Abstract
Rhizobia supply legumes with fixed nitrogen using a set of symbiosis genes. These can cross rhizobium species boundaries, but it is unclear how many other genes show similar mobility. Here, we investigate inter-species introgression using de novo assembly of 196 Rhizobium leguminosarum sv. trifolii genomes. The 196 strains constituted a five-species complex, and we calculated introgression scores based on gene-tree traversal to identify 171 genes that frequently cross species boundaries. Rather than relying on the gene order of a single reference strain, we clustered the introgressing genes into four blocks based on population structure-corrected linkage disequilibrium patterns. The two largest blocks comprised 125 genes and included the symbiosis genes, a smaller block contained 43 mainly chromosomal genes, and the last block consisted of three genes with variable genomic location. All introgression events were likely mediated by conjugation, but only the genes in the symbiosis linkage blocks displayed overrepresentation of distinct, high-frequency haplotypes. The three genes in the last block were core genes essential for symbiosis that had, in some cases, been mobilized on symbiosis plasmids. Inter-species introgression is thus not limited to symbiosis genes and plasmids, but other cases are infrequent and show distinct selection signatures.
Keywords: rhizobia, white clover, genome assembly, introgression, conjugation, symbiosis
Data Summary
The genomic data that support the findings of this study were deposited in the INSDC database under BioProject ID PRJNA510726 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA510726/). Accessions numbers are from SAMN10617942 to SAMN10618137 consecutively and are also provided in Table S10 (available in the online version of this article).
Orthologous gene alignments and SNP matrices are available on FigShare (file Data.zip): https://doi.org/10.6084/m9.figshare.11568894.v5
The source code of the present analyses is available at https://github.com/izabelcavassim/Rhizobium_analysis
Impact Statement.
Bacteria are notorious for horizontal gene transfer, and it has often been asserted that this process will blur the boundaries between species to the extent that species cannot be clearly defined. Our study provides strong evidence that this is not necessarily so. Using 196 newly sequenced genomes of the Rhizobium leguminosarum species complex, we find five clearly distinct genospecies that occur in sympatry but show little evidence of recent between-species gene transfer affecting either the core or the accessory genome, except for a few highly mobile genetic regions. These findings are based on a global analysis of gene introgression, where we develop and apply novel methods for detecting introgression events and for grouping introgressed genes based on patterns of intergenic linkage disequilibrium corrected for population structure. This study provides a significant advance in our understanding of gene-transfer rate and mechanisms among sympatric bacterial species.
Introduction
Mutation and meiotic recombination are the main sources of genetic variation in eukaryotes. In contrast, prokaryotes can rapidly diverge through other types of genetic exchange collectively known as horizontal gene transfer (HGT). These include transformation (through the cell membrane), transduction (through a vector) and conjugation (through cell-to-cell contact) [1, 2]. These processes can introgress adaptive genes to distantly related species, creating specific regions of high genetic similarity.
It has often been suggested that HGT would blur the boundaries between species to the extent that species phylogenies would be better represented by a net-like pattern than a tree [3]. This notion may have arisen when the methods for studying prokaryotic evolution and species delineation were still rudimentary, making it challenging to accurately evaluate the rate of HGT [4]. With ever-increasing numbers of bacterial whole-genome sequences (WGS), it has become possible to re-evaluate bacterial-species classification [5, 6]. HGT, or introgression, events can be inferred using parametric or phylogenetic methods. Parametric methods rely on comparing gene features such as nucleotide composition or k-mer frequencies to a genomic average in order to detect outliers, which may be associated with HGT [7, 8]. Explicit phylogenetic methods are based on comparisons of gene and species trees aimed at detecting topological differences, whereas implicit phylogenetic methods rely on detecting aberrant distances to an outgroup reference [9].
Whether sympatric species frequently exchange genetic material through HGT is still an open question and the nitrogen-fixing symbiont of legumes, Rhizobium leguminosarum , is a useful model for investigating inter-species introgression through HGT. There is extensive literature documenting the sharing of symbiosis-related genes among distinct, and sometimes distant, species of rhizobia [10–12]. This occurs whether the genes are on plasmids [13–18] or on conjugative chromosomal islands [19, 20]. It has been previously observed that R. leguminosarum can be divided into distinct genospecies (gsA, gsB, gsC, gsD and gsE), but the host-specific symbiovars that nodulate white clover ( R. leguminosarum sv. trifolii) and vetch ( R. leguminosarum sv. viciae) are not confined to distinct genospecies [21]. This provides another example of symbiosis-gene transfer between sympatric rhizobia.
Symbiosis genes are known to increase the fitness of both symbiont and plant host (reviewed by [22]), but it is still unclear if their frequent introgression represents a special case, or if HGT is common for a wide range of genes in sympatric rhizobia. To address this question and obtain a more general understanding of introgression characteristics and mechanisms among sibling bacterial species, we assembled 196 R . leguminosarum genome sequences and carried out an unbiased introgression analysis.
Methods
Rhizobium sampling and isolation
White clover (Trifolium repens) roots were collected from three breeding trial sites in the United Kingdom (UK), Denmark (DK) and France (F) (Fig. S1a, available in the online version of the article), and 50 Danish organic fields (DKO) (Fig. S1b). Roots were sampled from 40 plots from each trial site. The total number of plots was 170. The samples were stored at ambient temperature for 1–2 days and in the cold room (2 °C) for 2–5 days prior to processing. Pink nodules were collected from all samples, and a single bacterial strain was isolated from each nodule as described in [23]. From each plot, one to four independent isolates were sampled. In total, 249 strains were isolated from T. repens nodules. For each site the clover varieties were known, and representative soil samples from clover-free patches were collected and sent for chemical analysis. Furthermore, latitude and longitude data were collected (Table S1).
Genome assembly
A set of 196 strains was subjected to whole-genome shotgun sequencing using 2×250 bp Illumina (Illumina, USA) paired-end reads by MicrobesNG (https://microbesng.uk/, IMI – School of Biosciences, University of Birmingham, UK). In addition, 8 out of the 196 strains were re-sequenced using PacBio (Pacific Biosciences of California, USA) sequencing technology (Table S2; Figs S3–S5). Analysis of 16S rDNA confirmed that all 196 of the strains were Rhizobium leguminosarum .
Genomes were assembled using SPAdes (v. 3.6.2) [24]. SPAdes contigs were cleaned and assembled further, one strain at a time, using a custom Python script (Jigome, available at https://github.com/izabelcavassim/Rhizobium_analysis/tree/master/Jigome). First, low-coverage contigs were discarded because they were mostly contaminants from other genomes sequenced in the same Illumina run. The criterion for exclusion was a SPAdes k-mer coverage less than 30% of the median coverage of putative single-copy contigs (those >10 kb). Next, putative chromosomal contigs were identified by the presence of conserved genes that represent the syntenic chromosomal backbone common to all R. leguminosarum genospecies. A list of 3215 genes that were present, in the same order, in the chromosomal unitigs of all eight of the PacBio assemblies was used to query the Illumina assemblies using blastn (≥90 % identity over ≥90% of the query length). In addition, contigs carrying repABC plasmid replication genes were identified using a set of RepA protein sequences representing the twenty distinct plasmid groups found in these genomes (tblastn search requiring ≥95% identity over ≥90 % of the query length). A ‘contig graph’ of possible links between neighbouring contigs was created by identifying overlaps of complete sequence identity between the ends of contigs. The overlaps created by SPAdes were usually 127 nt, although overlaps down to 91 nt were accepted. Contigs were flagged as ‘unique’ if they had no more than one connection at either end, or if they were >10 kb in length. Other contigs were treated as potential repeats. The final source of information used for scaffolding by Jigome was a reference set of R. leguminosarum genome assemblies that included the eight PacBio assemblies and 39 genomes publicly available in GenBank. A 500 nt tag near each end of each contig, excluding the terminal overlap, was used to search this database by blastn; high-scoring matches to the same reference sequence, with the correct spacing and orientation, were subsequently used to choose the most probable connections through repeat contigs. Scaffolding was initiated by placing all the chromosomal backbone contigs in the correct order and orientation, based on the conserved genes that they carried, and extending each of them in both directions, using the contig graph and the pool of remaining non-plasmid contigs, until the next backbone contig was reached or no unambiguous extension was possible. Then each contig carrying an identified plasmid origin was similarly extended as far as possible until the scaffold became circular or no further extension was justified, and unique contigs that remained unconnected to chromosomal or plasmid scaffolds were extended. Finally, scaffolds were connected if their ends had appropriately spaced matches in the reference genomes. Scaffold sequences were assembled using overlap sequences to splice adjacent contigs exactly, or inserting an arbitrary spacer of 20 ‘N’ symbols if adjacent contigs did not overlap. The dnaA gene (which was the first gene in the chromosomal backbone set and is normally close to the chromosomal origin of replication) was located in the first chromosomal scaffold, and this scaffold was split in two, with chromosome-01 starting 127 nt upstream of the ATG of dnaA and chromosome-00 ending immediately before the ATG. The remaining chromosomal scaffolds were numbered consecutively, corresponding to their position in the chromosome. Plasmid scaffolds were labelled with the identifier of the repA gene that they carried. Scaffolds that could not be assigned to the chromosome or a specific plasmid were labelled ‘fragment’ and numbered in order of decreasing size. Subsequent analysis revealed large exact repeats in a few assemblies. These were either internal inverted repeats in the contigs created by SPAdes (five instances) or large contigs used more than once in Jigome assemblies (18 instances). They were presumed to be artifacts and removed individually. Assembly statistics were generated with quast (v 4.6.3, default parameters) [25] (Fig. S3). Genes were predicted using prokka (v 1.12) [26]. In summary, genomes were assembled into 10 to 96 scaffolds, with total lengths of 6 966 649 to 8 355 366 bp containing 6,642 to 8,074 annotated genes, indicating that we have produced assemblies of reasonable quality, which comprehensively captured the gene content of the sequenced strains (Tables S2 and S3).
Orthologous gene prediction
Orthologous gene groups were identified among a total of 1 468 264 predicted coding sequences present across all (196) strains. We used two software packages for orthologue identification: Proteinortho [27] and Syntenizer3000 (https://github.com/izabelcavassim/Rhizobium_analysis/tree/master/Syntenizer3000). The software Proteinortho (v5.16b), was executed with default parameters and the synteny flag enabled, to predict homologous genes while taking into account their physical location. For the analysis in this paper, we were only interested in orthologues and not paralogues. Paralogous genes predicted by Proteinortho were filtered out by analysing the synteny of homologous genes surrounded by a 40-gene neighbourhood (see Synteny section). After this filtering step, the orthologous gene groups were aligned using clustalo ([28], v. 1.2.0). Each gene sequence was translated into its corresponding amino acid sequence before alignment and back-translated to the original nucleotides. Each gap was replaced by three gaps, resulting in a codon-aware nucleotide alignment.
Pan-genome analysis
Pan-genome analysis was based on a total of 22 115 orthologous genes across all (196) strains. Genomes were randomly added one at a time and core (set of shared genes) and pan (set of all unique gene families observed) genomes were estimated. This procedure was repeated 50 times and estimates were plotted (Fig. S8).
Synteny
First, gene groups were aligned with their neighbourhoods (20 genes each side) using a modified version of the Needleman–Wunsch algorithm [29]. We counted the number of neighbouring genes that were syntenic across strains before a collinearity break. We used this score to disambiguate gene groups that contain paralogues. Paralogues are the result of gene duplication, and, as such, one of the paralogues is the original, and the rest are copies. Based on similarity, we kept the least divergent gene inside of the original homology group while removing the copied paralogues, if possible into a new gene group designated group name "-2". Orphan genes, that were present in only one strain, were removed from the analysis.
Variant calling
Codon-aware alignments were used in order to detect single nucleotide polymorphisms (SNPs). For a given gene alignment and position, we first counted the number of unique nucleotides (A, C, T, G). Sites containing two unique nucleotides were considered variable sites (bi-allelic SNPs). After finding variable sites, SNP matrices were encoded as follows: major alleles were encoded as 1 and minor alleles as 0. Gaps were replaced by the site mean. Later steps were executed in order to filter out unreliable SNPs. PCA (Fig. S7b, c) and ANI (Fig. 1a) analyses were restricted to genes found in at least 100 strains. By looking at the variants and their codon context, we excluded multi-allelic SNPs and SNPs placed in codons containing gaps or more than one SNP. Based on these criteria we ended up with 6529 out of 22 115 genes and 441 287 SNPs. Scripts and pipelines are available at a GitHub repository (https://github.com/izabelcavassim/Rhizobium_analysis/).
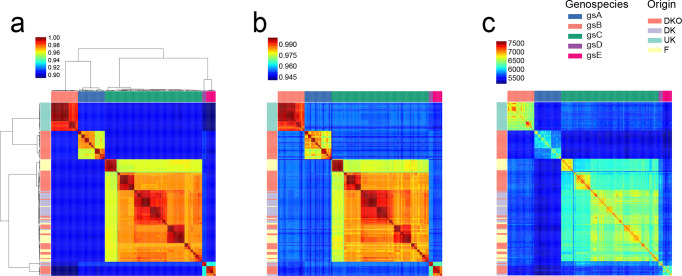
Fig. 1.
Genetic divergence across 196 rhizobium strains. Pairwise comparisons of genetic diversity were analysed at different levels. (a) Proportion of shared SNPs in genes that were present in at least 100 strains and that passed filtering criteria (6529 genes, 441 287 SNPs). Clusters of strains with SNP identity above 96 % were recognized as five genospecies: gsA (blue), gsB (salmon), gsC (green), gsD (purple), gsE (pink) as indicated in the legend. (b) Average nucleotide identity for concatenated sequences of 305 housekeeping genes. (c) Number of shared genes. Strain origins are indicated by coloured bars at the left (DKO in red, DK in purple, F in yellow, and the UK in green). Strains were ordered by clustering of the SNP data.
Plasmid replicon groups
Plasmid replication genes (repABC operons) were located in the genome assemblies by tblastn, initially using the RepA protein sequences of the reference strain 3841 (see Data bibliography) as queries [30]. Hits covering ≥70 % of the query length were accepted as repA genes, and those with ≥90 % amino acid identity were considered to belong to the same replication group (putative plasmid compatibility group). Hits with lower identity were used to define reference sequences for additional groups, using sequences from published Rhizobium genomes when available, or from strains in this study. Groups were numbered (Rh01, etc) in order of decreasing abundance in the genome set. RepB and RepC sequences corresponding to the same operons as the RepA references were used to check whether the full repABC operon was present at each location, requiring ≥85 % amino acid identity.
Presence of symbiosis genes in all strains
Since all sequenced strains were isolated from white-clover nodules, they are expected to carry the canonical symbiosis genes. One strain, SM168B, carried no symbiosis genes. Subsequent nodulation tests showed that the strain could colonize white clover and produce pink nodules, suggesting that the genes were lost during the pre-sequencing processing. On the other hand, strains SM165B and SM95 were found to have duplicated symbiosis regions.
Population genetic analysis
Population genetic parameters (Tajima’s D, nucleotide diversity, average pairwise differences and number of segregating sites) were estimated using the python library dendropy [31].
Introgression score
Despite the clear grouping of the 196 strains into distinct species, there was still extensive cross-species sequence conservation, allowing the construction of high-quality orthologous gene groups (Table S5). We took advantage of these for detecting introgression events by generating and traversing gene trees for each of the gene groups. Individual gene trees were first constructed using the neighbour-joining clustering method (software RapidNJ version 2.3.2) [32]. Each tree was traversed based on a depth-first traversal algorithm [33] by visiting each node after visiting its left child and before visiting its right child, searching deeper in the tree whenever possible. When the leaf of the tree was reached, the strain number and its genospecies origin were extracted. A list containing the genospecies was stored for the entire tree. The introgression score was computed as follows:
Introgression score=number of shifts -set(genospecies)+1.
The introgression score evaluates the number of times a shift (from one genospecies to another) is observed in a branch. The minimum possible is the total number of genospecies −1 shifts. A tree congruent to the species tree would have an introgression score equal to zero (Fig. S9).
Intergenic linkage disequilibrium corrected for population structure
Sample structure or relatedness between genotyped individuals leads to biased estimates of linkage disequilibrium (LD) and increase of type-I error. In order to correct for the population structure present in this data, the genotype matrix (coded as 0s and 1s) was adjusted as exemplified in [34] and [35].
The covariance between individuals was calculated as follows:
Let denote the total number of individuals and the total number of markers, the full genotype matrix () has dimensions with genotypes encoded as 0s and 1s. For simplicity, each SNP information is looked at as vectors, .
The first step of the calculations was to apply a Z-score normalization on the SNP vectors by subtracting each vector by its mean and dividing it by its standard deviation .
We then computed the covariance matrix between individuals as follows:
, can also be computed by the dot product of the genotype matrix:
The result is an matrix, where N is the number of strains. This matrix is also known as the genomic relationship matrix (GRM) [36]. We then decomposed the GRM matrix using the linalg function of scipy (python library).
Then the ‘decorrelation’ of the genotype matrix was achieved by multiplying by the inverse of the square root of as follows:
is therefore the pseudo SNP matrix, which is corrected for population structure.
The correlation between gene matrices (intergenic LD) was obtained by applying the Mantel test on the GRM (genetic distances) between pairs of genes:
For a data set composed of a distance matrix of gene X () and a genetic distance matrix of gene Y (), the scalar product of these matrices was computed, adjusted by the means and variances [var (x) and var (y)] of matrices X and Y:
The standardized Mantel test is actually the Pearson correlation between the elements of genes X and Y.
Filtering criteria for top introgressed genes
In order to identify genes that had trustable signals of introgression we used stringent filtering criteria as follows: number of sequences >50; number of segregating sites >10; average pairwise differences >10, ANI>0.7, introgression score >=10.
Results
Five distinct species constitute an R. leguminosarum species complex
Previous work has shown the existence of five distinct R. leguminosarum genospecies within one square meter of soil [21]. To acquire a broader diversity sample from a wider geographical area, we isolated 196 rhizobium strains from white-clover root nodules harvested in Denmark, France and the UK (Figs S1–S2; Table S1). We then sequenced and de novo assembled the genomes of all 196 strains, followed by genome annotation and construction of orthologous gene groups (Figs S3–S5; Table S2). To determine the relationship of our 196 strains with the previously identified genospecies, we constructed a phylogenetic tree containing rpoB sequences from known representatives of the five genospecies in addition to those from our 196 strains. This allowed us to assign all of our strains to a specific genospecies based on their position in the tree (Fig. S6). Since our extended sampling did not result in identification of additional genospecies, the five genospecies, gsA–E, likely represent a large part of northern European R. leguminosarum diversity.
The 196 strains shared a total of 4204 core gene groups, which had a higher median GC content than the 17 911 accessory gene groups (Fig. S7). This species complex has an open pan genome, since the total number of accessory genes identified increased indefinitely with the inclusion of more genomes. In contrast, the core genome gene count was stable at 4204 after inclusion of more than 100 genomes (Fig. S8).
We calculated average nucleotide identity (ANI) based on 305 conserved genes (Table S3) and on 6529 genes present in at least 100 strains, and clustered the strains based on pairwise ANI (Fig. 1a, b). The strains were collected from different countries and field-management regimes, but they clustered mainly by genospecies, although substructure related to geographic origin was also evident (Fig. 1a, b). These patterns were similar when clustering was carried out based on shared gene content (Fig. 1c; Fig. S7). In conjunction with earlier evidence that the standard whole-genome measure of ANI was lower than 0.95 for inter-genospecies comparisons [21], these results confirm that the genospecies should be considered genuinely distinct species constituting an R. leguminosarum species complex (Fig. 1a).
Plasmids are not genospecies specific
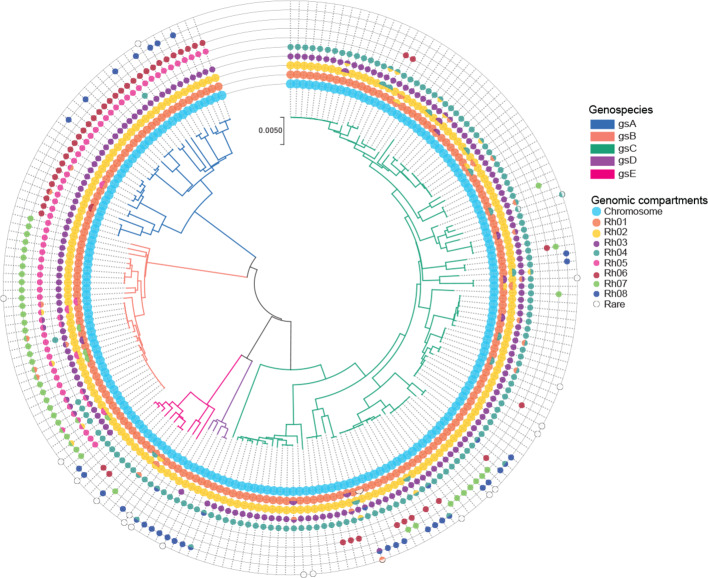
The genome of R. leguminosarum consists of a chromosome and a variable number of low-copy-number plasmids, including two that can be defined as chromids due to their size, ubiquitous presence across strains, and core-gene content [21, 30, 37]. In order to characterize the plasmid diversity within this species complex, we examined the sequence variation of a plasmid partitioning gene (repA) that is essential for stable maintenance of nearly all plasmids in Rhizobium . From all 196 genomes, 24 distinct repA sequence groups were identified. However, four of these correspond to isolated repA-like genes that are not part of repABC operons, and 12 others were rare (in no more than four genomes), so eight repA types account for nearly all plasmids identified (Fig. 2; Table S4). We numbered them Rh01 to Rh08 in order of decreasing frequency in the set of genomes. Of these, Rh01 and Rh02 correspond to the two chromids pRL12 and pRL11 of the reference strain 3841 [30] and are present in every genome. The distribution of the other plasmids shows some dependence on genospecies, but none are confined to a single genospecies. For example, Rh03 is present in all strains of gsA, gsB and gsC, but absent from gsE and in just one gsD strain, while Rh05 is universal in gsA and gsB but absent elsewhere (Fig. 2; Table S5).
Fig. 2.
Characterization of plasmid diversity. Species phylogeny based on the concatenation of 305 core genes using the neighbour-joining method. Branches are coloured by genospecies. Circles represent the genomic compartments observed in each strain. Chromids (Rh01 and Rh02) and plasmids (Rh03, Rh04, Rh05, Rh06, Rh07, Rh08) were defined based on the genetic similarity of the RepA plasmid partitioning protein.
Identification of introgression events based on gene trees
To evaluate the general rate of HGT within the present R. leguminosarum species complex, we developed a method to detect and quantify introgression (see Methods). For a given gene, present in all five genospecies, traversal of the gene phylogeny should encounter only four inter-species transitions if no introgression had occurred, yielding an introgression score of 0. All transitions in addition to the four expected would indicate introgression events, adding to the introgression score (Fig. S9). Most gene groups (55%) displayed very low introgression scores of 0 and 1 (Fig. 3a). On the other hand, only 2.43 % of the genes had an introgression score above or equal to 10, showing that introgression events were generally rare. After stringent filtering, we identified 171 genes with an introgression score above or equal to 10, indicating that they relatively frequently cross species boundaries (Fig. 3b).
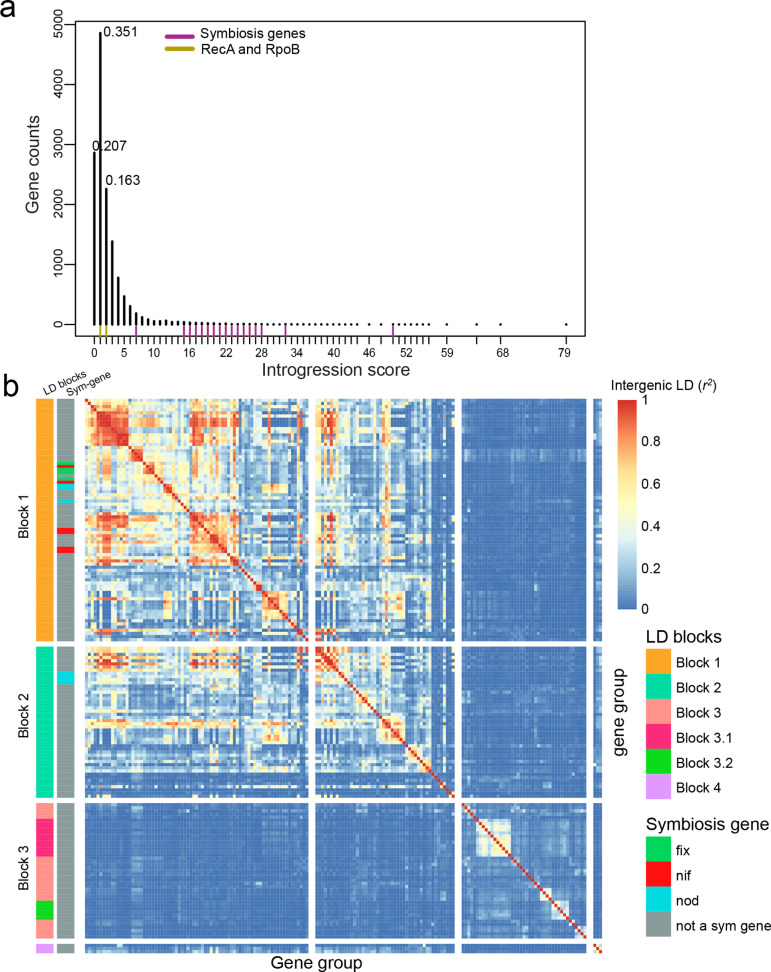
Fig. 3.
Introgression and LD analysis. (a) Distribution of introgression scores based on genes present in at least two genospecies (13 843), the frequencies of the top three introgression scores are shown. (b) Pairwise intergenic LD across highly introgressed genes (171 genes). The x and y axes represent genes ordered by LD clustering (complete method) rather than physical position. Genes were classified by LD blocks and by their contribution to symbiosis (left columns). The warmer the colour the greater the intergenic correlation (r 2). Chromosomal genes are found in LD block 3, while plasmid-borne genes are clustered in the first two blocks.
Clustering genes using population structure-corrected LD
Gene order was variable across the species complex (Fig. S10). To understand the nature of the genes displaying introgression, we therefore grouped them by linkage disequilibrium patterns rather than relying on the gene order of a single reference strain. The Mantel test is used to compare pairs of distance matrices, and here we used it to calculate intergenic LD by comparing genetic relationship matrices [36]. However, when population structure exists, this approach suffers from inflation [38], and we observed this effect in our data as unexpectedly high levels of LD between plasmid-borne symbiosis genes and chromosomal core genes (Fig. S11a). To address this issue, we calculated a genetic relationship matrix based on all SNPs and used it to generate pseudo-SNPs corrected for population structure for every gene (see Methods [34]). We then compared the gene pseudo-SNP genetic relationship matrices using the Mantel test in order to calculate intergenic LD. After this correction for population structure, symbiosis genes and chromosomal core genes no longer appeared to be in LD (Fig. S11b). We then proceeded to cluster the 171 genes that frequently crossed species boundaries based on their LD patterns (Fig. S12). The genes separated into four clusters, where LD blocks 1 and 2 comprised the plasmid-borne symbiosis genes, LD block 3 contained mainly chromosomal genes and block 4 comprised three genes with a distinct LD pattern (Fig. 3b; Fig. S13). It is worth noting that, because of our stringent criteria, the LD blocks detected by our method are just a representation of some of the introgressed genes within each LD block region (Fig. 4a–c).
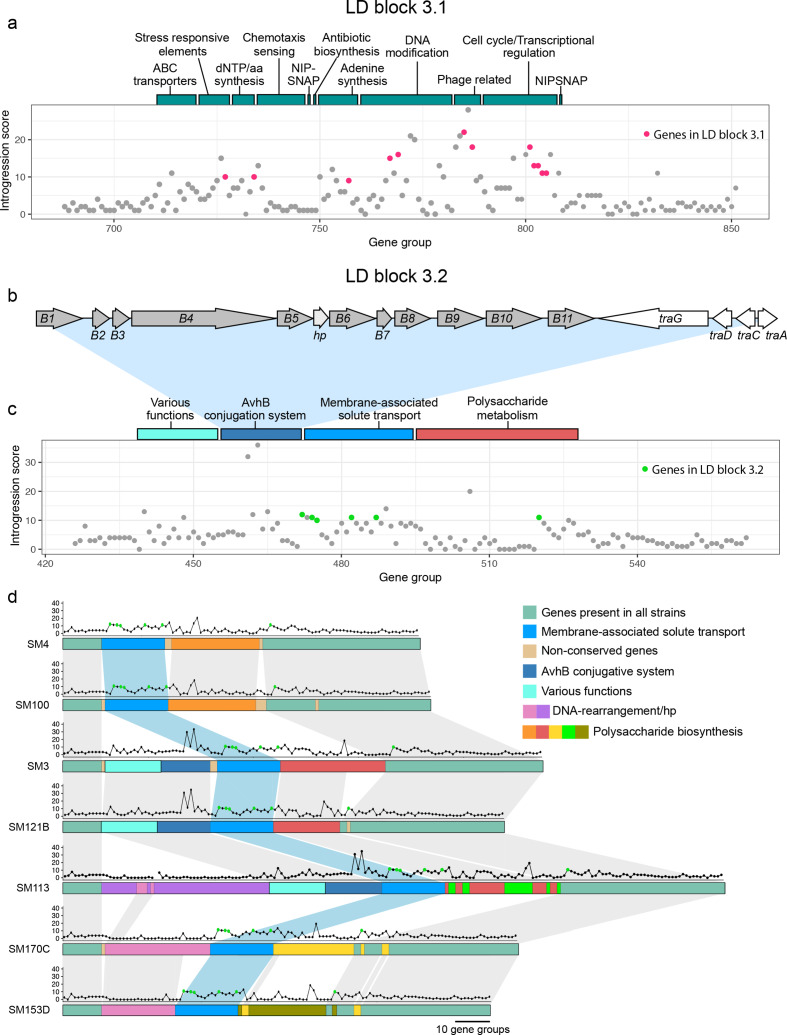
Fig. 4.
Functionality of chromosomal islands. (a) LD block 3.1 in strain SM3. Bars above the chart represent the classification of gene groups found in the region. (b) Gene organization of the avhB/tra type-IV secretion system from SM3. (c) Distribution of introgression scores for LD block 3.2. Coloured bars above the chart represent the classification of gene groups found in the area. (d) Illustration of synteny between gene groups in LD block 3.2 for strains lacking an insert (SM4, SM100), with the avhB/Tra conjugative system (SM3, SM121B), with a DNA rearrangement gene cluster (SM170C, SM153D), and one strain with both inserts (SM113). Dot plots above the gene group lines represent the introgression score for each gene in the gene group. Green dots represent the genes found in LD block 3.2.
Chromosomal introgression depends on specialized transfer systems
There was a clear substructure in the LD patterns among the genes in the chromosomal cluster (Fig. 3b, LD block 3), and we examined the larger LD blocks in greater detail. The largest block comprised 12 genes (Fig. 3b, LD block 3.1), most of which were present in nearly all of the 196 strains. Cluster 3.1 included a number of hypothetical proteins, a NIPSNAP family containing protein, a phage shock protein PspA and others (Table S9; Fig. 4a). We also observed toxin-antitoxin (VapC/YefM) genes (group696 and group697) in LD with this cluster (Table S7). However, we did not find genes that could directly explain the mobility of this introgressed region.
The second largest cluster (Fig. 3b, LD block 3.2) comprised six genes including a LysR family transcriptional regulator, an antibiotic biosynthesis monooxygenase, an exopolyphosphatase, TPR repeat-containing protein and an ABC transporter ATP-binding protein. To check whether the cluster could be in LD with genes that may explain its mobility, but which had not been detected by the stringently filtered introgression analysis, we extracted the genes in strongest LD with the six genes in the cluster 3.2 (Table S7). Three genes appeared to be in strong LD with at least one type-IV secretion protein. The introgressing genes were found in different genomic contexts, and are likely chromosomal core genes that have been mobilized by different types of transfer systems (Fig. 4b–d). In SM3 and SM121B the introgressing genes were downstream of a complete type-IV secretion system, which resembles the Agrobacterium tumefaciens AvhB system [39] (Fig. 4b–d). In SM170C and SM153D, another type of mobility system containing mostly hypothetical proteins along with some DNA-rearrangement genes and integrases neighboured the introgressed genes (Fig. 4d; Table S7). In SM4 and SM100 the same core genes are present, but the transfer system has likely been lost.
Symbiosis-gene introgression is driven by a few conjugative plasmids
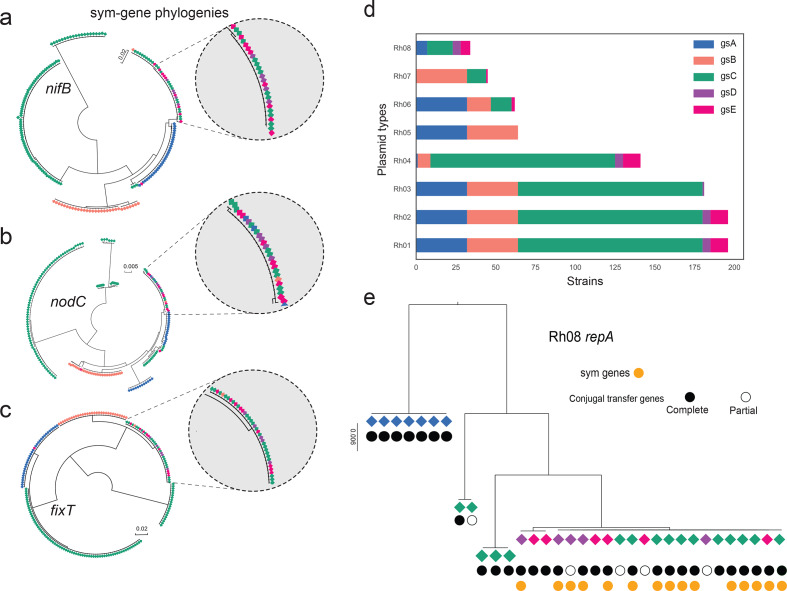
Symbiosis genes were in the tail of the introgression score distribution (Fig. 3a), and a detailed analysis of three symbiosis genes (nifB, nodC and fixT) confirmed these patterns of HGT (Fig. 5a–c). We also observed a complex LD pattern for the clusters comprising the symbiosis genes (Fig. 3b; LD block 1–2), which is consistent with the presence of multiple accessory genes in distinct symbiosis plasmids within the species complex. To understand the mechanisms behind sym-gene introgression, we investigated the symbiosis plasmids further. Where the assembly was complete enough to assign symbiosis genes to a specific plasmid, there was a clear pattern. Genospecies A symbiosis plasmids are all Rh06, in gsB they are Rh07, gsC has mostly Rh04 but some Rh07 and Rh08, gsD has Rh08, gsE has mostly Rh08 but some Rh06 and Rh07 (Fig. 2; Fig. 5d; Table S5). There are striking differences in the apparent mobility of these plasmids. Conjugal transfer genes (tra and trb) are present in some Rh06 plasmids and in all Rh07 and Rh08 plasmids, including those that are symbiosis plasmids. These transfer genes are all located together immediately upstream of the repABC replication and partitioning operon, in the same arrangement as in the plasmid p42a of R. etli CFN42, which has been classified as a class I, group I conjugation system [40]. Some repA sequences of sym-plasmids from strains of different genospecies are identical or almost identical in sequence (Fig. 5e and Fig. S14). The phylogenies of the corresponding conjugal transfer genes (e.g. traA, trbB and traG) show the same pattern (Fig. S15), indicating that symbiosis plasmids have crossed genospecies boundaries through conjugation. Rh08 is the most striking example (Fig. 5e), since all strains containing a Rh08 sym-plasmid were found in an introgressed clade (Fig. S15). We investigated the impact of this plasmid on introgression by repeating the introgression analysis in the absence of strains carrying Rh08. The mean introgression scores of all LD blocks decreased as a result of removing Rh08, but did not fully drop to background levels (Table 1). By randomly excluding the same number of strains and excluding them from the alignments we observed a slight decrease from 16.81 to 14.97 in the average introgression score across the 171 genes (Table S9). When we excluded all strains found in the fixT introgressed clade (Fig. 5c; Fig. S16), which includes strains carrying Rh08 or Rh07, the introgression scores of the plasmid-borne LD blocks (Fig. 3b, LD blocks 1 and 2) decreased greatly, whereas the chromosomal genes (Fig. 3b, LD block 3) were less affected (Table 1).
Fig. 5.
Evidence of horizontal symbiosis gene transfer between genospecies. (a)–(c) Examples of symbiosis-gene phylogenies, with insets showing clades in which identical alleles are shared across genospecies. (d) Distribution of plasmid groups, which were defined based on the genetic similarity of the RepA plasmid partitioning protein. (e) Phylogenetic analysis of the repA gene of plasmid type Rh08. A complete set of conjugal transfer genes has the following genes upstream of repA: traI,trbBCDEJKLFGHI,traRMHBFACDG, with the origin of transfer (oriT) between traA and traC. Partial sets are broken by the end of the scaffold, mostly after traM. The colour of the diamond indicates the genospecies origin with reference to (d).
Table 1.
Mean introgression scores
|
LD block |
Introgression score all strains |
Introgression score without Rh08 strains |
Introgression score without introgressed clade |
|---|---|---|---|
|
LD block 1 |
17.46 |
7.49 |
3.10 |
|
LD block 2 |
17.47 |
7.65 |
3.00 |
|
LD block 3 |
13.18 |
9.02 |
8.93 |
|
LD block 4 |
24.00 |
13.66 |
6.00 |
|
Symbiosis genes |
20.81 |
9.43 |
3.00 |
Symbiosis genes show a unique selection signature
The chromosomal and plasmid-borne genes that exhibited introgression were not in LD and their mobility appeared to depend on different transfer systems. We wanted to investigate if the differences between the two classes of genes displaying introgression extended to selection signatures. We therefore calculated Tajima’s D, which detects deviations from the expected level of nucleotide diversity based on the number of segregating sites and pairwise differences within each gene group. Across all 196 strains, only relatively few genes showed high Tajima’s D values (Table S5) indicating deviations from neutral evolution. The genes within the symbiosis clusters (LD blocks 1–2) were prominent among these, making up to 57 out of the genes with the top 250 Tajima’s D scores. Since Rh08 appeared to have spread rapidly with very limited accumulation of diversity, this plasmid could be the driver of the high Tajima’s D observed for the symbiosis genes. Again, we evaluated this by excluding Rh08-bearing strains from the analysis and re-calculating Tajima’s D (Table 2; Table S9). We found that plasmid LD blocks (LD blocks 1 and 2) showed decreased Tajima’s D values on exclusion of Rh08 strains, while Tajima’s D values for chromosomal genes (LD block 3) were generally unaffected (Table 2; Table S9). We then calculated Tajima’s D values exclusively for strains found in the introgressed clade (fixT, Fig. 5c), which includes both Rh08 and Rh07 carrying strains. The resulting Tajima's D values for the symbiosis genes were negative, consistent with fewer haplotypes than expected based on the number of segregating sites (Table 2).
Table 2.
Mean Tajima’s D values
|
LD block |
Tajima’s D all strains |
Tajima’s D without Rh08 strains |
Tajima’s D without introgressed clade |
Tajima’s D of only introgressed clade |
|---|---|---|---|---|
|
LD block 1 |
2.40 |
1.00 |
1.13 |
−0.81 |
|
LD block 2 |
1.90 |
0.66 |
0.51 |
0.08 |
|
LD block 3 |
0.45 |
0.42 |
0.47 |
0.54 |
|
LD block 4 |
−0.15 |
−0.32 |
−0.001 |
0.24 |
|
Symbiosis genes |
2.58 |
2.49 |
2.75 |
−0.62 |
Interestingly, after excluding all Rh08 strains or the clade of introgressed strains (Rh08 and some Rh07), symbiosis genes still retained high Tajima’s D values. This indicates that multiple symbiosis-gene haplotypes are also maintained at intermediate frequencies in the set of strains that does not exhibit symbiosis-gene introgression. Therefore, the elevated Tajima’s D values cannot be attributed solely to the existence of distinct versions of mobile symbiosis plasmids that have spread rapidly through the species complex.
Although the known symbiosis genes showed Tajima’s D patterns that were distinct from the average behaviour of the genes in the plasmid-borne LD blocks, there were other genes in these blocks that showed similar patterns (Table S8), suggesting that they may be either under direct selection, e.g. having unknown roles in symbiosis, or might be hitchhiking with symbiosis genes under selection.
Some fix genes show variation with respect to replicon location
Our LD analysis also singled out a small group of three genes that were in strong LD with each other, showed no LD with the chromosomal cluster and limited LD with the symbiosis cluster (Fig. 3b, LD block 4). These include fixH, fixG and a gene encoding an FNR-like protein, which are usually associated with a larger cluster of genes, fixNOQPGHIS, that are essential for symbiotic nitrogen fixation [30] However, they are atypical in several ways, as they have a high GC content similar to that of the core genome, they do not show the high Tajima’s D values we found typical of the main symbiosis genes, and they show variation with respect to the replicon they are associated with. In some strains, they are placed on symbiosis plasmids, in others they are located in the chromosome; other strains have two copies of the gene placed in two different genomic compartments (Table S5). The introgression signal is greatly reduced when the fixT introgressed clade is removed (Table 1), implying that most of the introgression of LD block 4 is mediated by the mobile Rh08 and Rh07 symbiosis plasmids.
Discussion
Robust detection of introgression events based on gene-tree traversal
HGT or introgression events in bacteria are often inferred using parametric or phylogenetic methods. Parametric methods [41–43] are most well suited for detecting introgression events between distantly related species, where introgression results in markedly different genomic signatures, such as abrupt changes in GC content. Detection of introgression between more closely related species, such as the members of the R. leguminosarum species complex described here, requires the use of phylogenetic methods that rely on gene trees derived from carefully constructed groups of orthologous genes. Because of the clear grouping of our strains into five distinct species (Fig. 1), we chose a simplified phylogenetic tree-traversal approach. Counting the number of transitions between genospecies on traversal proved to be a robust method for detecting introgression events, as we detected the symbiosis genes, which were candidates a priori. In addition, the method frequently detected groups of physically co-located and genetically linked genes, although the genes were analysed independently (Fig. 3b). The method is mainly limited by the accuracy of the gene trees and the level of differentiation between the species for each gene group, but we found that filtering away genes with too few segregating sites was efficient in controlling the false-positive rate. Another limitation is that our approach requires gene groups of a certain size, meaning that it cannot be used to detect introgression of accessory genes present at low frequency within the population. Here, we focused the introgression analysis on groups with more than 50 members, but we found no indication that the low-frequency genes showed extensive introgression either. The few genes present in groups with less than 50 members, which did show high introgression scores, were mostly annotated as transposases or associated with the introgressing chromosomal clusters (Table S6). The large proportion of singletons and low-frequency genes that were only present in a single genospecies could by definition not show introgression in our analysis, but have likely been acquired by HGT from strains outside the species complex.
Analysis of intergenic LD helps to resolve distinct introgression events
Within the R. leguminosarum species complex, the symbiosis genes are carried by different plasmid types (Fig. 2), and variation in gene order and content create complex syntenic relationships (Fig. S10). LD analysis is therefore a convenient way of understanding which introgressed genes travel together. The Mantel test has frequently been used in the comparison of genetic divergence with geographical distances [44]. In the present study, we have applied it to calculate the genetic correlations (LD) among genes by comparing their GRMs [36]. When autocorrelation of the GRM elements exists, possibly driven by population structure, then a relatively high false-positive rate is observed [45, 46]. Aware of this effect, we used the method proposed by [34] and corrected the bias due to population and phylogenetic structure. This approach is also frequently used for population structure correction in genome-wide association studies [47, 48]. To our knowledge, this is the first example of using the Mantel test combined with population structure-corrected pseudo-SNPs for estimation of intergenic LD. We found that calculating LD using this procedure resolved the LD inflation problem (Fig. S11), allowing us to reliably cluster the introgressed genes based on their LD patterns.
Introgression within the R. leguminosarum species complex is rare
Our introgression analysis clearly showed that genes travel across species boundaries within the species complex. Perhaps the most surprising finding was that the vast majority of genes showed no evidence of HGT, indicating that introgression events are rare. These results are in line with previous comparative genomic studies of sympatric isolates of rhizobium associated with common bean [16] and Medicago truncatula [49]. Based on the ratio of shared polymorphisms to fixed differences between 12 Sinorhizobium medicae and 32 S. meliloti strains, few HGT events (97 genes out of 3986 genes examined) were observed [49]. Nearly all of the HGT events involved plasmid-localized genes. Similar conclusions were drawn by Pérez-Carrascal et al [16]. Based on samples from a single agricultural field, Rhizobium species ( R. phaseoli , R. etli , R. leguminosarum , R. vallis and R. mesoamericanum ) were observed to co-exist in sympatry with low genetic recombination across their core genomes, but extremely similar symbiosis plasmids [16]. Another study, by Tian et al. [50], also examined recombination among core genes based on genomes of multiple rhizobium species, but reported a very high incidence of recombination because, by the Shimodaira–Hasegawa (SH) test [51], 247 out of 295 individual core-gene phylogenies were incongruent with the consensus phylogeny. At first sight, this is inconsistent with the findings of low HGT by Epstein et al., [49] Pérez-Carrascal et al [16]. and our present study, but Tian et al [50] included multiple strains of some species, so this incongruence may have reflected intraspecific recombination rather than interspecific HGT.
The sympatric, closely related species examined here were thus well-separated with respect to gene flow, and specialized, conjugative transfer mechanisms appear to be required for genes to cross species barriers. We found that one of the chromosomal introgressed regions (LD block 3.2) likely represented an integrative conjugative element (ICE). The avhB gene cassette and the traG gene of the type-IV secretion system of this putative ICE resembles a conjugative transfer system encoded by the virB/traG of the plasmid pSymA of S. meliloti [52, 53], and the virB/virD4 of Bartonella tribocorum [54]. However, both T4SSs in A. tumefaciens [55] and S. meliloti (AvhB and VirB, respectively) mediate the transfer of whole plasmids, whereas we are proposing that the T4SS encoded in LD block 3.2 mediates the transfer of an ICE. Other ICEs have been observed in the rhizobial genera (Mesorhizobium loti: [56]; Azorhizobium caulinodans : [57], Sinorhizobium : [58]) and in other species ( Streptococcus agalactiae : [59], Bacillus subtilis : [60], V. cholerae: [61]). Likewise, symbiosis plasmid transfer appears to require that the plasmids harbour a functional conjugal transfer system (traI,trbBCDEJKLFGHI,traRMHBFACDG), which is the case for all strains in the introgressed clade (Fig. 5; Fig S14–S16).
Symbiosis-gene transfer is mediated by conjugative plasmids
The occurrence of HGT of symbiosis genes within and between distant rhizobial genera ( Rhizobium , Bradyrhizobium , Sinorhizobium , Azorhizobium and Mesorhizobium ), nodulating different legume species, has been widely reported [10, 12, 16, 62, 63]. This shows that symbiosis-gene transfer is not restricted by genetic divergence and in many cases is not species specific [64, 65].
Here, we have shown that species-specific clades still exist even among symbiosis genes (Fig. 5a–c). In most species-specific clades, the genes were carried on a non-mobile symbiosis plasmid (Rh04) (Fig. S16), suggesting that, in this species complex, symbiosis-gene introgression was only observed when the strain had a plasmid with a conjugation apparatus. We verified this by characterizing the plasmid diversity within the strain pool. Symbiosis plasmids belong to a number of plasmid types (Rh04, Rh06, Rh07 and Rh08), and phylogenetic evidence indicated that some of them (Rh07 and Rh08) have been transferred through conjugation between different genospecies (Fig. 5e; Fig. S14). These transfers are likely recent since many of the sequences (repA and tra genes) have not yet diverged. Because conjugation requires cell-to-cell contact, plasmid transfer is not just constrained by genetic similarity [16, 66], but also by the requirement that the donor and recipient are found at the same location. This does not necessarily imply that the introgression events observed in this work occurred only among strains that are currently found at the same site. For instance, although most of the strains with introgressed sym genes were from Denmark, they were from a number of different farms (Table S1). It seems that the geographic mobility of these strains has a similar or shorter timescale than that of introgression.
This study, like previous work [21], has shown that distinct genospecies co-exist at one site, and also that the same genospecies are found in other regions where the local conditions (environment, soil type, rhizosphere) must be substantially different. Whether one genospecies is more adaptable to a multiplicity of environments than others is still an open question.
There is introgression of fix genes that vary in genomic location
The genes that displayed introgression and were on symbiosis plasmids (LD blocks 1 and 2) were not in LD with the introgressing chromosomal genes (LD block 3) (Fig. 3) and they displayed different selection signatures (Table 2), indicating that chromosomal and plasmid-associated introgression events are independent. LD block 4 was atypical, because it contained putative symbiosis genes that showed variation with respect to replicon location and were conspicuously absent from the immobile symbiosis plasmid Rh04 (Table S5). These genes are part of the fixNOQPGHIS cluster, and it is known that this set of genes is essential for symbiotic nitrogen fixation, but that a single copy is sufficient [30]. Nevertheless, their high GC content and frequent chromosomal location indicates that these are core genes that have been co-opted into a symbiotic role. Consistently, they show introgression when a copy has been acquired by one of the mobile types of symbiosis plasmid. This suggests that they have been mobilized as a consequence of their symbiotic function, perhaps because they confer an advantage when transferred to a recipient that does not have an optimal fixNOQPGHIS cluster for symbiosis.
Intermediate frequency symbiosis-gene haplotypes co-exist in sympatry
Just as the five genospecies co-exist, so do different symbiosis-gene haplotypes and plasmids. The symbiosis genes had strikingly high Tajima’s D values, indicating an excess of intermediate-frequency haplotypes (Table S8). The fixT gene is the gene with the fewest haplotypes, presenting only five haplotypes in total (Fig. S16). Four of these are present in the Danish organic fields, and the haplotype characteristic of the introgressed clade was found at trial sites in Denmark, France and the UK as well as in Danish organic fields (Table S5).
The presence of distinct groups of haplotypes at intermediate frequency could be a result of negative frequency-dependent selection [67–70]. This type of balancing selection could actively maintain symbiont diversity by increasing the fitness advantage of strains when they are rare. An alternative, not necessarily mutually exclusive hypothesis, is that distinct symbiosis haplotypes are maintained by host specialization. If the selective optimum between rhizobium and its host changes over time, symbiosis-gene alleles that contribute to the interaction will experience repeated partial sweeps, increasing the frequency of different adaptive alleles in different parts of the allelic range. Under balancing selection, these partial local sweeps can create elevated differentiation among allelic haplotypes and reduce nucleotide and haplotype diversity in the regions flanking each selected locus [71, 72].
The 196 strains characterized here were all collected from clover root nodules, and the colonization of nodules is a bottleneck that imposes strong selection. We see that certain haplotypes of symbiosis-related genes have introgressed across multiple genospecies, implying that these genes provide a fitness benefit that is largely independent of the genomic background. However, this pattern of selection appears to be exceptional, because the number of other genes that showed a similarly high introgression signal was very limited. Judging from our results, the high mobility of symbiosis genes, extensively documented in the literature, is not typical of the accessory genome in general, at least not for the accessory genes present at a frequency higher than 25 % within across the strains examined.
Conclusions
Using new methods for detection of introgression events and intergenic LD analysis, we carried out an unbiased investigation of introgression within an R. leguminosarum species complex. We found that introgression was generally very limited, with most genes displaying genetically distinct, species-specific variants. Striking exceptions are the genes located on symbiosis plasmids, especially the symbiosis genes, and a limited number of chromosomal islands, which appear to travel across species boundaries using conjugative transfer systems. The plasmid and chromosomal introgression events are independent and subject to different selective pressures, and some genes appear to move both between species and between replicons.
Data Bibliography
1. Genome assemblies: NCBI BioProject PRJNA510726: (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA510726/)
2. NCBI GenBank reference sequence (R. leguminosarum sv. viciae 3841: (https://www.ncbi.nlm.nih.gov/assembly/GCF_000009265.1/)
3. Orthologous gene alignments and SNP matrices: Data.zip on https://doi.org/10.6084/m9.figshare.11568894.v5
4. RepA types: representatives of repA types; Rh classification and nucleotide sequences: Supplementary tables (Table S4) on https://doi.org/10.6084/m9.figshare.11568894.v5
Supplementary Data
Funding information
This work was funded by grant no. 4105-00007A from Innovation Fund Denmark (S.U.A.). Genome sequencing was provided by MicrobesNG, which is supported by the BBSRC (grant number BB/L024209/1).
Acknowledgements
The authors would also like to thank industrial partners DLF Trifolium, SEGES and Legume Technology Ltd. for their contribution to the field trials.
Author contributions
Conceptualization: M.I.A.C., J.P.W.Y., S.M., M.H.S. and S.U.A.; Methodology: M.I.A.C., J.P.W.Y. and S.M.; Software: M.I.A.C., A.B., B.V., J.P.W.Y. and C.M.; Validation: M.I.A.C., C.M., S.M., J.P.W.Y.; Formal Analysis: M.I.A.C., J.P.W.Y., C.M., S.M., A.B., B.V. and B.F.; Investigation: S.M.; Resources: S.U.A., J.P.W.Y. and M.H.S.; Data curation: M.I.A.C., C.M., J.P.W.Y., S.M., S.U.A. and M.H.S.; Writing - Original Draft: M.I.A.C.; Writing - Review and Editing: M.I.A.C., J.P.W.Y., S.U.A., M.H.S., S.M., B.V.; Visualization: M.I.A.C., S.M., J.P.W.Y.; Supervision: S.U.A., J.P.W.Y., M.H.S; Project administration: S.U.A.; Funding acquisition: S.U.A.
Conflicts of interest
The authors declare that they have no competing interests.
Footnotes
Abbreviations: ANI, average nucleotide identity; GRM, genetic relationship matrix; HGT, horizontal gene transfer; ICE, integrative conjugative element; INSDC, International Nucleotide Sequence Database Collaboration; LD, linkage disequilibrium; PCA, principal component analysis; SNP, single nucleotide polymorphism; T4SS, type IV secretion system; WGS, whole genome sequencing.
The data that support the findings of this study are available in the INSDC databases under Study/BioProject ID PRJNA510726 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA510726/). Accessions numbers are from SAMN10617942 to SAMN10618137 consecutively.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Sixteen supplementary figures and ten supplementary tables are available with the online version of this article.
References
- 1.Hanage WP. Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb Perspect Biol. 2016;8:a018069. doi: 10.1101/cshperspect.a018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–M8. doi: 10.1016/S0962-8924(99)01664-5. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S, et al. High throughput ani analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 9.Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews M, De Meyer S, James EK, Stępkowski T, Hodge S, et al. Horizontal transfer of symbiosis genes within and between rhizobial genera: occurrence and importance. Genes. 2018;9:321. doi: 10.3390/genes9070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remigi P, Zhu J, Young JPW, Masson-Boivin C, et al. Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol. 2016;24:63–75. doi: 10.1016/j.tim.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Rogel MA, Ormeño-Orrillo E, Martinez Romero E, Romero EM. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol. 2011;34:96–104. doi: 10.1016/j.syapm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Cervantes L, Bustos P, Girard L, Santamaría RI, Dávila G, et al. The conjugative plasmid of a bean-nodulating Sinorhizobium fredii strain is assembled from sequences of two Rhizobium plasmids and the chromosome of a Sinorhizobium strain. BMC Microbiol. 2011;11:149. doi: 10.1186/1471-2180-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haukka K, Lindström K, Young JPW. Three phylogenetic groups of nodA and nifHGenes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl Environ Microbiol. 1998;64:419–426. doi: 10.1128/AEM.64.2.419-426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, et al. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2001;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- 16.Pérez Carrascal OM, VanInsberghe D, Juárez S, Polz MF, Vinuesa P, et al. Population genomics of the symbiotic plasmids of sympatric nitrogen-fixing Rhizobium species associated with Phaseolus vulgaris . Environ Microbiol. 2016;18:2660–2676. doi: 10.1111/1462-2920.13415. [DOI] [PubMed] [Google Scholar]
- 17.Segovia L, Young JP, Martínez-Romero E. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int J Syst Bacteriol. 1993;43:374–377. doi: 10.1099/00207713-43-2-374. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandasena KG, O'hara GW, Tiwari RP, Howieson JG. Rapid in situ evolution of nodulating strains for Biserrula pelecinus L. through lateral transfer of a symbiosis island from the original mesorhizobial inoculant. Appl Environ Microbiol. 2006;72:7365–7367. doi: 10.1128/AEM.00889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW, et al. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci U S A. 1995;92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar N, Lad G, Giuntini E, Kaye ME, Udomwong P, et al. Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum . Open Biol. 2015;5:140133. doi: 10.1098/rsob.140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesen ML. Widespread fitness alignment in the legume-rhizobium symbiosis. New Phytol. 2012;194:1096–1111. doi: 10.1111/j.1469-8137.2012.04099.x. [DOI] [PubMed] [Google Scholar]
- 23.Bailly X, Giuntini E, Sexton MC, Lower RPJ, Harrison PW, et al. Population genomics of Sinorhizobium medicae based on low-coverage sequencing of sympatric isolates. ISME J. 2011;5:1722. doi: 10.1038/ismej.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurevich A, Saveliev V, Vyahhi N, Tesler G, et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 27.Lechner M, Hernandez-Rosales M, Doerr D, Wieseke N, Thévenin A, et al. Orthology detection combining clustering and synteny for very large datasets. PLoS One. 2014;9:e105015. doi: 10.1371/journal.pone.0105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 30.Young JPW, Crossman LC, Johnston AWB, Thomson NR, Ghazoui ZF, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sukumaran J, Holder MT. DendroPy: a python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 32.Simonsen M, Pedersen CNS. Proceedings of the 2011 ACM Symposium on Applied Computing. ACM; 2011. Rapid computation of distance estimators from nucleotide and amino acid alignments; pp. 89–93. [Google Scholar]
- 33.Tarjan R. Depth-First search and linear graph algorithms. SIAM J Comput. 1972;1:146–160. doi: 10.1137/0201010. [DOI] [Google Scholar]
- 34.Mangin B, Siberchicot A, Nicolas S, Doligez A, This P, et al. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity. 2012;108:285. doi: 10.1038/hdy.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45:884. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 37.Harrison PW, Lower RPJ, Kim NKD, Young JPW, et al. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 2010;18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Guillot G, Rousset F. Dismantling the Mantel tests. Methods Ecol Evol. 2013;4:336–344. doi: 10.1111/2041-210x.12018. [DOI] [Google Scholar]
- 39.Chen L, Chen Y, Wood DW, Nester EW. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens . J Bacteriol. 2002;184:4838–4845. doi: 10.1128/JB.184.17.4838-4845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wetzel ME, Olsen GJ, Chakravartty V, Farrand SK. The repABC plasmids with Quorum-Regulated transfer systems in members of the Rhizobiales divide into two structurally and separately evolving groups. Genome Biol Evol. 2015;7:3337–3357. doi: 10.1093/gbe/evv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azad RK, Lawrence JG. Detecting laterally transferred genes: use of entropic clustering methods and genome position. Nucleic Acids Res. 2007;35:4629–4639. doi: 10.1093/nar/gkm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence JG, Ochman H. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 2002;10:1–4. doi: 10.1016/S0966-842X(01)02282-X. [DOI] [PubMed] [Google Scholar]
- 43.van Passel MWJ, Bart A, Thygesen HH, Luyf ACM, van Kampen AHC, et al. An acquisition account of genomic islands based on genome signature comparisons. BMC Genomics. 2005;6:163. doi: 10.1186/1471-2164-6-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, et al. Mantel test in population genetics. Genet Mol Biol. 2013;36:475–485. doi: 10.1590/S1415-47572013000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harmon LJ, Glor RE. Poor statistical performance of the Mantel test in phylogenetic comparative analyses. Evolution. 2010;64:2173–2178. doi: 10.1111/j.1558-5646.2010.00973.x. [DOI] [PubMed] [Google Scholar]
- 46.Rousset F. Partial Mantel tests: reply to Castellano and Balletto. Evolution. 2002;56:1874–1875. doi: 10.1111/j.0014-3820.2002.tb00204.x. [DOI] [Google Scholar]
- 47.Mamidi S, Lee RK, Goos JR, McClean PE. Genome-wide association studies identifies seven major regions responsible for iron deficiency chlorosis in soybean (Glycine max) PLoS One. 2014;9:e107469. doi: 10.1371/journal.pone.0107469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauvage C, Segura V, Bauchet G, Stevens R, Do PT, et al. Genome-Wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 2014;165:1120–1132. doi: 10.1104/pp.114.241521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Epstein B, Branca A, Mudge J, Bharti AK, Briskine R, et al. Population genomics of the facultatively mutualistic bacteria Sinorhizobium meliloti and S. medicae. PLoS Genet. 2012;8:e1002868. doi: 10.1371/journal.pgen.1002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A. 2012;109:8629–8634. doi: 10.1073/pnas.1120436109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimodaira H, Hasegawa M. Multiple comparisons of Log-Likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 52.Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, et al. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc Natl Acad Sci U S A. 2001;98:9883–9888. doi: 10.1073/pnas.161294798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galibert F, Finan TM, Long SR, Puhler A, Abola P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti . Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 54.Schulein R, Dehio C. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol Microbiol. 2002;46:1053–1067. doi: 10.1046/j.1365-2958.2002.03208.x. [DOI] [PubMed] [Google Scholar]
- 55.Alt-Mörbe J, Stryker JL, Fuqua C, Li PL, Farrand SK, et al. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmids is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid Vir genes. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/JB.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling J, Wang H, Wu P, Li T, Tang Y, et al. Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc Natl Acad Sci U S A. 2016;113:13875–13880. doi: 10.1073/pnas.1615121113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao R, Liu LX, Zhang YZ, Jiao J, Cui WJ, et al. Adaptive evolution of rhizobial symbiotic compatibility mediated by co-evolved insertion sequences. Isme J. 2018;12:101. doi: 10.1038/ismej.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae . Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 60.Merkl R. SIGI: score-based identification of genomic islands. BMC Bioinformatics. 2004;5:22. doi: 10.1186/1471-2105-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, et al. Dna sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirsch PR, Van Montagu M, Johnston AWB, Brewin NJ, Schell J, et al. Physical identification of bacteriocinogenic, nodulation and other plasmids in strains of Rhizobium leguminosarm. Microbiology. 1980;120:403–412. doi: 10.1099/00221287-120-2-403. [DOI] [Google Scholar]
- 63.Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, et al. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the core Cape subregion (South Africa) FEMS Microbiol Ecol. 2015;91:1–17. doi: 10.1093/femsec/fiu024. [DOI] [PubMed] [Google Scholar]
- 64.Greenlon A, Chang PL, Damtew ZM, Muleta A, Carrasquilla-Garcia N, et al. Global-level population genomics reveals differential effects of geography and phylogeny on horizontal gene transfer in soil bacteria. Proc Natl Acad Sci U S A. 2019;116:15200–15209. doi: 10.1073/pnas.1900056116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Provorov NA, Andronov EE, Onishchuk OP. Forms of natural selection controlling the genomic evolution in nodule bacteria. Russ J Genet. 2017;53:411–419. doi: 10.1134/S1022795417040123. [DOI] [Google Scholar]
- 66.Silva C, Vinuesa P, Eguiarte LE, Martínez-Romero E, Souza V, et al. Rhizobium etli and Rhizobium gallicum nodulate common bean (Phaseolus vulgaris) in a traditionally managed Milpa plot in Mexico: population genetics and biogeographic implications. Appl Environ Microbiol. 2003;69:884–893. doi: 10.1128/AEM.69.2.884-893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amarger N, Lobreau JP. Quantitative study of nodulation competitiveness in Rhizobium strains. Appl Environ Microbiol. 1982;44:583–588. doi: 10.1128/AEM.44.3.583-588.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bever JD. Dynamics within mutualism and the maintenance of diversity: inference from a model of interguild frequency dependence. Ecol Lett. 1999;2:52–61. doi: 10.1046/j.1461-0248.1999.21050.x. [DOI] [Google Scholar]
- 69.Provorov NA, Vorobyov NI. Interplay of Darwinian and frequency-dependent selection in the host-associated microbial populations. Theor Popul Biol. 2006;70:262–272. doi: 10.1016/j.tpb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 70.Provorov NA, Vorobyov NI. Population genetics of rhizobia: construction and analysis of an "Infection and Release" model. J Theor Biol. 2000;205:105–119. doi: 10.1006/jtbi.2000.2051. [DOI] [PubMed] [Google Scholar]
- 71.Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 2015;11:e1005004. doi: 10.1371/journal.pgen.1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoder JB, Stanton-Geddes J, Zhou P, Briskine R, Young ND, et al. Genomic signature of adaptation to climate in Medicago truncatula. Genetics. 2014;196:1263–1275. doi: 10.1534/genetics.113.159319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.