., Yao J., Lu L., Kim L. Mediastinal lymph node detection and station mapping on chest CT using spatial priors and random forest: mediastinal lymph node detection and station mapping. Med Phys. 2016;43:4362–4374. doi: 10.1118/1.4954009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feulner J., Zhou S.K., Hammon M., Hornegger J., Comaniciu D. Lymph node detection and segmentation in chest CT data using discriminative learning and a spatial prior. Med Image Anal. 2012;17 doi: 10.1016/j.media.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Cherry K.M., Wang S., Turkbey E.B., Summers R.M. In: Aylward S., Hadjiiski L.M., editors. 2014. p. 90351G. San Diego, California, USA. [DOI] [Google Scholar]
- 16.Barbu A., Suehling M., Xu Xun, Liu D., Zhou S.K., Comaniciu D. Automatic detection and segmentation of lymph nodes from CT data. IEEE Trans Med Imaging. 2012;31:240–250. doi: 10.1109/TMI.2011.2168234. [DOI] [PubMed] [Google Scholar]
- 17.Yan J., Zhuang T., Zhao B., Schwartz L. Lymph node segmentation from CT images using fast marching method. Comput Med Imaging Graph Off J Comput Med Imaging Soc. 2004;28:33–38. doi: 10.1016/j.compmedimag.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. University of Iowa; MS: 2010. Graph-based segmentation of lymph nodes in CT data. [DOI] [Google Scholar]
- 19.Dornheim J., Seim H., Preim B., Hertel I., Strauss G. Segmentation of neck lymph nodes in CT datasets with stable 3D mass-spring models. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv. 2006;9:904–911. doi: 10.1007/11866763_111. [DOI] [PubMed] [Google Scholar]
- 20.Unal G., Slabaugh G., Ess A., Yezzi A., Fang T., Tyan J. Proceedings of the IEEE International Conference on Image Processing. IEEE; Atlanta, GA: 2006. Semi-Automatic Lymph Node Segmentation in LN-MRI; pp. 77–80. [DOI] [Google Scholar]
- 21.Debats O.A., Litjens G.J.S., Barentsz J.O., Karssemeijer N., Huisman H.J. Automated 3-dimensional segmentation of pelvic lymph nodes in magnetic resonance images. Med Phys. 2011;38:6178–6187. doi: 10.1118/1.3654162. [DOI] [PubMed] [Google Scholar]
- 22.Heijnen L.A., Lambregts D.M.J., Mondal D., Martens M.H., Riedl R.G., Beets G.L. Diffusion-weighted MR imaging in primary rectal cancer staging demonstrates but does not characterise lymph nodes. Eur Radiol. 2013;23:3354–3360. doi: 10.1007/s00330-013-2952-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W., Li R., Deng H., Wang L., Lin W., Ji S. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L., Xia W., Zhang B., Qiu B., Gao X. MSFCN-multiple supervised fully convolutional networks for the osteosarcoma segmentation of CT images. Comput Methods Programs Biomed. 2017;143:67–74. doi: 10.1016/j.cmpb.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Jian J., Xiong F., Xia W., Zhang R., Gu J., Wu X. Fully convolutional networks (FCNs)-based segmentation method for colorectal tumors on T2-weighted magnetic resonance images. Australas Phys Eng Sci Med. 2018;41:393–401. doi: 10.1007/s13246-018-0636-9. [DOI] [PubMed] [Google Scholar]
- 26.Nam J.G., Park S., Hwang E.J., Lee J.H., Jin K.-.N., Lim K.Y. Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology. 2019;290:218–228. doi: 10.1148/radiol.2018180237. [DOI] [PubMed] [Google Scholar]
- 27.Huang X., Shan J., Vaidya V. Proceedings of the IEEE 14th International Symposium on Biomedical Imaging ISBI. IEEE; Melbourne, Australia: 2017. Lung nodule detection in CT using 3D convolutional neural networks; pp. 379–383. [DOI] [Google Scholar]
- 28.He K., Gkioxari G., Dollár P., Girshick R. Mask R-CNN. Proc. IEEE Int. Conf. Comput. Vis. 2017:2961–2969. doi: 10.1109/ICCV.2017.322. [DOI] [Google Scholar]
- 29.Meng X., Xia W., Xie P., Zhang R., Li W., Wang M. Preoperative radiomic signature based on multiparametric magnetic resonance imaging for noninvasive evaluation of biological characteristics in rectal cancer. Eur Radiol. 2019;29:3200–3209. doi: 10.1007/s00330-018-5763-x. [DOI] [PubMed] [Google Scholar]
- 30.Dao T., Gu A., Ratner AJ., Smith V., De Sa C., Ré C. A kernel theory of modern data augmentation. Proc Mach Learn Res. 2019;97:1528–1537. [PMC free article] [PubMed] [Google Scholar]
- 31.Lin T-Y, Dollár P., Girshick R., He K., Hariharan B., Belongie S. Feature pyramid networks for object detection. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017:2117–2125. [Google Scholar]
- 32.Ren S., He K., Girshick R., Sun J., Faster R-CNN. Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell. 2017:1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 33.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016:770–778. [Google Scholar]
- 34.Girshick R., Donahue J., Darrell T., Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2014:580–587. [Google Scholar]
- 35.Girshick R. Fast R-CNN. Proc. IEEE Int. Conf. Comput. Vis. 2015:1440–1448. [Google Scholar]
- 36.Kingma DP., BaAdam J. A Method for Stochastic Optimization. ArXiv14126980 Cs; 2014. [Google Scholar]
- 37.Lu Y., Yu Q., Gao Y., Zhou Y., Liu G., Dong Q. Identification of metastatic lymph nodes in MR imaging with faster region-based convolutional neural network. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-18-0494. canres.0494.2018. [DOI] [PubMed] [Google Scholar]
- 38.Muthusamy V.R., Chang K.J. Optimal methods for staging rectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2007;13:6877s–6884s. doi: 10.1158/1078-0432.CCR-07-1137. [DOI] [PubMed] [Google Scholar]
- 39.Al-Sukhni E., Messenger D.E., Charles Victor J., McLeod R.S., Kennedy E.D. Do MRI reports contain adequate preoperative staging information for end users to make appropriate treatment decisions for rectal cancer? Ann Surg Oncol. 2013;20:1148–1155. doi: 10.1245/s10434-012-2738-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.