Abstract
Positive sense (+) RNA viruses exploit membranes from a variety of cellular organelles to support the amplification of their genomes. This association concurs with the formation of vesicles whose main morphological feature is that of being wrapped by a double membrane. In the case of the SARS‐CoV virus, the outer membrane is not discrete for each vesicle, but seems to be continuous and shared between many individual vesicles, a difference with other +RNA viruses whose nature has remained elusive. I present morphological, biochemical and pharmacological arguments defending the striking analogy of this arrangement and that of entangled, nascent Lipid Droplets whose birth has been aborted by an excess of Phosphatidic Acid. Since Phosphatidic Acid can be targeted with therapeutical purposes, considering this working hypothesis may prove important in tackling SARS‐CoV infection.
Keywords: endoplasmic reticulum, lipid droplets, membrane subversion, phosphatidic acid, SARS‐CoV
Synopsis
Success in the replication of SARS‐CoV requires subversion of cellular membranes, which I propose could be based on the manipulation of Phosphatidic Acid, a powerful anionic lipid capable of altering membrane properties. (1) coronaviral proteins would tether ER membranes apposition and (2) subsequent lamellar‐to‐cubic ER transition. (3) Viral subversion of PA metabolism would support cubic ER cavities expansion and Lipid Droplets sequestration, (4) finally being supportive for viral amplification complexes. Modulation of Phosphatidic Acid levels may thus harbor therapeutic potential.
Abbreviations
- DMVs
double‐membraned vesicles
- ER
endoplasmic reticulum
- LD
lipid droplets
- MERS‐CoV
middle east respiratory syndrome‐coronavirus
- PA
phosphatidic acid
- SARS‐CoV
severe acute respiratory syndrome‐coronavirus
1.
Coronaviruses (CoVs), pathogenic in both men and animals, are enveloped positive sense RNA (+RNA) viruses whose genomes, of up to 32 kb, are considered the largest of their type. 1 Among them, some are classified as emergent, meaning that they are able to cross the species barrier to reach a new target species. SARS‐CoV emerged toward the end of 2002 in Southern China 2 and became a human threat. At present, SARS‐CoV‐2 is responsible for an unprecedented world‐wide sanitary crisis. Initial infection concurs with binding to and fusion with the host membranes through the spike protein. Subsequently, a major path for viral internalization relies on the endocytic pathway and culminates with the release of the viral RNA genome into the cytoplasm of the infected cell (reviewed in Reference 3). After this, +RNA viruses need to establish a complex capable of amplifying and further expressing their genome, the “replication/transcription complex”, as well as additional (termed “non‐structural”) proteins capable of coordinating accessory functions. All studied +RNA viruses hijack cytoplasmic cellular membrane compartments in order to achieve this step. 4 , 5 Although poorly characterized, this association presumably enhances RNA synthesis by concentrating viral macromolecules on a confined space and shields RNA molecules from nucleases and from the host cell's innate immune system. For different +RNA viruses, the targeted membrane source varies, including Golgi apparatus, lysosomes, late endosomes, autophagosomes, Endoplasmic Reticulum (ER) or mitochondria. 6 Even within the Coronaviridae family, where different studies have highlighted a better‐defined link with the ER, there is substantial morphological and probably etiological diversity in the sequestered membrane assemblies. 7 For example, gammacoronaviruses induce long stretches of parallel membranes that adopt round shapes at specific sites, 8 while betacoronaviruses as MERS‐ and SARS‐CoV concur with bona‐fide circular vesicles. 9 , 10 , 11 , 12 In any case, the exact nature of the membranes that SARS‐CoV sequesters in order to promote its multiplication remains enigmatic.
Nevertheless, efforts invested on performing careful electronic microscopy analyses at different times post‐SARS‐CoV‐infection have revealed recurrent, well‐confirmed features: the membranes emerge from or are closely apposed to the perinuclear ER, they are in tight and frequent contact with mitochondria and the infection‐specific vesicles that derive measure from 100 to 300 nm in diameter. 11 , 12 , 13 Since early stages of the infection (ie, 6 hours), some of these vesicles bear a single lipid layer and seem to be embedded in the ER. This description fully fits with that of nascent Lipid Droplets (LD). LD are membrane‐enclosed organelles that form by progressive nucleation of non‐polar lipids, mainly triacylglycerols and steryl esters, within the hydrophobic core of the ER bilayer. Sufficient nucleation drives the reorganization of an ER‐derived phospholipids monolayer around the neutral lipid core, progressive budding and final excision of the formed droplet into the cytoplasm. Along with a canonical role as a fat reservoir, the precedent years have highlighted many relevant roles for LD, including protection of lipids from oxidation, 14 depot of proteins with special regulation needs, signaling nodes and, as discussed later on, platforms for pathogens. 15 Of note, inspection of immunofluorescence against SARS‐CoV non‐structural proteins recurrently yields a pattern fully reminiscent of LD. 9 , 10 , 16 , 17 , 18 Later during infection (already at 9hours post‐infection), the interior of the SARS‐CoV‐induced vesicles becomes denser, filled with spider web‐like contents, and they appear as delimited by two membranes. In fact, during “classical” subversion of the membrane system upon infection by multiple + RNA viruses, the associated replication‐transcription vesicles are double‐membraned (DMV) 12 , 19 , 20 (Figure 1A, left panel). Since LD are by definition monolayer organelles, LD have been repeatedly dropped from consideration as the central structure being subverted by the virus during infection.
FIGURE 1.
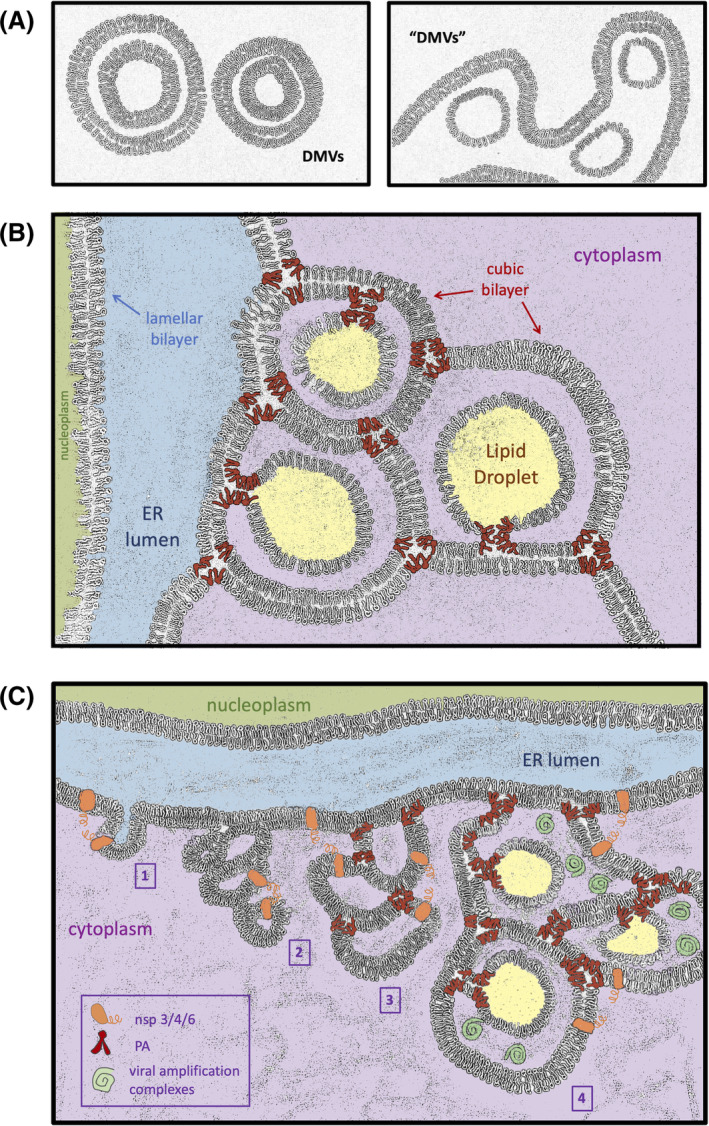
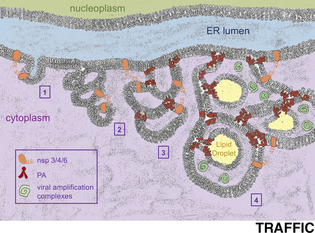
Schematics of double‐membraned vesicles origins. A, Left: Canonical shape of DMVs induced by +RNA viruses (as described in Reference 19). Vesicles are conformed by two concentric bilayers. Right: Morphology for DMVs‐like during SARS‐CoV infection (as adapted from Reference 11). The inner membrane of the vesicles is closed and appears as thinner, suggestive of a monolayer. The outer membrane is a bilayer common to all vesicles and corresponds to the endoplasmic reticulum membrane itself. B, Illustration of the ER displaying a canonical lamellar‐state bilayer (the inner nuclear membrane) and a cubic‐transitioned bilayer (the outer nuclear membrane). Each ER‐conformed cavity contains a filled, yet still anchored LD (in yellow). Both the ER cubic junctions and LD birth (and anchorage) are assisted and maintained thanks to PA moieties (in red). For clarity, all the three‐way junctions of the cubic structure have been drawn in the same plane, giving rise to three fully round DMVs‐like structures. The interior or the cavities has been colored as the cytoplasm, from which it derives. Yet, in a dense, 3D, cubic ER network, this space would remain substantially shielded from the cytoplasmic environment. C, Proposed kinetics of SARS‐CoV‐driven membrane subversion. Step 1: by interacting with each other through their cytosolic domains, coronaviral nsp3, nsp4 and/or nsp6 exploit their transmembrane segments to tether together and appose ER membranes. Step 2: repetitive tethering initiates the lamellar‐to‐cubic ER transition, which gives rise to the smaller, bilayered vesicles seen early upon infection. Step 3: viral subversion of PA metabolism may provide an increased pool of this lipid that supports size expansion of the cubic ER cavities and primes their filling with sequestered LD. Step 4: mature DMVs‐like structures are finally supportive for viral amplification complexes
Phosphatidic Acid (PA) is a central lipid with many assigned and interchangeable functions within the cell. First, it is a backbone for the subsequent creation of most phospholipid species. 21 Second, it can act as a signaling messenger. 22 , 23 Third, given its conical shape (meaning that its tail is wider than its headgroup) as well as its negatively charged head, it bears a fusogenic potential that translates into its ability to induce negative curvature on membranes. 24 , 25 In this context, deep characterizations combining cell imaging, biochemistry and genetics in the model organism S. cerevisiae have uncovered pathological situations in which an increase in the local concentration of PA provoke aberrant transitions at the perinuclear ER. Of relevance, these alterations are directly linked with the capacity of LD to correctly bud out from the ER. 26 , 27 , 28 , 29 , 30 , 31 In more detail, when the PA local concentration is elevated because of genetic deficiencies (such as lack of seipin) or by exposure to the combined presence of oleate and inositol, LD can form inside the leaflet of the ER outer membrane, but upon completion and acquisition of their own monolayer, are unable to emancipate into the cytoplasm. As a consequence, they remain anchored to the ER. 26 , 27 , 28 , 29 Independent biochemical and biophysical experiments in other model systems, including human cells, have validated the necessity of carefully modulating the surface tension at the ER by specific lipids 32 , 33 , 34 and by specific proteins 35 in order to permit correct LD budding. A relevant point when analyzing the morphological particularities of DMVs induced during SARS‐CoV infection is that, in contrast to those triggered by other +RNA viruses, the inner layer indeed constitutes a closed circle, but the outer one is continuous all along with that of the other DMVs, giving the impression that the whole is disconnected from the cytoplasm (Figure 1A, right panel). 11 This arrangement is strikingly similar to entangled LD, trapped within the ER as a consequence of a local excess in PA. 26 , 27 , 32 , 33 In this configuration, the inner membrane of the DMV would be the LD monolayer, while the outer membrane of the DMV, common to all DMVs, would be the ER membrane itself (Figure 1A, right panel). Yet, this does not assess how the ER bilayer manages to grow ahead of the LD monolayer.
Biological membranes are often conformed as lipid bilayers with planar geometry, termed lamellar symmetry. However, certain lipids have the potential, especially if possessing bulky hydrocarbon chains and small head groups, such as PA, of adopting a bilayered yet non‐lamellar status, named cubic phase. 36 In this configuration, the bilayer is no longer in a plane but bends and splits in space depicting curves whose overall mean curvature equals zero (for intuitive illustrations see 37 ). Canonical lamellar bilayers have the potential of evolving toward other membrane statuses through protein conformational changes or by increasing the presence of the appropriate lipids. 38 In particular, the cubic potential of the ER can be unlocked at a low or no energy cost by widespread homodimerization between membrane proteins 39 (Figure 1B). Crucially, the infections by both MERS‐ and SARS‐CoVs have been shown to promote the passage from lamellar to cubic bilayers in the ER. 37 , 40 Indeed, the sole expression of non‐structural nsp3, nsp4, and nsp6 viral proteins, all of which possess transmembrane and cytosolic domains, suffices to tether ER transitions yielding DMVs precursors capable of cubic behavior. 40 , 41 Yet, while structurally reminiscent, these bodies are smaller and less developed than the entities formed during a real infection. 41 I propose that nsp3‐4‐6 viral proteins serve to instruct the dimerization of membranes as to initiate the lamellar‐to‐cubic bilayer transition (Figure 1C, step 1). Subsequently, viral manipulation of host metabolism may support an increase in PA, whose ability to induce negative curvature in membranes would be crucial to permit the magnification of the cubic structure toward mature, bigger cavities (Figure 1C, step 2 to 3; Figure 1B). Last, PA would further sustain the growth yet the embedded state of nascent LD, whose presence inside the niches of the cubic bilayer would provide the final aspect of DMVs (Figure 1C, step 4; Figure 1B). This architecture explains how, despite not showing visible openings to the cytoplasm, the space between the outer and the inner membranes of the DMVs permits, at the same time, a shielded environment for amplification while easiness for viral genome release (Figure 1B and Figure 1C, step 4). Indeed, a solidly sealed DMV would represent a dead‐end for any RNA synthesized within, 7 while by turning the ER into a maze whose alleys are cytoplasmic by definition, yet kept apart from the cytoplasm, the problem is solved. I therefore propose that the membrane subversion step allowing SARS‐CoV replication may concur with the domestication of the LD birth process, by entangling them within cubic ER membranes and preventing their correct budding into the cytoplasm.
This scenario would represent an ideal niche for viral amplification and assembly. First of all, the LD represent a substrate for providing lipids that can fulfill structural, energetic, signaling and chemical chaperone functions. 15 , 42 This is the case during Hepatitis C Virus (HCV) 43 and Dengue virus infections. 44 Yet, mechanistically, while the biogenesis of LD is supported by the HCV itself as to keep on assisting the infection, 45 in the case of the Dengue virus the LD are exploited as a source of materials which lead to their eventual consumption. 46 Another example of RNA viruses whose success depends on LD include rotaviruses 47 and Bovine Viral Diarrhea virus. 48 Next, the LD harbor a very rich and dynamic proteome sitting on their surface, to a big extent inherited from ER membranes, that evolves depending on the metabolic and stress cues. 49 , 50 This makes of them a versatile platform for manipulation of the enzymatic and structural requirements the virus may have. Third, yet less well characterized, the LD represent an emerging substrate to assist nucleic acid‐related transactions. In this regard, elegant evidence comes from simplified models such as bacteria, where the genetic material is by definition harbored in the same compartment as the LD. In Rhodococcus jostii, DNA and LD interact through specialized proteins, and this contact is necessary to protect the DNA during harsh growing conditions, as well as to launch transcription factors that assist this growth. 51 In S. cerevisiae, LD forming inside the nucleus anchor factors, enriching them locally, thus regulating transcription. 52 In humans, mast cells have been found to bear RNA molecules both on top of and inside LD. 53 In the specific case of SARS‐CoV, only one explicit reference to LD has been made in the literature: the tyrosine‐based motif YXXΦ found in the viral protein 3a (where X can be any residue and Φ is a residue with a bulky hydrophobic side chain) is necessary for final targeting of the protein to the plasma membrane. Mutation in this motif maintains the protein in LD, and eventually may target it to lysosomes for destruction. 54 While authors interpret that the presence of viral proteins in LD is unscheduled and provoked by the mutation, it could also be that its presence in this organelle is part of normal viral physiology and that the mutation has just increased its residence time. In any case, the potential of LD to support nucleic acids metabolism makes of them an interesting viral assembly place in view of their multiplication requirements.
To reinforce the proposal, evidence that the virus alters local PA metabolism should be provided. For the time being, the only available data helping challenge that possibility come from a very recent work in which the viral proteins were tagged, expressed and their interacting proteome retrieved, identifying more than 300 high‐confidence SARS‐CoV‐2 interactors implicated in translation, transcription, ubiquitination and trafficking. 55 Different enzymes give rise to PA out of different initial substrates (Figure 2A), and whether an interaction is aimed at stimulating or at sequestering a protein cannot be said from such data. I searched for enzymes whose activity increases PA, namely Glycerol‐3‐phosphateacyltransferase (GPAT, four isoforms); acylglycerol‐3‐phosphate‐acyltransferase (AGPAT, five isoforms); phospholipase D (PLD, two isoforms); and diacylglycerolkinase (DGK, ten isoforms) (Figure 2A), as well as for Seipin, whose absence greatly promotes PA accumulation. 56 The results implicate five viral proteins, among which two of them interacting with PA‐yielding enzymes with a high fold‐enrichment in comparison to control (Figure 2B). These hits are the structural M protein, and the Orf9c protein, initially discarded as a false ORF 57 and as of today with unknown function. Importantly, regarding the host interactors, the pathway Glycerol‐3‐P > LysoPA > PA, whose consecutive steps are handled by GPATs and AGPATs, respectively, robustly emerged. The first step, conversion of 3‐Glycerol‐P to LysoPA, is catalyzed by GPATs, and one of the ER‐based isoforms, GPAT4, is a strong hit. Next, conversion of LysoPA to PA can be catalyzed by five different isoforms, two of which (AGPAT4 and AGPAT5) also solidly pop out in the analysis. AGPAT4, the best hit, exhibits a strong specificity for LysoPA 58 and has been found expressed in six tissues, among which, the lung. 58 , 59 At the subcellular level, and in agreement with the context of this proposal, it mainly localizes to the ER, followed by the mitochondria and the Golgi. 58 , 60 Each AGPAT isoform is responsible for generating a very specific pool of PA and, again of relevance for this proposal, the AGPAT4‐derived PA is reported to specifically mediate membrane remodeling. 60 Neither proteins related to the biology of LD, nor Seipin, were retrieved. Altogether, data reinforce the idea that, contrary to HCV or Dengue virus, it is not colonizing or hijacking the LD themselves, but to subvert the LD birth environment as a protective niche, that could benefit SARS‐CoVs replication.
FIGURE 2.
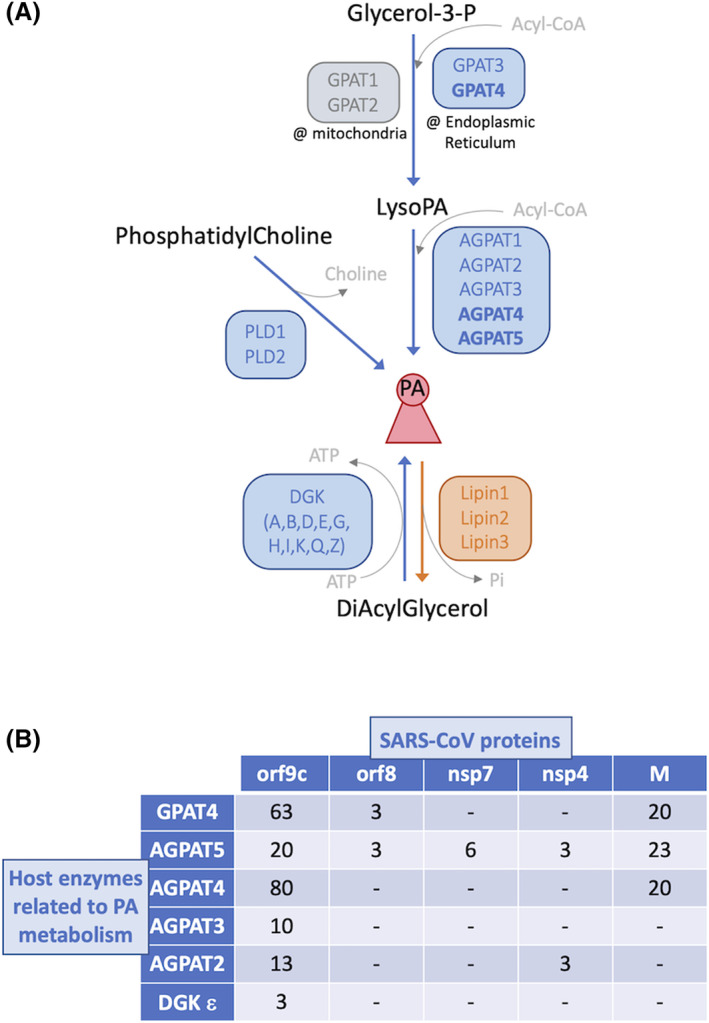
Metabolism of phosphatidic acid (PA) and its potential relationship with SARS‐CoV. A, Pathways yielding PA. PA acid is represented in the center of the scheme by a reddish conical structure. Enzymes and arrows of reactions favoring its accumulation are indicated in blue (or gray for those operating in mitochondria). Those leading to its consumption are indicated in orange. Enzymes emerging as strong interactors of SARS‐CoV‐2 proteins 55 are highlighted in bold. B, Two‐entry table illustrating the positive interactions between host PA‐yielding enzymes and viral proteins. The numbers indicate the fold‐enrichment compared to controls as reported by Reference 55. “‐” indicates lack of interaction
Experimental testing of the PA hypothesis would request visualization of the presumed entangled LD upon infection using appropriate LD and perinuclear ER lumen markers, such as for example BODIPY and Torsin A, respectively. Further, biosensor‐driven PA detection 61 should yield positive signals at perinuclear ER sites in the early hours after infection. Next, genetic manipulation of PA levels by ablation of the strongest hits, namely GPAT4 and AGPAT4, should both decrease LD entangling and, crucially, decrease the infection. Last, drugs lowering PA levels should provide similar results. In this sense, chloroquine, regularly used in the field of malaria, has shown antiviral efficiency against SARS‐CoV in vitro. 62 The strict mechanism by which chloroquine acts is not well‐defined and most probably implies different molecular activities, but it is since long established that chloroquine is a specific inhibitor of phospholipase D whose use decreases PA levels. 63 In addition, other phospholipase D inhibitors are recognized as powerful antiviral compounds. 64 These notions reinforce the possibility that a raise in PA is being exploited by the virus. Moreover, and in striking line with the inhibition of PA generation as a therapeutic strategy during SARS‐CoVs infection, an US patent deposited in 2015 claims the targeting of PA‐yielding enzymes as a therapy approach in a wide spectrum of viral infections, among which +RNA viruses. 65 While the authors argue that the rationale for inhibiting this pathway relies on the sole fact that PA is a basal phospholipid precursor, the patent provides evidence for the effectiveness of the treatment, thus indirectly supporting my proposal that PA may be key in membrane reorganization during infection. Last, it is worth mentioning that LD entangling as a consequence of excessive PA is exquisitely sensitive to diet, with high nutrient conditions, known to activate mTOR, favoring LD trapping. 26 , 27 This further agrees with the recent suggestion that mTOR inhibitors may be beneficial in treating this disease. 55 While the spectrum of consequences of using such drugs cannot be simplified to the only fact of imitating low nutrient intake, the effect on limiting PA accumulation would be straightforward. Last, targeting the coronaviral nsp6 protein, which permits the passage from lamellar to cubic ER bilayers presumably in synergy with PA (Figure 1C), fully abolishes viral replication. 66 Yet, this approach gave rise to resistant viral variants, 66 making of host PA levels modulation a preferred antiviral strategy in order to avoid resistance.
Most of the data we rely on nowadays have been obtained by studying the original SARS‐CoV, yet it is reasonable to expect a strong mechanistic conservation as to better understand and counteract SARS‐CoV‐2. In conclusion, I propose that a local increase in PA at perinuclear locations of LD birth and the cubic conversion of the ER may be considered as the membrane‐subverting mechanism exploited by SARS‐CoVs during their amplification. The morphological, biochemical and genetic available data make of it a plausible possibility. The metabolism of PA in the lung needs to be carefully regulated, 67 and LD are key in this organ for the correct synthesis and secretion of lung surfactant. 68 While awaiting direct experimental validation, the hypothesis is testable and the therapeutical perspective readily exploitable.
2. CONFLICT OF INTEREST
The author declares no competing interests.
ACKNOWLEDGMENTS
The author thanks Cécile Gauthier‐Rouvière and the anonymous reviewers for helpful insights improving this manuscript, Benjamin Pardo for help with the Artwork, and the ATIP‐Avenir program, La Ligue contre le Cancer and l'Institut National du Cancer (PLBIO19‐098 INCA_13832), France, for funding work in her laboratory.
Moriel‐Carretero M. The hypothetical role of phosphatidic acid in subverting ER membranes during SARS‐CoV infection. Traffic. 2020;21:545–551. 10.1111/tra.12738
Peer Review The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12738
[Correction added on 17 June 2020, after first online publication: Peer review history statement has been added.]
Funding information ATIP‐Avenir; Institut National du Cancer, France (INCa), Grant/Award Number: PLBIO19‐098 INCA_13832; La Ligue contre le Cancer
REFERENCES
- 1. Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967‐1976. [DOI] [PubMed] [Google Scholar]
- 3. Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci. 2020;16(10):1724‐1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6(11):967‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahlquist P. Parallels among positive‐strand RNA viruses, reverse‐transcribing viruses and double‐stranded RNA viruses. Nat Rev Microbiol. 2006;4(5):371‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denison MR. Seeking membranes: positive‐strand RNA virus replication complexes. PLoS Biol. 2008;6(10):e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neuman BW. How the double spherules of infectious bronchitis virus impact our understanding of RNA virus replicative organelles. mBio. 2013;4(6):e00987‐e00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maier HJ, Hawes PC, Cottam EM, et al. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4(5):e00801‐e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJ, de Haan CA. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol. 2007;81(22):12323‐12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stertz S, Reichelt M, Spiegel M, et al. The intracellular sites of early replication and budding of SARS‐coronavirus. Virology. 2007;361(2):304‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knoops K, Kikkert M, Worm SH, et al. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10(2):320‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Snijder EJ, van der Meer Y, Zevenhoven‐Dobbe J, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80(12):5927‐5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bailey AP, Koster G, Guillermier C, et al. Antioxidant role for lipid droplets in a stem cell niche of drosophila. Cell. 2015;163(2):340‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25(11):R470‐R481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harcourt BH, Jukneliene D, Kanjanahaluethai A, et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain‐like protease activity. J Virol. 2004;78(24):13600‐13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78(11):5619‐5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78(18):9977‐9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pedersen KW, van der Meer Y, Roos N, Snijder EJ. Open reading frame 1a‐encoded subunits of the arterivirus replicase induce endoplasmic reticulum‐derived double‐membrane vesicles which carry the viral replication complex. J Virol. 1999;73(3):2016‐2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double‐membrane vesicles. J Virol. 2002;76(8):3697‐3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carman GM, Han GS. Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009;50(Suppl):S69‐S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. English D, Cui Y, Siddiqui RA. Messenger functions of phosphatidic acid. Chem Phys Lipids. 1996;80(1–2):117‐132. [DOI] [PubMed] [Google Scholar]
- 23. Stace CL, Ktistakis NT. Phosphatidic acid‐ and phosphatidylserine‐binding proteins. Biochim Biophys Acta. 2006;1761(8):913‐926. [DOI] [PubMed] [Google Scholar]
- 24. Kooijman EE, Chupin V, de Kruijff B, Burger KN. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4(3):162‐174. [DOI] [PubMed] [Google Scholar]
- 25. Raben DM, Barber CN. Phosphatidic acid and neurotransmission. Adv Biol Regul. 2017;63:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124(Pt 22):3894‐3904. [DOI] [PubMed] [Google Scholar]
- 27. Wolinski H, Hofbauer HF, Hellauer K, et al. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim Biophys Acta. 2015;1851(11):1450‐1464. [DOI] [PubMed] [Google Scholar]
- 28. Han S, Binns DD, Chang YF, Goodman JM. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(DeltaNterm) only in combination with Ldb16p. BMC Cell Biol. 2015;16:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell. 2015;26(4):726‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fei W, Shui G, Gaeta B, et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180(3):473‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fei W, Shui G, Zhang Y, et al. A role for phosphatidic acid in the formation of "supersized" lipid droplets. PLoS Genet. 2011;7(7):e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choudhary V, Golani G, Joshi AS, et al. Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr Biol. 2018;28(6):915‐926. e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ben M'barek K, Ajjaji D, Chorlay A, Vanni S, Foret L, Thiam AR. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev Cell. 2017;41(6):591‐604. e597. [DOI] [PubMed] [Google Scholar]
- 34. Gao M, Huang X, Song BL, Yang H. The biogenesis of lipid droplets: lipids take center stage. Prog Lipid Res. 2019;75:100989. [DOI] [PubMed] [Google Scholar]
- 35. Salo VT, Li S, Vihinen H, et al. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev Cell. 2019;50(4):478‐493. e479. [DOI] [PubMed] [Google Scholar]
- 36. Mouritsen OG, Bagatolli LA. Life ‐ as a Matter of Fat: Lipids in a Membrane Biophysics Perspective. Switzerland: Springer Publishing Group; 2016. 10.10007/978-3-319-22614-9. [DOI] [Google Scholar]
- 37. Almsherqi ZA, McLachlan CS, Mossop P, Knoops K, Deng Y. Direct template matching reveals a host subcellular membrane gyroid cubic structure that is associated with SARS virus. Redox Rep. 2005;10(3):167‐171. [DOI] [PubMed] [Google Scholar]
- 38. Kinnunen PK. Fusion of lipid bilayers: a model involving mechanistic connection to HII phase forming lipids. Chem Phys Lipids. 1992;63(3):251‐258. [DOI] [PubMed] [Google Scholar]
- 39. Lingwood D, Schuck S, Ferguson C, Gerl MJ, Simons K. Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J Biol Chem. 2009;284(18):12041‐12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oudshoorn D, Rijs K, Limpens RWAL. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3‐4 polyprotein induce the formation of double‐membrane vesicles that mimic those associated with coronaviral rna replication. mBio. 2017;8(6). 10.1128/mbio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double‐membrane vesicles. mBio. 2013;4(4). 10.1128/mbio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moldavski O, Amen T, Levin‐Zaidman S, et al. Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev Cell. 2015;33(5):603‐610. [DOI] [PubMed] [Google Scholar]
- 43. Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med. 2015;21(1):34‐42. [DOI] [PubMed] [Google Scholar]
- 44. Samsa MM, Mondotte JA, Iglesias NG, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5(10):e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meyers NL, Fontaine KA, Kumar GR, Ott M. Entangled in a membranous web: ER and lipid droplet reorganization during hepatitis C virus infection. Curr Opin Cell Biol. 2016;41:117‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heaton NS, Randall G. Dengue virus‐induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8(5):422‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheung W, Gill M, Esposito A, et al. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84(13):6782‐6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Isken O, Langerwisch U, Schonherr R, et al. Functional characterization of bovine viral diarrhea virus nonstructural protein 5A by reverse genetic analysis and live cell imaging. J Virol. 2014;88(1):82‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bersuker K, Peterson CWH, To M , et al. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell. 2018;44(1):97‐112. e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cermelli S, Guo Y, Gross SP, Welte MA. The lipid‐droplet proteome reveals that droplets are a protein‐storage depot. Curr Biol. 2006;16(18):1783‐1795. [DOI] [PubMed] [Google Scholar]
- 51. Zhang C, Yang L, Ding Y, et al. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat Commun. 2017;8:15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Romanauska A, Kohler A. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell. 2018;174(3):700‐715. e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dvorak AM, Morgan ES, Weller PF. RNA is closely associated with human mast cell lipid bodies. Histol Histopathol. 2003;18(3):943‐968. [DOI] [PubMed] [Google Scholar]
- 54. Minakshi R, Padhan K. The YXXPhi motif within the severe acute respiratory syndrome coronavirus (SARS‐CoV) 3a protein is crucial for its intracellular transport. Virol J. 2014;11:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pagac M, Cooper DE, Qi Y, et al. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of Glycerol‐3‐phosphate acyltransferase. Cell Rep. 2016;17(6):1546‐1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Inberg A, Linial M. Evolutional insights on uncharacterized SARS coronavirus genes. FEBS Lett. 2004;577(1–2):159‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eto M, Shindou H, Shimizu T. A novel lysophosphatidic acid acyltransferase enzyme (LPAAT4) with a possible role for incorporating docosahexaenoic acid into brain glycerophospholipids. Biochem Biophys Res Commun. 2014;443(2):718‐724. [DOI] [PubMed] [Google Scholar]
- 59. Bradley RM, Duncan RE. The lysophosphatidic acid acyltransferases (acylglycerophosphate acyltransferases) family: one reaction, five enzymes, many roles. Curr Opin Lipidol. 2018;29(2):110‐115. [DOI] [PubMed] [Google Scholar]
- 60. Pagliuso A, Valente C, Giordano LL, et al. Golgi membrane fission requires the CtBP1‐S/BARS‐induced activation of lysophosphatidic acid acyltransferase delta. Nat Commun. 2016;7:12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Horchani H, de Saint‐Jean M, Barelli H, Antonny B. Interaction of the Spo20 membrane‐sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One. 2014;9(11):e113484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30(3):269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Madesh M, Balasubramanian KA. Effect of antimalarial drugs on rat enterocyte mitochondrial phospholipase D activity. Life Sci. 1998;62(2):177‐184. [DOI] [PubMed] [Google Scholar]
- 64. Brown HA, Thomas PG, Lindsley CW. Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat Rev Drug Discov. 2017;16(5):351‐367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu S, Rabinowitz JD, Shenk T. United States Patent US 9149445 B2; 2015.
- 66. Lundin A, Dijkman R, Bergstrom T, et al. Targeting membrane‐bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle east respiratory syndrome virus. PLoS Pathog. 2014;10(5):e1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cummings R, Parinandi N, Wang L, Usatyuk P, Natarajan V. Phospholipase D/phosphatidic acid signal transduction: role and physiological significance in lung. Mol Cell Biochem. 2002;234‐235(1‐2):99‐109. [PubMed] [Google Scholar]
- 68. McGowan SE, Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu Rev Physiol. 1997;59:43‐62. [DOI] [PubMed] [Google Scholar]


