Abstract
The transcription factor nuclear factor kappa B (NF-κB) is activated in hepatocytes in the pathogenesis of hepatic steatosis. However, the action mechanism of NF-κB remains to be established in the hepatic steatosis. In this study, the P50 subunit of NF-κB was found to promote the hepatic steatosis through regulation of histone deacetylase 1 (HDAC1) in hepatocytes. The activity was supported by the phenotypes of P50 knockout (P50-KO) mice and P65 knockout (P65-KO) mice. Hepatic steatosis was reduced in the P50-KO mice, but not in the P65-KO mice. The reduction was a result of inhibition of HDAC1 activity in the P50-KO cells. Knockdown of Hdac1 gene led to suppression of hepatocyte steatosis in HepG2 cells. A decrease in sterol-regulatory element binding protein 1c (SREBP1c) protein was observed in the liver of P50-KO mice and in cell with Hdac1 knockdown. The decrease was associated with an increase in succinylation of SREBP1c protein. The study suggests that P50 stabilizes HDAC1 to support the SREBP1c activity in hepatic steatosis in the pathophysiological condition. Interruption of this novel pathway in the P50-KO, but not the P65-KO mice, may account for the difference in hepatic phenotypes in the two lines of transgenic mice.
Key words: NF-κB, HDAC1, SREBP1, Succinylation, Hepatic steatosis
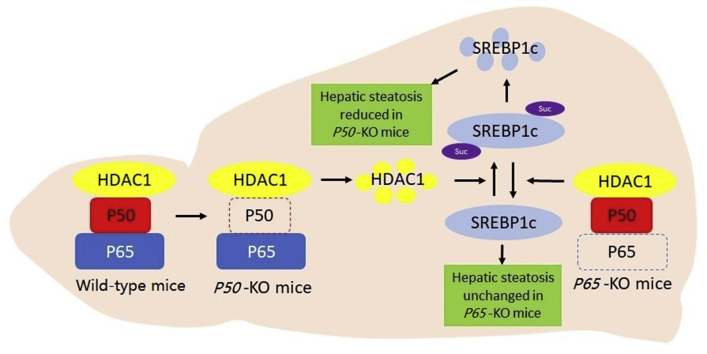
Graphical abstract
NF-κB promotes hepatic steatosis through an impact of P50 subunit in HDAC1 protein. P50 stabilizes HDAC1 protein to keep succinylation of SREBP1c under control in favor of lipogenesis in the liver of wild type or P65-KO mice. This pathway failed to operate in P50-KO mice leading to reduction of hepatic steatosis.
1. Introduction
Nuclear factor kappa B (NF-κB) was reported to promote lipid accumulation in hepatocytes in the progression of hepatic steatosis1,2. However, the exact mechanism of NF-κB action remains unknown. In adipocytes, NF-κB inhibits lipid accumulation through suppression of the transcriptional activity of peroxisome proliferator activated receptor gamma (PPARγ) in the lipogenic transcriptional network3. Sustained NF-κB activation leads to a reduction in lipid accumulation of adipocytes, which accounts for a resistance to obesity in the transgenic mice with overexpression of P65 (RelA) or inhibitor of kappa B kinase beta (IKKβ) in the adipocytes4,5. The lean phenotype together with improved insulin sensitivity6,7 is consistent with those observed in mice of global P50-KO8,9, which exhibit an increase in NF-κB activity. These studies suggest that NF-κB have opposite activities in the control of lipid accumulation between hepatocytes and adipocytes. The molecular basis is unknown for the opposite activities. NF-κB is generally formed by two different subunits, P65 and P50. The two subunits have different activities in the control of gene transcription, the P65 subunit triggers transactivation of target genes, and the P50 subunit binds to DNA to inhibit the P65 activity. The difference in the two subunits may provide a mechanism for the distinct activities of NF-κB in hepatocytes vs. adipocytes.
Histone deacetylase 1 (HDAC1) is a member of the class I histone deacetylase, and is located in the nucleus to catalyse the deacetylation reaction of histone proteins in the regulation of gene transcription. HDAC1 binds to NF-κB protein in the promoter DNA to perform the deacetylase activity. However, there is no literature about regulation of HDAC1 activity by NF-κB. HDAC1 activity has been reported in variety of systems. Global inactivation of Hdac1 gene results in embryonic lethality from severe developmental defects10, which is associated with a reduction in cell cycle progression. In the liver, HDAC1 is required for generation of new hepatocytes from the progenitor cells11. In the adipose tissue, HDAC1 controls differentiation of white adipocytes through an interaction with CCAAT-enhancer binding protein alpha (C/EBPα)12, or thermogenesis of brown adipocytes by suppression of uncoupling protein 1 (UCP-1) and PPARγ co-activator 1 (PGC-1) expression13. These studies suggest that HDAC1 regulates activities of multiple transcription factors in adipocytes. Unfortunately, there is no report about a feedback regulation of HDAC1 activity by the transcription factors.
Sterol-regulatory element binding protein 1c (SREBP1c) is one of the key transcription factors in the regulation of de novo lipogenesis in hepatocytes through induction of lipogenic gene expression14. SREBP1c activity is regulated at the transcriptional and post-translational levels. The transcriptional expression is well documented in the lipogenic effects of insulin. The posttranslational modification includes protease cleavage of SREBP1c for activation and modification by phosphorylation15 and acetylation16. Succinylation is one of the metabolism-associated post-translational modifications, whose activity is enhanced in non-alcoholic fatty liver17 and decreased by sirtuin 5 (SIRT5) through de-succinylate18. There is no report of SREBP1c regulation by succinylation.
In current study, we observed that HDAC1 protein decreased in the liver of P50-KO mice for a reduction in hepatic steatosis. HDAC1 may promote the hepatic steatosis through de-succinylation of SREBP1c protein in the nucleus. The study reveals a new mechanism of P50/HDAC1/SREBP1c for hepatic steatosis.
2. Materials and methods
2.1. Animals
The male P50-KO mice (strain name: B6.Cg-Nfkb1 tm Bal/J, Stock number: 006097) were purchased from the Jackson Laboratory at 4 weeks of age. The mice were crossed with wild-type (WT) C57BL/6J mice to generate heterozygous breeders. The male homozygous P50-KO mice (Experiment group) and wild-type littermates (Control group) were obtained by crossing the heterozygous mice. The male mice were fed on chow diet (5001, 13% calories in fat, LabDiet, Richmond, IN, USA) to 12 weeks of age, and then fed on a high-fat diet (HFD, D12331, 58% calories in fat, Research Diets, New Brunswick, NJ, USA) for 16 weeks to establish the fatty liver model. Tissue samples were collected from the mice at the end of HFD feeding. The male liver specific P65-KO mice were described in an early study6 and fed on HFD diet at 8 weeks of age for 14 weeks to induce the hepatic steatosis. The mice were housed at 4/cage with free access to water and diet. All procedures were performed in accordance with National Institutes of Health guidelines and approved by the Institute Animal Care and Use Committee (IACUC) at the Pennington Biomedical Research Center, Louisiana State University System, Baton Rouge, LA, USA.
2.2. Tissue collection
Liver tissues of mice were collected at the age of 9 months without fast. One part of liver was frozen in liquid nitrogen for protein and mRNA analysis and then kept at −80 °C, the other part of liver was treated with 4% formaldehyde for histological analysis and then kept at room temperature. The whole-body weight was collected at the same time.
2.3. Cell culture, reagents and treatment
HepG2 cells were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS at 37 °C in a 5% CO2 humidified atmosphere. MEF cells of P50-KO, IкBα-KO and Ikkβ-KO were described in the early studies3,19. The proteasome inhibitor MG-132 (SML1135, Sigma, St. Louis, MO, USA) was used at the final concentration of 50 μmol/L in the cell culture.
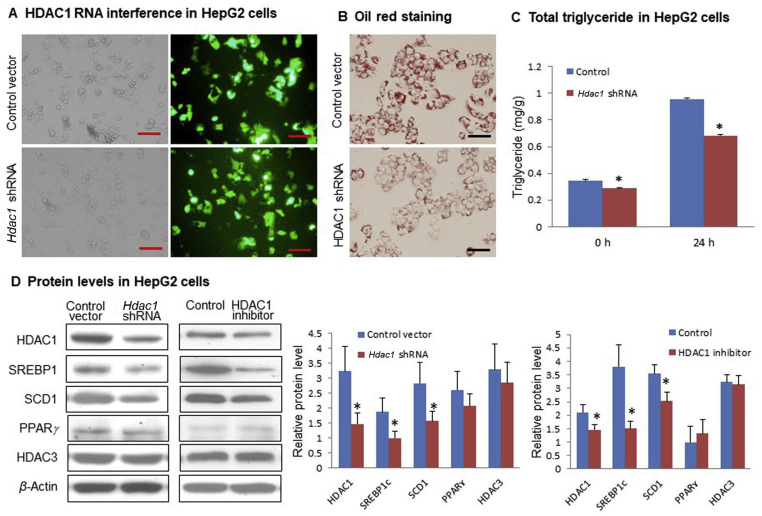
P50 shRNA. HepG2 cells were electrical transfected with mouse P50-shRNA expression vector (P/N: 9033, L/N: P04122008, Panomics, Redwood, CA, USA) through Amaxa Nucleofector™ II/2b Device, using cell line nucleofector Kit V (VCA-1003, Lonza cologne AG) with the program T-031. shRNA was used at 1–3 μg/well in the 6-well plate. The control cells were transfected with the empty vector (P/N: 8524, L/N: P04120901, Panomics). The transfected cells were cultured for 24 h and then treated with 0.3 mmol/L oleic acid (P5585, Sigma) for additional 24 h. The cells were harvested for Western blotting of SREBP1c, stearoyl-coenzyme A desaturase 1 (SCD1) and PPARγ and oil red O staining for triglyceride quantification.
Hdac1 shRNA. HepG2 cells were electrically transfected with Hdac1-shRNA expression vector through Amaxa Nucleofector™ II/2b Device, using the cell line nucleofector Kit V and program T-031. Control cells were transfected with the empty vector. The transfected cells were treated with 0.5 mmol/L oleic acid for 24 h Hdac1 expression vector together with the control vector were described elsewhere20.
HDAC1 inhibitor. HepG2 cells were pretreated with 0.4 mmol/L HDAC1 chemical inhibitor (SML0296, Sigma) for 30 min and then treated with 0.5 mmol/L oleic acid for 24 h to determine the impact on hepatic steatosis.
2.4. Oil red O staining
At the end of oleic acid treatment, HepG2 cells were fixed with 10% formalin for 30 min and stained oil red O dye (Fisher Biotech BP112-10, Fair Lawn, NJ, USA) as described elsewhere21.
2.5. Triglyceride assay
Liver tissues were homogenized in phosphate buffered saline (PBS; 1 g:20 mL) and the cell lysate was made with the cell lysis buffer on ice. The lipids were extracted from the liver tissue homogenization or cell lysate using a chloroform/methanol (2:1) mixture. Triglyceride was determined using the Serum Triglyceride Determination Kit (TR0100, Sigma) according to the manufacturer's instructions. The triglyceride concentration was also measured with the method of oil red O staining in HepG2 cells.
2.6. Western blot and immunoprecipitation (IP)
The whole cell lysate was prepared in a lysis buffer with sonication as described elsewhere21. In IP, the lysate protein (500 μg at 1 μg/μL) was mixed with 2 μg antibody in 100 μL protein G agarose (16–266, Millipore, Billerica, MA, USA) and rotated overnight at 4 °C. The agarose beads were washed with 800 μL washing buffer three times and collected as IP product after centrifugation at 13,000×g for 30 s. The succinylation signal was detected in the IP product after resolution in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Antibodies included: HDAC1 (H6287, Sigma), histone deacetylase 3 (HDAC3, ab7030; Abcam, Cambridge, UK), sirtuin 1 (SIRT1, ab12193; Abcam), SCD1 (ab19862, Abcam), SREBP1 (sc-8984, Santa Cruz, CA, USA), pan anti-succinyllysine polyclonal (106,768, NovoPro, shanghai, China), PPARγ (made in our laboratory), P50 (sc-8414, Santa Cruz, CA, USA), and β-actin (sc-47778, Santa Cruz, CA, USA).
2.7. Succinylation assay
Succinylation proteins were examined with the anti-succinyllysine antibody in the cell or tissue lysates. Succinylation of SREBP1c protein was determined in the IP product of SREBP1c antibody. The de-succinylation assay was conducted with the recombinant HDAC1 protein (BML-SE456, Enzo Life Sciences Inc., New York, NY, USA) in the IP product of SREBP1c. The succinylation signal of SREBP1c was determined with the anti-succinylation antibody in a Western blot.
2.8. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from frozen tissues using Tri-Reagent (T9424, Sigma) as described elsewhere22. The expression of mRNA was determined by one-step quantitative real-time PCR using TaqMan Master Mix (Applied Biosystems, Foster City, CA, USA). TaqMan primers and the probe were acquired from Applied Biosystems as followings: Hdac1 (Mm02391771_g1), Hdac3 (Mm00515916_m1), Sirt1 (Mm00493421_m1), fatty acid synthase (Fas) (Mm00662319_m1), Pparγ (Mm00440945_m1), Scd1 (Mm00772290_m1), Srebp1 (Mm00550338_m1), phosphodiesterase 3B (Pde3b, Mm00691635_m1), glucose-6-phosphatase (G6Pase, Mm00839363_m1), fatty acid translocase (Cd36, Mm00432403_m1), Pgc1α (Mm00447183_m1), phosphoenolpyruvate carboxykinase 1 (Pck1, Mn00440636_m1), tumor necrosis factor alpha (Tnf-α, Mm00443258_m1), interleukin 6 (Il-6, Mm00446190_ml), interleukin 1 beta (Il-1β, Mm00434228_ml). Mouse ribosome 18S rRNA_s1 (AIQJA2B) was used as an internal control.
2.9. Statistical analysis
The in vivo data were analyzed with two-way ANOVA. The in vitro experiments were conducted at least three times. Two-tailed, unpaired Student's t test was used in the statistical analysis of in vitro data. The data was presented as mean ± SE. Significance was considered at P < 0.05 in the statistical analysis.
3. Results
3.1. Hepatic steatosis was decreased in P50-KO mice
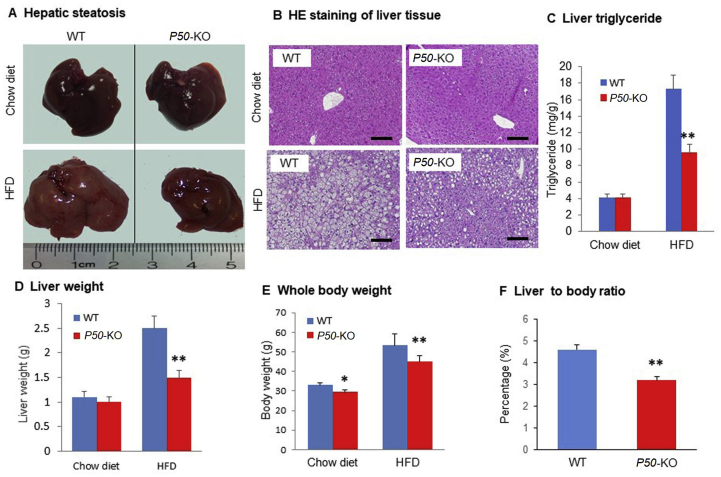
To explore the role of P50 in the control of hepatic steatosis, P50-KO mice were fed on HFD to establish the hepatic steatosis model. Compared to WT mice, the P50-KO mice exhibited a reduced degree in hepatic steatosis at the end of 4 months on HFD, which was observed in the liver appearance, liver weight and liver triglyceride content (Fig. 1A–D). The difference was observed in the mice on HFD, not on chow diet. The P50-KO mice exhibited a lower body weight on either chow diet or HFD with more significant difference on HFD (Fig. 1E). However, the liver to body ratio was significantly reduced in the P50-KO mice on HFD (Fig. 1F), suggesting that P50 has a liver-specific effect in the regulation of hepatic steatosis. The data suggest that P50-KO mice are resistant to the hepatic steatosis on HFD.
Figure 1.
Hepatic steatosis was decreased in P50-KO mice. Liver was examined in P50-KO and WT mice fed on HFD for 16 weeks. (A) Liver image. (B) Microscope image of liver tissue with H&E staining. Scale bar: 200 μm. (C) Triglyceride in liver tissue. (D) Liver weight. (E) Body weight of mice. (F) The liver weight to body weight ratio. In the bar figure, each data represents mean ± SE (n = 8). *P < 0.05, **P < 0.01 over WT mice.
3.2. Reduction of lipogenic gene expression in P50-KO mice
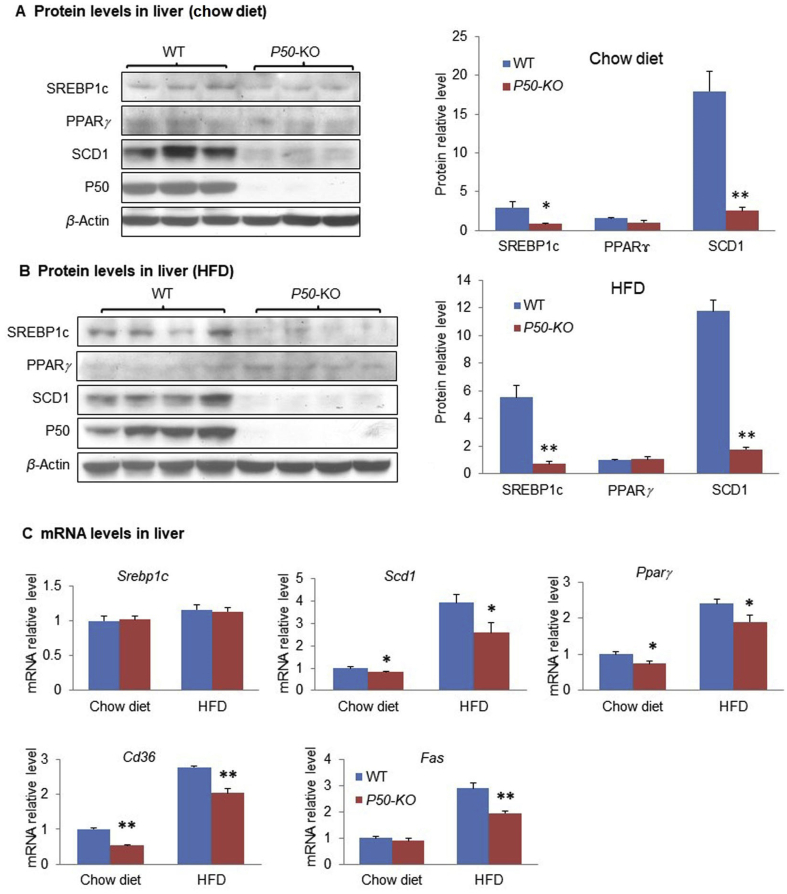
In order to understand the mechanism of reduced steatosis in P50-KO mice, lipogenesis capacity was examined in the liver by expression of lipogenic proteins including SREBP1c, PPARγ, and SCD1. A significant reduction was observed in SREBP1c and SCD1, but not PPARγ in the P50-KO mice on chow diet (Fig. 2A). The reduction status was maintained in the P50-KO mice on HFD (Fig. 2B). The SCD1 protein reduction suggests a decrease in de novo lipogenesis. Scd1 gene expression is subject to regulation by the transcription factor SREBP1c in the liver. The decreased abundance of SREBP1c protein provides a mechanism for the SCD1 reduction in P50-KO mice. The mRNA level of Scd1 was reduced in support of the mechanism (Fig. 2C). In addition, other lipogenic genes such as Fas and Cd36 also decreased in the liver of P50-KO mice (Fig. 2C). The mRNA level of Srebp1c was not reduced in the P50-KO mice (Fig. 2C). This group of data suggests that a reduction in lipogenesis occurs in the liver of P50-KO mice for the reduced steatosis.
Figure 2.
Expression of lipogenic genes was down-regulated in the liver of P50-KO mice. (A) Protein levels on chow diet. Western blot was performed for proteins of SREBP1, SCD1, PPARγ, and P50 in the liver tissue. (B) Protein level on HFD. Liver tissue was obtained at 16 weeks on HFD and used in the assay. (C) mRNA level of lipogenic genes. Srebp1, Scd1, Pparγ, Cd36 and Fas were examined by qRT-PCR in the liver tissue. Each experiment was repeated at least three times with consistent results. Representative blots were shown. In the bar figure, each data represents mean ± SE (n = 8). *P < 0.05, **P < 0.01.
3.3. Knockdown of P50 gene led to reduction of SREBP1c
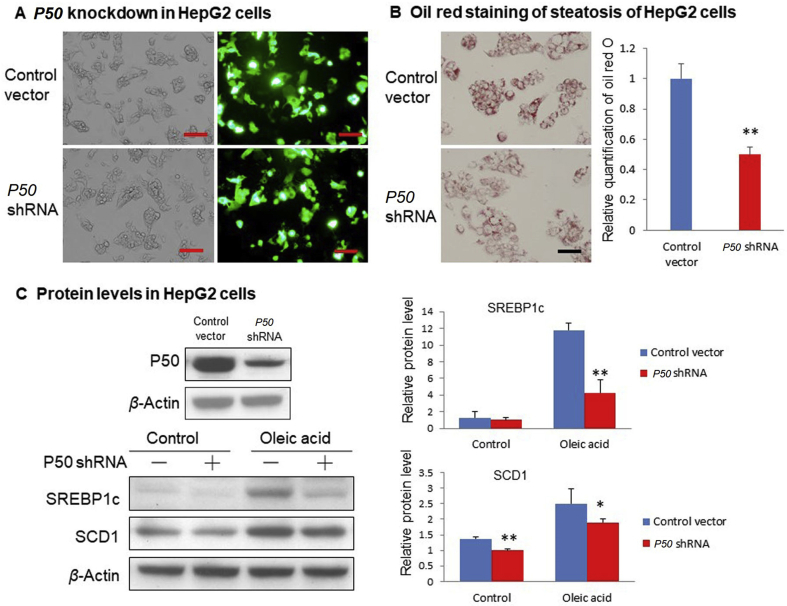
Above data suggest that the reduction in SREBP1c protein accounts for the attenuation of hepatic steatosis in the P50-KO mice. This led us to investigate the impact of P50 in SREBP1c protein in the cellular system with the knockdown strategy in HepG2 cells. The P50 knockdown was performed with an expression vector of P50 shRNA in transient transfection. shRNA was highly expressed in the cells following transfection (Fig. 3A), and P50 protein abundance was significantly reduced in the transfected cells (Fig. 3C). Steatosis model was established in the cells with the oleic acid treatment as indicated by the increased oil red O staining (Fig. 3C). In the model, the steatosis was reduced by P50 shRNA (Fig. 3B). Abundance of SREBP1c protein was significantly reduced in the knockdown cells, and the reduction was more significant in the steatosis condition (Fig. 3C). These data suggest that inhibition of P50 activity by gene knockdown reduces steatosis in HepG2 cells with a reduction in SREBP1c protein, which recapitulates the changes in the liver of P50-KO mice. The data supports a specific-connection of P50 and SREBP1c in the regulation of steatosis.
Figure 3.
Impact of P50 knockdown on lipogenic proteins in HepG2 cells. (A) HepG2 cells were electrically transfected with P50 shRNA expression plasmid using cell line nucleofector Kit L, program T031. Pictures were taken using a microscopy with 10 × object lenses under fluorescent light. Scale bar: 200 μm. (B) Oil red staining of triglyceride in transfected HepG2 cells after 0.3 mmol/L oleic acid treatment for 24 h. Relative triglyceride content was quantified in the HepG2 cells by measuring the oil red O amount in the samples. Scale bar: 100 μm. (C) Protein levels of P50, SREBP1, SCD1 and PPARγ were detected by Western blot in HepG2 cells. In the control group, HepG2 cells were transfected with P50 shRNA plasmid or control vector, and then cultured with DMEM only. In the oleic acid group, transfected cells were treated with 0.3 mmol/L oleic acid in DMEM for 24 h. In the bar figure, each data represents mean ± SE (n = 3). *P < 0.05, **P < 0.01.
3.4. P50-KO led to down-regulation of HDAC1 protein in hepatocytes
Above data suggest that SREBP1c protein reduction is not a result of a mRNA expression, suggesting a mechanism of protein level regulation. Given that stability of SREBP1c protein is regulated by acetylation16, we examined histone deacetylases including HDAC1, HDAC3 and SIRT1 in the liver for the SREBP1c reduction. HDAC1 protein was dramatically reduced in the KO mice relative to the WT mice, and the reduction was observed under both lean and obese conditions (Fig. 4A and B). HDAC3 and SIRT1 proteins were not changed in the same condition. Interestingly, Hdac1 mRNA was not decreased, but slightly increased in the P50-KO mice (Fig. 4C), suggesting that the HDAC1 reduction is unlikely a result of the transcriptional suppression. HDAC1 protein was elevated in the steatosis liver of WT mice together with SREBP1c (Fig. 4D), suggesting that HDAC1 may be required for the SREBP1c elevation in the hepatic steatosis. In contrast to HDAC1, SIRT1 protein was down-regulated and HDAC3 was not changed in the steatosis of WT mice (Fig. 4D). The association of HADC1 and SREBP1c suggests that HDAC1 may regulate SREBP1c in the hepatocytes.
Figure 4.
Reduction of HDAC1 protein in the liver of P50-KO mice. HDAC1 protein was decreased in the liver of P50-KO mice compared with that of WT mice. HDAC1 down-regulation was associated with the decrease in hepatic steatosis. (A) Nuclear protein in the liver tissue of P50-KO mice on chow diet. (B) Nuclear protein in the liver of P50-KO mice on HFD. (C) mRNA expression. mRNA level of HDAC1, HDAC3, SIRT1 was determined in the liver tissue of P50-KO mice on chow diet and HFD, respectively. (D) Proteins in liver tissue of WT mice on HFD. Western blot was performed for HDAC1, HDAC3, SIRT1, SCD1 and SREBP1 proteins in liver tissue of WT mice on HFD for 4 months. In the bar figure, each data represents mean ± SE (n = 8). *P < 0.05, **P < 0.01.
3.5. Knockdown of HDAC1 decreased SREBP1c in hepatocytes
To test above possibility, HDAC1 was knocked down in HepG2 cells by shRNA to mimic the liver condition of P50-KO mice. The HDAC1 protein was significantly reduced by the highly expressed shRNA in the transfected cells (Fig. 5A and D). The hepatic steatosis was decreased in the knockdown cells as indicated by the oil red O staining and total triglyceride quantification (Fig. 5B and C). A reduction in SREBP1c and SCD1 proteins was detected in the Hdac1 knockdown cells (Fig. 5D). PPARγ and HDAC3 were not changed in the cells, which enforces the relationship of HDAC1 and SREBP1c. A similar reduction in SREBP1c protein was observed with HDAC1 suppression by a chemical inhibitor (Fig. 5D). These data suggest that HDAC1 activity is required for the maintenance of SREBP1c protein abundance in hepatocytes.
Figure 5.
Inhibition of steatosis by Hdac1 knockdown in HepG2 cells. (A) Hdac1 shRNA transfection. HepG2 cells were electrical transfected with Hdac1 shRNA plasmid or control vector using cell line nucleofector Kit L, program T031. Pictures were taken using a microscopy with 10 × object lenses for transfection efficiency. Scale bar: 200 μm. (B) Impact of Hdac1 knockdown in steatosis. Oil red staining of triglyceride in transfected HepG2 cells after 0.5 mmol/L oleic acid treatment for 24 h. Scale bar: 100 μm. (C) Triglyceride quantification in the knockdown cells. (D) Suppression of lipogenic proteins by Hdac1 knockdown or chemical inhibitor. Western blot of HDAC1, HDAC3, SREBP1, SCD1, PPARγ protein in HepG2 cells after 0.5 mmol/L oleic acid treatment for 24 h. In the first column, Hdac1 was knocked down by shRNA. In the second column, HDAC1 was inhibited with HDAC1-specific inhibitor (0.4 mmol/L) for 30 min before oleic acid treatment in HepG2 cells. In the bar figure, each data represents mean ± SE (n = 3). *P < 0.05.
3.6. Succinylation of SREBP1c was increased in hepatic steatosis of P50-KO mice
Succinylation is a protein post-translational modification originally reported in 201118,23. It modifies proteins in both nucleus and cytoplasm24, 25, 26. Succinylation modification is extensive in hepatocytes, and an elevation is reported in some proteins under the hepatic steatosis27,28. However, the succinylation status was not known in the liver of P50-KO mice. To address this issue, we examined protein succinylation in the liver tissue with the anti-succinyllysine antibody in Western blot. The signal was observed in 3–4 major bands at the molecular weights between 130 and 55 kDa, which were significantly reduced by hepatic steatosis in the obese WT mice (Fig. 6A). However, the signal was significantly increased in the obese P50-KO mice (Fig. 6B). The enhanced signal was observed in all bands in the P50-KO mice. In the lean mice, the increase was observed only in the 130 kDa band in the P50-KO mice (Fig. 6C). The data suggest that the succinylation signals were increased in the liver of P50-KO mice.
Figure 6.
Succinylation of SREBP1c protein. Liver tissue was obtained from mice on HFD for 4 months. (A) Succinylation signal in the liver tissue of WT mice on chow diet or HFD, respectively. (B) Succinylation signal in the liver tissue of WT and P50-KO mice on HFD. (C) Succinylation signal in the liver tissue of WT and P50-KO mice on chow diet. (D) Succinylation of SREBP1c protein in the liver tissue of P50-KO mice on HFD. SREBP1c was isolated from the liver tissue by immunoprecipitation and resolved in SDS-PAGE gel. Succinylation was detected in the IP product with the succinylation antibody. (E) De-succinylation of SREBP1c protein by HDAC1. The de-succinylation assay was conducted with the recombinant HDAC1 in the IP product of SREBP1c of P50-KO mice. In the bar figure, each data represents mean + SE (n=3). *P < 0.05, **P < 0.01.
The SREBP1c protein has a molecular weight (125 kDa) close to the succinylation band of 130 kDa. Succinylation of SREBP1c protein was examined in the IP product of SREBP1 protein with the succinylation antibody. The succinylation signal was observed in SREBP1c, and an increase was observed in P50-KO mice (Fig. 6D). The increase was associated with HDAC1 reduction, suggesting a substrate/enzyme relationship for SREBP1c and HDAC1. To test the relationship, the HDAC1 enzyme assay was conducted for de-succinylation of SREBP1c using the recombinant protein. The de-succinylation reaction was catalysed by HDAC1 in a dose-dependent manner (Fig. 6E). The data suggest that SREBP1c protein is subjected to modification by succinylation, which is modulated by HDAC1 through de-succinylation. In the liver of P50-KO mice, the succinylation was increased due to the lack of HDAC1 activity.
3.7. Regulation of HDAC1 oscillation by NF-κB
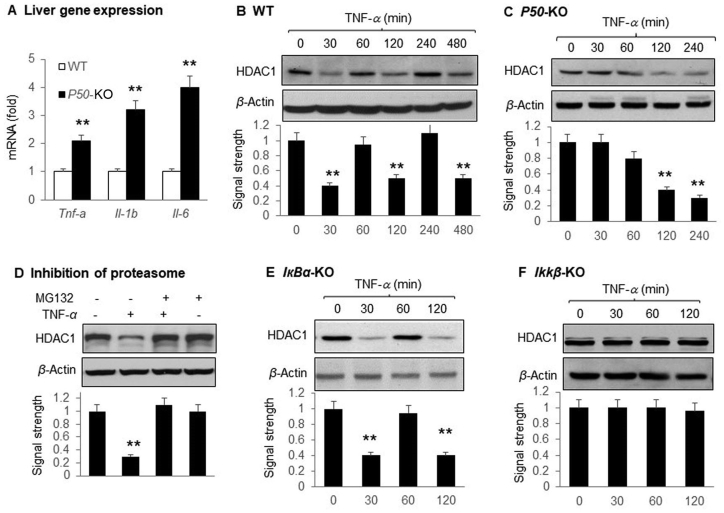
Above data suggest that regulation of HDAC1 by NF-κB is the key in the liver phenotype of P50-KO mice. However, the physiological relevance of the finding remains unknown. To address this issue, we tested HDAC1 response to TNF-α in the mouse embryonic fibroblast (MEF) cells. Tnf-α expression was elevated together with Il-1β and Il-6 in the liver of P50-KO mice (Fig. 7A), suggesting an enhanced inflammatory status. HDAC1 protein exhibited a pattern of oscillation in the wild-type MEF cells in response to NF-κB activation by TNF-α (Fig. 7B). HDAC1 was reduced by TNF-α at three time points (30, 120 and 480 min) with a recovery to the normal level between the reductions. MEF cells with different gene knockout in the NF-κB family were used to test the role of NF-κB in the TNF-α effect. In the P50-KO MEF cells, the HDAC1 oscillation disappeared, and a sustained HDAC1 reduction was observed under the TNF-α treatment (Fig. 7C). The HDAC1 reduction was blocked by the proteasome inhibitor MG-132 (Fig. 7D). In IкBα-KO cells in which NF-κB activation by TNF-α remains normal, the oscillation was not affected (Fig. 7E). In Ikkβ-KO cells, in which TNF-α could not activate NF-κB, TNF-α did not affect HDAC1 (Fig. 7F). The data suggest that NF-κB mediates the TNF-α signal in regulation of HDAC1, and P50 is required for the TNF-α effect. The HDAC1 protein degradation was dependent on proteasome in the absence of P50.
Figure 7.
Maintenance of HDAC1 protein level was dependent on P50. MEF cells with P50-KO, IκBα-KO and Ikkβ-KO were examined for HDAC1 after TNF-α (20 ng/mL) treatment at different times as indicated. The signal of HDAC1 protein was quantified and normalized with the signal of actin in the blot. (A) Liver gene expression. The expression was determined in the liver of P50-KO mice by qRT-PCR. (B) HDAC1 in the wild type MEF cells treated with TNF-α. (C) HDAC1 in the P50-KO MEF cells treated with TNF-α. (D) Blockage of HDAC1 reduction by proteasome inhibitor. HDAC1 protein was examined in P50-KO MEF cells that were pretreated with the proteasome inhibitor MG132 for 30 min followed by TNF-treatment for 120 min. (E) HDAC1 in the IκBα-KO MEF cells treated with TNF-α. (F) HDAC1 in the Ikkβ-KO MEF cells treated with TNF-α. Each experiment was repeated at least three times with consistent results. Representative blots were shown. In the bar figure, each data represents mean ± SE (n = 3). **P < 0.01.
3.8. Inactivation of NF-кB P65 did not alter the hepatic steatosis
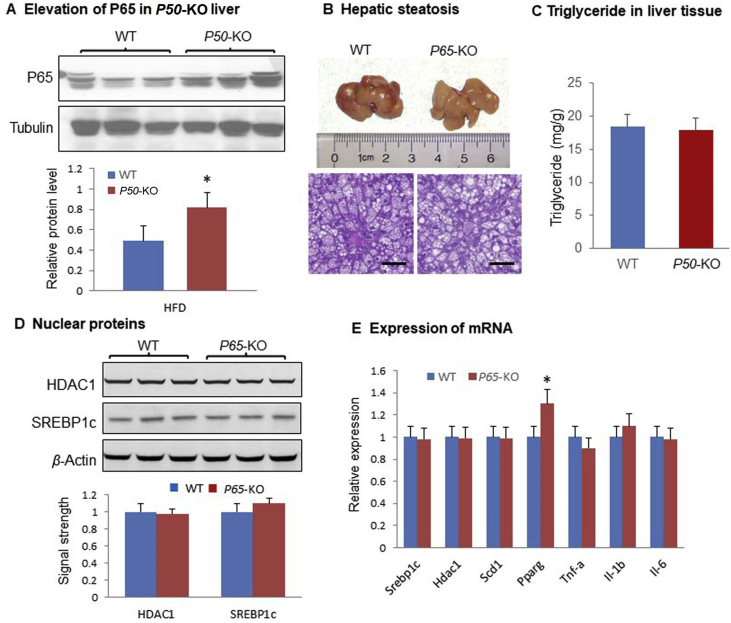
Above data suggests that the P50 subunit of NF-κB may regulate hepatic steatosis through an impact in the HDAC1/SREBP1c pathway. However, the role of P65 subunit remains unknown in the steatosis. To address this issue, P65 protein was examined in P50-KO mice and an elevation was observed the liver (Fig. 8A). The P65 activity was tested in the liver-specific P65-KO mice. The P65-KO mice did not exhibit any significant difference in the hepatic steatosis over the WT mice (Fig. 8B and C). A modest increase in the number of large lipid droplets was observed in the hepatocytes of P65-KO mice, but the difference did not lead to an increase in the triglyceride content (Fig. 8C). HDAC1 and SREBP1c did not show a difference in the liver between the P65-KO and WT mice (Fig. 8D). No difference was detected in mRNA of lipogenic genes and proinflammatory genes in the livers of P65-KO over WT mice except Pparγ (Fig. 8E). The mRNA of Pparγ was increased in the liver of P65-KO mice, which might account for the increased number of large lipid droplets. This group of data suggest that the P65 subunit did not play a direct role in the control of hepatic steatosis.
Figure 8.
Hepatic steatosis of P65-KO mice. (A) P65 protein in P50-KO liver. The P65 protein was examined in the liver of P50-KO mice on HFD. (B) Liver weight of P65-KO mice on HFD. The liver was collected from the mice at 14 weeks on HFD and the tissue image was taken under a microscope at 20 × from the tissue slide with H&E staining. Scale bar: 200 μm. (C) Triglyceride in liver tissue of mice on HFD. (D) Protein level in the liver tissues. (E) mRNA of lipogenic genes in the liver tissue. In the bar figure, each data represents mean ± SE (n = 7). *P < 0.05.
4. Discussion
Current study suggests that NF-κB may regulate hepatic steatosis through an impact in the HDAC1 activity by the P50 subunit. NF-κB is activated in response to multiple signals including TNF-α, endotoxin and reactive oxygen species, etc. It is known that the P50 subunit of NF-κB associates with HDAC1 on the promoter DNA of NF-κB target genes to regulate gene transcription. It is unknown if NF-κB regulates the abundance of HDAC1 protein in cells. Current study provides evidence that NF-κB indeed regulates the HDAC1 protein. The observation was made in the hepatocytes of P50-KO mice, in which HDAC1 protein was significantly decreased. The P50 impact on HDAC1 was confirmed in cells with P50 gene knockdown. The P50 impact was only observed in protein, but not mRNA of Hdac1, suggesting a role of post-translational modification in the control of HDAC1 protein by P50. The reduction is unlikely a result of suppression of protein synthesis as the liver development is normal in P50-KO mice. HDAC1 is required for normal differentiation of hepatocytes11. If HDAC1 protein synthesis was blocked, P50-KO liver should suffer a defect in development. Our data suggests that the HDAC1 reduction was a result of protein destabilization as the change was blocked by MG-132. The HDAC1 degradation is mediated by ubiquitination29. Other HDACs (HDAC3 and SIRT1) were not significantly altered in the liver of P50-KO mice, suggesting that P50 specifically regulates HDAC1. We speculate that P50 may stabilize HDAC1 through a protein–protein association to prevent HDAC1 degradation. However, the exact molecular event remains to be investigated for the HDAC1 degradation. The P65 subunit was reported to associate with HDAC130. However, the possibility is not supported by current study in P65-KO mice.
This study demonstrates that P65 acts in hepatocytes in the control of hepatic steatosis. Global P50 inactivation in mice was reported to generate several metabolic changes including resistance to diet-induced obesity and improvement in insulin resistance with an elevation in energy expenditure4,8,9,31. P50-KO mice exhibited an increase in energy expenditure and resistance to obesity as reported by this and other groups4,8,9. The reduction in hepatic steatosis may be a result of global increase in energy expenditure. However, the reduction in the liver to body weight ratio and the data of cellular models consistently support the direct role of P50 in hepatocytes. The hepatic effect may contribute to the global phenotype of P50-KO mice. The HDAC1 reduction is a potential mechanism for the reduced steatosis in P50-KO mice. Consistently, an increase in HDAC1 activity by gene overexpression enhances hepatic steatosis in mice although the mechanism was not clear32. In contracts to HDAC1, HDAC3 inhibits lipogenesis in the physiological conditions. The liver-specific Hdac3 knockout mice suffer severe hepatic steatosis33. HDAC3 may not contribute to the phenotype in this study for lack of HDAC3 alteration in the liver of P50-KO mice. In brown adipocytes, HDAC1 promotes lipid accumulation through inhibition of PGC-1α (peroxisome proliferator-activated receptor gamma co-activator 1alpha) activity13. Our data suggest that the P50 subunit of NF-κB promotes hepatic steatosis in the physiological condition by stabilization of HDAC1 protein in hepatocytes.
Our data suggests that HADC1 promotes hepatic steatosis through de-succinylation of SREBP1c protein in hepatocytes. There are two SREBP genes in mammalians. SREBP1 promotes fatty acid and triglyceride synthesis, and SREBP2 controls cholesterol synthesis. SREBP1c activity is induced by insulin through the PI3K/AKT pathway and by protein acetylation34. The activity of SREBP1c protein is inhibited by phosphorylation and ubiquitination-mediated protein degradation15,35. Succinylation was found as a new mechanism in the regulation of SREBP1c activity in current study. SREBP1c succinylation was found in P50-KO mice, and over modification of SREBP1c protein by succinylation was observed with the SREBP1c reduction. Succinylation occurs at the lysine residue in variety of proteins28. The succinylation status is regulated by SIRT5 through de-succinylation in mitochondria18,24. SIRT5 is located in the mitochondrion, and the identity of nuclear de-succinylation enzyme remains unknown. In current study, the succinylation signals were observed in the liver at molecules of 35–130 kDa, which was reduced by fatty liver in the WT mice. In P50-KO mice, the succinylation was increased under the none-obese and obese conditions. HDAC1 was found to de-succinylate SBREP1c protein in the enzyme assay, and lack of HDAC1 was responsible for the increased succinylation of SREBP1c in the P50-KO mice. The relationship among succinylation, acetylation and phosphorylation remains to be investigated in the SREBP1c protein in the future.
The P65 protein was elevated in the absence of P50 to induce expression of cytokines including TNF-α, IL-1 and IL-6 in the P50-KO liver. In the NF-κB dimer, P50 enhance the binding ability of P65 in the target gene DNA. However, the association reduces the transcriptional activity of P65 for lack of a transactivation domain in P50. In the absence of P50, the P50 homodimer binds to DNA with a low affinity, but higher transactivation ability as a result of doubled number of transactivation domain. This contributes to the elevated cytokine expression in the liver of P50-KO mice in current study. Kupffer cells are more active in the expression of the cytokines. When this happens in Kupffer cells and hepatocytes, more pro-inflammatory cytokines are produced. Current study suggests that the P65 subunit of NF-κB is not a major player in the pathogenesis of hepatic steatosis. In activation of P65 in the liver-specific P65-KO mice did not change the liver size, triglyceride content and body weight. The hepatic steatosis was not significantly altered in the P65-KO mice on HFD except the modest increase in the lipid droplet size, suggesting that P65 is not required for the hepatic steatosis.
5. Conclusions
This study demonstrates that the P50 subunit, not the P65 subunit, plays a direct role in hepatocytes in the regulation of steatosis. P50 protected HDAC1 activity through stabilization of the HDAC1 protein. The HDAC1 protein was reduced in the liver of P50-KO mice due to lack of the protection. HDAC1 preserved SREBP1c activity through de-succinylation, which was interrupted in the liver of P50-KO mice for super succinylation of SREBP1c and inhibition of hepatic steatosis. In the physiological condition, HDAC1 may stabilize SREBP1c protein through the de-succinylation activity to promote hepatic steatosis. These data suggest a novel signaling pathway of P50/HDAC1/SCREBP1c in hepatocytes in the control of hepatic steatosis. The pathway may apply to the pathogenesis of hepatic steatosis under endotoxin challenge or inflammatory status. However, this pathway remains to be explored further to identify the exact mechanisms for degradation of HDAC1 and SREBP1c proteins. A role of the gut microbiota, which influences hepatic steatosis36, remains to be investigated in the phenotype of P50-KO mice.
Acknowledgments
This study was funded by the National Key Research and Development Program of China (2018YFA0800603 to JY) the starting fund at Shanghai Jiao Tong University Affiliated Sixth People's Hospital to Jianping Ye. The animal study was conducted at the Pennington Biomedical Research Center, Louisiana State University, Baton Rouge, LA 70808, USA.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.02.005.
Author contributions
Yunwei Guo, Xiaoying Zhang, Zhiyun Zhao, Hongyun Lu, Bilun Ke and Xin Ye conducted the experiments and data analysis, involved in preparation of the figures and manuscript. Bin Wu and Jianping Ye designed the study and prepared the manuscript. All authors have read the manuscript and agreed to the conclusion.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang X.A., Zhang R., She Z.G., Zhang X.F., Jiang D.S., Wang T. Interferon regulatory factor 3 constrains IKKβ/NF-κB signaling to alleviate hepatic steatosis and insulin resistance. Hepatology. 2014;59:870–885. doi: 10.1002/hep.26751. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y., Sheng L., Xiong Y., Shen H., Liu Y., Rui L. Liver NF-κB-inducing kinase promotes liver steatosis and glucose counterregulation in male mice with obesity. Endocrinology. 2017;158:1207–1216. doi: 10.1210/en.2016-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Z., He Q., Peng B., Chiao P.J., Ye J. Regulation of nuclear translocation of HDAC3 by IκBα is required for tumor necrosis factor inhibition of peroxisome proliferator-activated receptor γ function. J Biol Chem. 2006;281:4540–4547. doi: 10.1074/jbc.M507784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang T., Zhang J., Yin J., Staszkiewicz J., Gawronska-Kozak B., Mynatt R. Uncoupling of inflammation and insulin resistance by NF-κB in transgenic mice through induction of energy expenditure. J Biol Chem. 2010;285:4637–4644. doi: 10.1074/jbc.M109.068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao P., Feng B., Ma J., Nie Y., Paul E., Li Y. Constitutive activation of IKKβ in adipose tissue prevents diet-induced obesity in mice. Endocrinology. 2012;153:154–165. doi: 10.1210/en.2011-1346. [DOI] [PubMed] [Google Scholar]
- 6.Ke B., Zhao Z., Ye X., Gao Z., Manganiello V., Wu B. Inactivation of NF-κB p65 (RalA) in liver improved insulin sensitivity and inhibited cAMP/PKA pathway. Diabetes. 2015;64:E496–E505. doi: 10.2337/db15-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Ibi D., Taniguchi K., Lee J., Herrema H., Akosman B. Inflammation improves glucose homeostasis through IKKβ–XBP1s interaction. Cell. 2016;167:1052–1066 e18. doi: 10.1016/j.cell.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minegishi Y., Haramizu S., Misawa K., Shimotoyodome A., Hase T., Murase T. Deletion of nuclear factor-κB p50 upregulates fatty acid utilization and contributes to an anti-obesity and high-endurance phenotype in mice. Am J Physiol Endocrinol Metab. 2015;309:E523–E533. doi: 10.1152/ajpendo.00071.2015. [DOI] [PubMed] [Google Scholar]
- 9.Bhatt B.A., Dedousis N., Sipula I.J., O'Doherty R.M. Elevated metabolic rate and skeletal muscle oxidative metabolism contribute to the reduced susceptibility of NF-κB p50 null mice to obesity. Phys Rep. 2018;6 doi: 10.14814/phy2.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagger G., O'Carroll D., Rembold M., Khier H., Tischler J., Weitzer G. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko S., Russell J.O., Tian J., Gao C., Kobayashi M., Feng R. HDAC1 regulates differentiation of bipotent liver progenitor cells during regeneration via SOX9B and CDK8. Gastroenterology. 2019;156:187–202. doi: 10.1053/j.gastro.2018.09.039. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiper-Bergeron N., Wu D., Pope L., Schild-Poulter C., Hache R.J. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 2003;22:2135–2145. doi: 10.1093/emboj/cdg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Wu R., Cui X., Zha L., Yu L., Shi H. Histone deacetylase 1 (HDAC1) negatively regulates thermogenic program in brown adipocytes via coordinated regulation of histone h3 lysine 27 (h3k27) deacetylation and methylation. J Biol Chem. 2016;291:4523–4536. doi: 10.1074/jbc.M115.677930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan L., Xiang W., Xie B., Yu L. Molecular mechanisms of fatty liver in obesity. Front Med. 2015;9:275–287. doi: 10.1007/s11684-015-0410-2. [DOI] [PubMed] [Google Scholar]
- 15.Sundqvist A., Bengoechea-Alonso M.T., Ye X., Lukiyanchuk V., Jin J., Harper J.W. Control of lipid metabolism by phosphorylation-dependent degradation of the srebp family of transcription factors by scf(fbw7) Cell Metabol. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Giandomenico V., Simonsson M., Gronroos E., Ericsson J. Coactivator-dependent acetylation stabilizes members of the srebp family of transcription factors. Mol Cell Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y., Hu H., Qu S., Wang J., Hua C., Zhang J. Sirt5 deacylates metabolism-related proteins and attenuates hepatic steatosis in ob/ob mice. EBioMedicine. 2018;36:347–357. doi: 10.1016/j.ebiom.2018.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M.J. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappab kinase complex. J Biol Chem. 2002;277:48115–48121. doi: 10.1074/jbc.M209459200. [DOI] [PubMed] [Google Scholar]
- 20.Gao Z., Chiao P., Zhang X., Zhang X., Lazar M.A., Seto E. Coactivators and corepressors of NF-κB in IκBα gene promoter. J Biol Chem. 2005;280:21091–21098. doi: 10.1074/jbc.M500754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu F., Gao Z., Zhang J., Rivera C.A., Yin J., Weng J. Lack of SIRT1 (mammalian sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le J., Zhang X., Jia W., Zhang Y., Luo J., Sun Y. Regulation of microbiota–GLP1 axis by sennoside a in diet-induced obese mice. Acta Pharm Sin B. 2019;9:758–768. doi: 10.1016/j.apsb.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M., Dai L. Sirt5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colak G., Xie Z., Zhu A.Y., Dai L., Lu Z., Zhang Y. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. 2013;12:3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rardin M.J., He W., Nishida Y., Newman J.C., Carrico C., Danielson S.R. Sirt5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metabol. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Y., Hou T., Ping J., Chen G., Chen J. Quantitative succinylome analysis in the liver of non-alcoholic fatty liver disease rat model. Proteome Sci. 2016;14:3. doi: 10.1186/s12953-016-0092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Yuan S., Tian X., Lin S., Wei S., Hu T. ABIN1 inhibits HDAC1 ubiquitination and protects it from both proteasome- and lysozyme-dependent degradation. J Cell Biochem. 2018;119:3030–3043. doi: 10.1002/jcb.26428. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.-K., Kim J.-H., Lee Y.C., Cheong J., Lee J.W. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappa B, and serum response factor. J Biol Chem. 2000;275:12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- 31.Gao Z., Yin J., Zhang J., He Q., McGuinness O.P., Ye J. Inactivation of NF-κB p50 leads to insulin sensitization in liver through post-translational inhibition of p70s6k. J Biol Chem. 2009;284:18368–18376. doi: 10.1074/jbc.M109.007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang A.G., Seo S.B., Moon H.B., Shin H.J., Kim D.H., Kim J.M. Hepatic steatosis in transgenic mice overexpressing human histone deacetylase 1. Biochem Biophys Res Commun. 2005;330:461–466. doi: 10.1016/j.bbrc.2005.02.179. [DOI] [PubMed] [Google Scholar]
- 33.Papazyan R., Sun Z., Kim Y.H., Titchenell P.M., Hill D.A., Lu W. Physiological suppression of lipotoxic liver damage by complementary actions of HDAC3 and SCAP/SREBP. Cell Metabol. 2016;24:863–874. doi: 10.1016/j.cmet.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You M., Liang X., Ajmo J.M., Ness G.C. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Xu S., Mihaylova M.M., Zheng B., Hou X., Jiang B. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metabol. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L., Pang Y., Wang X., Wu Q., Liu H., Liu B. Ablation of gut microbiota alleviates obesity-induced hepatic steatosis and glucose intolerance by modulating bile acid metabolism in hamsters. Acta Pharm Sin B. 2019;9:702–710. doi: 10.1016/j.apsb.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.