Abstract
The limited treatment options for the increasing occurrence of Lassa hemorrhagic fever in West Africa poses an urgent need for the discovery and development of novel therapeutics. Dietary supplements, especially natural products that are edible and safe for human use, are a good source of drug discovery with potential for uncovering novel applications. In this study, we tested 40 natural products of dietary supplements and identified capsaicin, a common dietary supplement abundant in chili peppers, as an inhibitor of Lassa virus (LASV) entry with EC50 of 6.9–10.0 μmol/L using an HIV based pseudovirus platform. Capsaicin inhibits the entry of five LASV strains but not against the Old World arenavirus lymphocytic choriomeningitis virus (LCMV), showing a preferential activity against LASV. Capsaicin inhibits LASV entry by blocking the pH dependent viral fusion through affecting the stable signal peptide (SSP)-GP2 transmembrane (GP2TM) region of the LASV surface glycoprotein. Mutational study revealed the key residues Ala25, Val431, Phe434 and Val435 in SSP-GP2TM region in capsaicin's antiviral effect. This study for the first time reveals a direct acting antiviral effect of capsaicin against the hemorrhagic fever causing LASV, providing detailed interaction hot spots in the unique SSP-GP2TM interface of LASV glycoprotein that is crucial in fusion inhibition, and offering a new strategy in discovering and developing antivirals from natural products that are safe for human use.
Key words: Lassa virus, Fusion inhibitor, Capsaicin, Dietary supplement, Natural product antivirals, Viral entry, Hemorrhagic fever, SSP-GP2TM region
Graphical abstract
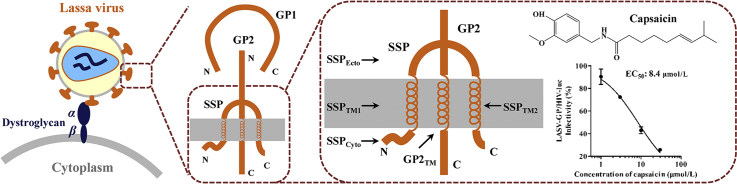
Capsaicin was identified as a Lassa virus (LASV) entry inhibitor. It blocks the LASV-GP mediated viral fusion through affecting the stable signal peptide-GP2 transmembrane region of LASV glycoprotein. Residues Ala25 in SSPTM1 and Val431, Phe434, Val435 in GP2TM are the key residues for the anti-LASV activity of capsaicin.
1. Introduction
Lassa virus (LASV) is an Old World arenavirus that causes Lassa fever, a severe and fatal viral hemorrhagic fever in human1. Endemic in West Africa, LASV causes an estimated 100,000 to 300,000 infections and 5000 deaths each year2. In the years of 2018 and 2019, Nigeria witnessed two unprecedented Lassa fever outbreaks, resulting in 633 and 810 confirmed cases with case fatalities of 27% and 20.6%, respectively3,4. According to World Health Organization (WHO)'s brochure on pandemic and epidemic diseases, LASV is listed as one of the top priority pathogens, and urgent research and development for diagnostic tests, antivirals and vaccines are underway1.
LASV is an enveloped single negative-strand RNA virus and the surface glycoprotein complex (GPC) is responsible for the entry of LASV into host cells5. LASV GPC is a trimer of three non-covalently associated subunits stable signal peptide (SSP), GP1 and GP2. The 58 amino-acid-long SSP is a twice membrane-spanning stable signal peptide that is crucial in GPC mediated fusion6. GP1 binds to α-dystroglycan (α-DG), the cell surface receptor of LASV, resulting in the virus–cell surface attachment7 followed by endocytosis. The fusion of LASV takes place in endosome, where LASV GP1 switches from α-DG to lysosome-associated membrane protein 1 (LAMP1) on the inner endosomal membrane8. GP2 is a class I fusion protein that drives viral-cell membrane fusion under high proton environment in the late endosomes9. Previous studies have shown that LASV GP2 exists in a non-covalent interaction with SSP10, 11, 12, 13, and that this interaction is indispensable in LASV GPC mediated membrane fusion6,14,15. Moreover, this interaction between GP2 and SSP is conserved among arenaviruses, providing a unique target for arenavirus fusion inhibition16.
There is currently no approved antivirals or vaccines against LASV. Clinical therapeutic treatment is limited to an off-label use of the broad-spectrum antiviral ribavirin17. Although a few small molecule LASV entry inhibitors have been reported18, 19, 20, 21, 22, 23, 24, LHF-535 is the only LASV entry inhibitor that is currently under clinical investigation25. Natural products have long been recognized as a valuable source of drug discovery, while the natural product in dietary supplement is one category among millions of natural products with the property of high safety besides of their structural diversity and biological significance26. In this study, we constructed a library of 40 natural products from dietary supplements and evaluated their anti-LASV entry activities. As a result, capsaicin, a well-known substance naturally abundant in chili peppers27 was identified as an inhibitor of LASV entry. As a predominant capsaicinoid, capsaicin is known as an agonist of the transient receptor potential vanilloid subtype 1 (TRPV1), or the capsaicin receptor28 resulting in the ‘hot’ and burning sensations associated with chili peppers as spice. This bioactivity was also applied clinically as an analgesic for the treatment of peripheral neuropathic pain, such as painful diabetic peripheral neuropathy, HIV-related neuropathic pain and postherpetic neuralgia29, 30, 31. This study for the first time disclosed capsaicin's activity against LASV entry, and it is the first report for this famous natural product as a direct acting antiviral.
2. Materials and methods
2.1. Cells and plasmids
HEK293T, A549, Vero and U-87MG cells were obtained from the China Infrastructure of Cell Line Resource (Beijing, China). Vero E6 cells were obtained from the American Type Culture Collection (ATCC). U-87 MG cells were cultured in MEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 μg/mL streptomycin and 100 IU/mL penicillin. All other cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FBS, 100 μg/mL streptomycin and 100 IU/mL penicillin. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
The codon-optimized GP genes of LASV lineage I strain LP, lineage II strain 803213, lineage III strain GA391 and lineage IV strain Josiah and LCMV strain Armstrong 53b were described previously32,33. The codon optimized GP gene of LASV lineage V strain AV (GenBank Accession No. AF246121.2) was synthesized and inserted into eukaryotic expression vector by TSINGKE Biotech (Beijing, China). Plasmids encoding chimeric LASV-GPs [i.e., LASV-GP-(SSP-GP2TM)LCMV, LASV-GP-(SSPCyto)LCMV, LASV-GP-(SSPTM1)LCMV, LASV-GP-(SSPEcto)LCMV and LASV-GP-(SSPTM2)LCMV] and LASV-GPs with single residue mutations were constructed as described previously33. The env-deficient HIV core plasmid (pNL4-3.Luc.R–E–) and vesicular stomatitis virus glycoprotein plasmid (VSV-G) were described previously32,33.
2.2. Compounds
The natural products were selected from an assembled compound library from dietary supplements (Cat. No. L6000, purity>95%, TargetMol, Boston, MA, USA; Cat. No. L1400, purity>95%, Selleck Chemicals, Houston, TX, USA). The capsaicin standard (purity>98%) was purchased from TargetMol (Cat. No. T1062). ST-193, F3406-2010, efavirenz and bafilomycin A1 were described previously32. Doxorubicin hydrochloride was purchased from TargetMol (Cat. No. T1020). All compounds were dissolved in dimethyl sulfoxide (DMSO, Cat. No. 34943, Sigma–Aldrich, St. Louis, MO, USA) and stored at −20 °C until use. DMSO (0.1%, v/v) was used as a solvent control in all experiments.
2.3. Cell viability assay
HEK293T and A549 cells were seeded in 96-well plates as 8 × 103 cells/well on the day before the assay. The cells were treated with the tested compound and incubated for 48 h. The cell viability was evaluated by using CellTiter-Glo® Luminescent Cell Viability Assay (Cat. No.G7571, Promega, Madison, WI, USA). Cells treated with 0.1% DMSO (v/v) served as the solvent control, which is the indicator of 100% cell viability. Cells with medium served as a negative control. Cells treated with doxorubicin served as the positive control. Cell viability (%) and selectivity index (SI) were calculated as Eqs. (1), (2):
| Cell viability (%) = Relative luminescence units (RLUs) of compound/RLUs of solvent control × 100 | (1) |
| SI = Half maximal cytotoxic concentration (CC50)/Half maximal effective concentration (EC50) | (2) |
2.4. Pseudovirus production
The pseudotyped-LASVs (lineages I–V) were generated as previously described33. Briefly, the plasmids encoding LASV-GP and the HIV vector (pNL4.3.Luc-R–E–) were co-transfected into HEK293T cells by using jetPRIME transfection reagent (Polyplus-transfection, New York, NY, USA). Forty-eight hours post-transfection, the LASV-GP/HIV-luc were collected and filtered through a 0.45 μm poresize filter (Millipore, Burlington, MA, USA). The pseudoviruses were layered onto 20% (w/v) sucrose and centrifuged at 300,000×g (L-100XP Ultracentrifuge, Beckman Coulter Inc., Brea, CA, USA) for 2 h at 4 °C. After that, the LASV-GP/HIV-luc were re-suspended in PBS, quantified by the HIV-1 p24 ELISA kit (Cat. No. KIT11695, Sino Biological Inc., Beijing, China) and stored at −80 °C until use. Pseudoviruses bearing the GP of LASV (strain AV) or chimeric LASV-GPs were generated under the same experimental condition.
2.5. Pseudovirus infection assay and compound detection in vitro
Pseudovirus infectivity was measured by the Luciferase Assay System (Cat. No. E1501, Promega) as described previously32, 33, 34. A549 cells were seeded into 24-well plates at a density of 4 × 104 cells/well the day before infection. The cells were incubated with reference compound or tested compounds 15 min prior to pseudoviron infection. Forty-eight hours post-infection, the cells were lysed, and luciferase activity was measured by a luciferase assay kit (Promega). All of these experiments were performed in triplicates.
2.6. Time-of-addition assay
This assay was performed as described previously32,33. Briefly, A549 cells were incubated with LASV pseudovirions at 4 °C for 2 h for the attachment. The supernatant containing the unattached virions was removed, and fresh medium was replenished. The cells were incubated at 37 °C for 48 h. Then, the cells were lysed, and luciferase activity was measured by the luciferase assay kit. Capsaicin (30 μmol/L), ST-193 (1 μmol/L) and bafilomycin A1 (3 nmol/L) were incubated with cells during LASV-GP/HIV attachment, post-attachment or throughout the entire process. The assay was performed in triplicates.
2.7. Virus binding assay
Capsaicin (300 μmol/L) was incubated with LASV-GP (strain Josiah)/HIV-luc or LCMV-GP/HIV-luc at 4 °C for 4 h. The pseudovirons were then layered onto 20% (w/v) sucrose and centrifuged at 300,000×g (L-100XP Ultracentrifuge, Beckman) for 2 h. The supernatant was removed, and the pseudovirons were resuspended in PBS. The harvested pseudoviruses were incubated with A549 cells for 48 h. The infected cells were lysed, and luciferase activity was measured by the Luciferase Assay System. The luciferase activity of the DMSO (0.1%, v/v) solvent control was used as the 100% infectivity indicator. ST-193 (0.1 μmol/L), F3406-2010 (10 μmol/L) and bafilomycin A1 (0.1 μmol/L) were used as the reference compounds. The assay was performed in triplicates.
2.8. Cell–cell fusion assay
The assay was performed as described previously32,33. Briefly, HEK293T cells were co-transfected with plasmids expressing LASV-GP (or LCMV-GP) and the enhanced green fluorescent protein (EGFP). The transfected cells were seeded into 48-well plates at a density of 1 × 105 cells/well. Twenty-four hours later, the medium was removed, and the cells were incubated with PBS (pH 4.7) for 20 min. Then the PBS was removed, and the fresh medium was added into the wells. The cells were incubated at 37 °C for 4 h, and syncytium formation was observed under a fluorescence microscope (IX71, Olympus Corp., Shinjuku, Tokyo, Japan). Capsaicin (50 μmol/L), bafilomycin A1 (10 nmol/L), ST-193 (0.1 μmol/L) or F3406-2010 (10 μmol/L) were added to the cells 4 h before low pH treatment, during low pH treatment or during the whole process, i.e., before and during low pH treatment.
2.9. Construction of chimeric LASV-GP and LCMV-GP plasmids
The genes of chimeric LCMV-GP [LCMV-GP-(SSP-GP2TM)LASV] and chimeric LASV-GP [LASV-GP-(SSP)LCMV] were created by overlapping PCR. The SSP (Met1 to Thr58) and GP2TM (Leu428 to Leu447) region of LASV-GP open reading frame were amplified by PCR and then were both used to replace the SSP (Met1 to Gly58) and GP2TM (Leu434 to Leu455) region of LCMV-GP to get LCMV-GP-(SSP-GP2TM)LASV. The LCMV-GP SSP (Met1 to Gly58) open reading frame were amplified by PCR and then were used to replace the SSP region (Met1 to Thr58) of LASV-GP to get LASV-GP-(SSP)LCMV. The chimeric gene sequences were inserted into pCMV3 vector and identified by DNA sequencing.
2.10. Statistical analysis
The mean values, standard deviation (SD), EC50, and CC50 values were calculated by GraphPad Prism software. The statistical analysis in this study was performed by using Student's t-test. The asterisks represent significant differences: *P < 0.05, **P < 0.01, and ***P < 0.001.
3. Results
3.1. Capsaicin is identified as a LASV specific entry inhibitor
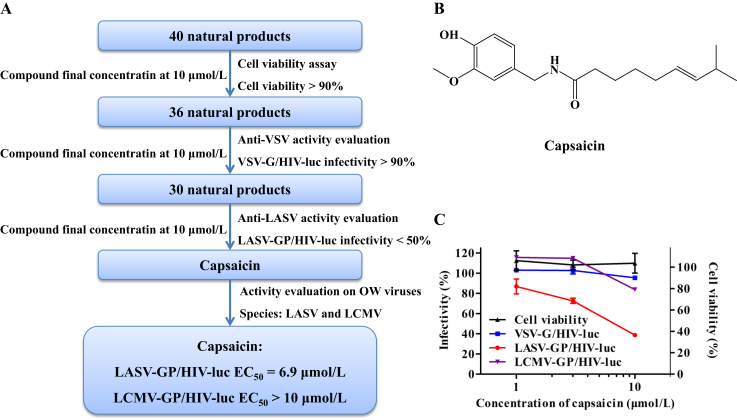
Dietary supplements of potential health benefits could be categorized into vitamins, minerals, proteins and amino acids, bodybuilding supplements, essential fatty acids, natural products, probiotics, etc. Among these categories, natural products, often the extracts from plants, animals, algae, fungi or lichens, are the source of biologically active compounds with wide structural diversity. In this study, we evaluated 40 natural products from an assembled library of labeled dietary supplements for their inhibitory activity against LASV-GP/HIV-luc (strain Josiah) infection (Fig. 1A and Supporting Information Table S1). These compounds were first tested for cytotoxicity on HEK293T cells by CellTiter-Glo assay at a final concentration of 10 μmol/L, and those resulting in less than 10% reduction of cell viability were selected for subsequent activity evaluations. The compounds were tested by VSV-G/HIV-luc pseudovirus infection to rule out compounds with any effect on VSV glycoprotein or HIV-1 replication, and compounds with less than 10% inhibitory activity on VSV-G/HIV-luc infection then proceed for evaluation against LASV-GP/HIV-luc infection. Compounds resulting in less than 50% infectivity of LASV-GP/HIV-luc were considered effective against LASV entry. Capsaicin was identified from this process with a specific inhibitory effect against LASV-GP/HIV-luc with an EC50 of 6.9 μmol/L (Fig. 1B and C). We tested the anti-LASV activity of capsaicin on A549, Vero E6, Vero and U-87MG cells to validate its anti-LASV activity, and our results show that the anti-LASV activity of capsaicin is not cell type specific (Supporting Information Table S2).
Figure 1.
Identification of capsaicin as a LASV entry inhibitor by screening a natural product library from dietary supplements. (A) A flow chart of screening 40 natural products against LASV-GP pseudotyped virus infection. (B) The chemical structure of capsaicin. (C) The dose–response curve demonstrating the inhibitory activity of capsaicin against LASV-GP/HIV-luc, LCMV-GP/HIV-luc and VSV-G/HIV-luc pseudovirus infection, as well as cell viability on HEK293T cells. Cells treated with 0.1% DMSO (v/v) served as the indicator of 100% cell viability. The data are represented as the mean ± SD (n = 3).
LASV belongs to Old World arenaviruses and is phylogenetically close to the lymphocytic choriomeningitis virus (LCMV, Supporting Information Fig. S1), which is a human pathogenic arenavirus of clinical significance35,36. We tested capsaicin against LCMV-GP/HIV-luc infection and found that capsaicin had no significant effect on LCMV infection (Fig. 1C and Table 1).
Table 1.
Effects of capsaicin on LASV-GP (lineages I–V) pseudotyped virus entry on A549 cells.
| Pseudovirus | Capsaicin |
ST-193 |
F3406-2010 |
||||
|---|---|---|---|---|---|---|---|
| EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | ||
| LASV-GP/HIV-luc | Lineage I (strain LP) | 15.4 | 10.9–21.8 | 0.013 | 0.0048–0.037 | >10 | / |
| Lineage II (strain 803213) | 15.9 | 10.0–25.1 | 0.0013 | 0.00058–0.0027 | >10 | / | |
| Lineage III (strain GA391) | 10.8 | 7.4–15.8 | 0.0013 | 0.00076–0.0022 | >10 | / | |
| Lineage IV (strain Josiah) | 8.4 | 6.9–10.2 | 0.0024 | 0.0016–0.0037 | >10 | / | |
| Lineage V (strain AV) | 6.5 | 5.4–7.8 | 0.00082 | 0.00055–0.0012 | >10 | / | |
| LCMV-GP/HIV-luc | (strain Armstrong 53b) | >30 | / | >10 | / | 0.15 | 0.14–0.16 |
95% CI, 95% confidence intervals.
/, not applicable.
The LASV species demonstrates a high genetic diversity, which could be further divided into 5 or 6 lineages, consistent to their geographic distribution37,38. In this study, in addition to the LASV strain Josiah (lineage IV) we used in the primary activity evaluation, we selected another four strains of LASV that are the genetic representative of each of their LASV lineages (Supporting Information Figs. S2 and S3), and tested capsaicin activity against the entry of these strains of LASV pseudotypes. Capsaicin showed a comparable activity towards 5 LASV strains with EC50s of 6.5–15.9 μmol/L (Table 1 and Supporting Information Fig. S4).
Taken together, capsaicin specifically inhibit LASV rather than LCMV among Old World arenaviruses, while displaying a comparable effect on various strains within the LASV species, indicating that capsaicin is a specific inhibitor of LASV entry.
3.2. Capsaicin blocks LASV entry by inhibiting LASV-GP mediated fusion
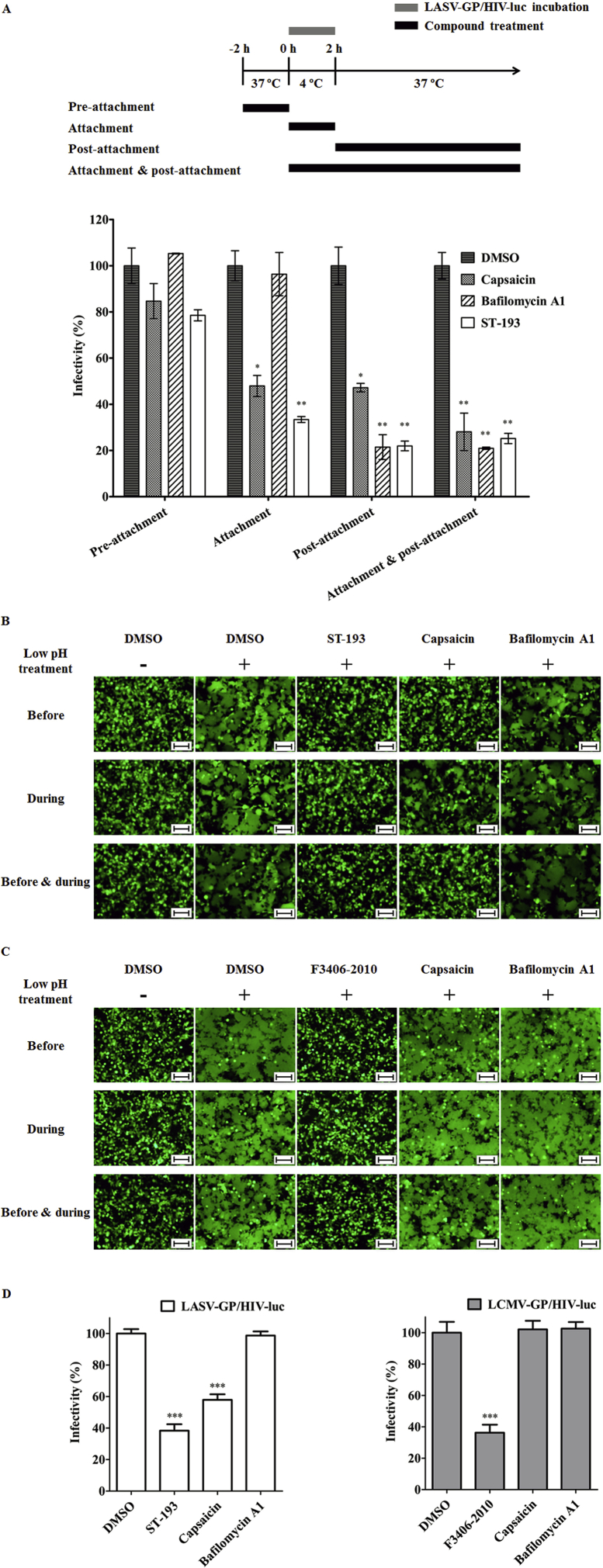
Many studies have delineated the stepwise fashion of LASV entry into host cells. LASV attaches to the cell surface through interaction between virus GP1 and cellular receptor α-DG39,40. Following endocytosis and endosomal acidification, LASV GP1 detaches from α-DG and the GPC to bind LAMP18, meanwhile LASV GP2 undergoes conformational changes and triggers viral-endosomal membrane fusion9, releasing virus genome into the cytoplasm. In this study, we first identify the step in which capsaicin exerts its effect by a time-of-addition assay32,33. Capsaicin as well as reference compounds bafilomycin A1 and ST-193 were incubated with A549 cells as depicted (Fig. 2). As shown in Fig. 2A, capsaicin is effective in both the attachment and the post-attachment phase, displaying a similar inhibitory pattern to ST-193, a known arenavirus fusion inhibitor that targets GPC,18 while the proton pump inhibitor bafilomycin A1 is only effective post-attachment, suggesting that capsaicin may interact directly with the LASV pseudovirions and that it may take effect at the viral fusion step.
Figure 2.
Capsaicin specifically inhibited low pH triggered LASV-GP mediated cell–cell fusion by binding directly to the virions. (A) Capsaicin affects the attachment and post-attachment, but not pre-attachment process of LASV entry. A549 cells were incubated with the test compounds or the same amount DMSO for 2 h at 37 °C (pre-attachment treatment) and then the supernatant was washed out. The LASV pseudovirions were added and incubated with the cells at 4 °C for 2 h (attachment). The supernatant which containing the unattached virions was removed and fresh medium was replenished. The cells were incubated at 37 °C for 48 h (post-attachment). The cells were lysed, and luciferase activity was measured by the luciferase assay kit. Capsaicin (30 μmol/L), ST-193 (1 μmol/L), bafilomycin A1 (3 nmol/L), or the same amount of DMSO was treated at pre-attachment, during the attachment, post-attachment or attachment + post-attachment. (B) and (C) Capsaicin inhibited low pH-triggered LASV-GP or LCMV-GP mediated cell–cell fusion. HEK293T cells were co-transfected with plasmids expressing LASV-GP (or LCMV-GP) and EGFP. The transfected cells were seeded into 48-well plates. Twenty-four hours later, the medium was removed, and the cells were incubated with PBS (pH 4.7) for 20 min. Then the low pH PBS was replaced with fresh medium and incubated for 4 h. Syncytium formation of the cells was observed via fluorescence microscopy. Capsaicin (50 μmol/L), bafilomycin A1 (10 nmol/L), ST-193 (0.1 μmol/L) or F3406-2010 (10 μmol/L) were added to the cells 4h before the low pH treatment, during low pH treatment or before + during low pH treatment. Scale bar, 100 μm. (D) Capsaicin blocks LASV entry by binding directly to LASV-GP/HIV-luc virions. LASV-GP/HIV-luc or LCMV-GP/HIV-luc was incubated with Capsaicin (300 μmol/L) at 4 °C for 4 h and then the supernatant was removed by ultracentrifugation. The pseudovirons were resuspended and used to infect A549 cells. Forty-eight hours post-infection, the cells were lysed, and luciferase activity was measured. The same amount of DMSO was used as the solvent control; ST-193 (0.1 μmol/L), F3406-2010 (10 μmol/L) and bafilomycin A1 (0.1 μmol/L) were used as the reference compounds. The luciferase activity of the solvent control was used as the 100% infectivity indicator. The data are represented as the mean ± SD (n = 3). Statistical significances between treatment group and DMSO group were calculated by Student's t-test using GraphPad Prism software, with asterisks representing significant differences: *P < 0.05, **P < 0.01, and ***P < 0.001.
We next used the established cell–cell fusion assay32,33 to confirm the effect of capsaicin on GPC mediated fusion. Capsaicin as well as reference compounds ST-193, F3406-201041, and bafilomycin A1 were added to cells expressing GPC and EGFP at specific time of low pH triggering as indicated in Fig. 2B and C, and cell–cell fusion was observed 4 h after low pH treatment as indicated by green fluorescence. The results showed that capsaicin inhibited LASV-GP mediated fusion, but it did not affect LCMV-GP mediated fusion, suggesting that capsaicin specifically targets the LASV fusion process.
A virus binding assay was also employed to explore whether there is an interaction between capsaicin and LASV virions. Capsaicin, along with reference compounds ST-193, F3046-2010 and bafilomycin A1 were incubated with LASV-GP/HIV-luc or LCMV-GP/HIV-luc. The compound-treated pseudoviruses were collected through ultracentrifugation to exclude un-bounded compound as well as supernatants. The harvested viruses were then used to infect A549 cells. The results showed that LASV-GP/HIV-luc treated with capsaicin had a low infectivity, similar to that of ST-193, indicating that capsaicin interacts directly with LASV-GP (Fig. 2D).
3.3. Capsaicin targets the SSP-GP2 interface of LASV glycoprotein
The arenavirus stable signal peptide (SSP) is a unique functional subunit in the GPC38. SSP forms a non-covalent interaction with GP2 near the transmembrane region which is essential to the LASV fusion process6. Compounds that interact with this complex directly will lead to a failure of the viral fusion. In this study, we investigated whether the specific inhibition of LASV-GP mediated fusion by capsaicin was associated with this interaction between SSP and GP2. To identity the region with which capsaicin interact, we established two chimeric GPC constructs where one is LASV-GP with its SSP and GP2TM substituted by LCMV SSP and GP2TM, termed LASV-GP-(SSP-GP2TM)LCMV, and another is LCMV-GP with its SSP and GP2TM substituted by the respective LASV SSP and GP2TM, termed LCMV-GP-(SSP-GP2TM)LASV (Table 2 and Fig. 3A). We tested the activity of capsaicin against the above chimeric GPC packed pseudoviruses, along with reference compounds ST-193 and F3406-2010. As shown in Table 2 and Supporting Information Fig. S5, the LASV fusion inhibitor ST-193 lost its activity against LASV-GP-(SSP-GP2TM)LCMV/HIV-luc while the LCMV specific inhibitor F3406-2010 was effective against LASV-GP-(SSP-GP2TM)LCMV/HIV-luc, which are consistent to their mode of action6,33. Capsaicin showed a comparable activity against LCMV-GP-(SSP-GP2TM)LASV/HIV-luc and LASV wild type (strain Josiah LASV pseudotype), while it completely lost its activity against LASV-GP-(SSP-GP2TM)LCMV/HIV-luc, indicating that capsaicin indeed targets the LASV SSP-GP2TM interface of GPC.
Table 2.
Effects of capsaicin on LASV-GP-(SSP-GP2TM)LCMV and LCMV-GP-(SSP-GP2TM)LASV mediated viral entry on A549 cells.
| Pseudovirus | Capsaicin |
ST-193 |
F3406-2010 |
|||
|---|---|---|---|---|---|---|
| EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | |
| LASV-GP/HIV-luc | 10.0 | 8.0–12.4 | 0.0024 | 0.0014–0.0043 | >10 | / |
| LCMV-GP/HIV-luc | >30 | / | >10 | / | 0.23 | 0.19–0.28 |
| LASV-GP-(SSP-GP2TM)LCMV/HIV-luc | >30 | / | >10 | / | 0.44 | 0.39–0.49 |
| LCMV-GP-(SSP-GP2TM)LASV/HIV-luc | 6.1 | 5.1–7.4 | 0.00089 | 0.00073–0.0011 | >10 | / |
95% CI, 95% confidence intervals.
/, not applicable.
Figure 3.
Fragment replacements and residue substitutions between LASV-GP (orange) and LCMV-GP (blue). (A) The domains of LASV-GP, LCMV-GP, LASV-GP-(SSP-GP2TM)LCMV and LCMV-GP-(SSP-GP2TM)LASV are showed in diagram. (B)–(D) Amino acid sequence alignment of SSP (B), SSPTM1 (C) and GP2TM region (D) between LASV-GP (strain Josiah) and LCMV-GP (strain Arm53b) along with their composition cartoons. The role of the replaced fragments and the residue substitutions which marked with black triangles is investigated in this study.
3.4. The N-terminal transmembrane region of SSP is a crucial region for the anti-LASV activity of capsaicin
SSP is a 58-residue long signal peptide that spans the membrane twice, with two cytoplasmic regions, two hydrophobic putative transmembrane regions and one ectodomain6. To further evaluate the specific regions in SSP-GP2TM and determine how they affect capsaicin activity, we divided LASV SSP into four segments (Table 3 and Fig. 3B) and replaced each segment with its LCMV SSP counterpart. Capsaicin was tested against infections of pseudoviruses bearing these chimeric LASV-GPs (Table 3 and Supporting Information Fig. S6) with ST-193 and F3406-2010 as references. Capsaicin showed a similar inhibitory activity on LASV-GP-(SSPCyto)LCMV/HIV-luc, LASV-GP-(SSPEcto)LCMV/HIV-luc and LASV-GP-(SSPTM2)LCMV/HIV-luc compared with wild type (strain Josiah LASV pseudotype), while it lost its activity on LASV-GP-(SSPTM1)LCMV/HIV-luc as well as LASV-GP-(SSP)LCMV/HIV-luc (Table 3), indicating that SSPTM1 is a key module in the anti-LASV activity of capsaicin.
Table 3.
Activities of capsaicin against LASV-GPs with fragment substitution in SSP mediated viral entry on A549 cells.
| Pseudovirus | Capsaicin |
ST-193 |
F3406-2010 |
|||
|---|---|---|---|---|---|---|
| EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | |
| LASV-GP/HIV-luc | 9.3 | 7.6–11.5 | 0.0024 | 0.0016–0.0037 | >10 | / |
| LASV-GP-(SSP)LCMV/HIV-luc | >30 | / | 0.0029 | 0.0021–0.0040 | 0.16 | 0.13–0.19 |
| LASV-GP-(SSPCyto)LCMV/HIV-luc | 8.3 | 6.4–10.7 | 0.0021 | 0.0012–0.0038 | 5.2 | 3.1–8.6 |
| LASV-GP-(SSPTM1)LCMV/HIV-luc | >30 | / | 0.19 | 0.089–0.41 | 1.1 | 0.47–2.5 |
| LASV-GP-(SSPEcto)LCMV/HIV-luc | 16.9 | 11.8–24.3 | 0.0057 | 0.0038–0.0085 | >10 | / |
| LASV-GP-(SSPTM2)LCMV/HIV-luc | 17.3 | 11.3–26.4 | 0.0054 | 0.0031–0.0093 | >10 | / |
95% CI, 95% confidence intervals.
/, not applicable.
Amino acid sequence alignment in the SSPTM1 region showed eight distinct residues between LASV and LCMV (Table 4 and Fig. 3C). To identify which residues affect the anti-LASV activity of capsaicin, we generated eight LASV-GP constructs, each bearing one of these single residues mutated to its LCMV counterpart (Supporting Information Fig. S7). We tested the activity of capsaicin against these LASV pseudoviruses with single residue mutations in SSPTM1 and found that capsaicin completely lost its activity on A25V mutant virus, and a slightly activity loss on S27I mutant virus with 2-fold EC50 increase, while displaying comparable activity on other single residue mutants (Table 4).
Table 4.
Activities of capsaicin against LASV-GP-SSPTM1 mutation mediated viral entry on A549 cells.
| Pseudovirus | Capsaicin |
ST-193 |
F3406-2010 |
|||
|---|---|---|---|---|---|---|
| EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | EC50(μmol/L) | 95% CI (μmol/L) | |
| LASV-GP/HIV-luc | 9.3 | 6.4–13.6 | 0.0044 | 0.0029–0.0067 | >10 | / |
| LASV-GPM19I/HIV-luc | 8.2 | 5.8–11.5 | 0.00040 | 0.00020–0.00078 | 3.7 | 3.2–4.2 |
| LASV-GPL23I/HIV-luc | 14.3 | 10.5–19.5 | 0.0096 | 0.0048–0.019 | 0.47 | 0.29–0.77 |
| LASV-GPA25V/HIV-luc | >30 | / | 0.0022 | 0.0011–0.0044 | 9.1 | 6.8–12.0 |
| LASV-GPS27I/HIV-luc | 21.0 | 13.7–32.1 | 0.090 | 0.032–0.25 | 9.5 | 5.5–16.3 |
| LASV-GPL29I/HIV-luc | 8.5 | 5.4–13.5 | 0.067 | 0.030–0.15 | 7.4 | 5.7–9.7 |
| LASV-GPA30T/HIV-luc | 8.3 | 6.2–11.2 | 0.022 | 0.014–0.034 | 8.5 | 3.8–18.7 |
| LASV-GPV31G/HIV-luc | 9.7 | 7.0–13.3 | 0.051 | 0.028–0.096 | 7.1 | 5.9–8.4 |
| LASV-GPL32I/HIV-luc | 11.4 | 7.6–17.1 | 0.032 | 0.019–0.052 | >10 | / |
95% CI, 95% confidence intervals.
/, not applicable.
Taken together, these results indicated that residue Ala25 in SSPTM1 is associated with the anti-LASV activity of capsaicin, and implied an interaction between the SSPTM1 and GP2 possibly mediated by the residue.
3.5. Val431, Phe434 and Val435 in the transmembrane region of GP2 involve in capsaicin activity
SSP is known to interact non-covalently with the fusion subunit GP2, participates in GPC mediated fusion and functions by stabilizing GP2 structure42. After identifying SSPTM1 as an important region in the SSP-GP2 interface, we next sought to identify residues in the respective GP2TM region that might associate with capsaicin activity. We inspected the amino acid sequences of LASV and LCMV in the GP2TM region and they revealed 6 distinct residues (Table 5 and Fig. 3D). We established 6 LASV pseudoviruses bearing these mutant LASV-GPs (Fig. S7), and tested capsaicin activity against their infection (Table 5). The results showed that capsaicin lost its activity against LASV pseudotypes bearing V431M, F434L or V435M mutation, indicating that Val431, Phe434, and Val435 in GP2TM of LASV are activity determinants of capsaicin, and might play important roles in SSP-GP2 interaction. The reference compounds ST-193 and F3406-2010 displayed a loss of activity consistent with previous reports18,41.
Table 5.
Effects of capsaicin on LASV-GP-GP2TM mutation mediated viral entry on A549 cells.
| Pseudovirus | Capsaicin |
ST-193 |
F3406-2010 |
|||
|---|---|---|---|---|---|---|
| EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | EC50 (μmol/L) | 95% CI (μmol/L) | |
| LASV-GP/HIV-luc | 8.3 | 6.9–10.0 | 0.0096 | 0.0051–0.018 | >10 | / |
| LASV-GPG429A/HIV-luc | 11.0 | 6.9–17.5 | 0.0076 | 0.0051–0.011 | >10 | / |
| LASV-GPV431M/HIV-luc | >30 | / | 0.73 | 0.52–1.0 | 1.5 | 1.2–1.9 |
| LASV-GPF434L/HIV-luc | >30 | / | 0.014 | 0.0060–0.031 | >10 | / |
| LASV-GPV435M/HIV-luc | >30 | / | 1.6 | 0.91–2.7 | >10 | / |
| LASV-GPF440A/HIV-luc | 14.0 | 10.0–19.7 | 0.0093 | 0.0032–0.027 | >10 | / |
| LASV-GPI443V/HIV-luc | 16.6 | 9.2–29.9 | 0.0095 | 0.0044–0.021 | >10 | / |
95% CI, 95% confidence intervals.
/, not applicable.
4. Discussion
In this study, we discovered capsaicin as a LASV entry inhibitor that functions by blocking the LASV-GP mediated viral fusion (Fig. 2B and C) and binding directly to the LASV pseudovirions (Fig. 2D). Further mechanism study showed that it targets the SSP–GP2 interface, a crucial functioning unit of LASV GPC mediated fusion (Table 2). Site-directed mutagenesis study revealed single residues in the SSPTM1 and GP2TM that affect anti-LASV activity of capsaicin (Table 3, Table 4, Table 5). These results implied a possible non-covalent interaction between residues Ala25 in SSPTM1 and Val431, Phe434, Val435 in GP2TM, and that a hydrophobic pocket may be formed. The two reference compounds we used in the mutational study, ST-193 and F3406-2010, showed a consistent pattern of activity variation with previous studies as in mutations V431M and V435M18,41, meanwhile displayed new sensitivities to mutations such as L23I and F434L. These results provided valuable information and new insight into the possible pattern of interaction between SSPTM1 and GP2TM when structural information regarding the SSP–GP2 interface is still unavailable. Taken together, these results not only provided support for the crucial role of SSPTM1 in the interaction between SSP and GP2TM, validated Val431 and Val435, two previously reported drug sensitive residues in the GP2TM, and identified two residues, Ala25 in SSPTM1 and Phe434 in GP2TM that are important for capsaicin activity, but also brought up valuable target information for novel arenavirus fusion inhibitors design or discovery based on the unique SSP–GP2 interface of arenavirus GPC.
Our study for the first time identified capsaicin, the famous natural product from dietary supplements, as a direct acting antiviral, which is notably distinct from its traditional biological effect and application. Capsaicin is the major active component of chili peppers, which is one of the most popular and historical foods, and is used widely as a spice in the world. It also has a long history in medical usage, as it was recorded as a medicine for gastrointestinal diseases in China since the 15th century. Modern pharmacology revealed that the traditional medical usage of capsaicin in pain relief is due to the stimulation and subsequent desensitization of TRPV128, a non-selective cation channel on neuron cells. Capsaicin was also investigated for a wide range of indications such as obesity43, osteoarthritis44, cancer complementary therapeutics45,46, pruritus47, and cannabinoid hyperemesis syndrome (CHS)48; however, the underlying mechanism of the effect of capsaicin on these non-neuropathic syndromes remains to be clarified. Historically peppers have been used as a dietary preservative for capsaicin's bactericidal properties49. There has been a report on the anti-herpes simplex virus activity of Capsicum annuum extract50; however, the anti-infective mechanism has been unclear. To the best of our knowledge, our study is the first report of the single molecular entity capsaicin as a direct acting antiviral.
The discovery of capsaicin as an antiviral agent not only offered a novel category of structural backbone in targeting LASV glycoprotein, but also disclosed a new potential medical usage for this historically used natural product and commonly known dietary supplement. There are two prominent advantages of developing therapeutic treatments from natural products over de novo drug design: compounds coming from natural products display a high level of structural diversity, which is not easily accomplished through small molecule design. Natural product components are often predisposed with biological significance through a long history of evolutionary selection and optimization51. In addition, natural products under the category of dietary supplements could also add safety to its existing advantages. According to the USA DSHEA act of 1994, dietary supplements are considered a subset of foods, and are regulated accordingly. There are strict regulations regarding the toxicity of dietary supplements that are for human ingestions, which gives them a high safety profile. In recent years, high-dose natural products have received much research attention because of their potential health effects26. In the field of antiviral discovery, the citrus peel extract tangeretin has been the first natural product reported with anti-LASV activity32. The discovery of capsaicin as yet another natural product with anti-LASV property suggests that compounds sourced from natural dietary supplements or traditional medicines could be a good start for the discovery of novel antiviral therapeutics.
Taken together, this study identified capsaicin as a LASV fusion inhibitor, providing a lead compound derived from natural dietary supplements with a distinct chemical scaffold that targets LASV glycoprotein. This study also highlighted the role of SSPTM1 in the interaction between SSP and GP2, and identified important residues in both SSPTM1 and GP2TM that affect antiviral activity, providing framework for the discovery and design of arenavirus fusion inhibitors based on the SSP–GP2 interface of mammarenavirus glycoprotein.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 81473256 and 81273561), the CAMS Innovation Fund for Medical Sciences (No. 2016-I2M-1-014, China), the Science and Technology Program of Beijing (No. Z151100000115008, China), the Beijing Key Laboratory of New Drug Mechanisms and Pharmacological Evaluation Study (No. BZ0150, China), and the Drug Innovation Major Project (Nos. 2015ZX09102-023 and 2018ZX09711001-003-002, China), the Disciplines Construction Project (No. 201920200802, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.02.014.
Author contributions
Ying Guo conceptualized and supervised this study. Ke Tang designed the experiments. Ke Tang and Xiaoyu Zhang carried out the experiments, performed data analysis, and drafted the manuscript. All authors revised the manuscript, and have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bhadelia N. Understanding Lassa fever. Science. 2019;363:30. doi: 10.1126/science.aav8958. [DOI] [PubMed] [Google Scholar]
- 2.Richmond J.K., Baglole D.J. Lassa fever: epidemiology, clinical features, and social consequences. BMJ. 2003;327:1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigeria Centre for Disease Control 2018 Lassa fever outbreak situation report [EB/OL] 31 December 2018. http://ncdc.gov.ng/diseases/sitreps Available from:
- 4.Nigeria Centre for Disease Control 2019 Lassa fever outbreak situation report [EB/OL] 15 December 2019. http://ncdc.gov.ng/diseases/sitreps Available from:
- 5.Hastie K.M., Saphire E.O. Lassa virus glycoprotein: stopping a moving target. Curr Opin Virol. 2018;31:52–58. doi: 10.1016/j.coviro.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messina E.L., York J., Nunberg J.H. Dissection of the role of the stable signal peptide of the arenavirus envelope glycoprotein in membrane fusion. J Virol. 2012;86:6138–6145. doi: 10.1128/JVI.07241-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunz S., Rojek J.M., Perez M., Spiropoulou C.F., Oldstone M.B. Characterization of the interaction of lassa fever virus with its cellular receptor alpha-dystroglycan. J Virol. 2005;79:5979–5987. doi: 10.1128/JVI.79.10.5979-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jae L.T., Raaben M., Herbert A.S., Kuehne A.I., Wirchnianski A.S., Soh T.K. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science. 2014;344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.York J., Romanowski V., Lu M., Nunberg J.H. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1–G2 complex. J Virol. 2004;78:10783–10792. doi: 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saunders A.A., Ting J.P., Meisner J., Neuman B.W., Perez M., de la Torre J.C. Mapping the landscape of the lymphocytic choriomeningitis virus stable signal peptide reveals novel functional domains. J Virol. 2007;81:5649–5657. doi: 10.1128/JVI.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.York J., Nunberg J.H. Distinct requirements for signal peptidase processing and function in the stable signal peptide subunit of the Junin virus envelope glycoprotein. Virology. 2007;359:72–81. doi: 10.1016/j.virol.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 13.Bederka L.H., Bonhomme C.J., Ling E.L., Buchmeier M.J. Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization. mBio. 2014;5 doi: 10.1128/mBio.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.York J., Nunberg J.H. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J Virol. 2009;83:4121–4126. doi: 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.York J., Nunberg J.H. Myristoylation of the arenavirus envelope glycoprotein stable signal peptide is critical for membrane fusion but dispensable for virion morphogenesis. J Virol. 2016;90:8341–8350. doi: 10.1128/JVI.01124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankar S., Whitby L.R., Casquilho-Gray H.E., York J., Boger D.L., Nunberg J.H. Small-molecule fusion inhibitors bind the pH-sensing stable signal peptide–GP2 subunit interface of the Lassa virus envelope glycoprotein. J Virol. 2016;90:6799–6807. doi: 10.1128/JVI.00597-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houlihan C., Behrens R. Lassa fever. BMJ. 2017;358:j2986. doi: 10.1136/bmj.j2986. [DOI] [PubMed] [Google Scholar]
- 18.Larson R.A., Dai D., Hosack V.T., Tan Y., Bolken T.C., Hruby D.E. Identification of a broad-spectrum arenavirus entry inhibitor. J Virol. 2008;82:10768–10775. doi: 10.1128/JVI.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A.M., Rojek J.M., Spiropoulou C.F., Gundersen A.T., Jin W., Shaginian A. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J Biol Chem. 2008;283:18734–18742. doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spence J.S., Melnik L.I., Badani H., Wimley W.C., Garry R.F. Inhibition of arenavirus infection by a glycoprotein-derived peptide with a novel mechanism. J Virol. 2014;88:8556–8564. doi: 10.1128/JVI.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrivastava-Ranjan P., Bergeron E., Chakrabarti A.K., Albarino C.G., Flint M., Nichol S.T. 25-Hydroxycholesterol inhibition of Lassa virus infection through aberrant GP1 glycosylation. mBio. 2016;7:e01808–e01816. doi: 10.1128/mBio.01808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M.K., Ren T., Liu H., Lim S.Y., Lee K., Honko A. Critical role for cholesterol in Lassa fever virus entry identified by a novel small molecule inhibitor targeting the viral receptor LAMP1. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P., Liu Y., Zhang G., Wang S., Guo J., Cao J. Screening and identification of Lassa virus entry inhibitors from an FDA-approved drug library. J Virol. 2018;92 doi: 10.1128/JVI.00954-18. e00954-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G., Cao J., Cai Y., Liu Y., Li Y., Wang P. Structure–activity relationship optimization for lassa virus fusion inhibitors targeting the transmembrane domain of GP2. Protein Cell. 2019;10:137–142. doi: 10.1007/s13238-018-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madu I.G., Files M., Gharaibeh D.N., Moore A.L., Jung K.H., Gowen B.B. A potent Lassa virus antiviral targets an arenavirus virulence determinant. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rautiainen S., Manson J.E., Lichtenstein A.H., Sesso H.D. Dietary supplements and disease prevention—a global overview. Nat Rev Endocrinol. 2016;12:407–420. doi: 10.1038/nrendo.2016.54. [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Salam O.M., Szolcsanyi J., Mozsik G. Capsaicin and the stomach. A review of experimental and clinical data. J Physiol Paris. 1997;91:151–171. doi: 10.1016/s0928-4257(97)89479-x. [DOI] [PubMed] [Google Scholar]
- 28.Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 29.Blair H.A. Capsaicin 8% dermal patch: a review in peripheral neuropathic pain. Drugs. 2018;78:1489–1500. doi: 10.1007/s40265-018-0982-7. [DOI] [PubMed] [Google Scholar]
- 30.Simpson D.M., Estanislao L., Brown S.J., Sampson J. An open-label pilot study of high-concentration capsaicin patch in painful HIV neuropathy. J Pain Symptom Manag. 2008;35:299–306. doi: 10.1016/j.jpainsymman.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Yong Y.L., Tan L.T., Ming L.C., Chan K.G., Lee L.H., Goh B.H. The Effectiveness and safety of topical capsaicin in postherpetic neuralgia: a systematic review and meta-analysis. Front Pharmacol. 2016;7:538. doi: 10.3389/fphar.2016.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang K., He S., Zhang X., Guo J., Chen Q., Yan F. Tangeretin, an extract from Citrus peels, blocks cellular entry of arenaviruses that cause viral hemorrhagic fever. Antivir Res. 2018;160:87–93. doi: 10.1016/j.antiviral.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Yan F., Tang K., Chen Q., Guo J., Zhu W. Identification of a clinical compound losmapimod that blocks Lassa virus entry. Antivir Res. 2019;167:68–77. doi: 10.1016/j.antiviral.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q., Tang K., Zhang X., Chen P., Guo Y. Establishment of pseudovirus infection mouse models for in vivo pharmacodynamics evaluation of filovirus entry inhibitors. Acta Pharm Sin B. 2018;8:200–208. doi: 10.1016/j.apsb.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labudova M., Pastorek J., Pastorekova S. Lymphocytic choriomeningitis virus: ways to establish and maintain non-cytolytic persistent infection. Acta Virol. 2016;60:15–26. doi: 10.4149/av_2016_01_15. [DOI] [PubMed] [Google Scholar]
- 36.Emonet S., Lemasson J.J., Gonzalez J.P., de Lamballerie X., Charrel R.N. Phylogeny and evolution of old world arenaviruses. Virology. 2006;350:251–257. doi: 10.1016/j.virol.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Bowen M.D., Rollin P.E., Ksiazek T.G., Hustad H.L., Bausch D.G., Demby A.H. Genetic diversity among Lassa virus strains. J Virol. 2000;74:6992–7004. doi: 10.1128/jvi.74.15.6992-7004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oloniniyi O.K., Unigwe U.S., Okada S., Kimura M., Koyano S., Miyazaki Y. Genetic characterization of Lassa virus strains isolated from 2012 to 2016 in southeastern Nigeria. PLoS Neglected Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunberg J.H., York J. The curious case of arenavirus entry, and its inhibition. Viruses. 2012;4:83–101. doi: 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torriani G., Galan-Navarro C., Kunz S. Lassa virus cell entry reveals new aspects of virus-host cell interaction. J Virol. 2017;91:e01902–e01916. doi: 10.1128/JVI.01902-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngo N., Henthorn K.S., Cisneros M.I., Cubitt B., Iwasaki M., de la Torre J.C. Identification and mechanism of action of a novel small-molecule inhibitor of arenavirus multiplication. J Virol. 2015;89:10924–10933. doi: 10.1128/JVI.01587-15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Wang W., Zhou Z., Zhang L., Wang S., Xiao G. Structure–function relationship of the mammarenavirus envelope glycoprotein. Virol Sin. 2016;31:380–394. doi: 10.1007/s12250-016-3815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanati S., Razavi B.M., Hosseinzadeh H. A review of the effects of Capsicum annuum L. and its constituent, capsaicin, in metabolic syndrome. Iran J Basic Med Sci. 2018;21:439–448. doi: 10.22038/IJBMS.2018.25200.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guedes V., Castro J.P., Brito I. Topical capsaicin for pain in osteoarthritis: a literature review. Reumatol Clínica. 2018;14:40–45. doi: 10.1016/j.reuma.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Cho S.C., Lee H., Choi B.Y. An updated review on molecular mechanisms underlying the anticancer effects of capsaicin. Food Sci Biotechnol. 2017;26:1–13. doi: 10.1007/s10068-017-0001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patowary P., Pathak M.P., Zaman K., Raju P.S., Chattopadhyay P. Research progress of capsaicin responses to various pharmacological challenges. Biomed Pharmacother. 2017;96:1501–1512. doi: 10.1016/j.biopha.2017.11.124. [DOI] [PubMed] [Google Scholar]
- 47.Gooding S.M., Canter P.H., Coelho H.F., Boddy K., Ernst E. Systematic review of topical capsaicin in the treatment of pruritus. Int J Dermatol. 2010;49:858–865. doi: 10.1111/j.1365-4632.2010.04537.x. [DOI] [PubMed] [Google Scholar]
- 48.McConachie S.M., Caputo R.A., Wilhelm S.M., Kale-Pradhan P.B. Efficacy of capsaicin for the treatment of cannabinoid hyperemesis syndrome: a systematic review. Ann Pharmacother. 2019;53:1145–1152. doi: 10.1177/1060028019852601. [DOI] [PubMed] [Google Scholar]
- 49.Marini E., Magi G., Mingoia M., Pugnaloni A., Facinelli B. Antimicrobial and anti-virulence activity of capsaicin against erythromycin-resistant, cell-invasive group a Streptococci. Front Microbiol. 2015;6:1281. doi: 10.3389/fmicb.2015.01281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafiz T., Mubaraki M., Dkhil M., Al-Quraishy S. Antiviral activities of Capsicum annuum methanolic extract against herpes simplex virus 1 and 2. Pakistan J Zool. 2017;49:251–255. [Google Scholar]
- 51.Martinez J.P., Sasse F., Bronstrup M., Diez J., Meyerhans A. Antiviral drug discovery: broad-spectrum drugs from nature. Nat Prod Rep. 2015;32:29–48. doi: 10.1039/c4np00085d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.