Abstract
The availability of next-generation sequencing techniques provides an unprecedented opportunity for the assignment of gene function. Streptococcus equi subspecies equi is the causative agent of strangles in horses, one of the most prevalent and important diseases of equids worldwide. However, the live attenuated vaccines that are utilized to control this disease cause adverse reactions in some animals. Here, we employ transposon-directed insertion-site sequencing (TraDIS) to identify genes that are required for the fitness of S. equi in whole equine blood or in the presence of H2O2 to model selective pressures exerted by the equine immune response during infection. We report the fitness values of 1503 and 1471 genes, representing 94.5 and 92.5 % of non-essential genes in S. equi , following incubation in whole blood and in the presence of H2O2, respectively. Of these genes, 36 and 15 were identified as being important to the fitness of S. equi in whole blood or H2O2, respectively, with 14 genes being important in both conditions. Allelic replacement mutants were generated to validate the fitness results. Our data identify genes that are important for S. equi to resist aspects of the immune response in vitro, which can be exploited for the development of safer live attenuated vaccines to prevent strangles.
Keywords: Streptococcus equi, transposon-directed insertion-site sequencing, whole blood, hydrogen peroxide
Data Summary
The DNA sequences generated in this study have been deposited in the European Nucleotide Archive under the accession number PRJNA578912.
Impact Statement.
Strangles, caused by Streptococcus equi subsp . equi , is one of the most frequently identified infectious diseases of horses worldwide, but the available live attenuated vaccines can survive for too long at the site of administration, leading to the development of adverse reactions. We applied a whole-genome approach to identify genes that are required in order for S. equi to grow in whole equine blood or in the presence of hydrogen peroxide, which simulate pressures exerted by the equine immune response. The simultaneous identification of every gene encoded within the DNA of S. equi that contributes to its ability to evade immune responses provides novel information about this important pathogen, and opens up exciting new opportunities for the design of safer and more effective vaccines with which to prevent strangles.
Introduction
Strangles, caused by Streptococcus equi subspecies equi , is one of the most frequently diagnosed infectious diseases of equids worldwide, and is responsible for considerable economic and welfare cost to the horse industry [1]. Following entry via the nasopharyngeal or oral routes, S. equi subsp. equi binds to and invades the mucosal epithelium before transitioning to the lymph nodes of the head and neck, where it can be identified within 3 h [2]. The presence of S. equi subsp. equi within lymph nodes induces infiltration of polymorphonuclear leukocytes, leading to swelling and abscessation, which may cause dysphagia, lending to this disease's common name of strangles [3].
The earliest vaccines against strangles used heat-killed bacteria, but whilst conferring significant levels of protection, these vaccines led to severe adverse reactions [4–8]. Cell-free-extract vaccines have also been developed, but published data suggested that the protection conferred by such vaccines was short-lived at best and that adverse reactions at the injection site remained a problem [9]. Recombinant subunit vaccines benefit from enhanced safety, and a new multi-component vaccine, Strangvac 4, has been shown to confer significant levels of protection against S. equi subsp. equi at 2 weeks post-combined intranasal and subcutaneous vaccination [10, 11]. However, Strangvac 4 is not yet available for use in horses.
Two live attenuated vaccines are available commercially for the prevention of strangles: Pinnacle IN [12] and Equilis StrepE [13]. These vaccines confer protection against challenge with S. equi subsp. equi , but the attenuated vaccine strains can cause adverse reactions and even strangles in some vaccinated animals [14–17]. A prototype live attenuated vaccine containing deletions in six genes conferred high levels of protection at 2 months post-second vaccination, but also led to adverse reactions when administered via the intramuscular route [18]. S. equi subsp. equi resists the equine immune system by producing known factors such as streptolysin S, immunoglobulin cleaving enzymes, a factor H-binding protein, SeM, fibronectin-binding proteins and a protective hyaluronic acid capsule [18–26]. However, it is likely that several other factors, which remain unidentified, are also employed to resist the equine immune system. The identification and attenuation of such factors provides an opportunity to enhance the safety of live attenuated vaccines.
The increased accessibility of functional genomics techniques has facilitated the development of a variety of transposon-insertion sequencing methods, which combine dense random mutant libraries and next-generation sequencing to identify essential bacterial genomes and assign gene function [27–33]. Exposure of mutant libraries to varying experimental conditions enables the relative fitness and conditional essentiality of each gene to be determined either in vitro [27, 34–36] or in vivo [37–42].
We developed a novel barcoded transposon-directed insertion-site sequencing (TraDIS) technique in S. equi subsp. equi utilizing pGh9:ISS1, which produces random, dense and stable transposon libraries [32]. To identify any novel genes involved in the survival of S. equi subsp. equi in the face of the equine immune system, S. equi mutant libraries were exposed to two conditions: whole equine blood and Todd–Hewitt broth (THB) containing hydrogen peroxide (H2O2). To validate the findings, a panel of six allelic replacement mutants were exposed to whole equine blood and H2O2, and the impact on their viability was measured.
Methods
Bacterial strains, DNA isolation and primers
S. equi subsp. equi strain 4047 (Se4047) was grown in THB at 37 °C in a humidified atmosphere containing 5 % CO2, unless otherwise stated. The Escherichia coli strain TG1 repA+ was used for the replication of the pGhost9 ISS1 plasmids at 37 °C. S. equi subsp. equi genomic DNA was extracted using GenElute spin columns (Sigma Aldrich), according to the manufacturer’s instructions. A list of all primers used in this study is available in Table S1 (available with the online version of this article).
Minimum inhibitory concentration (MIC) of hydrogen peroxide (H2O2)
To determine the concentration of H2O2 required to exert a selective pressure on S. equi subsp . equi , the MIC of H2O2 in THB was determined. An overnight culture of Se4047 was diluted 40-fold and incubated until OD600 0.3 was reached (containing approximately 2×108 c.f.u. ml−1). In a conical bottom 0.2 ml 96-well plate, the culture was diluted such that each well contained 4×105 c.f.u. ml−1 and doubling dilutions of THB containing H2O2, ranging from 1.5 to 0.00046 % (Sigma Aldrich). Wells containing ddH2O, instead of H2O2, were included as a control. The MIC was defined as the concentration of H2O2 in THB at which no growth of Se4047 occurred after incubation for 12 h at 37 °C in a humidified atmosphere containing 5 % CO2. The experiment was conducted in triplicate and repeated on three independent occasions.
TraDIS in whole horse blood, H2O2 or THB
Three transposon libraries, AC, CT and GA described in our previous work [32], were each generated using a different modified ISS1 transposon, such that the bases AC, CT or GA were located three bases downstream of the ISS1 inverted repeat and adjacent to the genome sequence of S. equi subsp. equi within each transposon mutant. As each barcoded library was generated independently, each served as an experimental replicate. One millilitre of each stored S. equi subsp. equi barcoded transposon library was added to 39 ml of pre-warmed and pre-gassed THB containing 0.5 µg erythromycin ml−1 (THBE), resulting in cultures of approximately 0.05–0.08 OD600. Cultures were grown at 37 °C in a humidified atmosphere containing 5 % CO2 for approximately 3 h until OD600 0.3 was reached. Five millilitres of the OD600 0.3 cultures were centrifuged at 10000 rpm for 5 min, generating pellets representing the input population of mutants. The supernatants were removed, and the pellets frozen for DNA extraction. One hundred microlitres of each OD600 0.3 culture was added to 50 ml of freshly drawn whole blood from the same Welsh mountain pony, THB containing 0.0004 % H2O2 (4×105 c.f.u. ml−1) or THB (control). This number of bacteria was equivalent to approximately 66 c.f.u. and 72 c.f.u. of each mutant in the whole equine blood/THB control and H2O2 pools, respectively. The cultures were incubated for 2 h (whole blood and 0.0004 % H2O2) or overnight (THB control) at 37 °C in a humidified atmosphere containing 5 % CO2 with rotation (30 r.p.m.). An incubation time of 2 h in whole blood has been used previously to demonstrate the effects of deletion of the hasA gene [18]. To ensure output pools contained viable bacteria, mutants were recovered after incubation with whole blood or H2O2 by plating 300 µl onto 20 Todd–Hewitt agar (THA) Petri dishes containing 0.5 µg erythromycin ml−1 (THAE) and 0.03 µg hyaluronidase ml−1, followed by overnight incubation at 37 °C in a humidified atmosphere containing 5 % CO2. Hyaluronidase was included to facilitate the recovery of distinct colonies of surviving mutants. Mutant colonies were washed off the Petri dishes using THB containing 50 % glycerol (v/v) for direct storage at −20 °C. Two millilitres of the recovered mutants were centrifuged at 10 000 r.p.m. for 5 min, the supernatants removed and the pellets frozen for DNA extraction. Five millilitres of the overnight control cultures in THB were centrifuged at 10 000 r.p.m. for 5 min, the supernatants removed and the pellets frozen for DNA extraction. The input pool of mutants for the whole blood analysis also served as the input pool for comparison with the overnight THB control culture.
DNA was extracted from the input pellets of each of the three independent ISS1 libraries and from each recovered library, which served as experimental replicates (three input and three output libraries). Purified DNA was sequenced by TraDIS, as previously described [32]. In brief, DNA was fragmented to approximately 600–800 bp, the fragment ends were repaired, and A-tailed and Y-adaptors ligated to the fragments. DNA was then digested with SmaI to minimize plasmid sequencing, and amplified by PCR with a specific ISS1 primer and a unique indexing PCR primer for each of the six samples (indexing primers AHT 6, 7, 15, 16, 21 and 32 in Table S1). PCR products were purified, and size selected using AMPure XP beads. Libraries were quantified using the Kapa Biosystems library qPCR quantification kit and gel electrophoresis. Each prepared library was diluted to 2 nM and combined in equal concentrations to form a pool of the six uniquely indexed samples. PhiX (Illumina) was also diluted to 2 nM. The library pool and PhiX were denatured and neutralized, combined in the ratio of 3:2 and sequenced on an Illumina MiSeq DNA sequencer, as previously described [32].
The TraDIS sequencing data from the triplicate output and input samples were analysed as previously described using the BioTraDIS pipeline [32, 43] and the Se4047 reference genome [25]. Stringent mapping criteria of 100 % read match were imposed on the dataset resulting in between 37.8 and 57.6 % of reads mapping to the Se4047 reference genome (Table S2). Reads mapping to the final 10 % of each gene were discounted to prevent false negatives of gene fitness and function as the transposition of ISS1 into this region of a gene can lead to no functional effect. Three genes (SEQ0285, SEQ0882 and SEQ1147) that were over-represented in the input pools due to the prevalence of a few specific ISS1 mutants were also removed from the dataset. Read counts per gene were normalized between the input libraries to facilitate data comparison. Eighty-five genes that contained <10 reads mapping to them, in any one of the three normalized input libraries, were removed to ensure each gene was sufficiently represented to minimize the effects of stochastic loss. Five-hundred and seventy-five genes previously identified as essential, ambiguous or not defined for survival in THB were also removed from the analysis [32]. These criteria permitted the inclusion of 1503 genes in the whole blood and THB overnight control analyses, which represent 94.5 % of non-essential genes in S. equi subsp . equi . A total of 1471 genes met the same inclusion criteria in the H2O2 analysis, which represented 92.5 % of non-essential genes in S. equi subsp. equi [32]. The three input pools contained on average 17 and 16 different ISS1 mutants in each of the 1503 and 1471 genes that passed filtering, respectively. All genes removed from the input data were similarly removed from the output data before the read counts per gene were normalized between the output libraries. The script tradis_comparison was then used to compare each of the three sets of three output libraries to the three input libraries, on a sequencing reads per gene basis, generating a fitness value [log2 fold change (FC)], P and q value for each gene. Genes were considered to have decreased fitness if they exhibited a log2FC value of less than −2 compared to the input control and had a q value of<0.05.
Validation of TraDIS whole equine blood and H2O2 fitness results
To confirm the reduced fitness of some genes as reported by TraDIS, allelic replacement mutants in Se4047 were generated lacking the genes pyrP (SEQ1316), mnmE (SEQ1365), addA (SEQ0953) and recG (SEQ0454). Strains of Se4047 lacking hasA (SEQ0269) and eqbE (SEQ1242) were also utilized, both of which have been described previously [18, 44]. The ΔhasA strain was used as a positive control in the whole equine blood assay, as it is known to be attenuated under this condition [18]. The ΔeqbE strain was used as a negative control, as TraDIS data showed that fitness in whole blood was not altered upon insertion of ISS1.
Deletion mutants were generated using an allelic replacement mutagenesis technique, as previously described for the generation of a ΔprtM mutant [45]. Briefly, 500 bp regions of S. equi subsp. equi DNA flanking the target gene were amplified using the primers listed in Table S1, ligated together and cloned into the pGhost9 plasmid [45]. Constructs were used individually to transform competent Se4047 cells, which were grown on THAE at 28 °C (the plasmid permissive temperature) for 2 days. Single colonies were inoculated into THBE overnight at 28 °C and then transferred to 37 °C for 3 h to induce chromosomal integration of the construct. Integrants were selected on THAE overnight at 37 °C; then, they were grown overnight at 37 °C in THBE, followed by dilution into THB and incubation at 28 °C for 48 h to excise pGhost9 and the target gene from the chromosome. Excised bacteria were grown on THA overnight at 37 °C to ensure free plasmid was lost. To confirm plasmid loss and, therefore, loss of erythromycin resistance, deletion strains were grown on both THAE and THA. Mutant alleles were confirmed by PCR using the appropriate P1 and P4 primers (suffixed with the gene name in Table S1) and sequencing on an ABI3100 DNA sequencer using BigDye fluorescent terminators. Deletion strains were stored in 25 % glycerol (v/v) at −80 °C.
Growth curves of validation strains
A single colony of each deletion mutant and Se4047 were inoculated into 10 ml THB in triplicate and grown for 16 h. Cultures were then diluted to approximately OD600 0.08 in pre-warmed and pre-gassed THB, and grown until stationary phase was reached. The mean OD600 across the three replicates of each strain and their se values were calculated. The doubling time of each replicate of each strain was calculated from exponential phase data and used to identify significant differences in growth rates of mutants compared to wild-type Se4047 using a two-tailed Student’s t-test.
Whole equine blood and H2O2 validation assays
Three overnight cultures for each deletion strain and six overnight cultures of Se4047 were generated by inoculating 10 ml THB with a single colony. Cultures were grown for 16 h, then diluted to OD600 0.08 in pre-warmed and pre-gassed THB. Cultures were grown to OD600 0.3 and then diluted to 1×105 c.f.u. ml−1 in THB. One hundred microlitres was then added to 10 ml freshly drawn blood from the same Welsh mountain pony that was used in the TraDIS study or THB containing 0.0004 % H2O2 and incubated for 2 h with rotation (30 r.p.m.). Immediately after adding the strains, 50 µl was plated neat, in triplicate, onto Columbia CNA staph/strep selective agar (Oxoid) [time point 0 (T0)] to enumerate the initial concentration of S. equi subsp. equi cells. Surviving cells were enumerated at 1 (T1), 2 (T2) and 3 h (T3) by plating various dilutions in PBS onto CNA agar in triplicate. Colony counts for each set of triplicate Petri dishes were converted into a mean c.f.u. ml−1 for each time point for each replicate. Mean c.f.u. ml−1 data from T1, T2 and T3 were transformed into a percentage relative to T0 within each replicate to normalize the data, as the T0 c.f.u. ml−1 varied slightly between experiments. The three values of transformed data per deletion strain at each time point were used to calculate the doubling time of each replicate of each strain, which was compared to wild-type Se4047 using a two-tailed Student’s t-test.
Results
Identification of genes important for fitness in whole equine blood
The three barcoded ISS1 libraries designated AC, CT and GA, described in our previous work [32], were grown to an OD600 of 0.3 immediately before use and re-sequenced to accurately identify input pool composition. Post-filtering, input libraries contained 26 381 unique mutants in library AC, 24 353 unique mutants in library CT and 28 128 in library GA (Table S3), representing 1503 (94.5 %) of the 1590 non-essential genes previously identified in S. equi .
The barcoded mutant libraries were each exposed to whole equine blood and the genes contributing to fitness in this environment were identified by TraDIS. The three barcoded libraries recovered from whole equine blood contained, on average, 9.4±3.5 % (sem) fewer unique mutants than were present in the input libraries (Table S3). The log2FC was calculated for all 1503 genes included in the analysis (Fig. 1a). Thirty-six genes were significantly reduced in fitness in whole equine blood (Fig. 1a, blue and red dots, Tables 1 and S4). The remaining 1466 genes exhibited no growth defects in whole equine blood as a result of ISS1 insertion (Fig. 1a, grey dots and green dot). Cluster of orthologous groups (COG) analysis of the 36 fitness genes (Fig. 1b) identified that the most prevalent categories included genes involved in replication, recombination and repair (n=4), transcription (n=4), and energy production and conversion (n=4). Five genes (14 % of fitness genes) did not belong to a defined COG category.
Fig. 1.
Fitness scores and COG categories of S. equi subsp. equi genes required for survival in whole equine blood. (a) Fitness scores (log2FC) per gene of S. equi subsp. equi ISS1 mutants post-incubation in whole equine blood, as determined by TraDIS. Blue dots, genes with significantly reduced fitness (log2FC < −2 and q<0.05); red dots, genes significantly reduced in fitness of which deletion mutants were made and retested to confirm TraDIS results; green dot, eqbE exhibiting no fitness effect that was used as a control for validation; grey dots, genes exhibiting no significant fitness effect. (b) Functional COG categories of the 36 fitness genes identified in whole equine blood. L, Replication, recombination and repair; K, transcription; C, energy production and conversion; T, signal transduction mechanisms; R, general function prediction only; F, nucleotide transport and metabolism; V, defence mechanisms; N, cell motility; M, cell wall/membrane/envelope biogenesis; G, carbohydrate transport and metabolism; D, cell cycle control, cell division, chromosome partitioning; U, intracellular trafficking, secretion and vesicular transport; O, posttranslational modification, protein turnover, chaperones; J, translation, ribosomal structure and biogenesis; E, amino acid transport and metabolism.
Table 1.
S. equi subsp. equi genes with reduced fitness in equine whole blood as a result of ISS1 insertion as identified by TraDIS. Genes highlighted in grey were deleted by allelic replacement mutagenesis and deletion strains incubated in whole equine blood to validate TraDIS results. An ΔeqbE deletion strain was used as a negative control
|
Gene |
Locus tag |
Function |
log2FC |
q value |
|---|---|---|---|---|
|
ackA |
SEQ0118 |
Acetate kinase |
−2.7 |
0.042 |
|
SEQ0231 |
SEQ0231 |
Putative Mga-like regulatory protein |
−2.9 |
<0.0005 |
|
hasA |
SEQ0269 |
Hyaluronan synthase |
−2.4 |
0.046 |
|
hasB |
SEQ0270 |
UDP-glucose 6-dehydrogenase |
−2.2 |
<0.0005 |
|
SEQ0306 |
SEQ0306 |
Putative ssDNA-binding protein |
−8.5 |
<0.0005 |
|
pepX |
SEQ0383 |
Xaa-Pro dipeptidyl-peptidase |
−2.3 |
0.017 |
|
recG |
SEQ0454 |
ATP-dependent DNA helicase |
−3.6 |
0.001 |
|
SEQ0492 |
SEQ0492 |
Putative mannose-specific phosphotransferase system (PTS), IID component |
−3.3 |
0.042 |
|
SEQ0494 |
SEQ0494 |
Putative mannose-specific phosphotransferase system (PTS), IIAB component |
−3.7 |
0.017 |
|
pptA/ecsA |
SEQ0506 |
ABC transporter ATP-binding protein |
−3.4 |
0.021 |
|
pptB/ecsB |
SEQ0507 |
ABC transporter protein |
−2.8 |
0.002 |
|
SEQ0562 |
SEQ0562 |
Exodeoxyribonuclease |
−2.7 |
0.022 |
|
bipA/typA |
SEQ0615 |
GTPase |
−4.5 |
0.007 |
|
pyrD |
SEQ0655 |
Putative dihydroorotate dehydrogenase |
−3.0 |
0.007 |
|
ppc |
SEQ0776 |
Putative phosphoenolpyruvate carboxylase |
−5.9 |
<0.0005 |
|
addA |
SEQ0953 |
Putative ATP-dependent exonuclease subunit A |
−9.2 |
<0.0005 |
|
SEQ1028 |
SEQ1028 |
GntR family regulatory protein |
−4.2 |
0.004 |
|
SEQ1073 |
SEQ1073 |
Putative phosphopantothenoylcysteine decarboxylase |
−7.9 |
<0.0005 |
|
SEQ1112 |
SEQ1112 |
Putative exported protein |
−4.5 |
0.001 |
|
SEQ1146 |
SEQ1146 |
Putative phosphate acetyltransferase |
−5.1 |
<0.0005 |
|
ldh |
SEQ1169 |
l-Lactate dehydrogenase |
−5.1 |
<0.0005 |
|
SEQ1180 |
SEQ1180 |
Putative DNA-binding protein |
−4.5 |
0.003 |
|
SEQ1181 |
SEQ1181 |
GntR family regulatory protein |
−8.0 |
<0.0005 |
|
SEQ1304 |
SEQ1304 |
Pyridine nucleotide-disulphide oxidoreductase family protein |
−6.4 |
<0.0005 |
|
pyrP |
SEQ1316 |
Uracil permease |
−4.6 |
<0.0005 |
|
mnmE |
SEQ1365 |
tRNA modification GTPase |
−5.2 |
<0.0005 |
|
SEQ1540 |
SEQ1540 |
Putative membrane protein |
−4.5 |
0.003 |
|
smc |
SEQ1566 |
Putative chromosome partition protein |
−3.8 |
<0.0005 |
|
ccpA |
SEQ1596 |
Catabolite control protein A |
−4.3 |
0.011 |
|
pepQ |
SEQ1597 |
Putative Xaa-Pro dipeptidase |
−5.4 |
<0.0005 |
|
SEQ1800 |
SEQ1800 |
Putative exported protein |
−8.3 |
<0.0005 |
|
scpA |
SEQ1863 |
Segregation and condensation protein A |
−4.5 |
<0.0005 |
|
greA |
SEQ1879 |
Transcription elongation factor |
−8.2 |
<0.0005 |
|
csrS |
SEQ1889 |
Sensor histidine kinase |
−6.1 |
<0.0005 |
|
yqeK |
SEQ1909 |
Hydrolase, HD family |
−4.5 |
0.002 |
|
pyrG |
SEQ1945 |
Putative CTP synthase |
−2.3 |
<0.0005 |
|
eqbE |
SEQ1242 |
Equibactin nonribosomal peptide synthase protein |
0.6 |
1 |
Identification of genes important for fitness in hydrogen peroxide
After filtering, input libraries contained 24 372 unique mutants in library AC, 22 734 unique mutants in library CT and 26 226 unique mutants in library GA (Table S5), which represented 92.5 % of the non-essential genes in S. equi subsp. equi [32]. The three output libraries recovered after H2O2 treatment contained, on average, 2.1±6.3 % (sem) fewer unique mutants than were present in the input libraries (Table S5). The effect of incubation with H2O2 on the fitness of ISS1 mutants was determined by calculating the log2FC for all 1471 genes passing the inclusion criteria (Fig. 2a). Fifteen genes were significantly reduced in fitness (log2FC < −2 and q<0.05; Fig. 2a, blue and red dots, Tables 2 and S6), with the remaining 1456 genes exhibiting no growth defect in the presence of H2O2 as a result of ISS1 insertion (Fig. 2a, grey and green dots). COG analysis of the 15 fitness genes (Fig. 2b) identified that the most prevalent categories included genes involved in energy production and conversion (n=4), and replication, recombination and repair (n=3). Fourteen of the fifteen genes identified in the H2O2 TraDIS screen were also implicated in survival in whole equine blood (Table 2, Fig. 3). One gene, ctsR, was uniquely identified in the H2O2 TraDIS screen (Table 2, highlighted in blue).
Fig. 2.
Fitness scores and COG categories of S. equi subsp. equi genes required for survival in hydrogen peroxide (H2O2). (a) Fitness scores (log2FC) per gene of S. equi subsp. equi ISS1 mutants post-incubation in H2O2, as determined by TraDIS. Blue dots, genes with significantly reduced fitness (log2FC < −2 and q<0.05); red dots, genes significantly reduced in fitness of which deletion mutants were made and retested to confirm TraDIS results; green dots, genes exhibiting no fitness effect that acted as negative controls for validation; grey dots, genes exhibiting no significant fitness effect. (b) Functional COG categories of the fitness genes identified in H2O2. C, Energy production and conversion; L, replication, recombination and repair; R, general function prediction only; E, amino acid transport and metabolism; D, cell cycle control, cell division, chromosome partitioning; O, posttranslational modification, protein turnover, chaperones; K, transcription; J, translation, ribosomal structure and biogenesis.
Table 2.
S. equi subsp. equi genes with reduced fitness in the presence of hydrogen peroxide (H2O2) as a result of ISS1 insertion, as identified by TraDIS. One gene highlighted in blue was uniquely identified in the presence of H2O2 when compared to genes identified as reduced in fitness in whole equine blood. The remaining genes were similarly identified as required in whole equine blood. The genes highlighted in grey were deleted by allelic replacement mutagenesis and deletion strains incubated in THB containing H2O2 to validate TraDIS results. The ΔeqbE deletion strain was used as a control.
|
Gene |
Locus tag |
Function |
log2FC |
q value |
|---|---|---|---|---|
|
SEQ0118 |
SEQ0118 |
Acetate kinase |
−4.1 |
0.0021 |
|
ctsR |
SEQ0200 |
Transcriptional regulator |
−3.9 |
0.0221 |
|
SEQ0306 |
SEQ0306 |
Putative ssDNA-binding protein |
−4.9 |
<0.0005 |
|
recG |
SEQ0454 |
ATP-dependent DNA helicase |
−5.1 |
<0.0005 |
|
SEQ0562 |
SEQ0562 |
Exodeoxyribonuclease |
−9.5 |
<0.0005 |
|
ppc |
SEQ0776 |
Putative phosphoenolpyruvate carboxylase |
−3.8 |
0.0221 |
|
addA |
SEQ0953 |
Putative ATP-dependent exonuclease subunit A |
−6.9 |
<0.0005 |
|
SEQ1028 |
SEQ1028 |
GntR family regulatory protein |
−3.8 |
0.0071 |
|
SEQ1146 |
SEQ1146 |
Putative phosphate acetyltransferase |
−8.5 |
<0.0005 |
|
ldh |
SEQ1169 |
l-Lactate dehydrogenase |
−4.5 |
0.0015 |
|
SEQ1304 |
SEQ1304 |
Pyridine nucleotide-disulphide oxidoreductase family protein |
−5.7 |
<0.0005 |
|
mnmE |
SEQ1365 |
tRNA modification GTPase |
−4.4 |
<0.0005 |
|
smc |
SEQ1566 |
Putative chromosome partition protein |
−3.6 |
<0.0005 |
|
pepQ |
SEQ1597 |
Putative Xaa-Pro dipeptidase |
−4.5 |
<0.0005 |
|
yqeK |
SEQ1909 |
Hydrolase, HD family |
−4.5 |
0.0009 |
|
hasA |
SEQ0269 |
Hyaluronan synthase |
0.6 |
1 |
|
pyrP |
SEQ1316 |
Uracil permease |
−0.7 |
1 |
|
eqbE |
SEQ1242 |
Equibactin nonribosomal peptide synthase protein |
0.5 |
1 |
Fig. 3.
Venn diagram of the 36 genes required for the survival of S. equi subsp. equi in whole equine blood compared to the 15 genes required for survival in hydrogen peroxide. The genes that were deleted by allelic replacement mutagenesis to validate the results are indicated.
Generation of allelic replacement mutants in putative fitness genes
To validate the findings of the TraDIS fitness screen, we generated four allelic replacement mutants lacking the genes recG, addA, pyrP and mnmE to confirm single mutant fitness effects in selective conditions. addA was selected for validation as it was the gene most negatively affected in whole equine blood as a result of ISS1 insertion. recG was selected because it was one of the least affected of the genes required for fitness in whole equine blood; in addition, a recA deletion has been incorporated previously into a live attenuated vaccine strain [18]. pyrP and mnmE were selected as these represented a middle ground in the affected genes. The ΔhasA and ΔeqbE mutants were generated previously. The ΔhasA mutant was utilized as it is known that this mutant has reduced fitness in whole equine blood [18], whilst the ΔeqbE allelic replacement mutant [44] was utilized as a negative control as ISS1 mutants in this gene exhibited no attenuation in whole equine blood or H2O2.
Deletion strains were grown in THB and optical densities measured over time to determine their growth characteristics (Fig. 4). The ΔaddA (P=0.0002), ΔrecG (P=0.0005) and ΔmnmE (P<0.0001) deletion strains grew significantly more slowly than Se4047 in THB, despite these genes being identified previously as non-essential [32].
Fig. 4.
Growth curves of the parental S. equi subsp. equi strain Se4047 and ΔpyrP, ΔhasA, ΔeqbE, ΔaddA, ΔrecG and ΔmnmE deletion mutation strains grown in THB at 37 °C in a humidified atmosphere in the presence of 5 % CO2. Error bars indicate the se. The significance of changes in doubling time using a two-sided Student’s t-test are indicated.
To determine whether the identification of genes that were important for the fitness of S. equi subsp. equi in the presence of whole equine blood or H2O2 by TraDIS was due to a generalized fitness defect from the increased time that mutation strains were grown in THB during these experiments, we compared the mutant populations of the three barcoded libraries recovered post-overnight incubation in THB with the input mutant pool immediately prior to growth in whole equine blood. Six genes (pyrD, sufB, sufD, SEQ1930, SEQ2142 and SEQ2146) were identified as having significantly reduced fitness in THB (Table S7). One of these genes, pyrD had indeed been identified as having reduced fitness in whole equine blood.
Survival of key mutation strains in whole blood
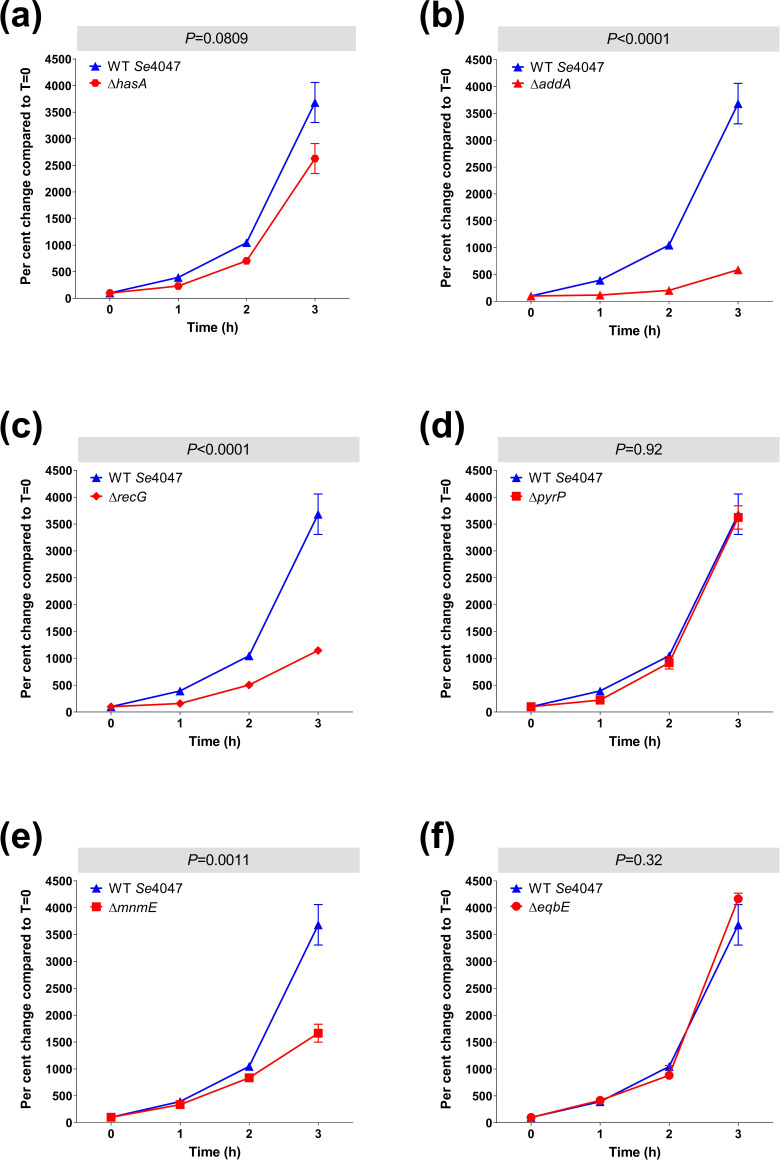
The six deletion strains were incubated in whole equine blood with reduced bacterial loads to more closely reflect the proportion of attenuated S. equi subsp. equi ISS1 mutants present in the original TraDIS assay. Validation assays were also incubated for an additional hour. The growth of each deletion strain in whole equine blood was measured over time and compared statistically to wild-type Se4047 (Fig. 5). The ΔhasA (P<0.0001) strain was highly attenuated in whole equine blood with a significantly longer doubling time (Fig. 5a), which is in agreement with published data on this strain [18]. The doubling times of the ΔaddA (P=0.0008), ΔrecG (P=0.0069) and ΔpyrP (P=0.019) strains were significantly longer than wild-type Se4047 in whole equine blood (Fig. 5b–d). However, the ΔmnmE strain (P=0.38) did not have a significantly longer doubling time than Se4047 in whole equine blood (Fig. 5e). The ΔeqbE control strain grew at the same rate as Se4047 (Fig. 5f).
Fig. 5.
Validation of the S. equi subsp. equi TraDIS screen in whole equine blood. Strains with deletion mutations of whole equine blood fitness genes, as identified by TraDIS, were incubated in blood for 3 h and their survival measured each hour. (a) ΔhasA, (b) ΔaddA, (c) ΔrecG, (d) ΔpyrP, (e) ΔmnmE and (f) ΔeqbE deletion mutation strains compared to the wild-type parental strain, Se4047. Error bars indicate the se. The significance of changes in doubling time using a two-sided Student’s t-test are indicated.
Survival of mutation strains in H2O2
The growth of each allelic replacement mutation strain in the presence of sub-MIC concentrations of H2O2 was measured over time and compared statistically to the growth of Se4047 (Fig. 6). The doubling times for the ΔaddA (P<0.0001), ΔrecG (P<0.0001) and ΔmnmE (P=0.0011) strains in the presence of H2O2 were significantly longer than that of Se4047 (Fig. 6b, c and e). The insertion of ISS1 into hasA, pyrP or eqbE did not confer any fitness defects in H2O2 (Table 2). In agreement with these data, the ΔhasA, ΔpyrP and ΔeqbE mutants grew at similar rates to Se4047 in the presence of H2O2(Fig. 6a, d and f).
Fig. 6.
Validation of the S. equi subsp. equi TraDIS screen in THB containing hydrogen peroxide. Strains with deletion mutations in H2O2 fitness genes, as identified by TraDIS, were incubated in THB containing H2O2 for 3 h and their survival measured each hour. (a) ΔhasA, (b) ΔaddA, (c) ΔrecG, (d) ΔpyrP, (e) ΔmnmE and (f) ΔeqbE deletion mutation strains compared to the wild-type parental strain, Se4047. Error bars indicate the se. The significance of changes in doubling time using a two-sided Student’s t-test are indicated.
Discussion
Here, we describe the genome-wide identification of genes required by S. equi subsp. equi for survival in the presence of whole equine blood and H2O2, conditions that mimic an interaction with the equine immune response. ISS1 mutants in 36 and 15 genes were significantly reduced in fitness upon exposure to whole equine blood or H2O2, respectively. Fourteen genes were required for fitness in both of these conditions. Four novel allelic replacement mutants lacking addA, recG, pyrP or mnmE were generated and tested to determine whether TraDIS had indeed identified novel genes that contribute to fitness in the presence of whole blood or H2O2. Two control mutants were also tested, a capsule deletion mutant, ΔhasA [18], and a ΔeqbE mutant [44].
addAB (also known as rexAB) encodes a major component of the homologous recombination process that repairs double-strand breaks by catalysing the unwinding of DNA [46–48]. In S. equi , addB was essential for growth in THB (insertion index=0.03, essential genes<0.034 [32]). Although, addA was not essential for growth in vitro or for fitness on overnight culture of the mutant libraries in THB, the ΔaddA mutant grew more slowly than the wild-type strain (Fig. 4). This slow growth phenotype was also observed in Streptococcus pneumoniae ΔaddA and ΔaddB mutants [47]. The S. equi subsp. equi ΔaddA deletion mutant was confirmed to be significantly attenuated in whole equine blood and H2O2. However, further investigation of the role of AddA is required as this result could, potentially, be related to the slow-growth phenotype of mutants lacking production of AddA.
recG encodes an ATP-dependent DNA helicase that is thought to be important for efficient recombination and DNA repair. RecG promotes the resolution of Holliday junctions by catalysing the conversion of junction intermediates to mature products by branch migration [49]. RecG is also thought to remove RNA from R-loops by unwinding the RNA–DNA hybrid [50, 51]. Although recG was non-essential for growth in vitro or for fitness on overnight culture of the mutant libraries in THB, the ΔrecG mutant was found to have a slower growth rate (Fig. 4). recG ISS1 mutants were more significantly attenuated in H2O2 compared to whole equine blood and this result was confirmed using the ΔrecG mutant (Figs 5 and 6). This result could, in part, be related to the slow-growth phenotype of mutants lacking production of RecG.
A membrane-bound uracil permease encoded by pyrP scavenges uracil from the environment for pyrimidine biosynthesis [52]. pyrP was required for fitness in whole equine blood, but not in H2O2 (Tables 1 and 2). The ΔpyrP deletion mutant had a similar growth rate to the wild-type Se4047 strain in THB. In agreement with the TraDIS data, the ΔpyrP strain had a significantly longer doubling time in whole equine blood, but a similar doubling time to Se4047 in the presence of H2O2. Interestingly, pyrD and pyrG, which are involved in the downstream biosynthetic pyrimidine pathway, were also required by S. equi subsp. equi for fitness in the presence of whole equine blood, but not H2O2 in vitro. Although ISS1 mutants in pyrD had significantly reduced fitness following overnight culture in THB (Table S7).
mnmE (also known as trmE) is predicted to encode a tRNA modification enzyme that forms a heterotetrameric complex with MnmG (also known as GidA) [53, 54]. The MnmEG complex catalyses two different GTP- and FAD-dependent reactions, generating 5-aminomethyluridine and 5-carboxymethylaminomethyluridine, utilizing ammonium and glycine as substrates, respectively [53]. GTP hydrolysis by MnmE causes structural rearrangements within the MnmEG complex, which is necessary for subsequent tRNA modification, in E. coli [55]. In S. equi , MnmG was found to be essential for growth in vitro [32], and it was critical for survival in Streptococcus pyogenes and Streptococcus agalactiae [32, 56, 57]. S. pyogenes ΔmnmE and ΔmnmG deletion mutants had decreased production of known virulence factors including streptolysin O, M-protein, mitogenic factor and NAD-glycohydrolase [58]. Deletion of mnmE or mnmG also decreased biofilm production by 50 % in Streptococcus mutans [59]. The growth rate of the S. equi subsp. equi ΔmnmE deletion mutation strain in THB was significantly decreased relative to Se4047 and ΔmnmE was the slowest growing of the allelic replacement mutants generated in this study (Fig. 4). ISS1 mutants of mnmE had reduced fitness in both whole equine blood and H2O2. However, the ΔmnmE deletion mutation strain was only found to have a significantly longer doubling time in the presence of H2O2.
Streptococcal capsule mutants have long been known to be susceptible to killing in both in vitro and in vivo [60–63]. Disrupting the capsule, exposes the bacterial surface, rendering the cells more susceptible to immune attack. The S. equi subsp. equi ΔhasA (hyaluronan synthase) mutant is known to be highly susceptible to killing in equine blood, and so it was expected that this gene would be identified as being required for fitness in whole blood using TraDIS (Fig. 1a, Table 1) and in the validation experiment (Fig. 5). Interestingly, the log2FC for hasA determined in the TraDIS screen of −2.4 (q=0.046) was close to the threshold of −2 used to determine attenuation; yet in isolation, the ΔhasA mutant was dramatically reduced in fitness in whole equine blood. One explanation for this result is that acapsular mutants benefit from the retained capsule of neighbouring mutants during fitness studies. Such a bystander effect could explain the recovery of acapsular mutants from the guttural pouches of persistently infected horses [64]. Mutants in the gene encoding UDP-glucose 6-dehydrogenase, hasB, were also found to meet the fitness threshold in the whole blood TraDIS screen (log2FC=−2.2, q=<0.0005). However, the analysis of hasC was confounded by the presence of two copies of this gene in the genome of Se4047 [25]. Although important for fitness in whole blood, acapsular ISS1 mutants in hasA had no fitness cost when exposed to H2O2 (log2FC=0.6, q=1). Our data suggest that at least some of the other 21 genes that were identified as being required for fitness in whole blood, but not H2O2, might similarly play a role in the evasion of phagocytosis, highlighting the application of TraDIS as a whole-genome functional genomics tool.
Other genes that were identified as being required for fitness in the presence of whole blood, but not H2O2, included ccpA, which putatively encodes catabolite control protein A. In Streptococcus suis , CcpA regulates many genes, primarily targeting those involved in carbohydrate metabolism and amino acid transporters, such as PTS uptake systems [65, 66]. Two PTS genes putatively required for mannose import, SEQ_0492 and SEQ_0494, and ldh, encoding lactate dehydrogenase, were also identified as important for survival in whole equine blood. Ldh was regulated by CcpA in S. suis [65, 66] and so the role of these genes in conferring fitness in the presence of whole equine blood may be interlinked.
The genes pptAB (also known as ecsAB) were required for the fitness of S. equi subsp. equi in whole blood, but not in the presence of H2O2. The pptAB genes encode ABC transporter proteins that export the quorum sensing peptides, SHP2 and SHP3, into the extracellular environment [67]. A pptAB deletion mutant of Staphylococcus aureus caused milder synovitis and reduced bone erosions in a murine model of arthritis [68]. The further study of pptAB to determine the role of quorum sensing to the virulence of S. equi subsp. equi is now warranted.
The transcriptional regulator CtsR was identified as being important for fitness in H2O2, but not whole equine blood. However, closer inspection of our data revealed that very few ISS1 mutants existed in the three input libraries and that these were represented by few reads; in library AC, 2 mutants were represented by 14 reads, in CT 4 mutants were represented by 17 reads and in GA 2 mutants were represented by 22 reads (Table S6). Therefore, although this gene met our inclusion criteria, we believe that this hit may be a false positive. In support of this hypothesis, a ΔctsR mutant of Lactobacillus plantarum was not significantly more susceptible to H2O2 than the wild-type parental strain [69].
With the exception of the capsule synthesis genes, none of the other genes that were previously linked to the survival of S. equi in whole blood were identified by TraDIS. The immunoglobulin-cleaving enzymes IdeE, IdeE2 [22, 23] and the Factor X-binding protein Se18.9 [21] are secreted and ISS1-associated defects are likely to be complemented by surrounding mutants. Unfortunately, the gene encoding the antiphagocytic protein SeM [19, 20] was removed from our analysis, as it was classed an essential gene in Se4047 [32]. None of the four fibronectin-binding proteins encoded by S. equi [24] were identified by TraDIS, which suggests that they may be functionally redundant in this system.
Conclusions
The TraDIS screens described herein have identified some interesting and novel genes, with many of them commonly identified between conditions. Two of the genes included in the validation panel, recG and addA, were attenuated in both conditions, but exhibited a slow growth phenotype compared to the wild-type parental strain, which restricts their usefulness as future targets for the development of live-attenuated vaccines as, for manufacturing purposes, vaccine strains should not be attenuated for growth in vitro. The capsule mutant hasA was confirmed to be required for survival in whole equine blood, but not H2O2. However, the reduced fitness of mnmE mutants in the TraDIS screen was only recapitulated in H2O2. The pyrP mutant had a normal growth rate in vitro and a slower growth rate in whole equine blood, suggesting that the deletion of pyrP may be useful for the development of safer live attenuated vaccines. Therefore, further validation of the genes identified by TraDIS is warranted to demonstrate their importance in the absence of competing strains, prior to the development of new live attenuated vaccines.
Data Bibliography
Charbonneau ARL, Taylor E, Mitchell CJ, Robinson C, Cain AK, Leigh JA, Maskell DJ, Waller AS.
Raw Illumina fastq files have been made available in Genbank at the Sequence Read Archive (SRA accession number: PRJNA578912). 2020.
Supplementary Data
Funding information
This work was funded through a grant from the Horse Trust (reference no. G4104). A.R.L.C. received a stipend from the University of Cambridge Doctoral Training Partnership scheme, which is funded by the BBSRC (Biotechnology and Biological Sciences Research Council) (reference no. 1503883). A.K.C was supported by an ARC (Australian Research Council) DECRA (Discovery Early Career Researcher Award) Fellowship (DE180100929).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Blood was collected under the auspices of a Home Office Project License, and following ethical review and approval by the Animal Health Trust’s Animal Welfare and Ethical Review Body (RPP 01_12).
Footnotes
Abbreviations: COG, cluster of orthologous groups; FC, fold change; MIC, minimum inhibitory concentration; TraDIS, transposon-directed insertion-site sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables are available with the online version of this article.
References
- 1.Boyle AG, Timoney JF, Newton JR, Hines MT, Waller AS, et al. Streptococcus equi infections in horses: guidelines for treatment, control, and prevention of strangles – revised consensus statement. J Vet Intern Med. 2018;32:633–647. doi: 10.1111/jvim.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Timoney JF, Kumar P. Early pathogenesis of equine Streptococcus equi infection (strangles) Equine Vet J. 2008;40:637–642. doi: 10.2746/042516408X322120. [DOI] [PubMed] [Google Scholar]
- 3.Harrington DJ, Sutcliffe IC, Chanter N. The molecular basis of Streptococcus equi infection and disease. Microbes Infect. 2002;4:501–510. doi: 10.1016/S1286-4579(02)01565-4. [DOI] [PubMed] [Google Scholar]
- 4.Bazeley PL, Battle J. Studies with equine streptococci 1. Aust Vet J. 1940;16:140–146. doi: 10.1111/j.1751-0813.1940.tb01317.x. [DOI] [Google Scholar]
- 5.Bazeley PL. Studies with equine streptococci 2. Aust Vet J. 1940;16:243–259. doi: 10.1111/j.1751-0813.1940.tb06315.x. [DOI] [Google Scholar]
- 6.Bazeley PL. Studies with equine streptococci 3. Aust Vet J. 1942;18:141–155. doi: 10.1111/j.1751-0813.1942.tb06344.x. [DOI] [Google Scholar]
- 7.Bazeley PL. Studies with equine streptococci 4. Aust Vet J. 1942;18:189–194. doi: 10.1111/j.1751-0813.1942.tb06359.x. [DOI] [Google Scholar]
- 8.Bazeley PL. Studies with equine streptococci 5. Aust Vet J. 1943;19:62–85. doi: 10.1111/j.1751-0813.1943.tb04318.x. [DOI] [Google Scholar]
- 9.Hoffman AM, Staempfli HR, Prescott JF, Viel L. Field evaluation of a commercial M-protein vaccine against Streptococcus equi infection in foals. Am J Vet Res. 1991;52:589–592. [PubMed] [Google Scholar]
- 10.Guss B, Flock M, Frykberg L, Waller AS, Robinson C, et al. Getting to grips with strangles: an effective multi-component recombinant vaccine for the protection of horses from Streptococcus equi infection. PLoS Pathog. 2009;5:e1000584. doi: 10.1371/journal.ppat.1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson C, Frykberg L, Flock M, Guss B, Waller AS, et al. Strangvac: a recombinant fusion protein vaccine that protects against strangles, caused by Streptococcus equi . Vaccine. 2018;36:1484–1490. doi: 10.1016/j.vaccine.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Walker JA, Timoney JF. Construction of a stable non-mucoid deletion mutant of the Streptococcus equi Pinnacle vaccine strain. Vet Microbiol. 2002;89:311–321. doi: 10.1016/S0378-1135(02)00205-5. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs AA, Goovaerts D, Nuijten PJ, Theelen RP, Hartford OM, et al. Investigations towards an efficacious and safe strangles vaccine: submucosal vaccination with a live attenuated Streptococcus equi . Vet Rec. 2000;147:563–567. doi: 10.1136/vr.147.20.563. [DOI] [PubMed] [Google Scholar]
- 14.Cursons R, Patty O, Steward KF, Waller AS. Strangles in horses can be caused by vaccination with Pinnacle I. N. Vaccine. 2015;33:3440–3443. doi: 10.1016/j.vaccine.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Kelly C, Bugg M, Robinson C, Mitchell Z, Davis-Poynter N, et al. Sequence variation of the SeM gene of Streptococcus equi allows discrimination of the source of strangles outbreaks. J Clin Microbiol. 2006;44:480–486. doi: 10.1128/JCM.44.2.480-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp-Symonds J, Kemble T, Waller A. Modified live Streptococcus equi ('strangles') vaccination followed by clinically adverse reactions associated with bacterial replication. Equine Vet J. 2007;39:284–286. doi: 10.2746/042516407X195961. [DOI] [PubMed] [Google Scholar]
- 17.Livengood JL, Lanka S, Maddox C, Tewari D. Detection and differentiation of wild-type and a vaccine strain of Streptococcus equi ssp. equi using pyrosequencing. Vaccine. 2016;34:3935–3937. doi: 10.1016/j.vaccine.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 18.Robinson C, Heather Z, Slater J, Potts N, Steward KF, et al. Vaccination with a live multi-gene deletion strain protects horses against virulent challenge with Streptococcus equi . Vaccine. 2015;33:1160–1167. doi: 10.1016/j.vaccine.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Boschwitz JS, Timoney JF. Inhibition of C3 deposition on Streptococcus equi subsp. equi by M protein: a mechanism for survival in equine blood. Infect Immun. 1994;62:3515–3520. doi: 10.1128/IAI.62.8.3515-3520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galán JE, Timoney JF. Molecular analysis of the M protein of Streptococcus equi and cloning and expression of the M protein gene in Escherichia coli . Infect Immun. 1987;55:3181–3187. doi: 10.1128/IAI.55.12.3181-3187.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiwari R, Qin A, Artiushin S, Timoney JF. Se18.9, an anti-phagocytic factor H binding protein of Streptococcus equi . Vet Microbiol. 2007;121:105–115. doi: 10.1016/j.vetmic.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Flock M, Frykberg L, Sköld M, Guss B, Flock J-I. Antiphagocytic function of an IgG glycosyl hydrolase from Streptococcus equi subsp. equi and its use as a vaccine component. Infect Immun. 2012;80:2914–2919. doi: 10.1128/IAI.06083-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulting G, Flock M, Frykberg L, Lannergård J, Flock J-I, et al. Two novel IgG endopeptidases of Streptococcus equi . FEMS Microbiol Lett. 2009;298:44–50. doi: 10.1111/j.1574-6968.2009.01698.x. [DOI] [PubMed] [Google Scholar]
- 24.Lannergård J, Flock M, Johansson S, Flock J-I, Guss B. Studies of fibronectin-binding proteins of Streptococcus equi. Infect Immun. 2005;73:7243–7251. doi: 10.1128/IAI.73.11.7243-7251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holden MTG, Heather Z, Paillot R, Steward KF, Webb K, et al. Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Beek C, Waern I, Eriksson J, Melo FR, Robinson C, et al. Streptococcal sagA activates a proinflammatory response in mast cells by a sublytic mechanism. Cell Microbiol. 2019;21:e13064. doi: 10.1111/cmi.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchard AM, Egan SA, Emes RD, Warry A, Leigh JA. PIMMS (Pragmatic Insertional Mutation Mapping System) laboratory methodology a readily accessible tool for identification of essential genes in Streptococcus . Front Microbiol. 2016;7:1645. doi: 10.3389/fmicb.2016.01645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charbonneau ARL, Forman OP, Cain AK, Newland G, Robinson C, et al. Defining the ABC of gene essentiality in streptococci. BMC Genomics. 2017;18:426. doi: 10.1186/s12864-017-3794-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barquist L, Boinett CJ, Cain AK. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 2013;10:1161–1169. doi: 10.4161/rna.24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile . mBio. 2015;6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L, Charbonneau ARL, Waller AS, Olsen RJ, Beres SB, et al. Novel genes required for the fitness of Streptococcus pyogenes in human saliva. mSphere. 2017;2:e00460-17. doi: 10.1128/mSphereDirect.00460-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, et al. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 2013;9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, et al. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere. 2016;1:e00013-15. doi: 10.1128/mSphere.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant AJ, Oshota O, Chaudhuri RR, Mayho M, Peters SE, et al. Genes required for the fitness of Salmonella enterica serovar Typhimurium during infection of immunodeficient gp91-/- phox mice. Infect Immun. 2016;84:989–997. doi: 10.1128/IAI.01423-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moule MG, Spink N, Willcocks S, Lim J, Guerra-Assunção JA, et al. Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei . Infect Immun. 2015;84:701–710. doi: 10.1128/IAI.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez MG, Yoder-Himes DR, Warawa JM. Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front Cell Infect Microbiol. 2015;5:78. doi: 10.3389/fcimb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L, Olsen R, Beres S, Eraso J, Saavedra MO, et al. Gene fitness landscape of group A Streptococcus during necrotizing myositis. J Clin Investig. 2019;129:887–901. doi: 10.1172/JCI124994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, et al. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics. 2016;32:1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heather Z, Holden MTG, Steward KF, Parkhill J, Song L, et al. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol Microbiol. 2008;70:1274–1292. doi: 10.1111/j.1365-2958.2008.06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton A, Robinson C, Sutcliffe IC, Slater J, Maskell DJ, et al. Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infect Immun. 2006;74:6907–6919. doi: 10.1128/IAI.01116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chédin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol Microbiol. 2002;43:823–834. doi: 10.1046/j.1365-2958.2002.02785.x. [DOI] [PubMed] [Google Scholar]
- 47.Halpern D, Gruss A, Claverys J-P, El Karoui M. rexAB mutants in Streptococcus pneumoniae . Microbiology. 2004;150:2409–2414. doi: 10.1099/mic.0.27106-0. [DOI] [PubMed] [Google Scholar]
- 48.Yeeles JTP, Gwynn EJ, Webb MR, Dillingham MS. The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single superfamily 1A motor domain. Nucleic Acids Res. 2011;39:2271–2285. doi: 10.1093/nar/gkq1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitby MC, Vincent SD, Lloyd RG. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. Embo J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. Embo J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent SD, Mahdi AA, Lloyd RG. The RecG branch migration protein of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 52.Martinussen J, Schallert J, Andersen B, Hammer K. The pyrimidine operon pyrRPB-carA from Lactococcus lactis . J Bacteriol. 2001;183:2785–2794. doi: 10.1128/JB.183.9.2785-2794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moukadiri I, Prado S, Piera J, Velázquez-Campoy A, Björk GR, et al. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yim L, Moukadiri I, Björk GR, Armengod M-E. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli . Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prado S, Villarroya M, Medina M, Armengod M-E. The tRNA-modifying function of MnmE is controlled by post-hydrolysis steps of its GTPase cycle. Nucleic Acids Res. 2013;41:6190–6208. doi: 10.1093/nar/gkt320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Breton Y, Belew AT, Valdes KM, Islam E, Curry P, et al. Essential genes in the core genome of the human pathogen Streptococcus pyogenes . Sci Rep. 2015;5:9838. doi: 10.1038/srep09838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hooven TA, Catomeris AJ, Akabas LH, Randis TM, Maskell DJ, et al. The essential genome of Streptococcus agalactiae . BMC Genomics. 2016;17:406. doi: 10.1186/s12864-016-2741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho KH, Caparon MG. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun. 2008;76:3176–3186. doi: 10.1128/IAI.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D, Shibata Y, Takeshita T, Yamashita Y. A novel gene involved in the survival of Streptococcus mutans under stress conditions. Appl Environ Microbiol. 2014;80:97–103. doi: 10.1128/AEM.02549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson AT. The relative importance of the capsule and the M-antigen in determining colony form of group A streptococci. J Exp Med. 1959;109:257–270. doi: 10.1084/jem.109.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolcock JB. The capsule of Streptococcus equi . J Gen Microbiol. 1974;85:372–375. doi: 10.1099/00221287-85-2-372. [DOI] [PubMed] [Google Scholar]
- 62.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dale JB, Washburn RG, Marques MB, Wessels MR. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect Immun. 1996;64:1495–1501. doi: 10.1128/IAI.64.5.1495-1501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris SR, Robinson C, Steward KF, Webb KS, Paillot R, et al. Genome specialization and decay of the strangles pathogen, Streptococcus equi, is driven by persistent infection. Genome Res. 2015;25:1360–1371. doi: 10.1101/gr.189803.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, et al. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis . Microbiology. 2011;157:1823–1833. doi: 10.1099/mic.0.046417-0. [DOI] [PubMed] [Google Scholar]
- 66.Willenborg J, de Greeff A, Jarek M, Valentin-Weigand P, Goethe R. The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol Microbiol. 2014;92:61–83. doi: 10.1111/mmi.12537. [DOI] [PubMed] [Google Scholar]
- 67.Chang JC, Federle MJ. PptAB exports Rgg quorum-sensing peptides in Streptococcus . PLoS One. 2016;11:e0168461. doi: 10.1371/journal.pone.0168461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonsson I-M, Juuti JT, François P, AlMajidi R, Pietiäinen M, et al. Inactivation of the Ecs ABC transporter of Staphylococcus aureus attenuates virulence by altering composition and function of bacterial wall. PLoS One. 2010;5:e14209. doi: 10.1371/journal.pone.0014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Bokhorst-van de Veen H, Bongers RS, Wels M, Bron PA, Kleerebezem M. Transcriptome signatures of class I and III stress response deregulation in Lactobacillus plantarum reveal pleiotropic adaptation. Microb Cell Fact. 2013;12:112. doi: 10.1186/1475-2859-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.