Key Points
Whole-genome bisulfite sequencing distinguishes between resistant and susceptible O. niloticus.
Promoter methylation regulates gene expression in immune-related genes.
Abstract
Streptococcus agalactiae is an important pathogenic bacterium causing great economic loss in Nile tilapia (Oreochromis niloticus) culture. Resistant and susceptible groups sharing the same genome showed significantly different resistance to S. agalactiae in the genetically improved farmed tilapia strain of Nile tilapia. The resistance mechanism is unclear. We determined genome-wide DNA methylation profiles in spleen of resistant and susceptible O. niloticus at 5 h postinfection with S. agalactiae using whole-genome bisulfite sequencing. The methylation status was higher in the spleen samples from resistant fish than in the susceptible group. A total of 10,177 differentially methylated regions were identified in the two groups, including 3725 differentially methylated genes (DMGs) (3129 hyper-DMGs and 596 hypo-DMGs). The RNA sequencing showed 2374 differentially expressed genes (DEGs), including 1483 upregulated and 891 downregulated. Integrated analysis showed 337 overlapping DEGs and DMGs and 82 overlapping DEGs and differentially methylated region promoters. By integrating promoter DNA methylation with gene expression, we revealed four immune-related genes (Arnt2, Nhr38, Pcdh10, and Ccdc158) as key factors in epigenetic mechanisms contributing to pathogen resistance. Our study provided systematic methylome maps to explore the epigenetic mechanism and reveal the methylation loci of pathogen resistance and identified methylation-regulated genes that are potentially involved in defense against pathogens.
Introduction
Tilapia show high adaptability, rapid growth, high yield, and palatable meat without intermuscular spines. It has become one of the world’s foremost cultured fish, being farmed in >100 countries and promoted worldwide by the Food and Agriculture Organization of the United Nations (1). In the 1950s, tilapia was introduced into China, which now leads the world with an annual production of ∼1.7 million metric tons (2). Tilapia farming areas are distributed in southern China, including the Guangdong, Guangxi, Hainan, Yunnan, and Fujian Provinces. The main species is the GIFT strain of Nile tilapia (3). At present, the chief problem in commercial tilapia culture is disease, with Streptococcus agalactiae the primary pathogen. The principal epidemic period is from June through September, with 40–90% mortality (4, 5). The disease is currently prevented and controlled by antibiotic drugs (6). But their long-term use contaminates the aquatic environment and poses a threat to consumer health. It is imperative to strengthen research on the selection of disease-resistant tilapia varieties to improve their ability to resist S. agalactiae.
S. agalactiae is a Gram-positive pathogenic bacterium shared by humans, livestock, and fish (7). Infection can cause meningitis, sepsis, and pneumonia. S. agalactiae is cosmopolitan and harmful to many fish species, including Letalurus punetaus, Trachinotus ovatus, and Oncorhynchus mykiss (8, 9). Tilapia is highly sensitive to S. agalactiae, and fish can be infected at varying ages with rapid propagation and high mortality. They are especially vulnerable to S. agalactiae when the water temperature is >30°C. The primary symptoms are floating at the water surface, loss of balance, intermittent turning or rolling, bleeding on the abdominal surface, and tail ulceration (10, 11). The pathogenic mechanism of S. agalactiae in tilapia is unclear.
DNA methylation is the major modification process of organisms from bacteria to eukaryotes, and its status has a profound effect on gene expression (6, 12, 13) and biological processes such as stabilization of genome and chromosome structure (14), X chromosome inactivation (15), genomic imprinting (16, 17), and gene silencing (18, 19). Increasing evidence has shown DNA methylation to play an important role in many common diseases. In humans, studies have shown low concordance in monozygotic twins of rates of diseases such as rheumatoid arthritis (20, 21) and multiple sclerosis (22), indicating environment or epigenetics involvement (23). In addition, research has shown differential DNA methylation related to breast, colorectal, and ovarian cancers (24–27). In flatfish, occurrence of liver tumors is related to DNA methylation (28). Global methylation was significantly reduced 1.8-fold in hepatocellular adenoma compared with noncancerous liver tissue, also supporting an epigenetic model of cancer (28). The global methylation profile of Danio rerio, Daphnia magna, Lepomis macrochirus, and Sebastiscus marmoratus following exposure to several environmental pollutants showed that epigenetic mechanisms were affected (29–34). This highlights the importance of DNA methylation in fish disease.
In a previous study, we bred S. agalactiae–resistant tilapia, but the mechanism of resistance is still unclear. We performed genome-wide DNA methylation and RNA sequencing (RNA-seq) to scan differentially methylated genes (DMGs) and differentially expressed genes (DEGs). We also identified the major genes involved in the resistance mechanism by determining the relationship between DMGs and DEGs. We further confirmed our data using bisulfite sequencing PCR and real-time PCR. Results showed that hypomethylation of promoters increased the expression of immune genes in tilapia resistant to S. agalactiae.
Materials and Methods
Fish and sample preparation
The GIFT strain Nile tilapia were obtained from the Guangxi Academy of Fishery Sciences. At 5 h postchallenge with S. agalactiae as previously described (35), spleen tissue was collected from 15 resistant and 15 susceptible fish. Each sample was divided into three parts, one preserved in RNALater (Applied Biosystems, Foster City, CA) and stored at −70°C for RNA extraction, the second fixed in 4% paraformaldehyde (pH 7.5) for 24 h and stored in 70% ethanol to detect the differences in microstructure by histology according to the described method (36), and a third preserved in ethanol for DNA extraction. Spleens of Nile tilapia with no pathogen exposure were collected and fixed in 4% paraformaldehyde (pH 7.5) as control. The RNA was extracted using the TRIzol method according to the manufacturer’s instruction. Genomic DNA was extracted using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China), including RNase A treatment, according the manufacture’s instruction. The DNA concentration and quality were detected by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and agarose gel electrophoresis.
Library construction and sequencing
Three replicate DNA samples from each resistant and susceptible group were used to prepare the library. Before bisulfite treatment, 25 ng of λ-DNA was added to 5 μg of genomic DNA from spleen. The mixed DNA was fragmented with a Sonicator (Sonics and Materials, Danbury, CT) to 450 bp. After blunting ends with addition of dA to 3′-end, Illumina methylated adapters were added according to the manufacturer’s instructions. The bisulfite conversion of DNA was carried out using ZYMO EZ DNA Methylation-Gold kit (ZYMO, Irvine, CA) and amplified by 12 cycles of PCR using the KAPA HiFi HotStart Uracil + ReadyMix (2X). Ultra-high throughput pair-end sequencing was carried out using the Illumina Hiseq 2500 platform according to manufacturer’s instructions. Raw Hiseq sequencing data were processed by Illumina base-calling pipeline (SolexaPipeline-1.0). Clean reads were obtained by 1) removal of 3′ adapter oligonucleotide, 2) removal of the unknown base <10% read, and 3) removal of low-quality reads with low-quality bases ≥50%. All clean bisulfite sequencing reads were mapped to the reference genome (37) using BSMAP aligner allowing up to two mismatches (38). The cytosine methylation levels were determined according to the uniquely mapped reads, as previously described (39). The library construction and RNA-seq were as described (36).
Identification of differentially methylated regions and DEGs
To identify methylated cytosine (mC) base in the Nile tilapia genome, the binomial test was used to assess whether the cytosine site can be considered an mC site as defined by a false discovery rate (FDR) <0.01 (40). Differentially methylated regions (DMRs) were detected with a 200-bp sliding window with a 50-bp step size. Genomic regions with levels of methylation greater than the cut-off value and a number of cytosines within the region greater than the cut-off value were considered DMRs. The DMRs were detected as follows. First, windows with an FDR value <0.05 and a >1.5-fold change in methylation level were submitted to further analysis. Second, the p value of each cytosine in the selected regions was calculated by Fisher exact test and considered differentially mC if the p value was ≥0.01 and showed fold changes ≥2 with an absolute methylation difference of 0.4, 0.2, and 0.1 for CG, CHG, and CHH, respectively. Only those regions containing at least seven differentially mCs were retained. Finally, neighboring DMRs were combined if the gap was ≤100 bp (41).
To identify DEGs, we set the criteria as FDR value <10−3 and an expression level differing at least 2-fold in the spleens from resistant and susceptible groups, as previously described (42).
Functional enrichment analysis
To understand the functions of the DMGs and DEGs, Gene Ontology (GO) functional enrichment was carried out using Goatools (https://github.com/tanghaibao/Goatools) with Fisher test (43). To control the false-positive rate, multiple tests (Bonferroni, Holm, Sidak, and FDR) were used to correct the p value. A p < 0.05 was interpreted as significant enrichment of the GO function. To confirm the relationship between the gene expression level and its methylation status, we analyzed the transcribed regions and promoter methylated levels.
Determination of association of methylation with gene expression
Previous reports have stated that promoter methylation inhibits gene expression (44). To elucidate the tilapia disease-resistance mechanism, the relationship between promoter methylation and gene expression were investigated. Four upregulated genes with promoter hypomethylation in the resistant group were selected: nuclear hormone receptor 38 (Nhr38), protocadherin 10 (Pcdh10), coiled-coil domain-containing protein 158 (Ccdc 158), and aryl hydrocarbon receptor nuclear translocator 2 (Arnt2). The primers were designed according to the reverse promoter sequence on both sides of the methylated island (Table I). The mixed DNA was treated using a DNA methylation kit (ZYMO) and subjected to PCR amplification following the manufacturer’s protocol. The PCR was conducted with the treated DNA as a reaction template. The PCR products were run on 2% agarose gel, and the purified fragment was ligated onto a pMD18-T Vector (Takara, Dalian, China). At least 15 positive clones from each group were sequenced. The difference in mean methylation level of the resistant and susceptible groups was analyzed by an independent sample t test using SPSS 19.0 software (IBM). The differences in the ratio of methylated to unmethylated CpG at each site were assessed by cross-tabulations followed by Fisher exact test. A p < 0.05 was considered significant.
Table I. Primers used in the study.
| Gene | Primer Sequence | Purpose | Product Size (bp) |
|---|---|---|---|
| Nhr38-BSP-F | 5′-TTTTGAGGTATTTATAAAAAGTTTGG-3′ | Identification of methylation level | 396 |
| Nhr38-BSP-R | 5′-CTTAAACACTTAAACCCTCTCTAAA-3′ | ||
| Pcdh10-BSP-F | 5′-TTTTTTTTGTAATAATATTGTTGTTG-3′ | Identification of methylation level | 321 |
| Pcdh10-BSP-R | 5′-TCCCTATCAAAAAATTACAAAC-3′ | ||
| Ccdc158-BSP-F | 5′-TTTATAGTTTTGGGGTTTAGAGG-3′ | Identification of methylation level | 428 |
| Ccdc158-BSP-R | 5′-ACAAATTCTAACTTTCACATCCA-3′ | ||
| Arnt2-BSP-F | 5′-TTTGATATAGAGGGGGTTATAAGT-3′ | Identification of methylation level | 264 |
| Arnt2-BSP-R | 5′-AAAATAACTTTTTATAACCCAATTTAT-3′ | ||
| Nhr38- F | 5′-AGGCCTTCAAGCTCGATGAT-3′ | qRT-PCR | 147 |
| Nhr38-R | 5′-AGCTTGGTGTTGGAGGAGAA-3′ | ||
| Pcdh10-F | 5′-TCACGATGACCTCAGACACC-3′ | qRT-PCR | 136 |
| Pcdh10-R | 5′-AGTCAGTTCCAACCGGCTAA-3′ | ||
| Ccdc 158-F | 5′-GTTCTGGAGAGATTTGGCGG-3′ | qRT-PCR | 145 |
| Ccdc 158-R | 5′-CCAGACCAAAGACCAAAGGC-3′ | ||
| Arnt2-F | 5′-ACCAGGCTATCTAGTGACGC-3′ | qRT-PCR | 140 |
| Arnt2-R | 5′-GGGCTGTTCTTAGTTCCCCT-3′ | ||
| Actin-F | 5′-TGGTGGGTATGGGTCAGAAAG-3′ | qRT-PCR | 217 |
| Actin-R | 5′-CTGTTGGCTTTGGGGTTCA-3′ |
qRT-PCR, quantitative RT-PCR.
To identify the association of methylation and gene expression, we extracted RNA from spleen of three resistant and three susceptible Nile tilapia. After the RNA reversed to cDNA, the relative expression of the selected gene was studied as previously described (3). Differences were determined using Fisher exact test. A p < 0.05 was regarded as significant.
Results
Splenic tissue analysis after S. agalactiae infection
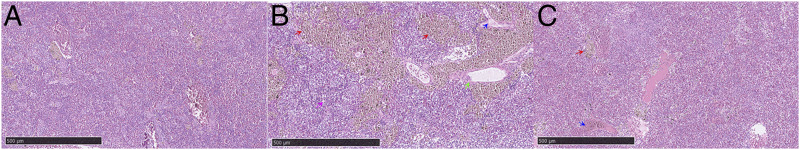
The control group showed no obvious pathologies (Fig. 1A). In the infected susceptible fish, multifocal hemosiderin deposition, congestion of blood vessels, large thrombi in blood vessels, and macrophages were observed. The infected resistant group showed multifocal hemosiderin deposition and large thrombi (Fig. 1B, 1C). We found a greater amount of multifocal hemosiderin deposition in the susceptible group than in resistant groups, suggesting that the susceptible fish were more severely affected by S. agalactiae (Table I).
FIGURE 1.
Tilapia spleen tissue section with H&E stain at 5 h after S. agalactiae infection. Red arrow shows multifocal hemosiderin deposition; green arrow shows the congestion of blood vessels; blue arrow shows large thrombus; pink arrow shows macrophages. (A) Control group without S. agalactiae infection; (B) susceptible group 5 h postinfection with S. agalactiae; (C) resistant group 5 h postinfection with S. agalactiae. Scale bars, 500 μm.
Genome-wide DNA methylation profiling and transcriptome sequencing in Nile tilapia spleen
To determine the difference in methylation level of the resistant and susceptible groups, we obtained 764,413,750 and 820,147,572 raw reads from 114.67G and 123.03G raw data, respectively (Table II). The string average depths were 9.18 (resistant) and 9.09 (susceptible). The data were deposited in the GenBank database under number SRP220470. All clean reads were assembled and mapped to the reference Nile tilapia genome (37). A mean of 75.43% (resistant) and 74.29% (susceptible) reads were mapped to the genome (Table II). A total of 24.54G and 22.26G raw data were generated from spleen of the resistant and susceptible groups, respectively. All raw data were deposited in the GenBank database SRP220463. An average of 50.74 and 46.46 million clean reads for resistant and susceptible groups, respectively, were analyzed (Table III). Approximately 90.57 and 89.94% clean reads in resistant and susceptible fish, respectively, were mapped to the reference genome data.
Table II. Summary of whole-genome bisulfite sequencing dataset.
| Sample | Raw Reads (n) | Raw Bases (G) | Clean Reads (n) | Clean Bases (G) | Mapped Reads (n) | Mapped Rate (%) | String Average Depth (X) |
|---|---|---|---|---|---|---|---|
| K5h-1 | 281,501,084 | 42.23 | 259,576,268 | 38.45 | 195,719,181 | 75.4 | 10.23 |
| K5h-2 | 215,580,902 | 32.34 | 197,787,930 | 29.29 | 147,034,221 | 74.34 | 7.45 |
| K5h-3 | 267,331,764 | 40.10 | 245,225,860 | 36.32 | 187,676,562 | 76.53 | 9.86 |
| Y5h-1 | 271,798,800 | 40.77 | 247,986,304 | 36.73 | 188,132,155 | 75.86 | 9.31 |
| Y5h-2 | 247,943,072 | 37.20 | 223,161,460 | 32.98 | 164,333,315 | 73.64 | 8.22 |
| Y5h-3 | 300,405,700 | 45.06 | 268,557,842 | 39.61 | 197,048,362 | 73.37 | 9.75 |
Table III. Summary of RNA-seq dataset.
| Sample | Raw Reads (n) | Raw Bases (G) | Clean Reads (n) | Clean Bases (G) | Clean Reads (n) | Mapped Reads (n) | Mapped Rate |
|---|---|---|---|---|---|---|---|
| K5h-1 | 56,893,102 | 8.53 | 53,211,176 | 7.88 | 53,211,176 | 48,352,995 | 90.87 |
| K5h-2 | 56,047,176 | 8.41 | 52,063,926 | 7.71 | 52,063,926 | 46,992,899 | 90.26 |
| K5h-3 | 50,665,376 | 7.60 | 46,959,588 | 6.95 | 46,959,588 | 42,531,298 | 90.57 |
| Y5h-1 | 40,506,268 | 6.08 | 37,853,660 | 5.60 | 37,853,660 | 33,591,337 | 88.74 |
| Y5h-2 | 54,235,004 | 8.14 | 51,153,314 | 7.58 | 51,153,314 | 46,498,362 | 90.90 |
| Y5h-3 | 53,627,686 | 8.04 | 50,382,204 | 7.47 | 50,382,204 | 45,429,633 | 90.17 |
DNA methylation patterns in spleen
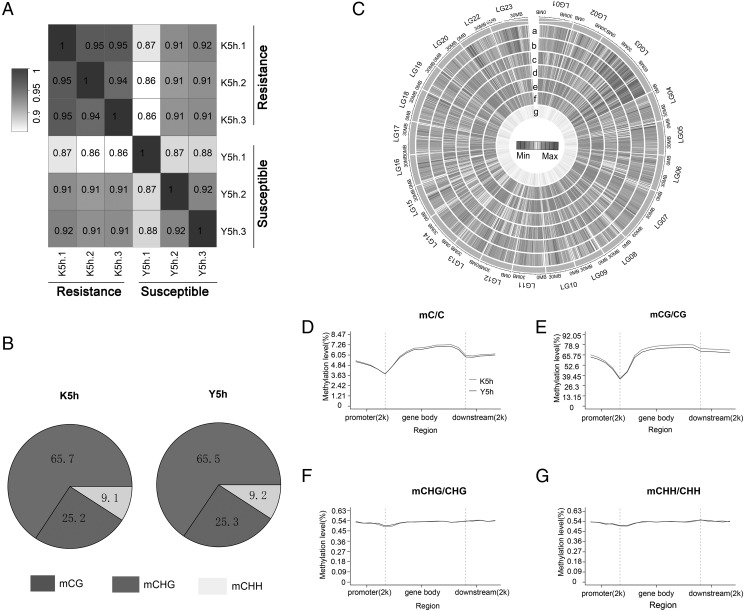
To assess the similarity of the six spleen samples, pairwise Pearson correlation coefficients were calculated between every two samples. The correlation coefficient was >0.94 in the resistant group and >0.87 in the susceptible group (Fig. 2A). An average of 196,730,852 and 181,819,451 mCs were detected in resistant and susceptible groups, respectively (Table IV). Most mCs were from mCG methylation (>92%) (Table IV). Three types of methylation were found in Nile tilapia: mCG, mCHG, and mCHH. The mCG type of methylation accounted for up to 65%, mCHG accounted for ∼25%, and mCHH accounted for ∼9% (Fig. 2B). To investigate the methylation distribution along the genome, methylation density was calculated in 300-kb windows across the genome (Fig. 2C). The maximum difference in the number of methylation sites of resistant and susceptible groups in mCG methylation was linkage group (LG) 14, and the minimum was LG17. In mCHG methylation, the maximum difference was LG14, and the minimum was LG16. In mCHH, maximum difference was LG14, and minimum was LG16. The difference in the methylation level of resistant and susceptible groups in different LGs was analyzed. In mCG methylation, the maximum difference was in LG14, and the minimum was in LG22. In mCHG, the maximum difference was in LG17, and the minimum was in LG14. In mCHH, the maximum difference was in LG17, and the minimum was in LG14. Methylation site number and level in mCHG and mCHH type were similar. To examine the methylation distribution in different genetic structure regions, we detected the methylation level in the gene body, promoter, and downstream region in mC, mCG, mCHG, and mCHH of resistant and susceptible groups. On genome-wide scale, a similar methylation profile was observed in resistant and susceptible groups with the resistant sample exhibiting a slightly higher methylation level (Fig. 2D). Distinct hypomethylation was observed in the region of transcription initiation site and the transcription termination sites. A similar profile was observed in CG methylation level (Fig. 2E). However, the CHG and CHH methylation profiles differed from that of CG. The CHG and CHH methylation profiles were similar, and methylation level did not differ in the gene body, promoter, and downstream region (Fig. 2F, 2G).
FIGURE 2.
Methylation profiles of spleen samples in resistant and susceptible tilapia. (A) Pearson correlation coefficient of genome-wide CpG methylation between the resistant and susceptible groups. (B) The proportion of mCs (mCG, mCHG, mCHH) in resistant and susceptible tilapia at 5 h after S. agalactiae infection. (C) Heatmap representation of average methylation densities in 300-kb windows independent of sequence context. a, mCG_K5h; b, mCG_Y5h; c, mCHH_aK5h; d, mCHH_Y5h; e, mCHG_K5h; f, mCHG_Y5h; g, gene density. The mC (D), mCG (E), CHG (F), and CHH (G) methylation levels in different sequence regions of resistant and susceptible groups.
Table IV. mCs in the spleen of S. agalactiae–resistant and S. agalactiae–susceptible groups.
| Sample | C (n [%]) | CG (n [%]) | CHG (n [%]) | CHH (n [%]) |
|---|---|---|---|---|
| K5h-1 | 210,462,338 (100) | 194,209,887 (92.29) | 4,216,077 (2.00) | 12,036,374 (5.71) |
| K5h-2 | 153,833,084 (100) | 142,783,771 (92.82) | 2,910,671 (1.89) | 8,138,642 (5.29) |
| K5h-3 | 225,897,136 (100) | 210,304,976 (93.01) | 4,182,918 (1.85) | 11,409,242 (5.05) |
| Y5h-1 | 186,761,315 (100) | 172,040,367 (92.12) | 3,866,816 (2.07) | 10,854,132 (5.81) |
| Y5h-2 | 165,841,768 (100) | 153,986,363 (92.85) | 3,139,123 (1.89) | 8,716,282 (5.26) |
| Y5h-3 | 192,855,270 (100) | 178,015,458 (92.31) | 3,941,047 (2.04) | 10,898,765 (5.65) |
Identification of DMRs and DEGs
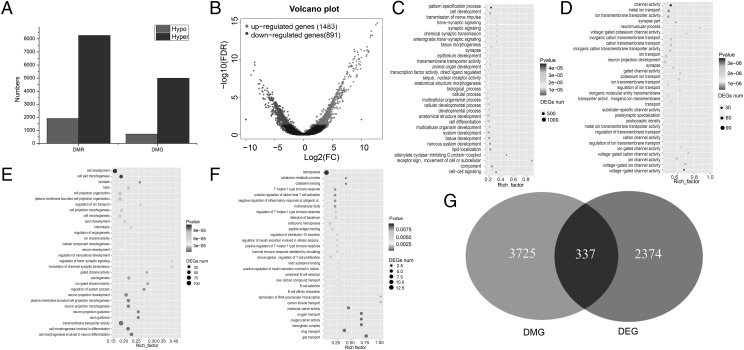
To identify the genomic regions with different levels of methylation in resistant and susceptible groups, 10,177 DMRs were detected, including 1913 hypo-DMRs and 8264 hyper-DMRs (Fig. 3A). A total of 717 hypo-DMRs were located in 596 gene body regions, and 1196 hypo-DMRs were located in 308 promoters. Additionally, 4991 hyper-DMRs were located in 3129 gene body regions, and 3273 hyper-DMRs were located in 528 promoters. To compare DEGs of resistant and susceptible groups, RNA-seq was carried out, revealing 2374 DEGs, including 1483 upregulated genes (resistant) and 891 downregulated genes (susceptible) (Fig. 3B).
FIGURE 3.
(A) Number of methylation regions and genes; (B) number of DEG; (C) GO terms enriched using hypermethylation genes; (D) GO terms enriched using hypomethylation genes; (E) GO terms enriched using upregulated genes; (F) GO terms enriched using downregulated genes; (G) overlap of DMG and DEG.
To better understand the mechanism of gene regulation networks of the susceptible groups, GO enrichment analysis was performed for the annotated DMGs and DEGs. The hyper-DMGs were significantly categorized as active in cellular processes, biological processes, and development processes (Fig. 3C). The hypo-DMGs were predominantly classified in ion transport, neuron projection development, and ion transmembrane transport (Fig. 3D). The upregulated genes were primarily involved in cell development, transmembrane transporter activity, and cell projection organization (Fig. 3E). The downregulated genes were significantly more active in in hemopoiesis, drug transport, and gas transport (Fig. 3F), activities closely related to the immune system.
Association of DNA methylation and RNA-seq data
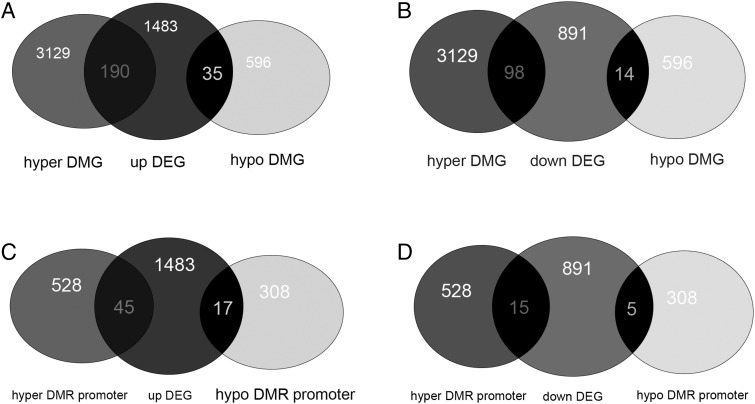
To better understand the mechanism of resistance in S. agalactiae, we analyzed the association of DNA methylation and gene expression. We found 337 overlapping genes in DEGs and DMGs, including 190 overlapping genes in hyper-DEG and hyper-DMG, 35 overlapping genes in upregulated DEG and hypo-DMG (Fig. 4A, Supplemental Table I), 98 overlapping genes in downregulated DEGs and hyper-DMGs, and 14 overlapping genes in downregulated DEGs and hypo-DMGs (Fig. 4B, Supplemental Table I). Eighty-two overlapping genes were observed in DEG and DMR promoters, including 45 in upregulated DEG and hyper-DMR promoter, 17 in upregulated DEG and hypo-DMR promoter (Fig. 4C, Supplemental Table I), 15 in downregulated DEG and hyper-DMR promoter, and five in downregulated DEG and hypo-DMR promoter (Fig. 4D, Supplemental Table I).
FIGURE 4.
Selection of condition-specific genes. (A) Venn diagram showing the number of overlapping genes of upregulated DEG and DMG. (B) Venn diagram showing the number of overlapping genes of downregulated DEG and DMG. (C) Venn diagram showing the number of overlapping genes of upregulated DEG and DMR promoter. (D) Venn diagram showing the number of overlapping genes of downregulated DEG and DMR promoter.
Relationship between DNA methylation and gene expression
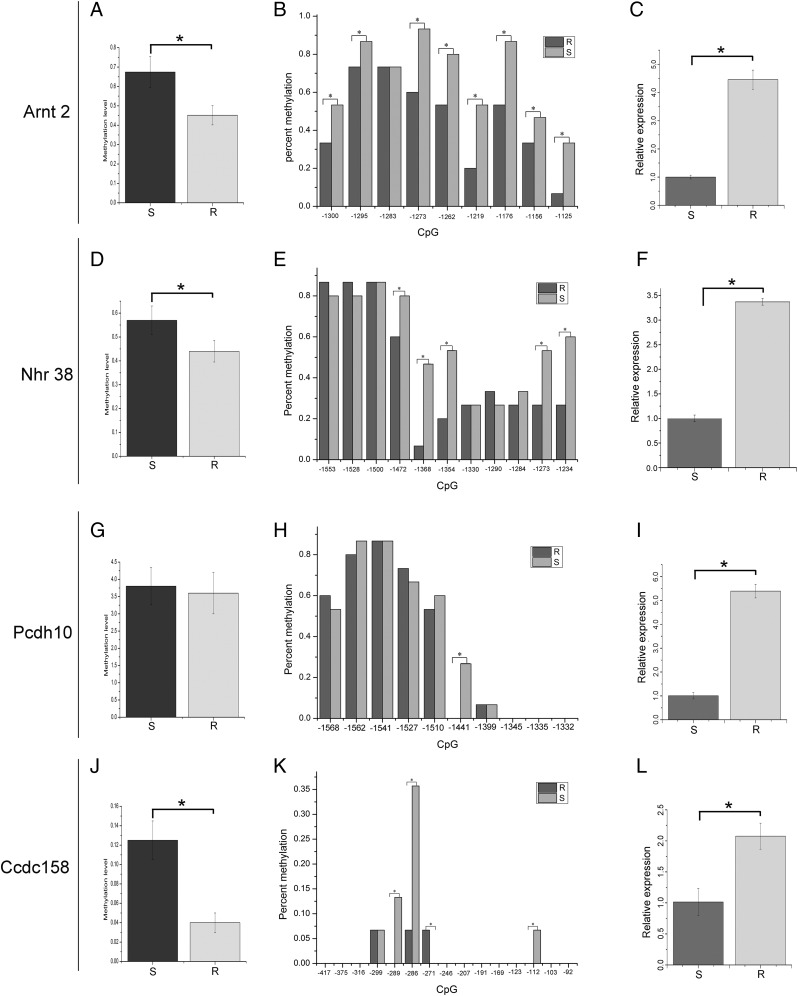
The gene promoter methylation level allowed characterization of the relationship of gene expression to methylation status. The CpG islands were determined from the genomic methylation sequences (Supplemental Table II). Results of bisulfite sequencing PCR showed that Arnt2, Nhr38, and Ccdc158 gene promoter methylation levels were significantly higher in the susceptible group than in resistant fish (Fig. 5A, 5D, 5J; p < 0.05; Supplemental Fig. 1). In Pcdh10 gene promoter, we also observed a high methylation level in the susceptible group but no significant difference with the resistant fish (Fig. 5G, p > 0.05; Supplemental Fig. 1). The CpG site methylation in the susceptible group was also higher than in resistant fish. In the Arnt2 promoter region, 8 out of 9 (88.9%) CpG sites showed methylation levels significantly higher in the susceptible group than in the resistant group (Fig. 5B, p < 0.05). In the Nhr38 gene promoter region, 5 out of 11 (45.5%) CpG sites showed methylation levels significantly higher in the susceptible group compared with the resistant group (Fig. 5E, p < 0.05). In the Pcdh10 gene promoter region, 1 out of 10 (10%) CpG sites presented a methylation level significantly higher in the susceptible group than the resistant group (Fig. 5H, p < 0.05). In the Ccdc158 gene promoter region, 3 out of 15 (20%) CpG sites showed significantly higher methylation levels in susceptible fish than in the resistant group (Fig. 5K, p < 0.05). We also found one CpG site (−271) with significantly higher methylation in the resistant group than in the susceptible group (Fig. 5K, p < 0.05). The expression of the corresponding genes showed that expression of all assessed genes in resistant tilapia was significantly higher than in susceptible fish (Fig. 5C, 5F, 5I, 5L). Gene expression was negatively associated with promoter methylation levels.
FIGURE 5.
Promoter methylation level and gene expression. (A) Difference in Arnt2 promoter methylation level of resistant (R) and susceptible (S) groups; (B) each CpG site methylation level difference in Arnt2 gene promoter regions of R and S groups; (C) difference in Arnt2 gene expression of R and S groups; (D) difference in Nhr38 promoter methylation level of resistant and susceptible groups; (E) CpG site methylation level difference in Nhr38 gene promoter regions of R and S groups; (F) difference in Nhr38 gene expression of R and S groups; (G) difference in Pcdh10 promoter methylation level of R and S groups; (H) each CpG site methylation level difference in Pcdh10 gene promoter region of R and S groups; (I) difference in Pcdh10 gene expression of R and S groups; (J) difference of Ccdc158 promoter methylation level between R and S groups; (K) CpG site methylation level difference in Ccdc158 gene promoter regions of R and S groups; (L) difference in Ccdc158 gene expression of R and S groups. *p < 0.05.
Discussion
Tilapia is one of most important fish species in freshwater aquaculture worldwide. However, tilapia are susceptible to bacterial infection, especially by S. agalactiae, an important pathogen of many vertebrates. Severe S. agalactiae infection may be fatal (45). Previous studies have shown the two main pathogenic bacteria infecting tilapia to be S. agalactiae and Streptococcus iniae. Because of rapid spread and high mortality, streptococcal infections have become the major bacterial disease in tilapia (46). Cases of cross-infection of S. agalactiae between humans and tilapia by direct contact have been reported (47). To prevent infection of tilapia and tilapia farm workers, we need to explore the molecular mechanisms of S. agalactiae infection. Varieties of tilapia resistant to S. agalactiae disease developed in an earlier study provide valuable research material for such study.
To identify pathological lesions in spleen (important immune tissue), histological examination was carried out postinfection of Nile tilapia with S. agalactiae. Fish susceptible to S. agalactiae showed more severe lesions than resistant fish and noninfected controls (Fig. 1), similar to observations in a previous study (10).
In this study, for the first time, to the best of our knowledge, the molecular mechanism of resistance was studied using the whole-genome-level profile of DNA methylation and RNA-seq to identify a set of genes that were significantly altered in DNA methylation status and RNA expression, and hence, potentially key genes in resistance. Overall, the gene bodies exhibited higher levels of CpG methylation relative to the promoter and downstream regions. This is consistent with reports in goat (48), human (49), and horse (50). We also observed higher methylation in the promoter region than in the gene body in other fish species (51).
Of the 10,177 DMRs, 3725 genes were annotated, including 3129 hyper-DMR and 596 hypo-DMR genes. Of 2374 DEGs, 1483 were upregulated, and 891 were downregulated. The GO function analysis showed that these DEGs were mainly enriched in immune response (ion transport, ion transmembrane transport) and development (biological process, development process, and neuron projection development). These results were similar to findings of previous studies (35) and showed the analyzed genes to play important roles in the resistance process. Because of the promoter methylation repression of gene expression, we combined the RNA-seq data and methylation data to analyze the overlapping gene with high expression and hypomethylation in promoter region. At last, we got 17 overlapping genes from the combination analysis. In data analysis, we set the susceptible group as the control to detect the DEGs and DMGs between two groups. If we sequence the uninfected sample as the control, the number of overlapping genes may be even less, and the accuracy may be increased. To ensure the accuracy of candidate genes, we selected four overlapped genes (Arnt2, Nhr38, Pcdh10, and Ccdc158) with hypomethylation in the promoter region and upregulated expression between the susceptible and resistant groups for further analysis. Arnt2 belongs to the basic helix-loop-helix-per-ARNT-sim (bHLH-PAS) superfamily and is an important transcription factor binding drug-metabolizing gene in vertebrates (52, 53). In humans and mice, ARNT gene spans 22 exons encoding multiple transcripts of different lengths, and each isoform has different functions (54). In humans, bHLH-PAS homology proteins regulated transcription factors involved in pathological function (55–57). However, this transcription factor must heterodimerize with Arnt2 to regulate gene expression (55). In rats, Arnt2 expression was detected mainly in brain, spinal cord, autonomic ganglia, and kidney by in situ hybridization of the whole body (58). After knockout of Arnt2 gene in mice, fumarylacetoacetate hydrolase activity was decreased, and perinatal lethal phenotype was displayed (59). Closer inspection showed that these mice also lack distinguishable paraventricular nucleus/supraoptic nucleus neurogenesis (60). Previous studies also show that ARNT2 regulated response to environmental stresses and resistance to toxic effects, such as those to 2,3,7,8-tetrachlorodibenzo-p-dioxin (54, 61). Nuclear hormone receptors (NHR) belong to a superfamily of ligand-activated transcription factors that regulate metabolic function and the pathophysiology of metabolic disease (62). Pcdh10 gene belongs to the protocadherin gene family, a subfamily of the cadherin superfamily. Expression of Pcdh10 is involved in inhibition of cell proliferation and induction of apoptosis (63, 64). In humans, Pcdh10 is a tumor suppressor gene in endometrial endometrioid carcinoma (EEC). Expression of Pcdh10 was significantly lower in EEC than in normal tissues or adjacent normal tissues in mRNA and protein level. In vitro, the apoptotic cells significantly increased in HEC-1 and KLE cells after transfecting with the Pcdh10 vector. Furthermore, Pcdh10 was found to regulate downstream target gene via Wnt/β-catenin signaling in EEC (65). In mice, the deficits were displayed with the expression of Pcdh10 reduction in amygdala function, including γ synchrony as well as increased density of immature filopodia-type spines (66). After deletions of Pcdh10 and regulatory region near Pcdh10, the rat phenotype shows similarly to autism spectrum disorders. This result suggested that Pcdh10 may play a key role in the pathophysiology of the disorder (67). Ccdc158 contained a conserved coiled-coil domain and was located on chromosome 4q21. Coiled-coil was a general domain in protein interaction and formation of protein skeleton. Special protein interaction of coiled-coil domain plays an important role in viral infection, molecular recognition, and signal transduction (68). In catfish, genome-wide association study was used to identify quantitative trait loci for providing an insight into the genomic basis of motile Aeromonas septicemia disease resistance. Three significant quantitative trait loci associated with motile Aeromonas septicemia were identified on LG 2,26,29 (69). Ccdc158 located on LG26 showed significant SNPs associated with Aeromonas septicemia resistance, suggesting Ccdc158 played a key role in Aeromonas septicemia resistance (69). In cattle, 15,552,968 markers from whole-genome sequencing were used to analyze the mastitis resistance genes by genome-wide association study approach. SNP marker in Ccdc158 gene was identified as most significantly associated SNP (70). The analyzed genes showed potential function in disease resistance. In Arnt2, Nhr38, and Ccdc158, promoter methylation levels were significantly higher in the susceptible group than in the resistant group, whereas Pcdh10 showed no significant difference. However, the methylation level of CpG site for the four genes was significantly higher in the resistant group compared with the susceptible group. The results were strongly consistent with the DNA methylation sequencing, although some sites showed no significant difference in methylation level. Higher expression was observed in the resistant fish than in the susceptible group. Gene expression was negatively correlated with methylation level and closely related to the pattern of higher expression associated with less severe infection. The results suggest that promoter methylation regulated the expression of the four genes to resist pathogen infection.
Supplementary Material
Acknowledgments
We thank the Biozeron company for help in data analysis.
This work was supported by the National Key Research and Development Program of China (2018YFD0900302-5), the Major Science and Technology Projects of Guangxi (AA17204080-2), the Guangxi Nature Fund Program (2018JJA130220), the Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture Program (GXIF2019A03-01), and the Major Science and Technology Projects of Nanning (20192242).
The sequence data presented in this article have been submitted to the GenBank under accession numbers SRP220470 and SRP220463.
The online version of this article contains supplemental material.
- Arnt2
- aryl hydrocarbon receptor nuclear translocator 2
- Ccdc158
- coiled-coil domain-containing protein 158
- DEG
- differentially expressed gene
- DMG
- differentially methylated gene
- DMR
- differentially methylated region
- EEC
- endometrial endometrioid carcinoma
- FDR
- false discovery rate
- GO
- Gene Ontology
- LG
- linkage group
- mC
- methylated cytosine
- Nhr38
- nuclear hormone receptor 38
- Pcdh10
- protocadherin 10
- RNA-seq
- RNA sequencing
- SNP
- single-nucleotide polymorphism.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Zhu J., Li C., Ao Q., Tan Y., Luo Y., Guo Y., Lan G., Jiang H., Gan X. 2015. Trancriptomic profiling revealed the signatures of acute immune response in tilapia (Oreochromis niloticus) following Streptococcus iniae challenge. Fish Shellfish Immunol. 46: 346–353. [DOI] [PubMed] [Google Scholar]
- 2.Yuan Y., Yuan Y. M., Dai Y. Y., Gong Y. C. 2017. Economic profitability of tilapia farming in China. Aquacult. Int. 25: 1253–1264. [Google Scholar]
- 3.Zhu J., Wei M., Wang Q., Ao Q., Tan Y., Luo Y., Wang H., Jiang H., Hu Q. 2019. Characterization and expression of galectin-3 after Streptococcus agalactiae and Aeromonas hydrophila challenge in GIFT strain Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 86: 974–980. [DOI] [PubMed] [Google Scholar]
- 4.Verner-Jeffreys D. W., Wallis T. J., Cano Cejas I., Ryder D., Haydon D. J., Domazoro J. F., Dontwi J., Field T. R., Adjei-Boteng D., Wood G., et al. 2018. Streptococcus agalactiae Multilocus sequence type 261 is associated with mortalities in the emerging Ghanaian tilapia industry. J. Fish Dis. 41: 175–179. [DOI] [PubMed] [Google Scholar]
- 5.Wang R., Chen M., Li C., Li L. P., Gan X., Huang J., Lei A. Y., Xu Z. H., Liang W. W. 2013. Identification of multiple genes and their expression profiles in four strains of Oreochromis spp. in response to Streptococcus iniae. J. Fish Biol. 82: 492–504. [DOI] [PubMed] [Google Scholar]
- 6.Yang B. T., Dayeh T. A., Volkov P. A., Kirkpatrick C. L., Malmgren S., Jing X., Renström E., Wollheim C. B., Nitert M. D., Ling C. 2012. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol. Endocrinol. 26: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira U. P., Mian G. F., Oliveira I. C. M., Benchetrit L. C., Costa G. M., Figueiredo H. C. 2010. Genotyping of Streptococcus agalactiae strains isolated from fish, human and cattle and their virulence potential in Nile tilapia. Vet. Microbiol. 140: 186–192. [DOI] [PubMed] [Google Scholar]
- 8.Amal M. N., Zamri-Saad M., Iftikhar A. R., Siti-Zahrah A., Aziel S., Fahmi S. 2012. An outbreak of Streptococcus agalactiae infection in cage-cultured golden pompano, Trachinotus blochii (Lacépède), in Malaysia. J. Fish Dis. 35: 849–852. [DOI] [PubMed] [Google Scholar]
- 9.Bowater R. O., Forbes-Faulkner J., Anderson I. G., Condon K., Robinson B., Kong F., Gilbert G. L., Reynolds A., Hyland S., McPherson G., et al. 2012. Natural outbreak of Streptococcus agalactiae (GBS) infection in wild giant Queensland grouper, Epinephelus lanceolatus (Bloch), and other wild fish in northern Queensland, Australia. J. Fish Dis. 35: 173–186. [DOI] [PubMed] [Google Scholar]
- 10.Laith A. A., Ambak M. A., Hassan M., Sheriff S. M., Nadirah M., Draman A. S., Wahab W., Ibrahim W. N., Aznan A. S., Jabar A., Najiah M. 2017. Molecular identification and histopathological study of natural Streptococcus agalactiae infection in hybrid tilapia (Oreochromis niloticus). Vet. World 10: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W., Gan X., Zhu J. J., Ao Q. W., Tan Y., Chen M., Luo Y. J. 2018. A pathological study of GIFT strain of Nile Tilapia (Oreochromis niloticus) infected by Streptococcus agalactiae. Agric. Biotechnol. 7: 136–142. [Google Scholar]
- 12.Curradi M., Izzo A., Badaracco G., Landsberger N. 2002. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 22: 3157–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baliga B. C., Kumar S. 2002. Role of Bcl-2 family of proteins in malignancy. Hematol. Oncol. 20: 63–74. [DOI] [PubMed] [Google Scholar]
- 14.Law J. A., Jacobsen S. E. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto I., Otte A. P., Allis C. D., Reinberg D., Heard E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. [DOI] [PubMed] [Google Scholar]
- 16.Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- 17.Trasler J. M. 2006. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod. Fertil. Dev. 18: 63–69. [DOI] [PubMed] [Google Scholar]
- 18.Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J. K. 2002. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y. Y., Sun L. X., Zhu J. J., Zhao Y., Wang H., Liu H. J., Ji X. S. 2017. Epigenetic control of cyp19a1a expression is critical for high temperature induced Nile tilapia masculinization. J. Therm. Biol. 69: 76–84. [DOI] [PubMed] [Google Scholar]
- 20.Aho K., Koskenvuo M., Tuominen J., Kaprio J. 1986. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 13: 899–902. [PubMed] [Google Scholar]
- 21.Bellamy N., Duffy D., Martin N., Mathews J. 1992. Rheumatoid arthritis in twins: a study of aetiopathogenesis based on the Australian Twin Registry. Ann. Rheum. Dis. 51: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bammer H., Schaltenbrand G., Solcher H. 1960. [Examinations of twins in multiple sclerosis]. Dtsch. Z. Nervenheilkd. 181: 261–279. [PubMed] [Google Scholar]
- 23.Robertson K. D. 2005. DNA methylation and human disease. Nat. Rev. Genet. 6: 597–610. [DOI] [PubMed] [Google Scholar]
- 24.McCann A. H., Miller N., O’Meara A., Pedersen I., Keogh K., Gorey T., Dervan P. A. 1996. Biallelic expression of the IGF2 gene in human breast disease. Hum. Mol. Genet. 5: 1123–1127. [DOI] [PubMed] [Google Scholar]
- 25.Cui H., Onyango P., Brandenburg S., Wu Y., Hsieh C. L., Feinberg A. P. 2002. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 62: 6442–6446. [PubMed] [Google Scholar]
- 26.Cui H., Cruz-Correa M., Giardiello F. M., Hutcheon D. F., Kafonek D. R., Brandenburg S., Wu Y., He X., Powe N. R., Feinberg A. P. 2003. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 299: 1753–1755. [DOI] [PubMed] [Google Scholar]
- 27.Murphy S. K., Huang Z., Wen Y., Spillman M. A., Whitaker R. S., Simel L. R., Nichols T. D., Marks J. R., Berchuck A. 2006. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 4: 283–292. [DOI] [PubMed] [Google Scholar]
- 28.Mirbahai L., Yin G., Bignell J. P., Li N., Williams T. D., Chipman J. K. 2011. DNA methylation in liver tumorigenesis in fish from the environment. Epigenetics 6: 1319–1333. [DOI] [PubMed] [Google Scholar]
- 29.Mirbahai L., Williams T. D., Zhan H., Gong Z., Chipman J. K. 2011. Comprehensive profiling of zebrafish hepatic proximal promoter CpG island methylation and its modification during chemical carcinogenesis. BMC Genomics 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandegehuchte M. B., Kyndt T., Vanholme B., Haegeman A., Gheysen G., Janssen C. R. 2009. Occurrence of DNA methylation in Daphnia magna and influence of multigeneration Cd exposure. Environ. Int. 35: 700–706. [DOI] [PubMed] [Google Scholar]
- 31.Vandegehuchte M. B., Lemière F., Janssen C. R. 2009. Quantitative DNA-methylation in Daphnia magna and effects of multigeneration Zn exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 150: 343–348. [DOI] [PubMed] [Google Scholar]
- 32.Vandegehuchte M. B., Lemière F., Vanhaecke L., Vanden Berghe W., Janssen C. R. 2010. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 151: 278–285. [DOI] [PubMed] [Google Scholar]
- 33.Shugart L. R. 1990. 5-Methyl deoxycytidine content of DNA from bluegill sunfish (Lepomis macrochirus) exposed to benzo[a]pyrene. Environ. Toxicol. Chem. 9: 205–208. [Google Scholar]
- 34.Wang Y., Wang C., Zhang J., Chen Y., Zuo Z. 2009. DNA hypomethylation induced by tributyltin, triphenyltin, and a mixture of these in Sebastiscus marmoratus liver. Aquat. Toxicol. 95: 93–98. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J., Fu Q., Ao Q., Tan Y., Luo Y., Jiang H., Li C., Gan X. 2017. Transcriptomic profiling analysis of tilapia (Oreochromis niloticus) following Streptococcus agalactiae challenge. Fish Shellfish Immunol. 62: 202–212. [DOI] [PubMed] [Google Scholar]
- 36.Hu Q. M., Tian H. F., Xiao H. B. 2019. Effects of temperature and sex steroids on sex ratio, growth, and growth-related gene expression in the Chinese giant salamander Andrias davidianus. Aquat. Biol. 28: 79–90. [Google Scholar]
- 37.Conte M. A., Gammerdinger W. J., Bartie K. L., Penman D. J., Kocher T. D. 2017. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi Y., Li W. 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lister R., O’Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., Millar A. H., Ecker J. R. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong S., Fei Z., Chen Y. R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J., et al. 2013. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159. [DOI] [PubMed] [Google Scholar]
- 41.Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Q., Tian H., Li W., Meng Y., Wang Q., Xiao H. 2019. Identification of critical sex-biased genes in Andrias davidianus by de novo transcriptome. Mol. Genet. Genomics 294: 287–299. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C., Pachter L., Salzberg S. L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffman A. R., Hu J. F. 2006. Directing DNA methylation to inhibit gene expression. Cell. Mol. Neurobiol. 26: 425–438. [DOI] [PubMed] [Google Scholar]
- 45.Ruane N. M., Collins E. M., Geary M., Swords D., Hickey C., Geoghegan F. 2013. Isolation of Streptococcus agalactiae and an aquatic birnavirus from doctor fish Garra rufa L. Ir. Vet. J. 66: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Harbi A. H. 2011. Molecular characterization of Streptococcus iniae isolated from hybrid tilapia (Oreochromis niloticus × Oreochromis aureus). Aquaculture 312: 15–18. [Google Scholar]
- 47.Lau S. K. P., Woo P. C. Y., Tse H., Leung K. W., Wong S. S. Y., Yuen K. Y. 2003. Invasive Streptococcus iniae infections outside North America. J. Clin. Microbiol. 41: 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C., Li Y., Zhou G., Gao Y., Ma S., Chen Y., Song J., Wang X. 2018. Whole-genome bisulfite sequencing of goat skins identifies signatures associated with hair cycling. BMC Genomics 19: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz M. D., He Y., Whitaker J. W., Hariharan M., Mukamel E. A., Leung D., Rajagopal N., Nery J. R., Urich M. A., Chen H., et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation [Published erratum appears in 2015 Nature 530: 242]. Nature 523: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J. R., Hong C. P., Moon J. W., Jung Y. D., Kim D. S., Kim T. H., Gim J. A., Bae J. H., Choi Y., Eo J., et al. 2014. Genome-wide analysis of DNA methylation patterns in horse. BMC Genomics 15: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao C., Li Q., Chen S., Zhang P., Lian J., Hu Q., Sun B., Jin L., Liu S., Wang Z., et al. 2014. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24: 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes H., Reisz-Porszasz S., Hankinson O. 1992. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science 256: 1193–1195. [DOI] [PubMed] [Google Scholar]
- 53.Gu Y. Z., Hogenesch J. B., Bradfield C. A. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40: 519–561. [DOI] [PubMed] [Google Scholar]
- 54.Abbott B. D., Schmid J. E., Brown J. G., Wood C. R., White R. D., Buckalew A. R., Held G. A. 1999. RT-PCR quantification of AHR, ARNT, GR, and CYP1A1 mRNA in craniofacial tissues of embryonic mice exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin and hydrocortisone. Toxicol. Sci. 47: 76–85. [DOI] [PubMed] [Google Scholar]
- 55.McIntosh B. E., Hogenesch J. B., Bradfield C. A. 2010. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72: 625–645. [DOI] [PubMed] [Google Scholar]
- 56.Semenza G. L. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furness S. G., Lees M. J., Whitelaw M. L. 2007. The dioxin (aryl hydrocarbon) receptor as a model for adaptive responses of bHLH/PAS transcription factors. FEBS Lett. 581: 3616–3625. [DOI] [PubMed] [Google Scholar]
- 58.Drutel G., Kathmann M., Heron A., Schwartz J. C., Arrang J. M. 1996. Cloning and selective expression in brain and kidney of ARNT2 homologous to the Ah receptor nuclear translocator (ARNT). Biochem. Biophys. Res. Commun. 225: 333–339. [DOI] [PubMed] [Google Scholar]
- 59.Grompe M., al-Dhalimy M., Finegold M., Ou C. N., Burlingame T., Kennaway N. G., Soriano P. 1993. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 7(12A): 2298–2307. [DOI] [PubMed] [Google Scholar]
- 60.Raimondo, A. 2011. Identification of downstream target genes and analysis of obesity-related variants of the bHLH/PAS transcription factor single-minded 1. Doctoral dissertation, University of Adelaide, Adelaide, Australia [Google Scholar]
- 61.Abbott B. D., Held G. A., Wood C. R., Buckalew A. R., Brown J. G., Schmid J. 1999. AhR, ARNT, and CYP1A1 mRNA quantitation in cultured human embryonic palates exposed to TCDD and comparison with mouse palate in vivo and in culture. Toxicol. Sci. 47: 62–75. [DOI] [PubMed] [Google Scholar]
- 62.Pearen M. A., Muscat G. E. O. 2010. Minireview: nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol. Endocrinol. 24: 1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami T., Hijikata T., Matsukawa M., Ishikawa H., Yorifuji H. 2006. Zebrafish protocadherin 10 is involved in paraxial mesoderm development and somitogenesis. Dev. Dyn. 235: 506–514. [DOI] [PubMed] [Google Scholar]
- 64.Nakao S., Platek A., Hirano S., Takeichi M. 2008. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J. Cell Biol. 182: 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y., Jiang Y., Jiang M., Zhang J., Yang B., She Y., Wang W., Deng Y., Ye Y. 2016. Protocadherin 10 inhibits cell proliferation and induces apoptosis via regulation of DEP domain containing 1 in endometrial endometrioid carcinoma. Exp. Mol. Pathol. 100: 344–352. [DOI] [PubMed] [Google Scholar]
- 66.Schoch, H. 2014. Molecular-genetic mechanisms of memory formation in mouse models of neurodevelopmental and neuropsychiatric disorders. Thesis, University of Pennsylvania, Philadelphia, PA. [Google Scholar]
- 67.Morrow E. M., Yoo S. Y., Flavell S. W., Kim T. K., Lin Y., Hill R. S., Mukaddes N. M., Balkhy S., Gascon G., Hashmi A., et al. 2008. Identifying autism loci and genes by tracing recent shared ancestry. Science 321: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mason J. M., Arndt K. M. 2004. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem 5: 170–176. [DOI] [PubMed] [Google Scholar]
- 69.Wang W., Tan S., Luo J., Shi H., Zhou T., Yang Y., Jin Y., Wang X., Niu D., Yuan Z., et al. 2019. GWAS analysis indicated importance of NF-κB signaling pathway in host resistance against motile Aeromonas septicemia disease in catfish. Mar. Biotechnol. (NY) 21: 335–347. [DOI] [PubMed] [Google Scholar]
- 70.Cai Z., Guldbrandtsen B., Lund M. S., Sahana G. 2018. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genomics 19: 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.