Abstract
Although most patients with COVID-19 pneumonia have a good prognosis, some patients develop to severe or critical illness, and the mortality of critical cases is up to 61.5%. However, specific molecular information about immune response in critical patients with COVID-19 is poorly understood. A total of 54 patients were enrolled and divided into three groups, among which 34 were common, 14 were severe, and 6 were critical. The constitution of peripheral blood mononuclear cells (PBMC) in patients was analyzed by CyTOF. The profile of cytokines was examined in plasma of patients using luminex. The IL-2 signaling pathway was investigated in the PBMC of patients by qRT-PCR. The count and percentage of lymphocytes were significantly decreased in critical patients compared to common and severe patients with COVID-19 pneumonia. The count of T cells, B cells, and NK cells was remarkably decreased in critical patients compared to normal controls. The percentage of CD8+ T cells was significantly lower in critical patients than that in common and severe patients with COVID-19 pneumonia. The expression of IL-2R, JAK1, and STAT5 decreased in PBMC of common, severe, and critical patients, but IL-2 level was elevated in severe patients and decreased in critical patients with COVID-19 pneumonia. The decrease of CD8+ T cells in critical patients with COVID-19 pneumonia may be related to the IL-2 signaling pathway. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia.
Subject terms: Interleukins, Viral infection
Introduction
In early December 2019, cases of unknown pneumonia were found in Wuhan, Hubei province in China, which were subsequently identified as an acute respiratory infectious disease caused by a novel coronavirus1–3. On 11 February 2020, the World Health Organization named these diseases as Corona Virus Disease 2019 (COVID-19). This infectious disease is highly contagious and threatens human life and health4–7.
The clinical spectrum of COVID-19 pneumonia includes mild, common, severe, and critical cases. Although most patients with COVID-19 pneumonia have a good prognosis, some patients develop to severe or critical illness, and the mortality of critical cases can reach up to 61.5%8. However, specific molecular information about immune responses in critical patients with COVID-19 remains poorly understood.
Reduction of mortality in critical patients is crucial for clinical response against COVID 19. In this study, we analyzed the potential molecular mechanism underlying the decrease of lymphocytes in critical patients with COVID-19 pneumonia, which may provide the new therapeutic targets for COVID-19.
Results
The decrease of lymphocytes in critical patients with COVID-19 pneumonia
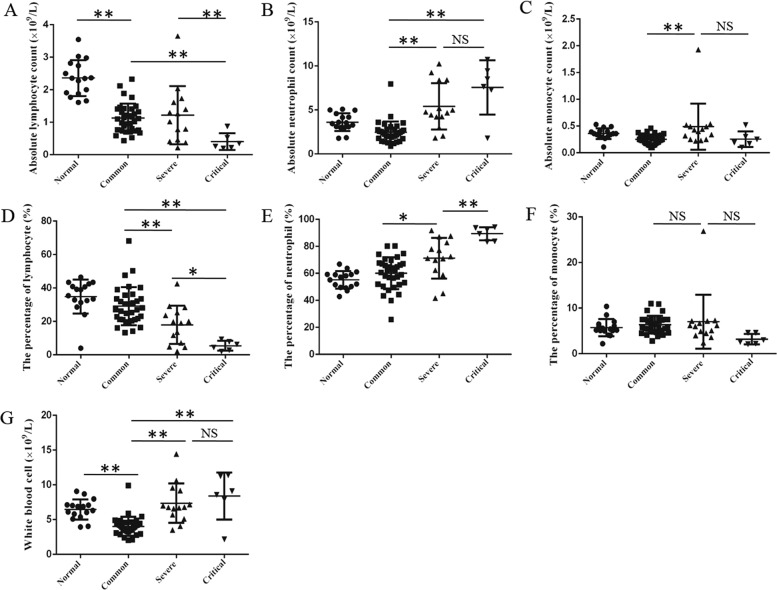
Patients with COVID-19 pneumonia were enrolled and divided into common, severe, and critical type by the guidance of the National Health Commission of China9 (Table 1). The absolute lymphocyte count in patients with COVID-19 pneumonia was remarkably decreased compared with normal controls (Fig. 1a). Notably, the absolute lymphocyte count in critical patients was significantly lower than that in common or severe patients, but there was no difference between common patients and severe patients (Fig. 1a). In addition, the percentage of lymphocyte in white blood cells (WBC) decreased in turn in common, severe, and critical patients, and the differences among them were statistically significant (Fig. 1d). Meanwhile, the WBC count increased in turn in common, severe, and critical patients, and there was a statistical difference between common and critical patients (Fig. 1g). The increase in WBC count was driven by an increase in absolute neutrophil count (Fig. 1b), which was consistent with the trend of C reactive protein and procalcitonin (Table 2). All in all, the count and percentage of lymphocyte were decreased in critical patients compared to common and severe patients with COVID-19 pneumonia.
Table 1.
Clinical categorization of the patients with COVID-19 pneumonia by National Health Commission of China.
| Common | Severe | Critical |
|---|---|---|
| Meet any of the followings: | Meet any of the followings: | |
| Fever | 1. Respiratory distress, RR ≥30/min | 1. Respiratory failure and mechanical ventilation |
| Respiratory symptoms | 2. Saturation of oxygen ≤93% at rest state | 2. Shock |
| Pneumonia on X-ray or CT | 3. Arterial partial pressure of oxygen (PaO2) or Fraction of inspiration O2 (FiO2) ≤300 mnHg (1 mmHg = 0.133 kPa) | 3. Other organ failure and ICU monitoring and treatment |
Fig. 1. The blood routine analysis in patients with COVID-19 pneumonia.
The patients were enrolled and divided into common, severe, and critical types. The absolute count of lymphocyte (a), neutrophil (b), monocyte (c), and the percentage of lymphocyte (d), neutrophil (e), monocyte (f), and white blood cell count (g) in patients and normal controls were acquired by full-automatic blood routine detector. One-way analysis of variance (ANOVA) followed by post hoc LSD was used to compare differences between groups. *P < 0.05 and **P < 0.01.
Table 2.
The clinical information of normal controls and patients with COVID-19 pneumonia.
| Normal controls (n = 16) |
Common patients (n = 34) |
Severe patients (n = 14) |
Critical patients (n = 6) |
P value | |
|---|---|---|---|---|---|
| Ages (years old) | 55 ± 13 | 50 ± 15 | 63 ± 5 | 75 ± 10 | − |
| Gender(male/female) | 6/10 | 14/20 | 6/8 | 2/4 | − |
| White blood cell (×109/L) | 6.46 ± 0.36 | 4.02 ± 0.23 | 7.36 ± 0.76 | 8.39 ± 1.38 | 0.000 |
| Neutrophil (×109/L) | 3.61 ± 0.25 | 2.47 ± 0.21 | 5.41 ± 0.71 | 7.57 ± 1.26 | 0.000 |
| Lymphocyte (×109/L) | 2.36 ± 0.14 | 1.13 ± 0.08 | 1.22 ± 0.24 | 0.41 ± 0.11 | 0.000 |
| Monocyte (×109/L) | 0.36 ± 0.02 | 0.25 ± 0.02 | 0.49 ± 0.11 | 0.26 ± 0.06 | 0.005 |
| Neutrophil (%) | 55.18 ± 1.63 | 60.07 ± 2.02 | 71.24 ± 4.03 | 89.38 ± 1.99 | 0.000 |
| Lymphocyte (%) | 34.86 ± 2.56 | 29.11 ± 1.95 | 17.98 ± 3.07 | 5.54 ± 1.18 | 0.000 |
| Monocyte (%) | 5.72 ± 0.47 | 6.37 ± 0.33 | 7.01 ± 1.58 | 3.18 ± 0.47 | 0.082 |
| Alanine aminotransferase (U/L) | 19.85 ± 2.33 | 49.53 ± 9.8 | 49.29 ± 9.1 | 49.17 ± 15.06 | 0.232 |
| Aspartate aminotransferase (U/L) | 22.23 ± 1.71 | 38.53 ± 5.41 | 39.14 ± 5.79 | 85.17 ± 31.05 | 0.003 |
| Total bilirubin (μmol/L) | 14.71 ± 1.29 | 9.73 ± 0.56 | 11.29 ± 2.16 | 18.40 ± 2.24 | 0.000 |
| Direct bilirubin (μmol/L) | 4.67 ± 0.47 | 1.91 ± 0.12 | 3.79 ± 1.81 | 6.52 ± 1.58 | 0.006 |
| Total protein (g/L) | 74.93 ± 1.02 | 73.52 ± 0.92 | 71.69 ± 2.02 | 69.52 ± 3.64 | 0.241 |
| Albumin (g/L) | 46.1 ± 0.54 | 36.70 ± 0.71 | 31.05 ± 0.96 | 30.70 ± 1.88 | 0.000 |
| Creatinine (μmol/L) | 56.19 ± 2.92 | 59.94 ± 2.76 | 56.43 ± 3.66 | 96.83 ± 25.72 | 0.002 |
| Glomerular filtration rate (mL/min/1.73 m2) | 126.59 ± 5.62 | 113.32 ± 5.62 | 101.19 ± 3.47 | 70.85 ± 14.15 | 0.000 |
| Procalcitonin (ng/mL) | 0.10 ± 0.001 | 0.13 ± 0.004 | 0.18 ± 0.04 | 3.42 ± 3.15 | 0.015 |
| C reactive protein (mg/L) | 2.50 ± 0.25 | 16.50 ± 2.73 | 46.34 ± 9.82 | 96.73 ± 21.72 | 0.000 |
| Saturation of oxygen (%) | 98.53 ± 1.21 | 96.83 ± 0.90 | 92.84 ± 0.59 | 87.03 ± 2.95 | 0.019 |
The constitution of PBMC in patients with COVID-19 pneumonia
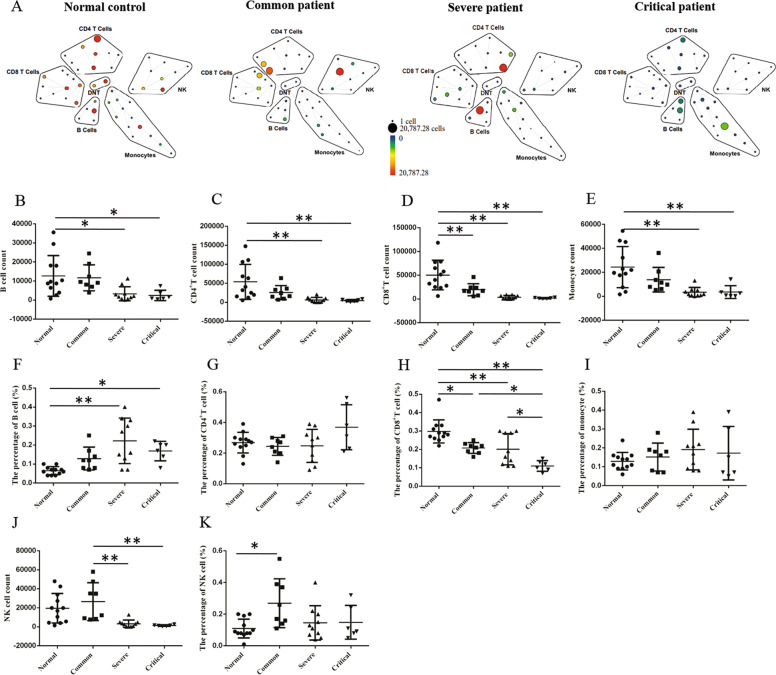
To investigate the cause of lymphocytic decrease, we analyzed the constitution of peripheral blood mononuclear cell (PBMC) in patients with COVID-19 pneumonia by CyTOF. As well as we know, PBMC consists largely of lymphocytes and lymphocyte includes T cell, B cell, and NK cell, and T cell is the main part. CyTOF revealed that the count of T cells, B cells, NK cells, and monocytes was remarkably decreased in critical patients and severe patients compared to normal controls (Fig. 2a–e, j). In addition, the percentage of B cells was increased in severe and critical patients compared to normal controls (Fig. 2f). The percentage of NK cells in common patients was higher than that in normal controls (Fig. 2k). The percentage of CD4+ T cells and monocytes had no statistical difference among the patients and normal controls (Fig. 2g, i). However, the percentage of CD8+ T cells was significantly lower in patients with COVID-19 pneumonia than that in normal controls, especially the percentage of CD8+ T cells in critical patients was decreased compared to common and severe patients with COVID-19 pneumonia (Fig. 2h).
Fig. 2. The CyTOF analysis in PBMC of patients with COVID-19 pneumonia.
The patients were enrolled and divided into common, severe, and critical types. The count and percentage of B cell (a, b, f), CD4+ T cell (a, c, g), CD8+ T cell (a, d, h), monocyte (a, e, i), and NK cell (a, j, k) in PBMC of patients and normal controls was analyzed by CyTOF. ANOVA followed by post hoc LSD was used to compare differences between groups. *P < 0.05 and **P < 0.01.
The profile of cytokines in patients with COVID-19 pneumonia
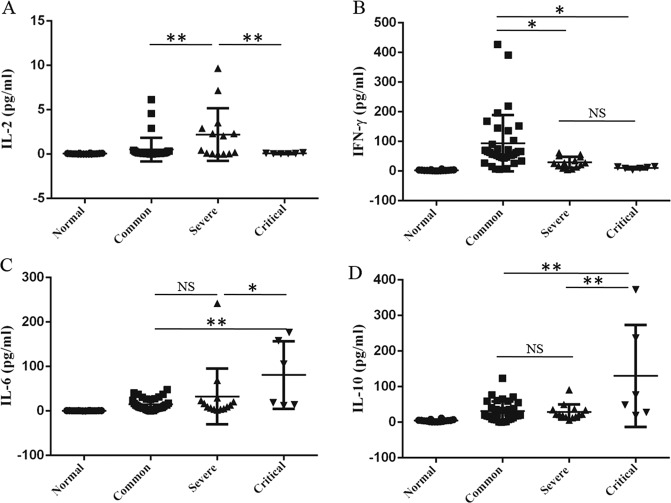
To make further investigation, we analyzed the profile of cytokines in plasma of patients with COVID-19 pneumonia using luminex. The level of interleukin-2 (IL-2) in critical patients was significantly lower than that in severe patients, whereas the level of IL-2 in severe patients was higher than that in normal controls and common patients (Fig. 3a). For interferon-γ (IFN-γ), its level in critical patients was significantly decreased compared to common patients, but its level in common patients was increased compared to the normal controls (Fig. 3b). Contrary to IL-2 and IFN-γ, the level of interleukin-6 (IL-6) and interleukin-10 (IL-10) in critical patients was higher than that in common and severe patients with COVID-19 pneumonia (Fig. 3c, d). Because IL-2 is required for T cells proliferation, differentiation, and activation10, we speculated that the decrease of CD8+ T cells in critical patients with COVID-19 pneumonia might be related to the IL-2 signaling pathway.
Fig. 3. The cytokine profile in plasma of patients with COVID-19 pneumonia.
The patients were enrolled and divided into common, severe, and critical types. The levels of II-2 (a), IFN-γ (b), IL-6 (c), and IL-10 (d) in plasma of patients and normal controls were analyzed by luminex. The experiment shown was replicated in the laboratory twice. ANOVA followed by post hoc LSD was used to compare differences between groups. *P < 0.05 and **P < 0.01.
The expression of IL2 receptor, JAK1, STAT5 in patients with COVID-19 pneumonia
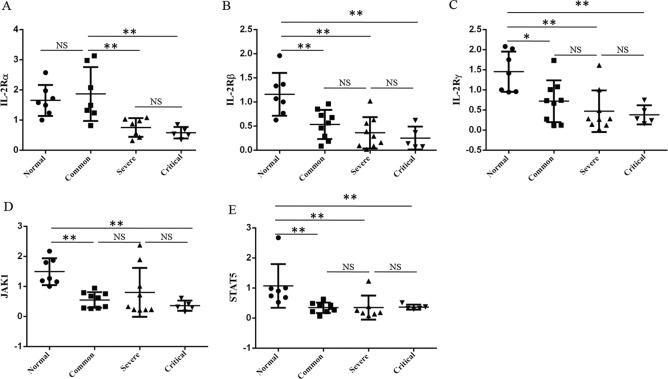
It has been reported that IL-2 and IL-2 receptor (IL-2R) provides an important signal for T cell activation through JAK1-STAT5 signaling pathway11. IL-2R has three subunits: IL-2Rα, IL-2Rβ, and IL-2Rγc, and the activated T cells can simultaneously express α, β, and γc chain and form a high-affinity receptor12. We found that the IL-2Rα expression in PBMC of severe and critical patients was significantly lower than that in common patients and normal controls (Fig. 4a). In addition, the expression of IL-2Rβ and IL-2Rγc in common, severe, and critical patients was significantly lower than that in normal controls, whereas there was no statistical difference among those patients with COVID-19 pneumonia (Fig. 4b, c). Accordingly, the expression of JAK1 and STAT5 showed similar trends as IL-2Rβ and IL-2Rγc in patients and normal controls (Fig. 4d, e). Thus, the decrease of T cells in patients with COVID-19 pneumonia was correlated to low expression of IL-2R and JAK1-STAT5, especially in severe and critical patients.
Fig. 4. The expression of IL-2 signaling pathway in PBMC of patients with COVID-19 pneumonia.
The patients were enrolled and divided into common, severe, and critical types. The expression of IL-2Rα (a), IL-2Rβ (b), IL-2Rγc (c), JAK1 (d), and STAT5 (e) in PBMC of patients and normal controls were detected by qRT-PCR. The experiment shown was replicated in the laboratory twice. ANOVA followed by post hoc LSD was used to compare differences between groups. *P < 0.05 and **P < 0.01.
Discussion
The most significant finding of this study was the low level of IL-2 in plasma and low expression of IL-2R in PBMC of critical patients, which may result in the remarkable decrease of CD8+ T cell and lymphocytes in critical patients with COVID-19 pneumonia. In addition, we found that total T cell, B cell, and NK cell counts were remarkably decreased in critical patients compared to normal controls.
At present, the rising epidemic situation in China has been effectively curbed through a series of preventive control and medical treatment. The new confirmed and suspected cases in China are basically cleared, and the treatment of remaining severe and critical cases has become the most important task. According to the latest guidelines for novel coronavirus pneumonia, the progressive decrease of peripheral blood lymphocytes is one of the clinical early warning indicators for adult patients with severe and critical illness. Similar to previous studies13, we also found that peripheral blood lymphocytes were decreased in the patients with COVID-19 pneumonia, especially in critical patients. Importantly, we found that CD8+ T cells were remarkably decreased, which was the same as another study14. Some studies have also shown that critical patients with COVID-19 pneumonia have an immune deficiency and hypoimmunity, which may also lead to serious infection and death15. Therefore, CD8+ T cell reduction may result in underlying increased mortality in critical patients.
The secretion of cytokines plays an important role in the development and differentiation of immune cells. Many studies have suggested that cytokine storms might be one of the causes of multiple organ failure and death in severe and critical patients with COVID-19 pneumonia16,17. Similarly, our study also found that some inflammatory cytokines such as IL-6 and IL-10 increased in critical patients with COVID-19 pneumonia. Differently, we found that IL-2 was elevated in severe patients but decreased in critical patients with COVID-19 pneumonia. IL-2, also known as the T cell growth factor, is mainly produced by activated CD4+ T cells and CD8+ T cells18–21. It has been demonstrated that IL-2 at high concentration enhances CD4+ T and CD8+ T cell activation by stimulating expansion and differentiation of conventional T cells and IL-2 at low concentration inhibits CD4+ T and CD8+ T cell activation by maintaining activity and survival of T regulatory cells (Treg)21. IL-2 is so specific and critical for T cells activation that we investigate the IL-2 signaling pathway. Consistent with IL-2 level in plasma, we found that the expression of IL-2R and JAK1-STAT5 in PBMC was decreased in critical patients with COVID-19 pneumonia. Therefore, the decrease of CD8+ T cells and lymphocytes in critical patients with COVID-19 pneumonia may be related to the inhibition of IL-2 signaling pathway.
In addition, we also observed that IL-10 and IL-6 was increased and IFN-γ was decreased in critical patients with COVID-19 pneumonia. Relative to IL-2, IL-10, IL-6, and IFN-γ have more versatile functions. IL-10 is considered a prototypical anti-inflammation cytokine22, but the recent findings indicate that high exogenous IL-10 level promotes the CD8+ T cell cytotoxicity and intermediate endogenous IL-10 level induces exhaustion of CD8+ T cells in tumor23,24. IL-10 can inhibit IL-2 secretion and has heterogenity on CD8+ T cell activation which may depend on other environmental cytokines24, so we speculate that it may take part in T cell activation through IL-2 pathway. IL-6 can induce the differentiation of T cells, whereas its level was increased in our study, so we think it plays an important role in secretion of acute phase proteins such as C reactive protein and inflammatory cytokines25. IFN-γ has a critical role in recognizing and eliminating pathogens, but it has no direct effects on CD8+ T cells activation26,27. Therefore, we focus on the IL-2 pathway to explore its possible effect on CD8+ T cells and lymphocytes.
This study is limited by sample size for critical patients with COVID-19 pneumonia, and clinical significance and power would be increased with more patients. However, our findings still provide evidences and clues for the diagnosis and treatment of critical patients with COVID-19 pneumonia. The progressive decrease of IL-2 in plasma may be a warning factor of disease deterioration in patients with COVID-19 pneumonia. For critical patients with COVID-19 pneumonia, appropriate IL-2 supplementation could be beneficial by improving the immune disorder so as to reduce mortality.
Materials and methods
Patients
All patients were enrolled from inpatients in Beijing Youan Hospital, Capital Medical University. The patients were confirmed by nucleic acid detection of novel coronavirus using the protocol as described previously6,28. A total of 54 patients were divided into three groups according to the guidance of the National Health Commission of China9 (Table 1), among which 34 were common type, 14 were severe type, and 6 were critical type. Sixteen normal controls were from the physical examination population in Beijing Youan Hospital, Capital Medical University. The clinical information of normal controls and patients with COVID-19 pneumonia was described in Table 2. The informed consent was obtained from all the patients. The study was approved by the Ethics Committee of Beijing Youan Hospital, Capital Medical University, China.
CyTOF analysis
Peripheral blood mononuclear cells (PBMC) were cultured with 2 μM cisplatin (195-Pt, Fluidigm) for 2 min before quenching with CSB (Fluidigm) to identify viability. A fix I (Fluidigm) buffer was then used to fix cells for 15 min at room temperature (RT). All metal-conjugated antibodies (Fluidigm) were titrated for optimal concentration before staining with cells. Cells were cultured with antibodies in a total 50 μL CSD (Fluidigm) for 30 min at RT, triple washed in CSB, and incubated with 0.125-μm intercalator in fix and perm buffer (Fluidigm) at 4 °C overnight. Prior to the acquisition, samples were resuspended in deionized water containing 10% EQ4 element beads (Fluidigm), and the concentration of cells was adjusted into 1 × 106 cell/ml. Data acquisition was performed on a Helios mass cytometer (Fluidigm). Data analysis was performed according to the previous paper29,30.
Luminex assay
Cytokines in human plasma were analyzed by luminex assay (Millipore, Billerica, USA). The diluted standard and quality control was added into appropriate wells. The assay buffer and sample was added into sample wells. The premixed beads were added into each well. The plate was sealed and incubated in plate shaker overnight at 4 °C. After washing the plate, the detection antibody was added into each well and incubated for 1 h at room temperature. The streptavidin–phycoerythrin was added into each well-containing detection antibody for 30 min at room temperature. After washing, sheath fluid was added into all wells, and the plate was read on FlexMAP3D.
qRT-PCR
Total RNA was extracted according to the instructions of the RNeasy Micro Kit (Qiagen, CA, USA), and 4 μl of total RNA was reverse transcribed into cDNA by SuperScript III First Strand cDNA synthesis kit (Invitrogen, CA, USA). After diluting the cDNA for 20-fold, quantitative real-time PCR was performed on a high-throughput qRT-PCR instrument (Fluidigm, USA) using 1 μl of cDNA and Applied Biosystems Gene Expression Arrays with Universal Taqman Fast Master Mix (Applied Biosystems, CA, USA). The PCR conditions were 2 min at 50 °C, then 2 min at 95 °C followed by 50 cycles at 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 50 s. A glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene was used as a reference control. The primers and probes of qRT-PCR were seen in Table 3.
Table 3.
The primer and probe of qRT-PCR.
| Primer and probe 5′−3′ | |
|---|---|
| IL2Rβ FORWARD | CGCATCCTTAAGCAGCAACC |
| IL2Rβ REVERSE | AGGCCTCTATCTCCAAGGCA |
| IL2Rβ PROBE | TCGCTGACCAGCTGCTTCACCAACCA |
| IL2Rα FORWARD | TTATCATTTCGTGGTGGGGCA |
| IL2RαREVERSE | CTTGTCTTCCCGTGGGTCAT |
| IL2Rα PROBE | ACAGAGGTCCTGCTGAGAGCGTCTGCA |
| IL2Rγ FORWARD | CCACTGTTTGGAGCACTTGG |
| IL2Rγ REVERSE | CCGAACACGAAACGTGTAGC |
| IL2Rγ PROBE | CCGGACTGACTGGGACCACAGCTGG |
| STAT5A FORWARD | AAGCCCCACTGGAATGATGG |
| STAT5A REVERSE | GGAGTCAAACTTCCAGGCGA |
| STAT5A PROBE | AAGCAACAGGCCCACGACCTGCTCA |
| JAK1 FORWARD | ACGAGTGTCTAGGGATGGCT |
| JAK1 REVERSE | CGCATCCTGGTGAGAAGGTT |
| JAK1 PROBE | TGCAGTTGCCAGAACTGCCCAAGGACA |
| GAPDH FORWARD | CAGCCTCAAGATCATCAGCA |
| GAPDH REVERSE | TGTGGTCATGAGTCCTTCCA |
| GAPDH PROBE | CAAGGTCATCCATGACAACTTTGG |
Statistical analysis
All data was analyzed using GraphPad 6.02. One-way analysis of variance (ANOVA) followed by post hoc LSD was used to compare differences between groups. Differences were considered statistically significant at confidence levels *P < 0.05 or **P < 0.01.
Supplementary information
Acknowledgements
This study was financially supported by COVID-19 Key Technology Research and Development Funding of Beijing Hospital Authority; The National Natural Science Foundation of China (81672026); and National Science and Technology Major Project of China (2018ZX10302205–005).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by H-U Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hongbo Shi, Wenjing Wang
Contributor Information
Ronghua Jin, Email: jin_eagle@sina.com.
Dexi Chen, Email: dexichen@ccmu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2636-4).
References
- 1.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;8:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel corona virus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–512. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan W, et al. A novel coronavirus genome identified in a cluster of pneumonia cases-Wuhan, China 2019-2020. China CDC Wkly. 2020;2:61–66. [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, et al. Early transmission dynamics in Wuhan, China, of novelcoronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J, et al. A familiar cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phan L, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;S2213:30079–30085. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The National Health Commission of China. Diagnosis and Treatment plan for 2019 Novel Coronavirus-Infected Pneumonia (trial version fourth). http://www.nhc.gov.cn/xcs/zhengcwj/202001/4294563ed35b43209b31739bd0785e67.shtml (2020).
- 10.Read KA, Powell MD, McDonald PW, Oestreich KJ. IL-2, IL-7, and IL-15: multistage regulators of CD4(+) T helper cell differentiation. Exp. Hematol. 2016;44:799–808. doi: 10.1016/j.exphem.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Fu X, et al. Enhanced interactionbetween SEC2 mutant and TCR Vβ induces MHC II-independent activation of T cells via PKCθ/NF-κB and IL-2R/STAT5 signaling pathways. J. Biol. Chem. 2018;293:19771–19784. doi: 10.1074/jbc.RA118.003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 13.Guan W, et al. China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan, S. et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv10.1101/2020.02.10.20021832 (2020).
- 16.Zhou, Y. et al. Aberrant pathogenic GM-CSF+T cells and inflammatory CD14+CD16+monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv10.1101/2020.02.12.945576 (2020).
- 17.Chen C, Zhang XR, Ju ZY, He WF. Advances in the research of cytokine storm mechanism induced by Corona Virus Disease 2019 and the corresponding immunotherapies. Zhonghua Shao Shang Za Zhi. 2020;36:E005. doi: 10.3760/cma.j.cn501120-20200224-00088. [DOI] [PubMed] [Google Scholar]
- 18.Wilson EB, Livingstone AM. Cutting edge: CD4+ T cell-derived IL-2 is essential for help-dependent primary CD8+ T cell responses. J. Immunol. 2008;181:7445–7448. doi: 10.4049/jimmunol.181.11.7445. [DOI] [PubMed] [Google Scholar]
- 19.Malek TR. The biology of interleukin-2. Annu Rev. Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 20.Bao Y, Cao X. The immune potential and immunopathology of cytokine-producing B cell subsets: a comprehensive review. J. Autoimmun. 2014;55:10–23. doi: 10.1016/j.jaut.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Abbas AK, Trotta E, Simeonov DR, Marson A, Bluestone JA. Revisiting IL-2: biology and therapeutic prospects. Sci. Immunol. 2018;3:eaat1482. doi: 10.1126/sciimmunol.aat1482. [DOI] [PubMed] [Google Scholar]
- 22.O’Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol. Rev. 2008;223:114–131. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]
- 23.Mannino MH, et al. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367:103–107. doi: 10.1016/j.canlet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Bedke T, Muscate F, Soukou S, Gagliani N, Huber S. Title: IL-10-producing T cells and their dual functions. Semin. Immunol. 2019;44:101335. doi: 10.1016/j.smim.2019.101335. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Narazaki M, Kishimoto T. Interleukin (IL-6) Immunotherapy. Cold Spring Harb. Perspect. Biol. 2018;10:a028456. doi: 10.1101/cshperspect.a028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kak G, Raza M, Tiwari BK. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts. 2018;9:64–79. doi: 10.1515/bmc-2018-0007. [DOI] [PubMed] [Google Scholar]
- 27.Zaidi MR. The interferon-gamma paradox in cancer. J. Interferon Cytokine Res. 2019;39:30–38. doi: 10.1089/jir.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van U, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 2017;8:1740. doi: 10.1038/s41467-017-01689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.