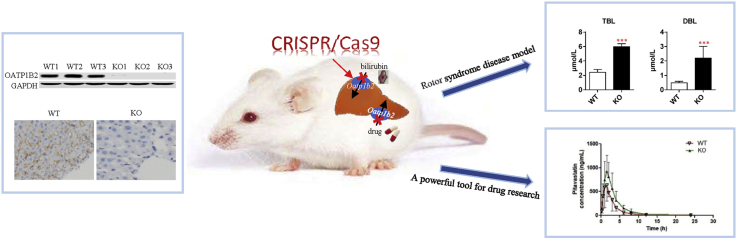
Abstract
Organic anion transporting polypeptide 1B1 and 1B3 (OATP1B1/3) as important uptake transporters play a fundamental role in the transportation of exogenous drugs and endogenous substances into cells. Rat OATP1B2, encoded by the Slco1b2 gene, is homologous to human OATP1B1/3. Although OATP1B1/3 is very important, few animal models can be used to study its properties. In this report, we successfully constructed the Slco1b2 knockout (KO) rat model via using the CRISPR/Cas9 technology for the first time. The novel rat model showed the absence of OATP1B2 protein expression, with no off-target effects as well as compensatory regulation of other transporters. Further pharmacokinetic study of pitavastatin, a typical substrate of OATP1B2, confirmed the OATP1B2 function was absent. Since bilirubin and bile acids are the substrates of OATP1B2, the contents of total bilirubin, direct bilirubin, indirect bilirubin, and total bile acids in serum are significantly higher in Slco1b2 KO rats than the data of wild-type rats. These results are consistent with the symptoms caused by the absence of OATP1B1/3 in Rotor syndrome. Therefore, this rat model is not only a powerful tool for the study of OATP1B2-mediated drug transportation, but also a good disease model to study hyperbilirubinemia-related diseases.
Key Words: OATP1B1/3, OATP1B2, CRISPR/Cas9, Transporter, Rat model
Abbreviations: ADRs, adverse drug reactions; ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; A/G, albumin/globulin ratio; AST, aspartate aminotransferase; AUC, the area under the time–plasma concentration curve; BUN, blood urea nitrogen; Chr, chromosome; CL/F, clearance/bioavailability; Cmax, peak concentration; CR, reatinine; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, mature CRISPR RNA; DAB, 3,3′-diaminobenzidine; DBL, direct bilirubin; DDI, drug–drug interaction; DMSO, dimethyl sulfoxide; FDA, the U.S. Food and Drug Administration; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GLB, globulin; GLU, glucose; HE, haemotoxylin and eosin; HCG, human chorionic gonadotropin; HDL-C, high density lipoprotein cholesterol; HMG, hydroxymethylglutaryl; HRP, horseradish peroxidase; HZ, heterozygous; IBIL, indirect bilirubin; IS, internal standard solution; KO, knockout; LDL-C, low density lipoprotein cholesterol; MC, methylcellulose; MRT, mean residence time; NC, nitrocellulose; OATPs, organic anion transporting polypeptides; OATP1B1/3, organic anion transporting polypeptide 1B1 and 1B3; PAM, protospacer adjacent motif; PMSG, pregnant mare serum gonadotropin; p.o., peroral; R-GT, γ-glutamyltranspeptidase; SD, Sprague–Dawley; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; sgRNA, single guide RNA; SLC, solute carrier; SNPs, single nucleotide polymorphisms; T7E I, T7 endonuclease I; TALEN, transcription activator-like effector nuclease; TBA, total bile acid; TBL, total bilirubin; TBST, Tris-buffered saline Tween 20; T-CH, total cholesterol; TG, triglyceride; Tmax, peak time; TP, total protein; UA, uric acid; Ugt1a1, UDP glucuronosyltransferase family 1 member A1; Vd/F, the apparent volume of distribution/bioavailability; WT, wild type; ZFN, zinc-finger nucleases
Graphical abstract
The Slco1b2 KO rat model was successfully constructed by CRISPR/Cas9. This rat model is not only a powerful tool for the study of Oatp1b2-mediated drug transportation, but also a good disease model to study hyperbilirubinemia-related diseases.
1. Introduction
Organic anion transporting polypeptides (OATPs), a family of 12 transmembrane domain glycoproteins, belong to the solute carrier (SLC) family, and transport important endogenous and exogenous substrates into cells1. The human OATP superfamily consists of 11 transporters subdivided into six families (OATP1–6), in which the OATP1B plays a crucial role2. The human OATP1B subfamily contains two members, OATP1B1 and OATP1B3, which are encoded by the SLCO1B1 and SLCO1B3 genes, respectively. The two proteins are not only mainly expressed in the sinusoidal membrane of the liver, but also are found in some tumor tissues3, 4, 5. The substrates of OATP1B1 and OATP1B3 cover a wide range, including endogenous substances such as bilirubin, bile acids and steroid hormones, as well as clinically used drugs like angiotensin II receptor blockers (sartans), hydroxymethylglutaryl (HMG)-CoA reductase inhibitors (statins), anticancer drugs (paclitaxel) and so on6, 7, 8.
Since most drugs are metabolized by hepatocytes, the hepatic uptake rate mediated by OATP1B1/3 is a determinant of total clearance9. Inhibition of the activities of OATP1B1/3 can alter the systemic drug exposure and lead to serious adverse reactions10. Several clinical drug–drug interaction (DDI) cases of cyclosporine combined with statins were reported, as cyclosporine inhibited the activities of OATP1B1/3 and resulted in severe muscular toxicity of statins11. The U.S. Food and Drug Administration (FDA) guidelines for “in vitro metabolism- and transporter-mediated drug–drug interaction studies” also point out that DDI potentials mediated by OATP1B1 and OATP1B3 must be considered in the development of novel drugs12. On the other hand, the activities of OATP1B1 and OATP1B3 are closely related to human health. For example, the biotransformation of bilirubin, the substrate of OATP1B1/3, depends on the function of OATP1B1/3 in vivo2. If the function of two transporters is decreased or lost, the concentration of bilirubin in the blood will increase and lead to hyperbilirubinemia13. Rotor syndrome is an autosomal recessive inheritance of hyperbilirubinemia, caused an increase in conjugated bilirubin, which is due to the loss of activity of OATP1B1/314.
Human OATP1B1 and OATP1B3 also play significant roles in the progression of tumors like prostate cancer. One study showed that the uptake capacity of androgen in prostate cancer cells correlated with the activities of OATP1B1 and OATP1B3, and overexpressing OATP1B1 and OATP1B3 could increase cell viability15. In clinical surveillance, patients with the loss-of-function SLCO1B3 334GG/699AA haplotype showed longer median survival and improved 10-year survival rate16. At the same time, SLCO1B3 may determine the progression time of the patients receiving androgen deprivation therapy for prostate cancer17. Therefore, OATP1B3 is a potential target for prostate cancer treatment, but there are no sufficient relevant studies at present. Thus, it is important to develop useful methods to study the functions of OATP1B1 and OATP1B3 in pathology and clinical medicine18.
Knockout (KO) animal models lacking the key transporters are specific and fit for preclinical studies, especially pharmacological research19. OATP1B2 (also named SLCO1B2 or LST-1), encoded by the Slco1b2 gene, has been identified in mice and rats, which is the closest ortholog of human OATP1B1 and OATP1B320. The Slco1b2 KO mouse model was generated in 2008 by homologous recombination21,22. In pharmacokinetic experiments, however, rats have many advantages that make them superior to mice, such as enough blood volume, strong tolerance and large sample size. In particular, rats are the preferred animal model to carry out hyperbilirubinemia research23,24. Therefore, we want to generate a Slco1b2 KO rat model.
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) technology as the third generation of gene editing technology after zinc-finger nucleases (ZFN) and transcription activator-like effector nuclease (TALEN), presents distinct advantages in editing multiple genes simultaneously in the same cell25. Its core component is Cas9 protein, which combines mature CRISPR RNA (crRNA) to recognize and cleave exogenous nucleic acid molecules26,27. The cleavage of the target gene will cause double-strand breaks, and then initiate the repair method of non-homologous terminal conjugation, which will result in deletion, replacement or insertion mutations of cleavage sites, thus complete gene editing28. Currently, CRISPR/Cas9 system has been identified as one breakthrough technology in the biomedical research29.
In this study, we knocked out the Slco1b2 gene in rats by using the CRISPR/Cas9 system, and successfully obtained the Slco1b2 KO rat model. This model can be applied to pharmacokinetics studies. At the same time, the model can be used as a disease model of hyperbilirubinemia–Rotor syndrome.
2. Materials and methods
2.1. Animals
Male and female Sprague–Dawley (SD) rats were purchased from National Rodent Laboratory Animal Resources (Shanghai, China). Animals were fed in a barrier free of specific pathogens and allowed to eat and drink freely. The barrier was a temperature and humidity controlled environment with 12-h light/dark cycles. All the animal experiment protocols in this study were approved by the Ethics Committee on Animal Experimentation of East China Normal University (Shanghai, China).
2.2. Chemicals and reagents
The oligos (60 bp, containing Slco1b2 knock out target-site) and all primers for PCR/Q-PCR were synthesized by Biosune Biotechnology Co., Ltd. (Shanghai, China). KOD-plus-Neo polymerase was purchased from Toyobo (Osaka, Japan). SYBR Premix Ex Taq and Prime Script RT Reagent Kit were bought from Takara (Dalian, China). T7 endonuclease I (T7E I) was purchased from New England Biolabs (Ipswich, MA, USA). Phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) was purchased from Amresco (Cleveland, OH, USA). Bicinchoninic acid kit was purchased from Thermo Scientific (Waltham, MA, USA). The agarose gel recovery kit was bought from Generay Biotech Co., Ltd. (Shanghai, China). A primary antibody for OATP1B2 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was purchased from Abcam (Cambridge, UK). The fluorescence-conjugated secondary antibody to rabbit IgG and mouse IgG were bought from Cell Signaling Technology (Boston, MA, USA). Pitavastatin was obtained from MedChemExpress (Monmouth Junction, NJ, USA).
2.3. Target-site selection
The Slco1b2 gene sequence of Rattus norvegicus (Norway rat) was obtained from NCBI (https://www.ncbi.nlm.nih.gov/pmc/), and exon regions were sequentially labeled (from 1st to 15th). In order to completely knock out the target gene, the selected target is at the first exon of the rat Slco1b2 gene. The first exon sequence was input into the online website (https://benchling.com/) to obtain an 18 bp target sequence followed by a protospacer adjacent motif (PAM) site (5′-NGG-3′) in the 3′ end. The target sequence is 5′-GCAAACAAGGTTCTGCGA-3′. PAM site is TGG.
2.4. In vitro single guide RNA (sgRNA) template synthesis and transcription
The 60 bp oligo fragment containing the T7 promoter and 18 bp target sequence (5′-GATCACTAATACGACTCACTATAGGGCAAACAAGGTTCTGCGAGTTTTAGAGCTAGAAAT-3′) was synthesized by Biosune Biotechnology Co., Ltd. (Shanghai, China). Using this product, the sgRNA double-stranded template was synthesized by overlapping PCR using KOD-plus-neo polymerase, and then transcribed by the T7 transcription kit. The sgRNA of Slco1b2 gene was extracted and purified by phenol/chloroform method, and then stored in −80 °C refrigerator for later experimentation.
2.5. Embryo microinjection
First, healthy adult male rats (over 8 weeks old) were sterilized and caged with healthy adult female rats (over 8 weeks old). The female rats with vaginal suppositories were used as pseudo-pregnant rats for subsequent experiments. Then, the normal adult female rats were injected with pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (HCG) for superovulation and then mated and conceived. Female rats that had been mated successfully were sacrificed, and the embryos were taken out and cultured in culture medium for 2 h in a 37 °C, 5% CO2 incubator. Finally, the sgRNA template (25 ng/μL) and Cas9 mRNA (50 ng/μL) were co-injected into the cytoplasm of the embryos by microinjection technique. The concentration ratio of sgRNA to Cas9 mRNA was 1:2. After several hours of culture, the surviving embryos were transplanted into the fallopian tube of the pseudo-pregnant rats. The pseudo-pregnant rats were returned to the cage for normal feeding.
2.6. Genotype identification
2.6.1. Genotype identification of F0 offspring rats
The newborn rats were cut the toes and randomly numbered when they grew to a week or so. The genomic DNA was extracted by phenol/chloroform method, and specifically amplified 600 bp sequences flanking the target site by the primers of Slco1b2 (Table 1) with Easy-Taq DNA polymerase. Gradient annealing of 10 μL PCR product was carried out under the condition that 95 °C was maintained for 5 min, and each cycle decreased by 1 °C for 60 cycles, each cycle lasted for 30 s. The annealed products were digested in 37 °C incubator for 30 min using 0.1 μL T7 endonuclease I (T7E I). T7E I could recognize and cleave the mismatched bases with insertion or deletion mutations, and then multiple bands can be seen when electrophoresed on a 1.5% agarose gel. The positive PCR products (showed multiple bands) were recovered and purified by the agarose gel recovery kit and ligated into pMD18-T vector according to the TA cloning kit, and then the clones were sequenced to determine the accurate gene editing results. Although the clones tested can only reflect part of the genetic editing in the rat genome, we mainly use them as a reference to select the rats needed for subsequent experiments. The mutation is available only if there were insertion or deletion of non-three-integer multiple bases occurred on the exon. In addition, in order to simplify future sequence analysis, gene editing with replacement mutations or confusion of sequencing maps was discarded.
Table 1.
Primers accurate information for target and off-target sites.
| Primer name | Primer sequence (5′–3′) | Product length (bp) | Temperature (°C) |
|---|---|---|---|
| Slco1b2-genotyping-S | CAGGGTAGACGACCATT | 631 | 56 |
| Slco1b2-genotyping-A | CTTTCTGCCAGGTGTTTT | ||
| Slco1b2-off 1-S | TTGACCATCACAACGGAATC | 475 | 60 |
| Slco1b2-off 1-A | CTGGGTGAATGAACTACTTGC | ||
| Slco1b2-off 2-S | TGTGGGGCCAGAGGACAG | 445 | 62 |
| Slco1b2-off 2-A | AGTTGGATGAAGATGAGGACTG | ||
| Slco1b2-off 3-S | GGGGAACCAAGATGTGAA | 410 | 60 |
| Slco1b2-off 3-A | GAGCAAATAAGAAAGTCAGGAA |
2.6.2. Genotype identification of F1 and F2 offspring rats
The F0 offspring, carrying the available mutations, were caged with wild type (WT) rats and the progeny obtained were F1 generation. The genomic DNA of F1 rats was sequenced and analyzed. The healthy adult F1 male and female rats with the same mutation were selected for mating and breeding to obtain F2 generation. The genomic DNA of F2 rats were performed by PCR amplified sequences flanking the target site, and data was analyzed by DNAman (Lynnon Biosoft, San Ramon, CA, USA).
2.7. Off-target site detection
The Slco1b2 gene target sequence was input into the online target prediction website (https://benchling.com/) to obtain potential off-target sites. Three sites (Table 2), with a relative high probability of off-target (score>2.0) were selected, and 300 bp sequences of upstream and downstream potential sites were found in NCBI (https://www.ncbi.nlm.nih.gov/pmc/), respectively. Based on the above sequences, primers (Table 1) for PCR were designed using Primer Premier. Three genome of KO rats were randomly selected to amplify the potential off-target sites using the primers in Table 1 and Easy-Taq DNA polymerase. The products were digested with T7E I and then performed by agarose gel electrophoresis (As described in Section 2.6.1). If there is off-target effect, two bands appear after electrophoresis.
Table 2.
Details for potential off-target sites examined.
| Match name | Location | Spacer + PAM | Off-target score |
|---|---|---|---|
| Slco1b2-sgRNA | Chr4-175819244 | GCAAACAAGGTTCTGCGATGG | 100.00 |
| Slco1b2-off-1 | Chr1-13544758 | GCAAACAAGGTTCTGCGGTAG | 19.60 |
| Slco1b2-off-2 | Chr14-3156777 | GCAAACAGAGTTCTGCGAAGG | 3.19 |
| Slco1b2-off-3 | Chr7-114058847 | GGAGACAGGGTTCTGCGAGGG | 2.67 |
Chr, chromosome.
2.8. Detection of protein OATP1B2 expression
Since OATP1B2 is almost specifically expressed in the liver except in some tumor tissues, the expression of OATP1B2 in the livers of WT and KO rats was detected by Western blot and immunohistochemistry, respectively. Rats (8 weeks old) were killed by CO2 asphyxiation and the livers were carefully separated and frozen in −80 °C for further processing.
2.8.1. Western blot analysis
Every unit of liver (0.1 g) was added with 400 μL lysate, containing RIPA, protease inhibitor, phosphatase inhibitor and phenylmethanesulfonyl fluoride. The sample was homogenized by Automatic Sample Rapid Grinding Instrument (JXFSTPRP-24, shanghai, China) and pyrolyzed at 4 °C for 2 h to extract total protein. Protein concentration was detected by using the Bicinchoninic acid Kit. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 60 μg protein sample per well, and then the protein was transferred to a nitrocellulose (NC) membrane and blocked with 5% skim milk powder in Tris-buffered saline Tween 20 (TBST) buffer for 1 h. After incubation of primary antibody, mouse anti-OATP1B2 (sc-376904, diluted 1:200) and rabbit anti-GAPDH (AB181602, diluted 1:10,000) at 4 °C overnight, the anti-mouse/rabbit secondary antibody (diluted 1:10,000) were incubated for 1 h in the dark, and the blots were scanned by Odyssey imager system (LI–COR, Lincoln, NE, USA). The GAPDH was used as a loading control.
2.8.2. Immunohistochemical analysis
The liver tissue was carefully cut into pieces (1 cm × 1 cm × 0.5 cm), soaked in 4% paraformaldehyde, and completed the preparation according to the standard paraffin section immunohistochemical process. The primary antibody, mouse anti-OATP1B2 (sc-376904, diluted 1:100) was incubated at 4 °C overnight, and the secondary antibody used horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (GB23301, diluted 1:200) was incubated at room temperature for 50 min. Color was developed using the immunohistochemistry kit 3,3-diaminobenzidine (DAB) chromogenic reagent, and the nucleus was counterstained with hematoxylin.
2.9. Detection of physiological condition
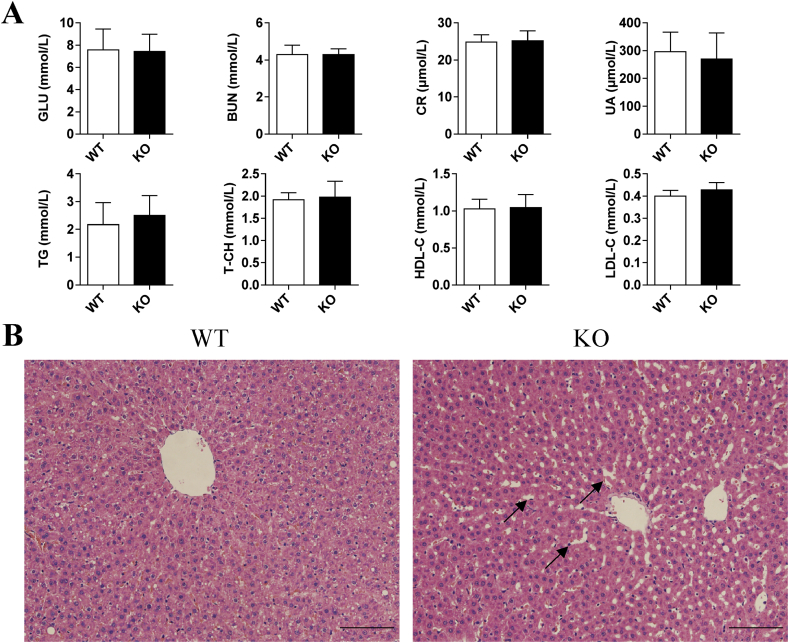
In order to detect whether the physiological condition changed in the Slco1b2 KO rat, blood samples from WT and KO female and male rats (4-week-old and 8-week-old) were taken by the tail vein, and the serum physiological indexes were detected by ADICON Clinical Laboratories (Shanghai, China). Liver function indicators include total protein (TP), albumin (ALB), globulin (GLB), albumin/globulin ratio (A/G), total bilirubin (TBL), direct bilirubin (DBL), Indirect bilirubin (IBIL), aspartate aminotransferase (AST), alanine aminotransferase (ALT), aspartate aminotransferase/alanine aminotransferase (AST/ALT), alkaline phosphatase(ALP), γ-glutamyltranspeptidase (R-GT) and total bile acid (TBA). Renal function indicators include blood urea nitrogen (BUN), creatinine (CR), uric acid (UA). Blood lipid indexes include triglyceride (TG), total cholesterol (T-CH), high density lipoprotein cholesterol (HDL-C) and low density lipoprotein cholesterol (LDL-C). Glucose (GLU), a commonly used indicator of physiological status, was also detected. In addition, Haemotoxylin and Eosin (HE) staining according to standard procedures was used to detect whether the liver was abnormal.
2.10. Detection of compensatory effects
Total RNA in the livers of WT and KO rats were extracted using the Trizol method according to the instructions. After measuring the concentration with nano-drop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), the RNA (total 2000 ng) was quantitatively reverse transcribed into cDNA using the Takara RT kit (RR036A). The primary efflux and absorption transporters were detected by real-time quantitative PCR was performed using Quant Studio 3 Real-Time PCR System (Thermo Fisher Scientific). The efflux transporters included Abcc1, Abcc2, Abcc3, Abcb1a, Abcb1b, Abcg2, Abcg5, Abcg8, and Abcb11, while the absorbing transporters included Slco1b2, Slco1a1, Slco2a1, Slco1a2, Slco2b1, Slc22a1, Slc22a3, Slc22a7, Slc22a8, and Slc10a1. Primers information is listed in the Supporting Information Table S1. The β-actin was set as the internal reference.
2.11. Pitavastatin pharmacokinetic studies in WT and KO rats
In order to determine whether the protein OATP1B2 lost its function in KO rats, pitavastatin, recommended by FDA, was used as the substrate of OATP1B2 to detect whether the pharmacokinetic behavior changed in 8-week-old KO male rats. Pitavastatin powder was dissolved with 5% dimethyl sulfoxide (DMSO) and 95% methylcellulose (MC) as solvents. The dosage was 5 mg/kg and the volume was 4 mL/kg, and the final concentration of pitavastatin was 1.25 mg/mL. The dosage was obtained according to the clinical dosage and the predicted experimental results30. After fasting for 12 h, pitavastatin was given to rats intragastrically. Blood samples were collected through tail vein at 0.17, 0.5, 1.0, 1.5, 2.0, 3.0, 4.0, 6.0, 8.0, 12.0, and 24.0 h after administration. Samples were centrifuged at 8000×g for 10 min, and the plasma was collected and stored at −20 °C for subsequent experiments.
2.12. Quantification of pitavastatin in plasma samples by LC–MS/MS
Before quantification, the plasma samples were thawed at room temperature and treated with liquid–liquid extraction. One hundred microliter of each plasma sample was added with 10 μL methanol, 10 μL of internal standard solution (IS, 100 ng/mL mebendazole in acetonitrile) and 800 μL of ethyl acetate. The mixture was mixed thoroughly and centrifuged at 14,000×g for 10 min. An aliquot of 600 μL upper organic layer was transferred to a new 1.5 mL tube afterwards, and dried with a gentle flow of hot nitrogen. The residue was then reconstituted with 100 μL methanol/water (1:1, v/v) and centrifuged at 14,000×g for 10 min. Finally 80 μL of supernatant was used for LC–MS/MS analysis.
The LC–MS/MS system consisted of an Agilent 1290 HPLC and a 6460 triple quadrupole mass spectrometer coupled with an Agilent Jet Stream ESI ion source (Agilent Technologies, Santa Clara, CA, USA). Chromatography separation was performed on a Zorbax Eclipse Plus C18 column (50 mm × 2.1 mm, 1.8 μm, Agilent Technologies), using a mobile phase system of water (A) and methanol (B) both containing 0.1% formic acid (v/v). The flow rate was 0.2 mL/min and the injection volume was 2 μL. The gradient elution program was used as follows: 0–0.5 min, 45% B; 0.5–3 min, 45%–95% B; 3–3.5 min, 95% B; 3.5–3.7 min, 95%–45% B; 3.7–5.5 min, 45% B. Pitavastatin and IS were both monitored in positive ESI mode, with the ion transition of 422.1 → 22.1I and 296.1 → 264.1, respectively.
2.13. Statistical data analysis
All data were presented as mean ± SD. All the graphs were plotted by GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA, USA). Statistical analysis between different groups was performed using the two-tailed t-test. The difference was considered to be statistically significant if P < 0.05. The pharmacokinetic parameters of pitavastatin were calculated by WinNonlin software version 5.2.1 (Pharsight Corporation, Mountain View, CA, USA) based on the non-compartmental model.
3. Results
3.1. Gene analysis of Slco1b2 KO rats
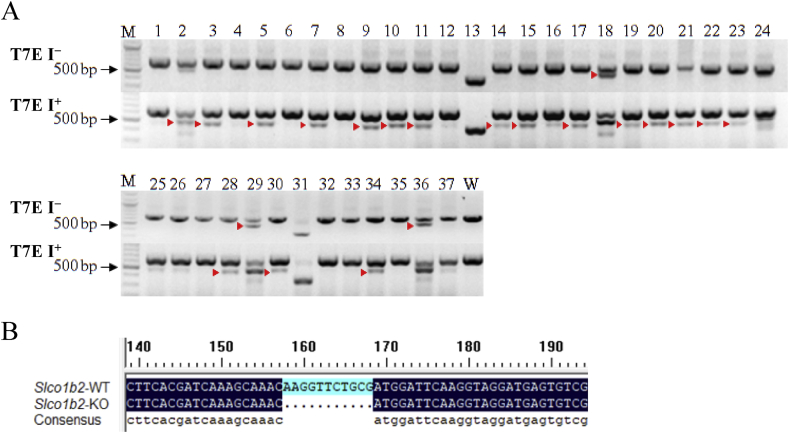
In the genetic identification of F0 generation rats, the T7E I assay and sequence analysis were used. As shown in Fig. 1A, in the absence of T7E I (T7E I−) cleavage, the bands of 13# and 31# rats were significantly lower than the WT, and 2#, 18#, 29#, and 36# showed obvious two bands, indicating that there may be a deletion of longer fragments. After sequencing analysis, it was confirmed that more than 100 bp base deletion occurred in the above rat genome. However, because of the occurrence of non-effective editing in 18# and 31#, and a replacement mutation occurred in 2#, 13#, 29#, and 36#, the above rats were abandoned. Sequence analysis of other genomes (3#, 5#, 7#, and etc.) revealed that 10#, 14#, 15#, 20#, 21#, 22#, and 30# contained effective editing. But in order to simplify the future workload, we randomly selected 10#, 15#, and 21#F0 rats for subsequent experiments. Part of the sequencing results are shown in the Supporting Information Table S2.
Figure 1.
Genotype identification of offspring. (A) The mutations in F0 generation were detected by T7E I digestion using PCR products amplified from F0 rat tail genomic DNA by Primer. T7E I−, before T7E I digestion. T7E I+, after T7E I digestion. " ", mutation band. WT, wild type as negative control. M, DNA molecular weight marker. (B) Gene sequence near the target in WT and Slco1b2 KO rats. "…", missing sequence.
", mutation band. WT, wild type as negative control. M, DNA molecular weight marker. (B) Gene sequence near the target in WT and Slco1b2 KO rats. "…", missing sequence.
The genomic DNA of F1 rats was sequenced and analyzed. The healthy adult F1 male and female rats with the same mutation were selected for mating and breeding to obtain F2 generation. The possible genotypes of the F2 generation are WT rats, homozygous KO rats, and heterozygous (HZ) rats. After specific amplification of the F2 generation rat genomic DNA, sequencing analysis was performed. As shown in Fig. 1B, individuals with a homozygous deletion of 11 bp were obtained.
3.2. Off-target analysis
Since CRISPR/Cas9 system allows one to three base mismatches when sgRNA paired with target, it may lead to the combination of sgRNA and non-target sequence, resulting in off-target effect. Therefore, it is necessary to detect the off target in gene-editing rats. As shown in Fig. 2, all the bands were single, bright and uniform in size, indicating that the Slco1b2 KO rats did not have off-target effects at the detected off-target sites.
Figure 2.
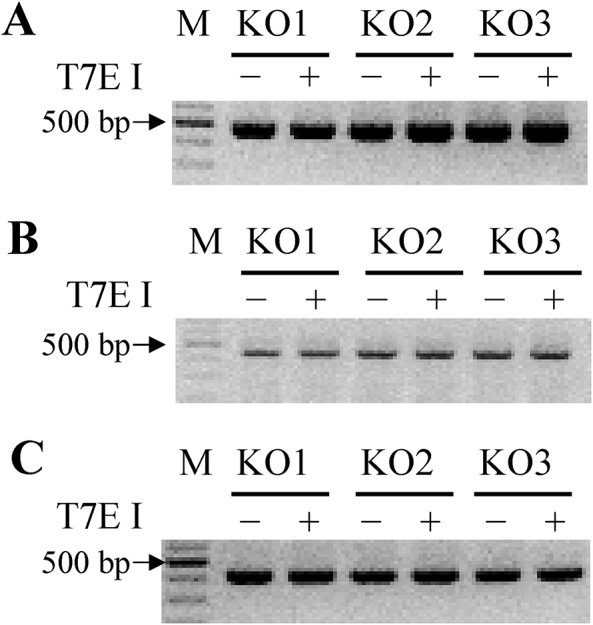
Off-target analysis in Slco1b2 KO rats. Three off target sites for Slco1b2 sgRNA were selected for T7E I enzyme cleavage. (A) Slco1b2-off target-1, (B) Slco1b2-off target-2, (C) Slco1b2-off target-3 analysis of KO rats (n = 3). T7E I−, before T7E I digestion; T7E I+, after T7E I digestion. M, DNA molecular weight marker.
3.3. Expression of OATP1B2 in WT and KO rats liver
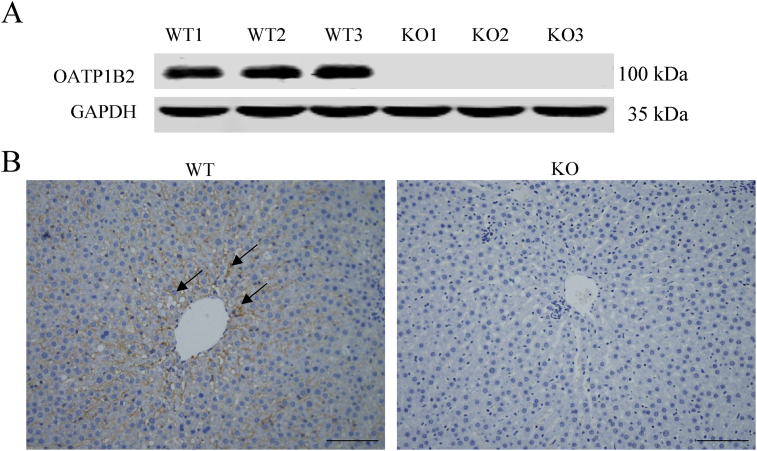
The expression of OATP1B2 in the liver of WT and KO rats was checked by Western blot and immunohistochemistry, respectively. As shown in Fig. 3A, the OATP1B2 protein was completely absent in the liver of KO rats. In Fig. 3B, in WT rats, OATP1B2 was highly expressed on the sinusoidal membrane of the liver (shown in brown), while there was almost no positive result in the liver of KO rats. These results confirmed that OATP1B2 protein was absent completely in KO rats, suggesting that the KO model was successfully constructed.
Figure 3.
Analysis of OATP1B2 protein expression in WT and KO rats. (A) Western blot analysis of OATP1B2 protein expression level in the livers of WT and KO rats (n = 3). The molecular weight of OATP1B2 is about 100 kDa. The molecular weight of GAPDH (the internal reference) is about 35 kDa. (B) Immunohistochemistry analysis of OATP1B2 protein expression in the livers of WT and KO rats. Positive results are shown in brown (↑). Scale bars, 200 μm in length.
3.4. Detection of physiological conditions of WT and KO rats
Since some important endogenous substances, such as bilirubin and bile acids, are substrates of OATP1B2, it is necessary to detect whether the physiological indexes of Slco1b2 KO rats change and whether the rats grow and breed normally. We detected the physiological conditions of WT and KO rats by serum analysis and HE staining.
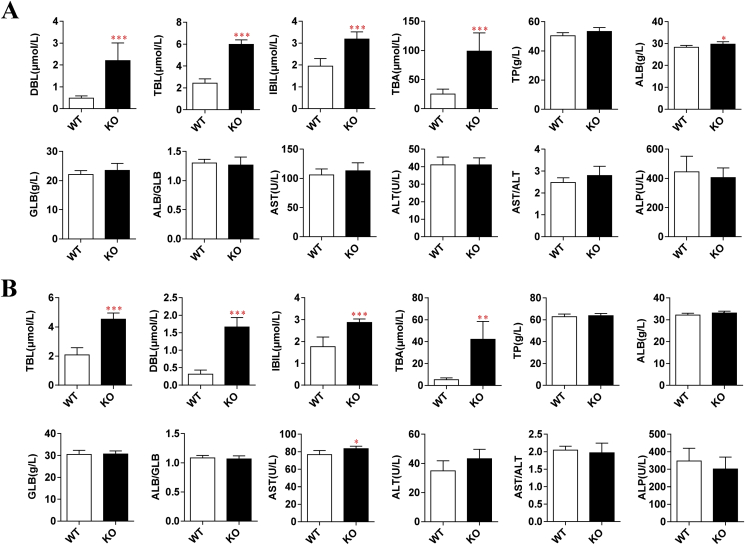
In addition, the Slco1b2 KO rats may be used as a disease model of Rotor syndrome, and the main disease characteristics are the increase of DBL and TBL in adolescents. Therefore, we randomly selected WT and KO rats (4-week-old and 8-week-old) to perform serum tests of liver function. As shown in Fig. 4, the TBL, DBL and IBIL increased significantly in male KO rats at both 4 (Fig. 4A) and 8 (Fig. 4B) weeks old compared with WT rats. Among them, the 4-week-old rats increased 2.46-, 4.55-, and 1.63-fold, respectively. And the 8-week-old rats increased 2.18-, 5.26-, and 1.62-fold, respectively. These results indicate that Slco1b2 KO rats can mimic Rotor syndrome with a phenotype similar to humans. In particular, the TBA increased significantly by 3.93- and 7.98-fold in 4- and 8-week old KO rats, respectively. Moreover, ALB in 4-week-old and AST in 8-week-old KO rats were showing statistically significant differences. However, these parameters are still within the normal range of the rat, and there is no clinical significance. Except for the above indexes, there was no significant difference in other liver function indexes between KO and WT rats. At the same time, as shown in Fig. 5A, there was no significant difference in renal function indications, blood lipid indications and glucose between KO and WT rats. Furthermore, we also tested female rats and the results were consistent with those of male rats (data not shown).
Figure 4.
Indicators of liver function in WT and KO male rats. Liver function indicators in (A) 4-week-old and (B) 8-week-old WT and KO male rats (n = 6). Indicators include TBL, DBL, IBIL, TBA, TP, ALB, GLB, ALB/GLB, AST, ALT, AST/ALT, and ALP. Values are presented as mean ± SD (n = 6). *P < 0.05, **P < 0.01 and ***P < 0.001 compared with WT rats.
Figure 5.
Physiological phenotype in WT and KO male rats. (A) Glucose, renal function and blood lipids indexes in 8-week-old WT and KO rats. Indicators include GLU, BUN, CR, UA, TG, T-CH, HDL-C, and LDL-C. Values are shown as mean ± SD (n = 6). (B) HE staining for liver sections in 8-week-old rats. The cytoplasm was stained mauve by eosin, and the cell nucleus was stained blue by hematoxylin. Scale bars, 100 μm in length.
HE staining was performed on the liver of 4-week-old and 8-week-old KO and WT rats to observe the morphology of hepatocytes. There was no significant difference in liver morphology between WT and KO rats at 4 weeks old (Supporting Information Fig. S1). However, in the slices of liver tissues of 8-week-old WT rats (Fig. 5B), the central lobular area showed a radial arrangement of hepatic cords from the central vein outward, with normal cell morphology, uniform cytoplasm and orderly arrangement. In contrast, the structure of hepatocyte cord in the 8-week-old KO rat was blurred and disordered, and the hepatic sinusoids were enlarged, suggesting slight damage to the liver, which may be related to the long-term high content of bilirubin and bile acid.
3.5. Compensatory expression of major transporters in WT and KO rats
In order to detect the compensatory expression of other major transporters in rats after Slco1b2 gene knockout, we detected the mRNA levels of major uptake and efflux transporters by real-time quantitative PCR. The data showed, except for the target gene Slco1b2, there was no significant difference in mRNA levels of other transporters (Supporting Information Fig. S2). Therefore, the transporters detected did not compensate at the transcriptional level in KO rats.
3.6. Pharmacokinetics of pitavastatin in WT and KO rats
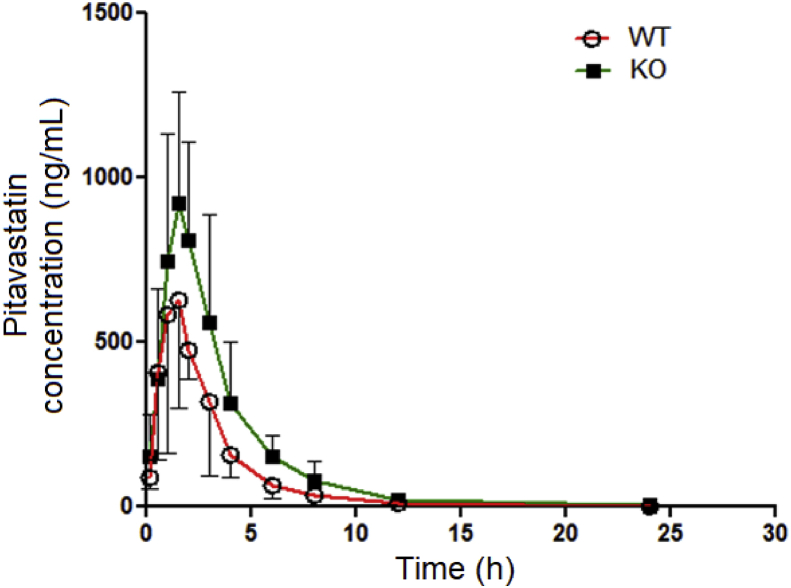
To further detect whether the function of OATP1B2 in KO rats is affected, we used pitavastatin, a specific OATP1B2 substrate recommended by FDA, to conduct pharmacokinetic experiments in WT and KO rats by oral administration (Fig. 6). The dose of pitavastatin (5 mg/kg) is based on clinical doses and preliminary experiments. Compared with WT rats, the area under the time–plasma concentration curve (AUC), both AUC0–24 h and AUC0–inf of KO rats were significantly increased by 68.8%, while the apparent volume of distribution/bioavailability (Vd/F) and clearance/bioavailability (CL/F) were significantly decreased by 39.8% and 38.5%, respectively (Table 3). Although peak time (Tmax), peak concentration (Cmax) and mean residence time (MRT) had no significant differences, they all had trends of increase. The results showed that the pharmacokinetic behavior of pitavastatin in KO rats was changed, especially a significant increase of in vivo exposure. Therefore, the function of OATP1B2 was absent in KO rats.
Figure 6.
Pharmacokinetics of pitavastatin in WT and KO rats. The concentrations of pitavastatin were determined by LC–MS/MS. Values are presented as mean ± SD (n = 6).
Table 3.
Pharmacokinetic parameters of pitavastatin after oral administration.
| Parameter | Unit | WT | KO |
|---|---|---|---|
| t1/2 | h | 2.98 ± 0.62 | 2.92 ± 0.43 |
| Tmax | h | 1.58 ± 0.86 | 2.08 ± 0.97 |
| Cmax | ng/mL | 757.70 ± 316.98 | 1001.32 ± 335.82 |
| AUC0–t | h·ng/mL | 2022.50 ± 393.63 | 3414.25 ± 1041.89* |
| AUC0–inf | h·ng/mL | 2037.19 ± 368.42 | 3439.51 ± 1051.67* |
| Vd/F | mL/kg | 10931.16 ± 3360.51 | 6580.87 ± 1897.02* |
| CL/F | mL/h/kg | 2539.70 ± 572.91 | 1562.04 ± 430.31** |
| MRT0–t | h | 3.01 ± 0.84 | 3.65 ± 0.89 |
The dose was 5 mg/kg (p.o.). Results were mean ± SD (n = 6). *P < 0.05, and **P < 0.01 compared to WT rats.
4. Discussion
In recent years, the studies on OATP1B1 and OATP1B3 have become more and more popular because of their transport function of important endogenous and exogenous substances11. The past research mainly focused on transporter-involved drug safety evaluation, drug resistance and disease progression9,31,32. The in vitro and in vivo models for studying the role of OATP1B in drug research are limited with many drawbacks. In vitro cell models mainly include primary cells and transfected cells. However, cell culture conditions are strict and lack regulatory pathways, which do not reflect the real situation of complex living organisms33,34. In vivo models include organ perfusion model, inhibitor inhibiting transporter activity and gene knockout model. However, both the organ perfusion model and the use of inhibitors have the disadvantages of low specificity of inhibitors and high cost35.
Gene knockout models have attracted more and more attention in recent years because they can better reflect the function of transporters under physiological conditions. In 2008, Oatp1b2-null mice were engineered by homologous recombination and their phenotype was characterized21,22. However, due to technical limitation such as the difficulty in obtaining and culturing rat embryonic stem cells, there has been no rat model with Slco1b2 gene knockout to date. Compared with mice, rats have many advantages in pharmacokinetic and toxicological studies, such as large size and easy to handling, adequate blood volume and strong tolerance. In particular, the rat model can more accurately and comprehensively reflect the absorption, distribution, metabolism and excretion of drugs or compounds in vivo36, 37, 38. In addition, current research on hyperbilirubinemia has found that the rats are the preferred animal model23,24. Therefore, Slco1b2 knockout rats can be used as the disease model of Rotor syndrome. The early gene knockout technology based on the principle of homologous recombination is very difficult to generate rat models because of the low efficiency and long experimental period. However, the emergence of CRISPR/Cas9 technology has brought light to the gene editing of rats25. CRISPR/Cas9 system is a new generation of gene editing technology. Compared with the traditional homologous recombination method, CRISPR/Cas9 has many advantages, such as easy operation, short cycle, low cost, high success rate and no species limitation39. Therefore, this study selected CRISPR/Cas9 technology to successfully generate the Slco1b2 KO rat model for the first time.
Compared with WT rats, there was no expression of OATP1B2 protein in the liver of KO rats by the Western blot and immunohistochemistry analysis (Fig. 3). We also detected the mRNA expression of other transporters to determine whether there were compensatory effects by using the screening method of Q-PCR. The results showed that the transporters detected did not compensate at the transcriptional level in KO rats. Furthermore, we used the OATP1B2 substrate pitavastatin to verify the change of OATP1B2 function in vivo. The suspension of pitavastatin in CMC-Na was used in the pharmacokinetic study via the oral administration, which might be the cause for large deviation in absorption and AUC. Despite of relatively large individual deviation, KO and WT rats still showed significant differences in the pharmacokinetic parameters. The AUC0–24 h and AUC0–inf of KO rats were significantly increased by both 68.8%, while the Vd/F and CL/F were significantly decreased by 39.8% and 38.5%, respectively (Table 3). In addition, Tmax, Cmax and MRT had trends of increase, while t1/2 was not significantly changed. In fact, pitavastatin was uptaken into the hepatocytes by OATP1B2, and then excreted into the bile by P-gp and BCRP in WT rat livers40. In contrast, pitavastatin uptake in the liver of KO rats was significantly reduced, and thus the drug molecules were more likely to be retained in the bloodstream, resulting in a smaller value of Vd. At the same time, hepatic efflux transporters were hardly saturated in KO rats because the hepatic pitavastatin influx rate was significantly lower. If the hepatic clearance of pitavastatin remained unchanged, the unchanged systemic elimination rate constant ke resulted in similar t1/2 of the WT and KO rats, as well as the lower CL/F in KO rats decided by smaller Vd. Based on the mentioned above, the Slco1b2 KO rat model was successfully constructed because of the loss of expression and function of Oatp1b2.
In the process of developing new drugs, we can compare the pharmacokinetic profiles of the tested compounds or specific substrates in WT and KO rats to determine whether the drugs are substrates, inhibitors or inducers of OATP1B2. Careful conclusions could be further extrapolated to human OATP1B1/3, especially OATP1B1/3-mediated drug transport and drug–drug interactions. Compared with cell models, the results from the rat model can provide more information about the significance of the influx transporter on systemic exposure of the drug at an integrate level. Although Oatp1b2 KO mice model was constructed, mRNA levels of other transporters such as Oatp1a4, Oat2 and Mrp3 were significantly changed22. Therefore, the Oatp1b2 KO rats were a better model to study the function of OATP1B2 than the mice model because of no changes of other transporters. Moreover, adverse drug reactions (ADRs) caused by inter-patient differences in drug responsiveness affect 2%–6% of hospitalized patients, mainly due to genetic polymorphism41. Actually, there are 10 single nucleotide polymorphisms (SNPs) of SLCO1B1 to decrease the activity of OATP1B1, among which the most common and influential are the c.512T > C and c.388A > G. The SLCO1B3 c.334T > G and c.699G > A SNPs decrease the transport capacity42,43. Therefore, the Slco1b2 KO rat model could be used for pre-clinical pharmacokinetic experiments to provide some support for personalized medication for patients with SLCO1B1 and SLCO1B3 SNPs.
On the other hand, this Slco1b2 KO model can be used in cancer-related research, because the OATP research has gradually associated with drug resistance and target therapy in recent years44. The low expression of OATP1B1 or OATP1B3 often leads to the decrease of intracellular drug concentrations, which results in drug resistance32. In contrast, overexpression of OATP1B1 or OATP1B3 in drug-resistant cells can reverse the resistance to a certain extent and enhance the therapeutic effects45. In addition, OATP1B1 and OATP1B3 are almost specifically expressed in the liver under normal conditions, but in some tumors they are not only expressed, but also expressed differently at different stages of tumors5,46,47. These specific mechanisms are not clear yet, but it can be used as a potential target to provide new ideas for cancer treatment46. Therefore, the Slco1b2 KO rat model is a powerful experimental tool to explore the relationship between OATP1B1/3 and tumors.
The Slco1b2 KO rats are capable of not only making contribution to drug evaluation48, but also serving as a disease model. For example, there are many causes of hyperbilirubinemia. At present, different animal models have been established to simulate hyperbilirubinemia, such as intraperitoneal or intravenous injection of bilirubin into neonatal rats, which results in the increase of bilirubin content in rats; or the construction of a UDP glucuronosyltransferase family 1 member A1 (Ugt1a1) gene KO mouse model to simulate neonatal jaundice49, 50, 51. But there is no disease model of Rotor syndrome to date. In this study, the Slco1b2 KO rat model presented the same pathogenesis and disease characteristics as clinical Rotor syndrome. For example, the total bilirubin (TBL) and direct bilirubin (DBL) in the serum of KO rats were significantly higher than those in WT rats (Fig. 4). These results are consistent with the symptoms caused by the absence of OATP1B1 and OATP1B3 in Rotor syndrome14. Therefore, the Slco1b2 KO rat model can be used as a disease model of Rotor syndrome and hyperbilirubinemia, as well as the hyperbilirubinemia-related diseases.
Since Rotor syndrome is a kind of jaundice in children and adolescents, we detected the physiological indexes of liver function in serum of 4-week-old KO rats and WT rats. The results showed that TBL, DBL, IBIL and TBA in KO rats were significantly higher than those in WT rats. But these phenomena still existed in 8-week-old rats. At the same time, HE staining showed that the structure of hepatocytes was blurred, the arrangement was disordered and the hepatic sinusoids were enlarged in the liver tissue sections of KO rats. These results indicated that hyperbilirubinemia has accompanied the growth of rats. Probably due to the long-term exposure of hyperbilirubinemia environment, liver cells were damaged to a certain extent. The total bile acid content in KO rats was significantly increased, because the bile acids were the substrate for Oatp1b252. The loss of Oatp1b2 function resulted in the accumulation of total bile acids in the blood. These results suggest the long-term monitoring of hepatic function in patients with Rotor syndrome is necessary, and the high risks of hepatocyte lesions as well as cholestasis should be noticed.
In summary, the Slco1b2 KO rat model was successfully constructed by CRISPR/Cas9 technology, with no off-target and compensatory effects. This rat model can be used in pharmacokinetic research. At the same time, this model presents the disease characteristics of hyperbilirubinemia-Rotor syndrome, especially caused by the absence of OATP1B1/3. Thus this model can also be used as a disease model to study hyperbilirubinemia-related diseases.
Acknowledgments
This work was supported in whole or part by grants from the National Natural Science Foundation of China (No. 81773808), the Science and Technology Commission of Shanghai Municipality (Nos. 17140901000, 17140901001 and 18430760400). This work was also supported from ECNU Multifunctional Platform for Innovation (011, China).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.11.007.
Author contributions
Xin Wang was responsible for the conception and design of the study. Xinrun Ma, Xuyang Shang, Xuan Qin, and Jian Lu performed experiments. Xinrun Ma, Xuyang Shang and Xin Wang analyzed results and interpreted data. Xinrun Ma, Xuyang Shang, Xuan Qin, and Jian Lu drafted the manuscript. Mingyao Liu and Xin Wang revised the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wolkoff A.W. Organic anion uptake by hepatocytes. Comp Physiol. 2014;4:1715–1735. doi: 10.1002/cphy.c140023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagenbuch B., Stieger B. The SLCO (former SLC21) superfamily of transporters. Mol Asp Med. 2013;34:396–412. doi: 10.1016/j.mam.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth M., Obaidat A., Hagenbuch B. OATPs, OATs and OCTs: the organic anion and cation transporters of the SLCO and SLC22A gene superfamilies. Br J Pharmacol. 2012;165:1260–1287. doi: 10.1111/j.1476-5381.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe T., Unno M., Onogawa T., Tokui T., Kondo T.N., Nakagomi R. LST-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology. 2001;120:1689–1699. doi: 10.1053/gast.2001.24804. [DOI] [PubMed] [Google Scholar]
- 5.Thakkar N., Lockhart A.C., Lee W. Role of organic anion-transporting polypeptides (OATPs) in cancer therapy. AAPS J. 2015;17:535–545. doi: 10.1208/s12248-015-9740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda K. Organic anion transporting polypeptide (OATP)1B1 and OATP1B3 as important regulators of the pharmacokinetics of substrate drugs. Biol Pharm Bull. 2015;38:155–168. doi: 10.1248/bpb.b14-00767. [DOI] [PubMed] [Google Scholar]
- 7.Stieger B., Hagenbuch B. Organic anion-transporting polypeptides. Curr Top Membr. 2014;73:205–232. doi: 10.1016/B978-0-12-800223-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sissung T.M., Reece K.M., Spencer S., Figg W.D. Contribution of the OATP1B subfamily to cancer biology and treatment. Clin Pharmacol Ther. 2012;92:658–660. doi: 10.1038/clpt.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlgren M., Vildhede A., Norinder U., Wisniewski J.R., Kimoto E., Lai Y. Classification of inhibitors of hepatic organic anion transporting polypeptides (OATPs): influence of protein expression on drug–drug interactions. J Med Chem. 2012;55:4740–4763. doi: 10.1021/jm300212s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shitara Y. Clinical importance of OATP1B1 and OATP1B3 in drug–drug interactions. Drug Metab Pharmacokinet. 2011;26:220–227. doi: 10.2133/dmpk.DMPK-10-RV-094. [DOI] [PubMed] [Google Scholar]
- 11.International Transporter Consortium, Giacomini K.M., Huang S.M., Tweedie D.J., Benet L.Z., Brouwer K.L. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.U.S. Food and Drug Administration Guidance for industry: in vitro metabolism and transporter mediated drug–drug interaction studies. https://www.federalregister.gov/documents/2017/10/25/2017-23102/in-vitro-metabolism--and-transporter-mediated-drug-drug-interaction-studies-and-clinical-drug Available from:
- 13.Dhumeaux D., Erlinger S. Hereditary conjugated hyperbilirubinaemia: 37 years later. J Hepatol. 2013;58:388–390. doi: 10.1016/j.jhep.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 14.van de Steeg E., Stránecký V., Hartmannová H., Nosková L., Hřebíček M., Wagenaar E. Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J Clin Investig. 2012;122:519–528. doi: 10.1172/JCI59526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green S.M., Kaipainen A., Bullock K., Zhang A., Lucas J.M., Matson C. Role of OATP transporters in steroid uptake by prostate cancer cells in vivo. Prostate Cancer Prostatic Dis. 2017;20:20–27. doi: 10.1038/pcan.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada A., Sissung T., Price D.K., Danesi R., Chau C.H., Sharifi N. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14:3312–3318. doi: 10.1158/1078-0432.CCR-07-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang M., Xie W., Mostaghel E., Nakabayashi M., Werner L., Sun T. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29:2565–2573. doi: 10.1200/JCO.2010.31.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Steeg E., Wagenaar E., van der Kruijssen C.M., Burggraaff J.E., de Waart D.R., Elferink R.P. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J Clin Investig. 2010;120:2942–2952. doi: 10.1172/JCI42168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez F.J., Fang Z.Z., Ma X. Transgenic mice and metabolomics for study of hepatic xenobiotic metabolism and toxicity. Expert Opin Drug Metabol Toxicol. 2015;11:869–881. doi: 10.1517/17425255.2015.1032245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagenbuch B., Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 21.Lu H., Choudhuri S., Ogura K., Csanaky I.L., Lei X., Cheng X. Characterization of organic anion transporting polypeptide 1b2-null mice: essential role in hepatic uptake/toxicity of phalloidin and microcystin-LR. Toxicol Sci. 2008;103:35–45. doi: 10.1093/toxsci/kfn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaher H., Meyer zu, Schwabedissen H.E., Tirona R.G., Cox M.L., Obert L.A., Agrawal N. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Mol Pharmacol. 2008;74:320–329. doi: 10.1124/mol.108.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro S.M. Bilirubin toxicity in the developing nervous system. Pediatr Neurol. 2003;29:410–421. doi: 10.1016/j.pediatrneurol.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Mikkaichi T., Nakai D., Yoshigae Y., Imaoka T., Okudaira N., Izumi T. Liver-selective distribution in rats supports the importance of active uptake into the liver via organic anion transporting polypeptides (OATPs) in humans. Drug Metab Pharmacokinet. 2015;30:334–340. doi: 10.1016/j.dmpk.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 28.Ceasar S.A., Rajan V., Prykhozhij S.V., Berman J.N., Ignacimuthu S. Insert, remove or replace: a highly advanced genome editing system using CRISPR/Cas9. Biochim Biophys Acta. 2016;1863:2333–2344. doi: 10.1016/j.bbamcr.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Liu X., Wu S., Xu J., Sui C., Wei J. Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B. 2017;7:292–302. doi: 10.1016/j.apsb.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Z., Zhang Y., Gu J., Feng P., Wang Y. Pharmacokinetic properties of single- and multiple-dose pitavastatin calcium tablets in healthy Chinese volunteers. Curr Ther Res Clin Exp. 2015;77:52–57. doi: 10.1016/j.curtheres.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemi M., Pasanen M.K., Neuvonen P.J. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63:157–181. doi: 10.1124/pr.110.002857. [DOI] [PubMed] [Google Scholar]
- 32.de Morrée E.S., Böttcher R., van Soest R.J., Aghai A., de Ridder C.M., Gibson A.A. Loss of SLCO1B3 drives taxane resistance in prostate cancer. Br J Canc. 2016;115:674–681. doi: 10.1038/bjc.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulvestad M., Darnell M., Molden E., Ellis E., Åsberg A., Andersson T.B. Evaluation of organic anion-transporting polypeptide 1B1 and CYP3A4 activities in primary human hepatocytes and HepaRG cells cultured in a dynamic three-dimensional bioreactor system. J Pharmacol Exp Ther. 2012;343:145–156. doi: 10.1124/jpet.112.195750. [DOI] [PubMed] [Google Scholar]
- 34.Qin X., Wang X. Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharm Sin B. 2019;9:1087–1098. doi: 10.1016/j.apsb.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Wang C., Liu Q., Meng Q., Cang J., Sun H. Pharmacokinetic interaction between JBP485 and cephalexin in rats. Drug Metab Dispos. 2010;38:930–938. doi: 10.1124/dmd.110.032060. [DOI] [PubMed] [Google Scholar]
- 36.Liang C., Zhao J., Lu J., Zhang Y., Ma X., Shang X. Development and characterization of MDR1 (Mdr1a/b) CRISPR/Cas9 knockout rat model. Drug Metab Dispos. 2019;47:71–79. doi: 10.1124/dmd.118.084277. [DOI] [PubMed] [Google Scholar]
- 37.Qin X., Lu J., Wang P., Xu P., Liu M., Wang X. Cytochrome P450 3A selectively affects the pharmacokinetic interaction between erlotinib and docetaxel in rats. Biochem Pharmacol. 2017;143:129–139. doi: 10.1016/j.bcp.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Tang Y., Lu J., Shao Y., Qin X., Li Y. Characterization of novel cytochrome P450 2E1 knockout rat model generated by CRISPR/Cas9. Biochem Pharmacol. 2016;105:80–90. doi: 10.1016/j.bcp.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Wang H.X., Li M., Lee C.M., Chakraborty S., Kim H.W., Bao G. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 40.Prueksaritanont T., Chu X., Evers R., Klopfer S.O., Caro L., Kothare P.A. Pitavastatin is a more sensitive and selective organic anion-transporting polypeptide 1B clinical probe than rosuvastatin. Br J Clin Pharmacol. 2014;78:587–598. doi: 10.1111/bcp.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott S.A. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13:987–995. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong I.Y., Kim R.B. Impact of genetic variation in OATP transporters to drug disposition and response. Drug Metab Pharmacokinet. 2013;28:4–18. doi: 10.2133/dmpk.dmpk-12-rv-099. [DOI] [PubMed] [Google Scholar]
- 43.Niemi M., Backman J.T., Kajosaari L.I., Leathart J.B., Neuvonen M., Daly A.K. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–478. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 44.König S.K., Herzog M., Theile D., Zembruski N., Haefeli W.E., Weiss J. Impact of drug transporters on cellular resistance towards saquinavir and darunavir. J Antimicrob Chemother. 2010;65:2319–2328. doi: 10.1093/jac/dkq324. [DOI] [PubMed] [Google Scholar]
- 45.Herraez E., Sanchez-Vicente L., Macias R.I.R., Briz O., Marin J.J.G. Usefulness of the MRP2 promoter to overcome the chemoresistance of gastrointestinal and liver tumors by enhancing the expression of the drug transporter OATP1B1. Oncotarget. 2017;8:34617–34629. doi: 10.18632/oncotarget.16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teft W.A., Welch S., Lenehan J., Parfitt J., Choi Y.H., Winquist E. OATP1B1 and tumour OATP1B3 modulate exposure, toxicity, and survival after irinotecan-based chemotherapy. Br J Canc. 2015;112:857–865. doi: 10.1038/bjc.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright J.L., Kwon E.M., Ostrander E.A., Montgomery R.B., Lin D.W., Vessella R. Expression of SLCO transport genes in castration-resistant prostate cancer and impact of genetic variation in SLCO1B3 and SLCO2B1 on prostate cancer outcomes. Cancer Epidemiol Biomark Prev. 2011;20:619–627. doi: 10.1158/1055-9965.EPI-10-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Qin X., Shang X., Liu M., Wang X. Organic anion transport polypeptide 1b2 selectively affects the pharmacokinetic interaction between paclitaxel and sorafenib in rats. Biochem Pharmacol. 2019;169:113612. doi: 10.1016/j.bcp.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 49.Rice A.C., Shapiro S.M. A new animal model of hemolytic hyperbilirubinemia-induced bilirubin encephalopathy (kernicterus) Pediatr Res. 2008;64:265–269. doi: 10.1203/PDR.0b013e31817d9be0. [DOI] [PubMed] [Google Scholar]
- 50.Hansen T., Tommarello S., Allen J. Subcellular localization of bilirubin in rat brain after in vivo i.v. administration of [3H]bilirubin. Pediatr Res. 2001;49:203–207. doi: 10.1203/00006450-200102000-00012. [DOI] [PubMed] [Google Scholar]
- 51.Porro F., Bockor L., De Caneva A., Bortolussi G., Muro A.F. Generation of Ugt1-deficient murine liver cell lines using TALEN technology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slijepcevic D., Roscam Abbing R.L.P., Katafuchi T., Blank A., Donkers J.M., van Hoppe S. Hepatic uptake of conjugated bile acids is mediated by both sodium taurocholate cotransporting polypeptide and organic anion transporting polypeptides and modulated by intestinal sensing of plasma bile acid levels in mice. Hepatology. 2017;66:1631–1643. doi: 10.1002/hep.29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.