Abstract
Haemophilus influenzae causes common and sometimes severe adult and pediatric disease including chronic obstructive respiratory disease, otitis media and infections of the central nervous system. Serotype b strains, with a b-type capsule, have been the historical cause of invasive disease, and the introduction of a serotype b-specific vaccine has led to their decline. However, unencapsulated or non-b-type H. influenzae infections are not prevented by the vaccine and appear to be increasing in frequency. Here we report two pediatric cases of severe central nervous system H. influenzae infection presenting to the same hospital in San Diego, California during the same week in January 2016. Due to good vaccine coverage in this part of the world, H. influenzae cases are normally rare and seeing two cases in the same week was unexpected. We thus suspected a recent transmission chain, and possible local outbreak. To test this hypothesis, we isolated and sequenced whole genomes from each patient and placed them in a phylogenetic tree spanning the known diversity of H. influenzae . Surprisingly, we found that the two isolates (SD2016_1 and SD2016_2) belonged to distantly related lineages, suggesting two independent transmission events and ruling out a local outbreak. Despite being distantly related, the two isolates belong to two different lineages that have exchanged capsule loci in the recent past. Therefore, as in other bacterial pathogens, capsule switching by horizontal gene transfer may be an important evolutionary mechanism of vaccine evasion in H. influenzae .
Keywords: Haemophilus influenzae, capsule, phylogenetics, epidemiology, meningitis, horizontal gene transfer
Data Summary
H. influenzae SD2016_1 and SD2016_2 genome sequences have been deposited in NCBI under BioProject PRJNA534512.
Outcome.
Two cases of severe central nervous system H. influenzae infection occurred during the same week in the same hospital in San Diego, California – a region where such infections are usually very rare due to vaccine coverage. We thus suspected a local outbreak of an H. influenzae clone not covered by the vaccine. Using whole-genome sequencing and phylogenetic analysis of two isolates (SD2016_1 and SD2016_2, one from each patient), we found that they were distantly related, rapidly ruling out a local outbreak and suggesting independent transmission events. In this case, SD2016_1 and SD2016_2 both encoded a-type capsules, whereas the vaccine targets b-type capsules. These results highlight the potential importance of horizontal gene transfer in the capsule locus in allowing H. influenzae to escape vaccine coverage.
Introduction
Haemophilus influenzae is traditionally classified into encapsulated or unencapsulated strains, with encapsulated strains being subdivided into serotypes (or types) a-f, each with a distinct type of polysaccharide capsule. Type-b has long been associated with invasive disease [1] and has thus been a major vaccine target. Since the introduction of the Hib vaccine against type-b H. influenzae , a dramatic decrease of severe cases has been observed [2]. However, this drop in severe type-b infections was followed by an increase of acute infections caused by non-b-type (i.e. a, c, d, e and f capsule types) and non-typeable (unencapsulated) strains, although not to the level of type-b infections pre-vaccine [3–6]. In certain regions and populations, more disease is now caused by type-a than type-b infections [7, 8]. In other populations, f-types and predominantly non-typeable strains are on the rise [9]. Non-typeable H. influenzae (NTHi) are associated with otitis media in children and chronic obstructive respiratory disease in adults [10–12] but cause much less invasive disease than type-b.
As a common surface antigen and vaccine target, the capsule is a target of diversifying selection, and the capsule locus is a hotspot of recombination in bacterial pathogens including Klebsiella pneumoniae [13], Streptococcus pneumoniae [14] and Neisseria meningitidis [15] – but has been less thoroughly studied in H. influenzae . Many (but not all) natural H. influenzae isolates are competent for DNA uptake [16], and are inferred to undergo relatively frequent recombination [17]. Capsule types tend to be associated with particular phylogenetic lineages of H. influenzae , leading to the assertion that capsule genes evolve clonally, with limited recombination [18]. On the other hand, the capsule locus can be deleted naturally by recombination [19], and the capsule locus is occassionally recombined among phylogenetically distant lineages [20]. Thus, capsular recombination in H. influenzae appears to be relatively rare [21], but its impact on the evolution and epidemiology of H. influenzae infections could be substantial. For example, capsular switching could in principle allow successful pathogenic lineages to evade vaccines and persist. Alternatively, if capsular recombination is limited, we would expect vaccine lineages to be replaced with other lineages, encoding different capsules or no capsule at all. Evidence to distinguish between these scenarios is currently lacking.
Here we describe two H. influenzae genomes, each isolated from a meningitis patient at Rady Children’s Hospital (CA, USA) within 1 week of each other in January of 2016. As such severe central nervous system infections are extremely rare in Western countries since the introduction of H. influenzae vaccines in the 1990s, the appearance of two cases in a such narrow geographic and time window leads us to address the following questions using a comparative genomic approach:
Are the two strains closely related, suggesting an outbreak of a particular H. influenzae clone – possibly a vaccine-escape mutant? By placing the two strains on a phylogeny of other sequenced H. influezae genomes, we found that the two strains were unrelated. This surprising result led us to ask a second question:
Do these two unrelated strains share particular genes that might have allowed them both to emerge at the same place and time?
Methods
Strain collection and patient characteristics
H. influenzae strains were isolated from the blood culture of two unrelated individuals (Table 1), both children under 5 years of age. They presented to Rady Children’s Hospital within 1 week of each other in January 2016. They were both treated with antibiotics and were eventually cured with no apparent complications. Blood culture was H. influenzae positive for both patients and showed that these strains were non-type b, but with an encapsulated appearance. Both strains were sent to the United States Centers for Disease Control and Prevention (CDC) for serotyping, which confirmed them both to be type-a. Further patient characteristics are given in Table 1. Strains isolated from patients 1 and 2 were, respectively, named SD2016_1 and SD2016_2 (also called ‘isolate 1’ and ‘isolate 2’, respectively, for short) in this study.
Table 1.
Patient characteristics
|
Patient 1 |
Patient 2 |
|
|---|---|---|
|
Positive culture |
Blood culture |
Blood and cerebrospinal fluid (CSF) culture |
|
CSF cell profile on first tap |
Protein 267 Glucose 47 Erythrocytes 112, Nucleated cells 619 |
Protein 68 Glucose 49 Erythrocytes 28 Nucleated cells 707 |
|
CNS complications |
Subdural empyema |
Seizure |
|
Antibiotic treatment |
Vancomycin and meropenem for 18 days |
Ceftriaxone for 10 days |
|
Time to fever resolution |
14 days |
1 day |
Ethical approval
This study was approved by the Internal Ethical Review Board and the Privacy Board of Rady Children's Hospital-San Diego (file 19005C). The study was deemed to be a case report, which does not involve a systematic investigation and therefore does not meet the definition of research as outlined in 45 CFR 46.102(d) and are not subject to the Human Subject Regulations (45 CFR 46). The privacy board concluded that no protected health information (PHI) is disclosed in this study.
DNA extraction and sequencing
H. influenzae strains were cultured overnight on chocolate agar plates (Thermo Fisher Sceintific), and DNA was extracted using the Bactozol DNA extraction kit (MRC). Extracted DNA was further purified using the PowerClean Pro DNA Clean-Up Kit (MOBIO Laboratories). Libraries were prepared using the Nextera XT kit (Illumina) following the standard Illumina protocol and library size was confirmed at approximately 1000 bp with a Qiaxcel Advanced System (QIAGEN). We performed paired end sequencing (2×300 bp) using the MiSeq reagent Kit V3 (Illumina) on the MiSeq system (Illumina) yielding a total of 1 128 523 paired-end reads for SD2016_1 and 1 708296 paired-end reads for SD2016_2.
Genome assembly, annotation and phylogenetics
Sequencing reads were trimmed with Trimmomatic v.0.33 [22] with default parameters. Trimmed reads were assembled into contigs using IDBA v1.1.3 [23]. We then compared the SD2016_1 and SD_2016_2 genomes with a dataset of high-quality H. influenzae genome assemblies from 591 non-typeable, 8 a-capsule, 52 b-capsule, 4 c-capsule, 1 d-capsule, 19 e-capsule and 7 f-capsule isolates available in NCBI as of 8 October 2018, previously reported by Watts and Holt [21], for a total of 684 genomes [see Table S1, (available in the online version of this article) for NCBI accession numbers]. Contigs were annotated using the rast v.0.1.1 server [24]. Translated gene predictions were clustered into 12 840 orthologous groups using Roary v3.11.2 [25], of which 9180 were present in >1 genome, and 502 core gene families were present in >99 % of isolates (Table S2). Each core gene family was aligned separately using mafft v.7 [26] and the concatenated alignment was used to infer a phylogeny using FastTreeMP v.2.1 [27].We rooted the tree by adding eight H. haemolyticus outgroup genomes (Table S1) to the alignment (yielding a smaller core genome alignment based on 104 genes), inferring the phylogeny with FastTreeMP, identifying the location of the root, and then placing the root on the tree inferred from the larger alignment. We also used FastTreeMP to reconstruct each individual gene tree (or gene fragments in the case of genes belonging to the capsule locus). Phylogenies were displayed in FigTree v.1.4 4 (http://tree.bio.ed.ac.uk/software/figtree/). Capsule genes were identified using the hicap software [21].
Horizontal gene transfer detection
To infer putative horizontal gene transfer (HGT) events in the phylogeny, we used the 'reconcile' mode of Notung v.2.9 [28] with all other parameters set as default. Rooted trees were used as inputs to resolve polytomies. Notung was able to reconcile 5698 of the 9180 gene families, with the other being too uncertain or computationally complex to yield a result. We only considered recent HGT among extant isolates, not internal branches, which could contain more phylogenetic uncertainty. In total, we identified exchanges involving 2167 gene families and 662 genomes, for a total of 11 138 exchanges (Table S3). We visualized this network of HGT in Cytoscape v.3.5.1 [29] using the edge-weighted spring-embedded layout based on the number of genes exchanged between each pair of strains.
Results
SD2016_1 and SD2016_2 genomes are distantly related
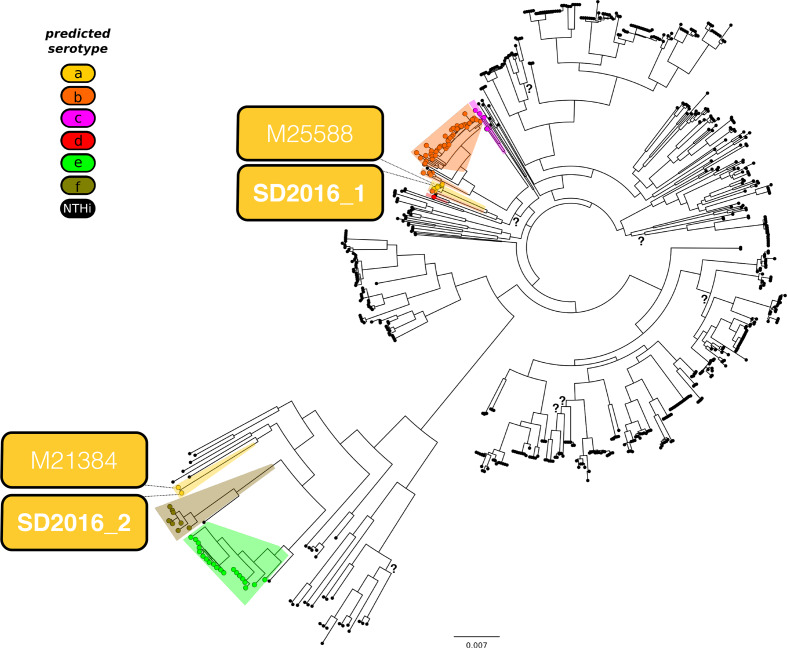
The core genome phylogeny shows that SD2016_1 and SD2016_2 belong to two distinct and distantly related lineages (Fig. 1, File S1). The closest sequenced relative of SD2016_1 is M25588, deposited in GenBank by the US CDC in 2018 (and thus of likely American origin, although this is not specified in the GenBank entry). Another close relative of isolate 1, NML_Hia1, was isolated in Canada in 2011, and also carries an a-type capsule locus. The nearest neighbouring clade of isolate 1 include NTHi and d-type strains, and the next closest clade includes b- and c-types (Fig. 1). Isolate 2 is nearly identical to M21384, another CDC genome recently inferred to have undergone a recombination event importing a-type capsule genes into an f-type ancestor [21]. Consistent with the f-type ancestry of this lineage, the next closest relatives to SD2016_2 and M21384 are f-type genomes, including Hi794 (Finland, date unknown) and KR494 [30]. As SD2016_1 and SD2016_2 are distantly related in the tree, they appear not to be epidemiologically linked and likely represent independent infection events.
Fig. 1.
Core genome phylogeny of H. influenzae shows the two San Diego 2016 isolates are distantly related. Serotypes are shown as coloured circles, with non-typeable (NTHi) isolates in black. Isolates SD2016_1 and 2 are indicated in coloured boxes (indicating serotype a), along with each of their nearest-neighour isolates. All branches are well-supported (bootrap support >0.8, unless noted with a question mark (?). H. haemolyticus was used as an outgroup to place the root of the tree (not shown). The scale bar is in units of nucleotide substitutions per site.
Detailed phylogenies and potential exchanges in the capsule locus
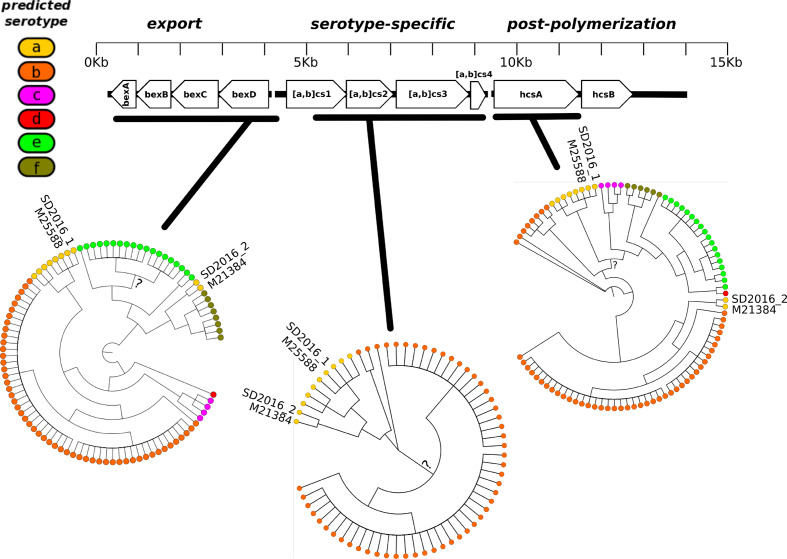
The presence of two a-type strains (SD2016_1 and SD2016_2) in two distantly related clades leads us to investigate in greater detail the evolution of capsule locus genes. By manually inspecting individual gene alignments at this locus (File S2), we found that SD2016_1 and SD2016_2 sequences were very similar – but not identical – spanning an ~5 kb region encompassing most of the serotype-specific genes (Fig. 2). Throughout the capsule locus, SD2016_1 and SD2016_2 each clustered with its respective nearest neighbour (M25588 or M21384) from the core-genome phylogeny. NML_Hia1, another a-type genome isolated in Canada in 2011, also shared a similar or identical sequence in the serotype-specific region, suggesting that the putative recombination event in this region occurred in 2011 or earlier. Upstream of the serotype-specific region, SD2016_2 was most similar to f-type strains, whereas downstream it was most similar to b-types (Fig. 2). This suggests that an f-type ancestor of SD2016_2 acquired ~5 kb of serotype-specific DNA from an a-type donor strain, resulting in a serotype switch, which could have been further affected by multiple recombination events downstream of the serotype-specific region. In summary, recent ancestors of SD2016_1 and SD2016_2 likely engaged in HGT at the capsule locus. However, SD2016_1 and SD2016_2 capsule loci are non-identical, notably in the acsC gene (six substitutions over 2400 bp) (Fig. 2, File S2), making it unlikely that the exchange occured in Rady Children's Hospital.
Fig. 2.
The two San Diego 2016 isolates have similar sequences in the serotype-specific capsule region, but are distinct in flanking regions. The schematic (top) shows the arrangement of genes within the capsule locus. The dendrograms (bottom) show the phylogenies inferred from the alignment of three separate regions of the locus. All branches are well-supported (bootrap support >0.8) unless noted with a question mark (?). Dendrograms show tree topology only; branch lengths are not to scale. Alignments and phylogenies inferred for individual genes and gene fragments are available in File S2.
Global analysis of HGT
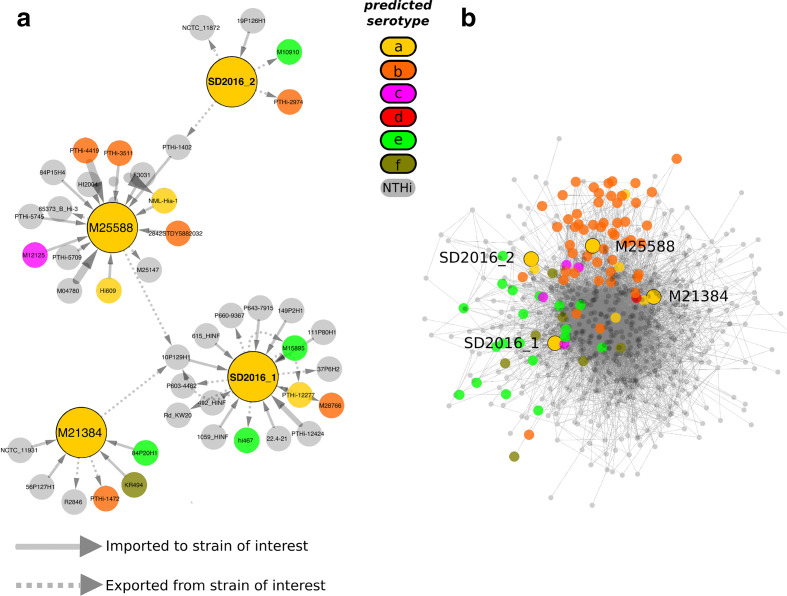
Despite being phylogenetically unrelated, SD2016_1 and SD2016_2 were both isolated during the same week from the same hospital. We thus hypothesized that they may have recently exchanged genes via HGT. Using a reconciliation method to detect HGTs (Methods) we inferred a total of 11 138 gene transfers (Table S3), but none directly between SD2016_1 and SD2016_2 (Fig. 3a). However, the two strains were indirectly connected via isolates M25588, 10SP129H1 (USA, 2005) and PTHi-1402 (Portugal, 1992). The dates and locations of these isolates does not suggest recent transfer. We note that this gene-by-gene analysis would not be expected to detect the similarities observed in the capsule locus because of the conflicting phylogenetic signal within genes (Fig. 2). A broad cross-species genomic comparison study recently found that HGT is more frequent in bacteria encording capsules [31]. In contrast, we found that the 'core' of the HGT network in H. influenzae is dominated by unencapsulated NTHi strains (Fig. 3b). Serotypes b and e form distinct clusters at the periphery of the network, suggesting a preference for HGT within, but less frequently between, these serotypes. Serotype a strains, including SD2016_1 and 2, although relatively less sampled, did not form a cohesive cluster in the HGT network, suggesting widespread and promiscuous HGT with NTHi and other serotypes. More comprehensive sampling will be needed to support this hypothesis.
Fig. 3.
Network of gene exchange among H. influenzae genomes. (a) Inferred HGT events involving SD2016_1, SD2016_2, or their closest relatives (large yellow circles). Arrows show gene exchange events between genomes (circles), with arrow width proportional to the number of genes exchanged. (b) Overall network of inferred gene exchanges. Serotypes are shown as larger coloured circles, and NTHi genomes are smaller grey circles. Edges show HGT events between genomes. Genomes sharing more genes are drawn closer together in the network by the edge-weighted spring-embedded layout. Genomes of intereset (SD2016_1 and 2) along with their phylogenetic nearest-neighbours are outlined in black around the circle.
Discussion
The appearance of two H. influenzae infections in the same hospital in the same week was highly unexpected because such infections are exceedingly rare in areas of high vaccine coverage. This raised concerns that the two cases were part of a local outbreak, involving H. influenzae transmission in the San Diego area. By sequencing the two isolate genomes (SD2016_1 and SD2016_2) and placing them on a phylogenetic tree encompassing the known genomic diversity of H. influenzae , we found that they belonged to distantly related clades, indicating that each infection was aquired independently and the two were not linked in a recent transmission chain. The entire analysis, from strain isolate to sequencing and phylogenetic analysis, could be performed in about a week, allowing us to rapidly rule out local transmission. Rather, the observation that SD2016_1 and SD2016_2 are distantly related reinforces the potential for rapid spread of different H. influenzae lineages. These results are broadly consistent with an earlier study that showed little or no evidence that lineages are constrained by geographic barriers [18].
Despite being distantly related, SD2016_1 and SD2016_2 share a very similar capsule locus, particularly in the serotype-specific region. Flanking the serotype-specific region and elsewhere in the genome, SD2016_2 (like its nearest phylogenetic neighbour M21384) is more similar to f-type strains. Together, these observations point to a scenario in which an f-type ancestor of SD2016_2 acquired an a-type capsule by recombination with a recent ancestor of SD2016_1. This scenario is also supported by another recent study, which inferred a transfer of a-type capsule genes into the previously f-type M21384, the nearest-neighbour of isolate 2 [21]. This recent study made the in silico prediction that this recombination event yielded an a-type capsule, but they did not confirm this with serotyping. Our serotyping results confirmed that the recombination event does indeed yield an a-type serological phenotype. Due to limited geographic and temporal information about these strains, we can only speculate about precisely where and when the recombination event occurred. However, strain 1 is in the same lineage as a Canadian type-a strain isolated in 2011, suggesting that the exchange happened in North America sometime in the past decade. Recent ancestors of the f-type acceptor lineage were found in Scandinavia as recently as 2011, suggesting the acceptor strain migrated from Europe to North America, but more thorough sampling will be required to confirm this scenario.
In our clinical experience at Rady Children's Hospital, type-a H. influenzae has never caused anywhere near the rate of invasive disease caused by type-b. Type-a infections typically occur once every 3 to 5 years. That both SD2016_1 and SD2016_2 were isolated at the same place and time appears to be a coincidence, not an outbreak in this case. Yet combined with broader surveys, it does suggest that these a-type strains could be evolving to fill the vacant ‘invasive disease’ niche left by b-type strains targeted by current vaccines. This is consistent with the increasing disease burden of a-type infections in certain populations [7, 8, 32], while in other populations f-types and predominantly non-typeable strains are on the rise [9, 33]. Our results also indicate that vaccines need not select for lineage replacement, but could allow multiple different lineages to adapt via acquisition of new capsule loci by HGT. We show that such a scenario is plausible, but further analysis of larger population genomic samples will be needed to assess the relative importance of lineage replacement vs. capsule HGT in the evolutionary response of H. influenzae to vaccine pressure.
Data Bibliography
1. H. influenzae SD2016_1 and SD2016_2 genome sequences have been deposited in NCBI under BioProject PRJNA534512.
2. The additional H. influenzae genome assemblies used in our analyses are available at: https://monash.figshare.com/articles hicap_validation_assembly_set/7562363
3. Alignments and phylogenies of individual capsule genes and gene fragments (File S2) are available on Figshare at doi: 10.6084/m9.figshare.11800785.
Supplementary Data
Funding information
This work was supported by the Canadian Institutes of Health Research and a Canada Research Chair to B.J.S.
Author contributions
Conceptualization: Y.T., L.F., J.B., B.J.S. Formal analysis: Y.T. Funding acquisition: B.J.S. Investigation: Y.T., N.T. Methodology: Y.T., B.J.S. Project administration: L.F., J.B., B.J.S. Software: Y.T. Visualization: Y.T. Writing – original draft: Y.T., B.J.S. Writing – review and editing: Y.T., L.F., J.B., N.T., B.J.S.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: HGT, horizontal gene transfer; NTHi, non-typeable H. influenzae.
All supporting data, code and protocols have been provided within the article or through supplementary data files.Three supplementary tables and two supplementary data files are available with the online version of this article.
References
- 1.Pittman M. Variation and type specificity in the bacterial species Hemophilus influenzae . J Exp Med. 1931;53:471–492. doi: 10.1084/jem.53.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peltola H. Worldwide Haemophilus influenzae type B disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev. 2000;13:302–317. doi: 10.1128/CMR.13.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giufrè M, Cardines R, Brigante G, Orecchioni F, Cerquetti M. Emergence of invasive Haemophilus influenzae type a disease in Italy. Clin Infect Dis. 2017;64:1626–1628. doi: 10.1093/cid/cix234. [DOI] [PubMed] [Google Scholar]
- 4.Headrick A, Schmit EO, Kimberlin DW. Fulminant Haemophilus influenzae type A infection in a 4-year-old with previously undiagnosed asplenic heterotaxy. Pediatr Infect Dis J. 2018;37:e108–e110. doi: 10.1097/INF.0000000000001758. [DOI] [PubMed] [Google Scholar]
- 5.Ladhani SN, Collins S, Vickers A, Litt DJ, Crawford C, et al. Invasive Haemophilus influenzae serotype E and F disease, England and Wales. Emerg Infect Dis. 2012;18:725–732. doi: 10.3201/eid1805.111738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang RSW, Ulanova M. The changing epidemiology of invasive Haemophilus influenzae disease: emergence and global presence of serotype a strains that may require a new vaccine for control. Vaccine. 2017;35:4270–4275. doi: 10.1016/j.vaccine.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Eton V, Schroeter A, Kelly L, Kirlew M, Tsang RSW, et al. Epidemiology of invasive pneumococcal and Haemophilus influenzae diseases in Northwestern Ontario, Canada, 2010-2015. Int J Infect Dis. 2017;65:27–33. doi: 10.1016/j.ijid.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Ulanova M. Global epidemiology of invasive Haemophilus influenzae type a disease: do we need a new vaccine? J Vaccines. 2013;14 [Google Scholar]
- 9.Resman F, Ristovski M, Ahl J, Forsgren A, Gilsdorf JR, et al. Invasive disease caused by Haemophilus influenzae in Sweden 1997–2009; evidence of increasing incidence and clinical burden of non-type B strains. Clin Microbiol Infect. 2011b;17:1638–1645. doi: 10.1111/j.1469-0691.2010.03417.x. [DOI] [PubMed] [Google Scholar]
- 10.Gkentzi D, Slack MPE, Ladhani SN. The burden of nonencapsulated Haemophilus influenzae in children and potential for prevention. Curr Opin Infect Dis. 2012;25:266–272. doi: 10.1097/QCO.0b013e32835310a4. [DOI] [PubMed] [Google Scholar]
- 11.Lim CT, Parasakthi N, Puthucheary SD. Neonatal meningitis due to non-encapsulated Haemophilus influenzae in a set of twins: A case report. Singapore Med J. 1994;35:104–105. [PubMed] [Google Scholar]
- 12.Moghaddam SJ, Ochoa CE, Sethi S, Dickey BF. Nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease and lung cancer. Int J Chron Obstruct Pulmon Dis. 2011;6:113–123. doi: 10.2147/COPD.S15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyres KL, Gorrie C, Edwards DJ, Wertheim HFL, Hsu LY, et al. Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol Evol. 2015;7:1267–1279. doi: 10.1093/gbe/evv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mostowy RJ, Croucher NJ, De Maio N, Chewapreecha C, Salter SJ, et al. Pneumococcal capsule synthesis locus CPS as evolutionary hotspot with potential to generate novel serotypes by recombination. Mol Biol Evol. 2017;34:2537–2554. doi: 10.1093/molbev/msx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartley SN, Mowlaboccus S, Mullally CA, Stubbs KA, Vrielink A, et al. Acquisition of the capsule locus by horizontal gene transfer in Neisseria meningitidis is often accompanied by the loss of UDP-GalNAc synthesis. Sci Rep. 2017;7:44442. doi: 10.1038/srep44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maughan H, Redfield RJ. Extensive variation in natural competence in Haemophilus influenzae. Evolution. 2009;63:1852–1866. doi: 10.1111/j.1558-5646.2009.00658.x. [DOI] [PubMed] [Google Scholar]
- 17.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 18.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, et al. Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A. 2014;111:5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroll JS, Hopkins I, Moxon ER. Capsule loss in H. influenzae type B occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell. 1988;53:347–356. doi: 10.1016/0092-8674(88)90155-9. [DOI] [PubMed] [Google Scholar]
- 20.Musser JM, Kroll JS, Moxon ER, Selander RK. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci U S A. 1988;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watts SC, Holt KE. hicap: in silico serotyping of the Haemophilus influenzae capsule locus. J Clin Microbiol. 2019;57:e00190–19. doi: 10.1128/JCM.00190-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 24.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, et al. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics. 2012;28:i409–i415. doi: 10.1093/bioinformatics/bts386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su Y-C, Hörhold F, Singh B, Riesbeck K. Complete genome sequence of encapsulated Haemophilus influenzae type F KR494, an invasive isolate that caused necrotizing myositis. Genome Announc. 2013;1:e00470–13. doi: 10.1128/genomeA.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rendueles O, de Sousa JAM, Bernheim A, Touchon M, Rocha EPC. Genetic exchanges are more frequent in bacteria encoding capsules. PLoS Genet. 2018;14:e1007862. doi: 10.1371/journal.pgen.1007862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyler K, Meehan M, Bennett D, Mulhall R, Harrison O, et al. Spontaneous capsule loss in Haemophilus influenzae serotype B associated with Hib conjugate vaccine failure and invasive disease. Clin Microbiol Infect. 2019;25:390–391. doi: 10.1016/j.cmi.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Resman F, Svensjö T, Ünal C, Cronqvist J, Brorson H, et al. Necrotizing myositis and septic shock caused by Haemophilus influenzae type F in a previously healthy man diagnosed with an IgG3 and a mannose-binding lectin deficiency. Scand J Infect Dis. 2011a;43:972–976. doi: 10.3109/00365548.2011.589079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.